Nutrigenomic Studies on the Ameliorative Effect of Enzyme-Digested Phycocyanin in Alzheimer’s Disease Model Mice
Abstract
:1. Introduction
2. Materials and Methods
2.1. PC Products and Aβ25–35
2.2. Animals
2.3. Y Maze Spontaneous Alternation Test
2.4. Total RNA Isolation
2.5. DNA Microarray Analysis
2.6. Bioinformatic Analysis of Microarray Data
2.7. Statistical Analysis
3. Results
3.1. Spontaneous Alternation
3.2. Gene Expression Profiles of the Hippocampi of EDPC- and PC-Administered Mice
3.2.1. Upregulated Aβ-Related Genes
3.2.2. Downregulated EDPC-Related Genes
3.2.3. Downregulated PC-Related Genes
3.2.4. Downregulated Aβ-Related Genes
3.2.5. Upregulated EDPC-Related Genes
3.2.6. Upregulated PC-Related Genes
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sanabria-Castro, A.; Alvarado-Echeverría, I. Molecular Pathogenesis of Alzheimer’s Disease: An Update. Ann. Neurosci. 2017, 24, 46–54. [Google Scholar] [CrossRef]
- Kim, H.Y.; Lee, D.K. Intracerebroventricular injection of amyloid-peptides in normal mice to acutely induce Alzheimer-like cognitive deficits. J. Vis. Exp. 2016, 109, e53308. [Google Scholar] [CrossRef] [Green Version]
- Schmid, S.; Jungwirth, B. Intracerebroventricular injection of beta-amyloid in mice is associated with long-term cognitive impairment in the modified hole-board test. Behav. Brain. Res. 2017, 324, 15–20. [Google Scholar] [CrossRef]
- McCarty, M.F.; DiNicolantonio, J.J. A Fundamental Role for Oxidants and Intracellular Calcium Signals in Alzheimer’s Pathogenesis-And How a Comprehensive Antioxidant Strategy May Aid Prevention of This Disorder. Int. J. Mol. Sci. 2021, 22, 2140. [Google Scholar] [CrossRef]
- Pentón-Rol, G.; Marín-Prida, J. C-Phycocyanin and phycocyanobilin as remyelination therapies for enhancing recovery in multiple sclerosis and ischemic stroke: A preclinical perspective. Behav. Sci. 2018, 8, 15. [Google Scholar] [CrossRef] [Green Version]
- Rimbau, V.; Camins, A. Protective effects of C-phycocyanin against kainic acid-induced neuronal damage in rat hippocampus. Neurosci. Lett. 1999, 276, 75–78. [Google Scholar] [CrossRef]
- Agrawa, M.; Peruma, Y. Phycocyanin alleviates ICV-STZ induced cognitive and molecular deficits via PI3-Kinase dependent pathway. Food Chem. Toxicol. 2020, 145, 111684. [Google Scholar] [CrossRef]
- Li, Z.; Gan, L. Effect of C-phycocyanin on HDAC3 and miRNA-335 in Alzheimer’s disease. Transl. Neurosci. 2020, 11, 161–172. [Google Scholar] [CrossRef]
- Vyas, Y.; Montgomery, J.M. Hippocampal Deficits in Amyloid-β-Related Rodent Models of Alzheimer’s Disease. Front. Neurosci. 2020, 14, 266. [Google Scholar] [CrossRef]
- Hosseinian, S.; Arefian, E. A meta-analysis of gene expression data highlights synaptic dysfunction in the hippocampus of brains with Alzheimer’s disease. Sci. Rep. 2020, 10, 8384. [Google Scholar] [CrossRef]
- Szczepanik, J.C.; Garcia, A.F. Protective effects against memory impairment induced by methylglyoxal in mice co-treated with FPS-ZM1, an advanced glycation end products receptor antagonist. Acta Neurobiol. Exp. 2020, 80, 364–374. [Google Scholar] [CrossRef]
- Rothman, S.M.; Tanis, K.Q. Human Alzheimer’s disease gene expression signatures and immune profile in APP mouse models: A discrete transcriptomic view of Aβ plaque pathology. J. Neuroinflammation 2018, 15, 256. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, L.; Zeng, L. Microarray Profile of Long Noncoding RNA and Messenger RNA Expression in a Model of Alzheimer’s Disease. Life 2020, 10, 64. [Google Scholar] [CrossRef] [PubMed]
- Dong, S.; Zhao, J. F-box protein complex FBXL19 regulates TGFβ1-induced E-cadherin down-regulation by mediating Rac3 ubiquitination and degradation. Mol. Cancer 2014, 13, 76. [Google Scholar] [CrossRef] [Green Version]
- Hol, F.A.; Geurds, M.P. PAX genes and human neural tube defects: An amino acid substitution in PAX1 in a patient with spina bifida. J. Med. Genet. 1996, 33, 655–660. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lipkin, S.M.; Näär, A.M. Identification of a novel zinc finger protein binding a conserved element critical for Pit-1-dependent growth hormone gene expression. Genes Dev. 1993, 7, 1674–1687. [Google Scholar] [CrossRef] [Green Version]
- Ciminelli, B.M.; Menduti, G. Polymorphic Genetic Markers of the GABA Catabolism Pathway in Alzheimer’s Disease. J. Alzheimers Dis. 2020, 77, 301–311. [Google Scholar] [CrossRef]
- Rossi, A.; Rigotto, G. Defective Mitochondrial Pyruvate Flux Affects Cell Bioenergetics in Alzheimer’s Disease-Related Models. Cell Rep. 2020, 30, 2332–2348.e10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bagyinszky, E.; Kang, M.J. Early-onset Alzheimer’s disease patient with prion (PRNP) p.Val180Ile mutation. Neuropsychiatr. Dis. Treat. 2019, 15, 2003–2013. [Google Scholar] [CrossRef] [Green Version]
- Jeong, J.K.; Park, S.Y. Neuroprotective effect of cellular prion protein (PrPC) is related with activation of alpha7 nicotinic acetylcholine receptor (α7nAchR)-mediated autophagy flux. Oncotarget 2015, 6, 24660–24674. [Google Scholar] [CrossRef]
- Wang, Y.H.; Ding, Z.Y. An Efficient Screen for Cell-Intrinsic Factors Identifies the Chaperonin CCT and Multiple Conserved Mechanisms as Mediating Dendrite Morphogenesis. Front. Cell. Neurosci. 2020, 14, 577315. [Google Scholar] [CrossRef]
- Mauceri, D.; Buchthal, B. Nasally delivered VEGFD mimetics mitigate stroke-induced dendrite loss and brain damage. Proc. Natl. Acad. Sci. USA 2020, 117, 8616–8623. [Google Scholar] [CrossRef]
- Jones, E.; Mead, S. Genetic risk factors for Creutzfeldt-Jakob disease. Neurobiol. Dis. 2020, 142, 104973. [Google Scholar] [CrossRef] [PubMed]
- Ferrer, I.; Garcia-Esparcia, P. Olfactory Receptors in Non-Chemosensory Organs: The Nervous System in Health and Disease. Front. Aging Neurosci. 2016, 8, 163. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guo, X.; Keenan, B.T. Age attenuates the transcriptional changes that occur with sleep in the medial prefrontal cortex. Aging Cell 2019, 18, e13021. [Google Scholar] [CrossRef] [Green Version]
- Venoux, M.; Delmouly, K. Gene organization, evolution and expression of the microtubule-associated protein ASAP (MAP9). BMC Genom. 2008, 9, 406. [Google Scholar] [CrossRef] [Green Version]
- Schmidt, C.; Frahm, C. Phosphoinositide 3-Kinase Restrains Neurotoxic Effects of Microglia After Focal Brain Ischemia. Mol. Neurobiol. 2016, 53, 5468–5479. [Google Scholar] [CrossRef]
- D’Andrea, I.; Fardella, V. Lack of kinase-independent activity of PI3Kγ in locus coeruleus induces ADHD symptoms through increased CREB signaling. EMBO Mol. Med. 2015, 7, 904–917. [Google Scholar] [CrossRef] [Green Version]
- Mohan, V.; Wade, S.D. Close Homolog of L1 Regulates Dendritic Spine Density in the Mouse Cerebral Cortex Through Semaphorin 3B. J. Neurosci. 2019, 39, 6233–6250. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, K.T.; Shann, Y.J. Identification of latent biomarkers in hepatocellular carcinoma by ultra-deep whole-transcriptome sequencing. Oncogene 2014, 33, 4786–4794. [Google Scholar] [CrossRef]
- Huang, J.; Teng, L. ZNF216 Is an A20-like and IkappaB kinase gamma-interacting inhibitor of NFkappaB activation. J. Biol. Cham. 2004, 279, 16847–16853. [Google Scholar] [CrossRef] [Green Version]
- Lee, D.; Takayama, S. ZFAND5/ZNF216 is an activator of the 26S proteasome that stimulates overall protein degradation. Proc. Natl. Acad. Sci. USA 2018, 115, E9550–E9559. [Google Scholar] [CrossRef] [Green Version]
- Zöller, T.; Attaai, A. Aged Mouse Cortical Microglia Display an Activation Profile Suggesting Immunotolerogenic Functions. Int. J. Mol. Sci. 2018, 19, 706. [Google Scholar] [CrossRef] [Green Version]
- Hiraoka, N.; Misra, A. Molecular cloning and expression of two distinct human N-acetylgalactosamine 4-O-sulfotransferases that transfer sulfate to GalNAc beta 1-->4GlcNAc beta 1-->R in both N- and O-glycans. Glycobiology 2001, 11, 495–504. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fujikawa, D.G. The role of excitotoxic programmed necrosis in acute brain injury. Comput. Struct. Biotechnol. J. 2015, 13, 212–221. [Google Scholar] [CrossRef] [Green Version]
- Teruya, R.; Ikejiri, A.T. Expression of oxidative stress and antioxidant defense genes in the kidney of inbred mice after intestinal ischemia and reperfusion. Acta Cir. Bras. 2013, 28, 848–855. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shin, J.; Berg, D.A. Single-Cell RNA-Seq with Waterfall Reveals Molecular Cascades underlying Adult Neurogenesis. Cell Stem Cell 2015, 17, 360–372. [Google Scholar] [CrossRef] [Green Version]
- Chiu, C.; Monat, C. SAPCD2 Controls Spindle Orientation and Asymmetric Divisions by Negatively Regulating the Gαi-LGN-NuMA Ternary Complex. Dev. Cell 2016, 36, 50–62. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ultanir, S.K.; Hertz, N.T. Chemical genetic identification of NDR1/2 kinase substrates AAK1 and Rabin8 Uncovers their roles in dendrite arborization and spine development. Neuron 2012, 73, 1127–1142. [Google Scholar] [CrossRef] [Green Version]
- Léger, H.; Santana, E. Ndr kinases regulate retinal interneuron proliferation and homeostasis. Sci. Rep. 2018, 8, 12544. [Google Scholar] [CrossRef]
- Klimek, C.; Jahnke, R. The Hippo network kinase STK38 contributes to protein homeostasis by inhibiting BAG3-mediated autophagy. Biochim. Biophys. Acta Mol. Cell Res. 2019, 1866, 1556–1566. [Google Scholar] [CrossRef] [PubMed]
- Kizuka, Y.; Kitazume, S. An aberrant sugar modification of BACE1 blocks its lysosomal targeting in Alzheimer’s disease. EMBO Mol. Med. 2015, 7, 175–189. [Google Scholar] [CrossRef] [PubMed]
- Shigeta, M.; Shibukawa, Y. beta1,4-N-Acetylglucosaminyltransferase III potentiates beta1 integrin-mediated neuritogenesis induced by serum deprivation in Neuro2a cells. Glycobiology 2006, 16, 564–571. [Google Scholar] [CrossRef] [PubMed]
- Alonso, A.D.; Grundke-Iqbal, I. Abnormal phosphorylation of tau and the mechanism of Alzheimer neurofibrillary degeneration: Sequestration of microtubule-associated proteins 1 and 2 and the disassembly of microtubules by the abnormal tau. Proc. Natl. Acad. Sci. USA 1997, 94, 298–303. [Google Scholar] [CrossRef] [Green Version]
- Rather, M.A.; Khan, A. Inflammation and Alzheimer’s Disease: Mechanisms and Therapeutic Implications by Natural Products. Mediators Inflamm. 2021, 2021, 9982954. [Google Scholar] [CrossRef]
- Chen, Y.Y.; Wang, M.C. Redox signaling and Alzheimer’s disease: From pathomechanism insights to biomarker discovery and therapy strategy. Biomark. Res. 2020, 8, 42. [Google Scholar] [CrossRef]
- Agrawal, I.; Jha, S. Mitochondrial Dysfunction and Alzheimer’s Disease: Role of Microglia. Front. Aging Neurosci. 2020, 12, 252. [Google Scholar] [CrossRef]
- D’Agostino, G.; Russo, R. Palmitoylethanolamide protects against the amyloid-25–35-induced learning and memory impairment in mice, an experimental model of Alzheimer disease. Neuropsychopharmacology 2012, 37, 1784–1792. [Google Scholar] [CrossRef] [Green Version]
- Farooq, S.M.; Boppana, N.B. C-phycocyanin confers protection against oxalate-mediated oxidative stress and mitochondrial dysfunctions in MDCK cells. PLoS ONE 2014, 9, e93056. [Google Scholar] [CrossRef]

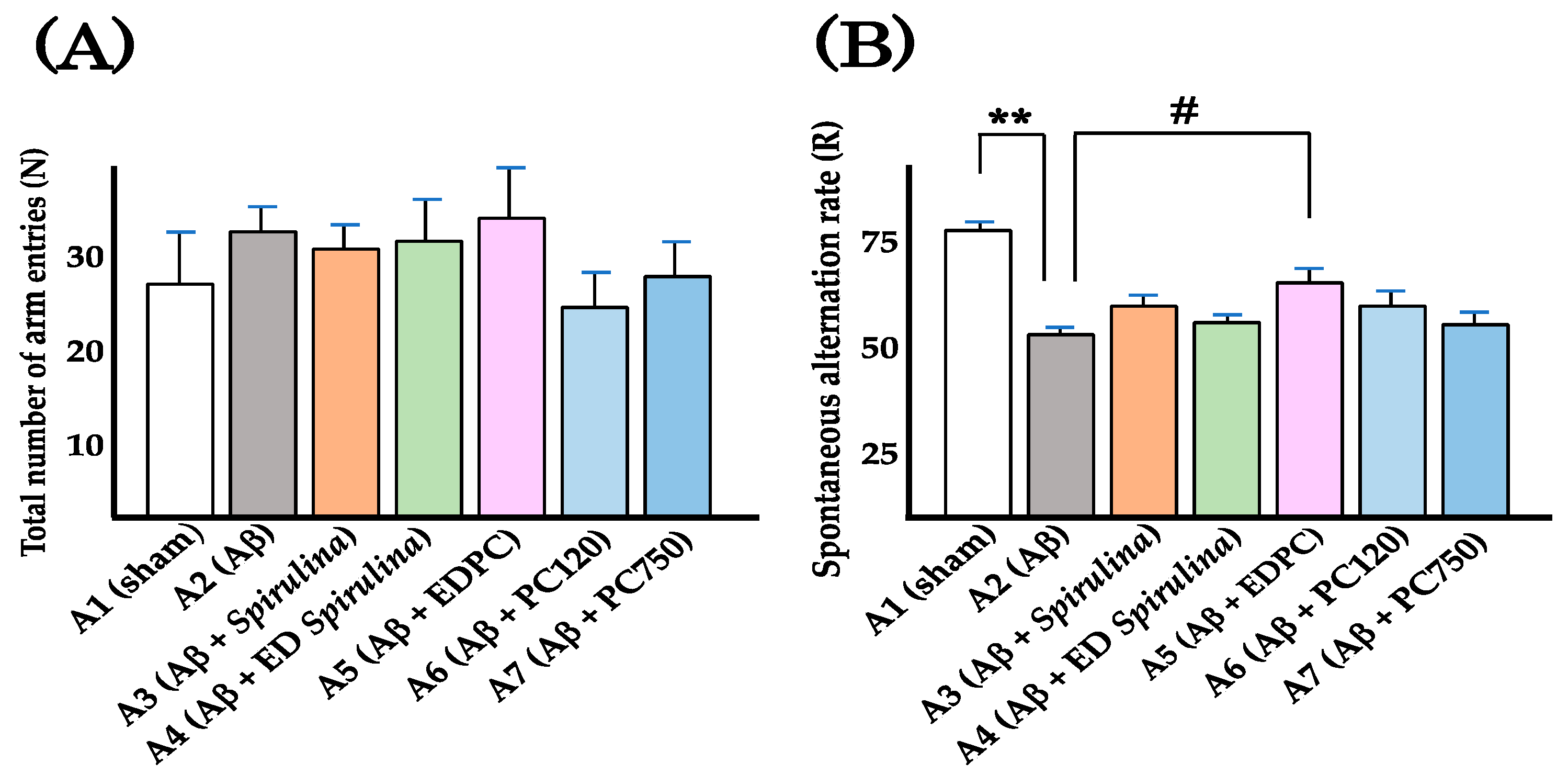

| Group | Oral Administration | I. C. V. Injection | Total Number of Arm Entries (N) | Number of Alternation (M) | R (%) 1 |
|---|---|---|---|---|---|
| A1 | Vehicle | Vehicle | 26.0 ± 5.7 | 17.8 ± 4.0 | 75.1 ± 2.5 |
| A2 | Vehicle | Aβ25–35 | 32.0 ± 2.5 | 15.3 ± 1.5 | 51.0 ± 2.1 2 |
| A3 | Spirulina | Aβ25–35 | 30.5 ± 2.5 | 16.5 ± 2.0 | 57.2 ± 3.7 |
| A4 | ED Spirulina | Aβ25–35 | 31.0 ± 3.4 | 16.0 ± 2.5 | 53.9 ± 2.5 |
| A5 | EDPC | Aβ25–35 | 33.4 ± 5.7 | 19.2 ± 3.3 | 61.8 ± 3.7 3 |
| A6 | PC (120 mg/kg) | Aβ25–35 | 24.5 ± 3.2 | 13.0 ± 2.4 | 56.6 ± 4.5 |
| A7 | PC (750 mg/kg) | Aβ25–35 | 26.8 ± 3.9 | 12.7 ± 2.2 | 51.6 ± 3.8 |
| Genes | Gene Product | Fold Change 1 | Reference | ||
|---|---|---|---|---|---|
| Aβ 2 | Aβ + EDPC 3 | Aβ + PC 4 | |||
| Cd9 | Cd9 antigen | 3.0 | 3.2 | 4.2 | [12] |
| Ctsz | Cathepsin Z | 2.1 | 2.4 | 2.6 | [12] |
| Trem2 | Triggering receptor expressed on myeloid cells 2 | 3.1 | 2.7 | 3.8 | [12] |
| Igf1 | Insulin-like growth factor 1 | 3.5 | 2.3 | 3.6 | [13] |
| Serpine1 | Serine (or cysteine) peptidase inhibitor, clade E, member 1 | 4.2 | 4.7 | 3.2 | [13] |
| Egfr | Epidermal growth factor receptor | 3.2 | 2.6 | 3.1 | [13] |
| Nfe2l2 | Nuclear factor, erythroid derived, like 2 (also known as Nrf2) | 3.0 | 2.3 | 3.7 | [13] |
| Gene | Product/Description | Fold Change | Function [Reference] | RPKM 1 | ||||
|---|---|---|---|---|---|---|---|---|
| Aβ | Aβ + EDPC | Aβ + PC | CNS E11.5 | CNS E14 | CNS E18 | |||
| Fbxl19 | F-box and leucine-rich repeat protein19 | 2.2 | 1.0 | 1.0 | Ubiquitination [14] | 26.6 | 25.5 | 23.7 |
| Pax1 | Paired box gene 1 | 2.9 | 1.2 | 1.4 | Regulation of transcription [15] | 2.4 | 0.4 | 0.0 |
| Zfp292 | Zinc finger protein 292 | 14.9 | 2.5 | 1.5 | Regulation of transcription [16] | 6.0 | 5.7 | 4.2 |
| Gene | Product/Description | Aβ | Aβ + EDPC | Aβ + PC | Function [Reference] |
|---|---|---|---|---|---|
| Abat * | 4-Aminobutyrate aminotransferase | 4.0 | 2.9 | 1.8 | Salvage reaction Regulation of GABA [17] |
| Brp44 (Mpc2) * | Mitochondrial pyruvate carrier 2 | 9.1 | 5.5 | 3.5 | TCA cycle [18] |
| Gpr123 (Adgra1) | G protein-coupled receptor 123 | 7.2 | 4.7 | 3.2 | Excitatory neuron |
| Bsph1 | Binder of sperm protein homolog 1 | 2.0 | 1.5 | 0.9 | Sperm |
| Butr1 (Btnl10) | Butyrophilin related 1 | 2.4 | 1.6 | 1.0 | Putative ligand |
| Casp8ap2 | Caspase 8 associated protein 2 | 2.3 | 1.5 | 1.1 | Apoptosis |
| G6pc3 | Glucose 6 phosphatase, catalytic, 3 | 2.3 | 1.4 | 1.0 | Gluconeogenesis |
| Gucy2e | Guanylate cyclase 2e | 2.3 | 1.5 | 1.0 | Generation of cGMP |
| Irf2bpl | Interferon regulatory factor 2 binding protein-like | 2.1 | 1.4 | 0.8 | Neuronal network |
| Mnt | Max binding protein | 2.1 | 1.5 | 1.0 | Regulation of transcription |
| Pxk (MONaKA) | PX domain containing serine/threonine kinase | 2.9 | 1.8 | 1.3 | Transport of Na+ and K+ |
| Slx4ip | SLX4 interacting protein | 3.9 | 2.9 | 1.9 | DNA repair |
| Zcchc24 | Zinc finger, CCHC domain containing 24 | 2.1 | 2.0 | 0.6 | Regulation of transcription |
| Gene | Product/Description | Aβ | Aβ + EDPC | Aβ + PC | Function [Reference] |
|---|---|---|---|---|---|
| Prnp | Prion protein | 0.2 | 1.6 | 2.1 | Prion protein [19] Autophagy [20] |
| Cct4 | Chaperonin containing Tcp1, subunit 4 | 0.2 | 1.5 | 2.1 | Protein folding Dendrite morphogenesis [21] |
| Figf (Vegfd) | c-Fos induced growth factor | 0.02 | 0.04 | 0.02 | Angiogenesis Protection of neurons [22] |
| Gal3st1 (Cst) | Galactose-3-O-sulfotransferase | 0.03 | 0.11 | 0.03 | Myelin membrane fluidity Risk factor of CJD [23] |
| Olfr181 | Olfactory receptor 181 | 0.29 | 0.73 | 0.54 | Olfactory receptor [24] |
| Olfr847 | Olfactory receptor 847 | 0.05 | 0.11 | 0.05 | Olfactory receptor [24] |
| Olfr859 | Olfactory receptor 859 | 0.2 | 1.2 | 1.6 | Olfactory receptor [24] |
| Olfr963 | Olfactory receptor 963 | 0.04 | 0.10 | 0.04 | Olfactory receptor [24] |
| Gene | Product/Description | Aβ | Aβ + EDPC | Aβ +PC | Function [Reference] |
|---|---|---|---|---|---|
| Ccd163 | Coiled-coil domain containing 163 | 0.4 | 1.5 | 1.9 | Wake [25] |
| Kcnj12 | Potassium inwardly rectifying channel, subfamily J, number 12 | 0.2 | 0.8 | 0.7 | Action potential |
| Lamb4 | Laminin subunit beta 4 | 0.3 | 0.8 | 0.6 | Laminin |
| Mtap9 (Map9) | Microtubule-associated protein 9 | 0.3 | 1.5 | 2.3 | Microtubule Cell cycle [26] |
| Pik3cg | Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit gamma | 0.1 | 0.4 | 0.3 | Microglia [27] Induction of ER stress, synaptic plasticity [28] |
| Sema3b | SEMA domain, immunoglobulin domain (Ig), short basic domain, secreted 3B | 0.3 | 0.7 | 0.5 | Removal of spines of dendrites [29] |
| Tmem82 | Transmembrane protein 82 | 0.4 | 1.1 | 0.9 | Cell proliferation [30] |
| Zfand5 | Zinc finger, AN1-type domain 5 | 0.2 | 0.7 | 0.3 | Inflammation [31] Apoptosis [32] |
| Gm 14439 | Predicted gene 14439 | 0.1 | 0.1 | 0.2 | Ribosomal protein |
| LOC100503859 | RIKEN cDNA 1110015O18 gene | 0.3 | 1.0 | 0.8 | Unknown |
| D330050I16Rik | RIKEN cDNA D330050 gene | 0.3 | 0.7 | 0.6 | Unknown |
| Gene | Product/Description | Aβ | Aβ + EDPC | Aβ +PC | Function [Reference] |
|---|---|---|---|---|---|
| Cd244 | CD244 natural killer receptor 2B4 | 4.0 | 2.9 | 1.8 | NK cells Microglia [33] |
| Chst9 | Carbohydrate (N-acetylgalactosamine4-O) sulfotransferase 9 | 0.03 | 0.11 | 0.05 | Modification of pro-opiomelanocortin [34] |
| Endog | Endonuclease G | 0.38 | 0.86 | 0.58 | Apoptosis/Necrosis [35] |
| Hbq1a | Hemoglobin theta 1A | 0.03 | 0.09 | 0.04 | Oxidative stress [36] |
| Hopx | HOP homeobox | 0.48 | 0.97 | 0.66 | Neural stem cells [37] |
| LOC547349 | H2-K1 | 0.09 | 0.23 | 0.12 | Self-recognition |
| Proser2 | Proline and serine rich 2 | 0.37 | 1.04 | 0.70 | Unknown (Tumor marker) |
| Rnmt | RNA (guanine-7-)methyltransferase | 0.12 | 0.24 | 0.16 | 5′-Cap structure of mRNA |
| Sapcd2 | Suppressor APC domain containing 2 | 0.03 | 0.10 | 0.05 | Migration Spindle [38] |
| Stk38 | Serine/threonine kinase 38 | 0.04 | 0.10 | 0.04 | Spines of dendrites [39] Retinal neurons [40] Mitophagy [41] |
| Trhde | TRH degrading enzyme | 0.03 | 0.11 | 0.05 | Degradation of TRH |
| Vmn1r62 | Vomeronasal 1 receptor 62 | 0.03 | 0.11 | 0.04 | Pheromone receptor |
| U90926 | Putative incRNA | 0.04 | 0.10 | 0.03 | Putative incRNA |
| Gene | Product/Description | Aβ | Aβ + EDPC | Aβ + PC | Function [Reference] |
|---|---|---|---|---|---|
| Mgat3 * | Mannoside acetylglucosaminyltransferase 3 (GnT-III) | 0.01 | 0.01 | 0.04 | Modification of BACE1 [42] Neuritogenesis [43] |
| Cela3b | Chymotrypsin-like elastase family, member 3B | 0.48 | 0.70 | 1.0 | Protease |
| Cipc | CLOCK interacting protein, circadian | 0.37 | 0.54 | 1.0 | Cell cycle |
| Ybx1 | Y-box binding protein 1/DNA binding protein B | 0.39 | 0.72 | 0.79 | Transcription and translation |
| G430095P16 | Long non-coding RNA | 0.08 | 0.13 | 0.17 | Non-coding RNA |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Imai, Y.; Koseki, Y.; Hirano, M.; Nakamura, S. Nutrigenomic Studies on the Ameliorative Effect of Enzyme-Digested Phycocyanin in Alzheimer’s Disease Model Mice. Nutrients 2021, 13, 4431. https://doi.org/10.3390/nu13124431
Imai Y, Koseki Y, Hirano M, Nakamura S. Nutrigenomic Studies on the Ameliorative Effect of Enzyme-Digested Phycocyanin in Alzheimer’s Disease Model Mice. Nutrients. 2021; 13(12):4431. https://doi.org/10.3390/nu13124431
Chicago/Turabian StyleImai, Yasuyuki, Yurino Koseki, Makoto Hirano, and Shin Nakamura. 2021. "Nutrigenomic Studies on the Ameliorative Effect of Enzyme-Digested Phycocyanin in Alzheimer’s Disease Model Mice" Nutrients 13, no. 12: 4431. https://doi.org/10.3390/nu13124431
APA StyleImai, Y., Koseki, Y., Hirano, M., & Nakamura, S. (2021). Nutrigenomic Studies on the Ameliorative Effect of Enzyme-Digested Phycocyanin in Alzheimer’s Disease Model Mice. Nutrients, 13(12), 4431. https://doi.org/10.3390/nu13124431





