Associations of Arachidonic Acid Synthesis with Cardiovascular Risk Factors and Relation to Ischemic Heart Disease and Stroke: A Univariable and Multivariable Mendelian Randomization Study
Abstract
:1. Introduction
2. Materials and Methods
2.1. Genetic Instruments for AA Synthesis
2.2. Genetic Associations with Risk Factors for CVD
2.3. Genetic Associations with IHD and Ischemic Stroke
2.4. Statistical Analysis
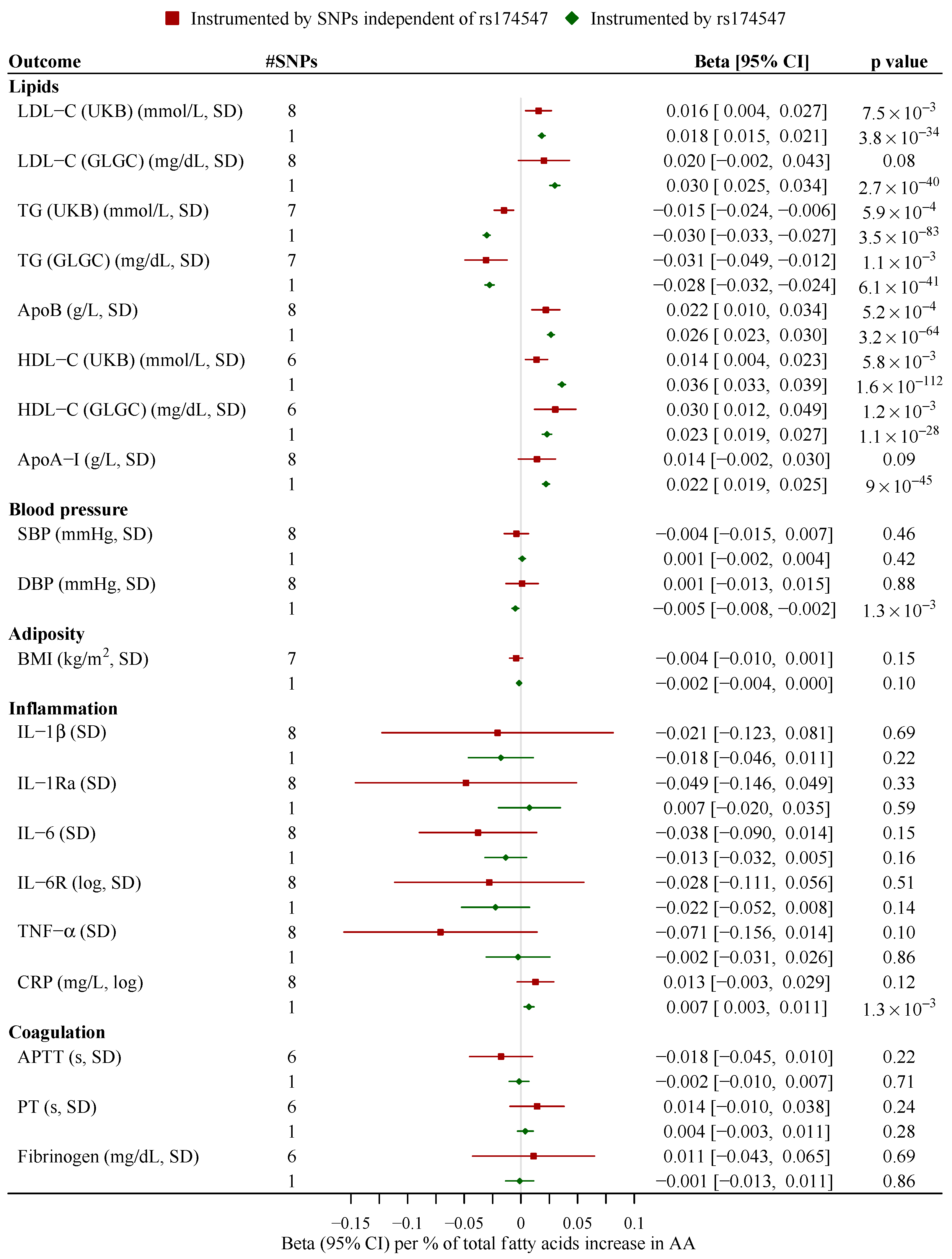
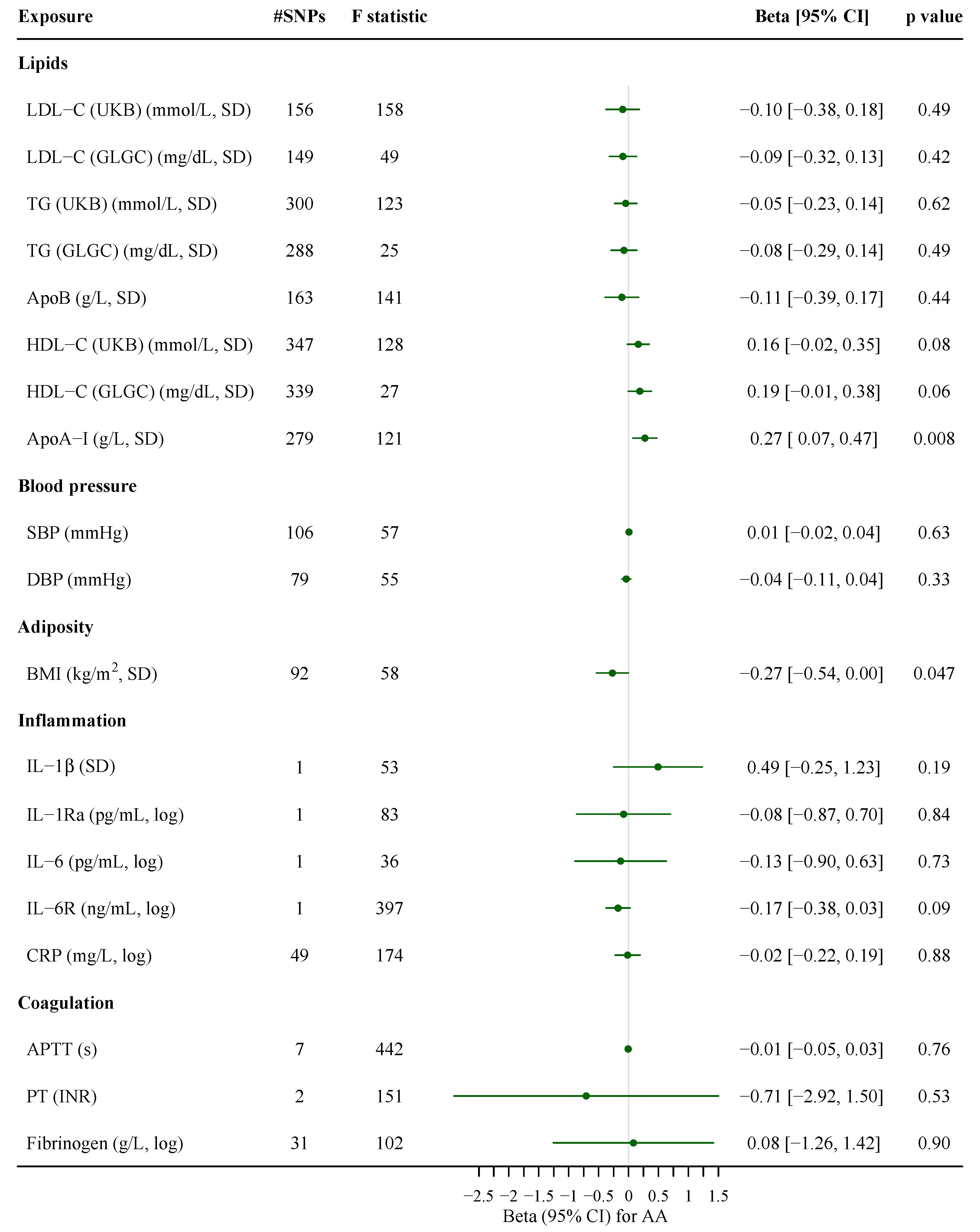
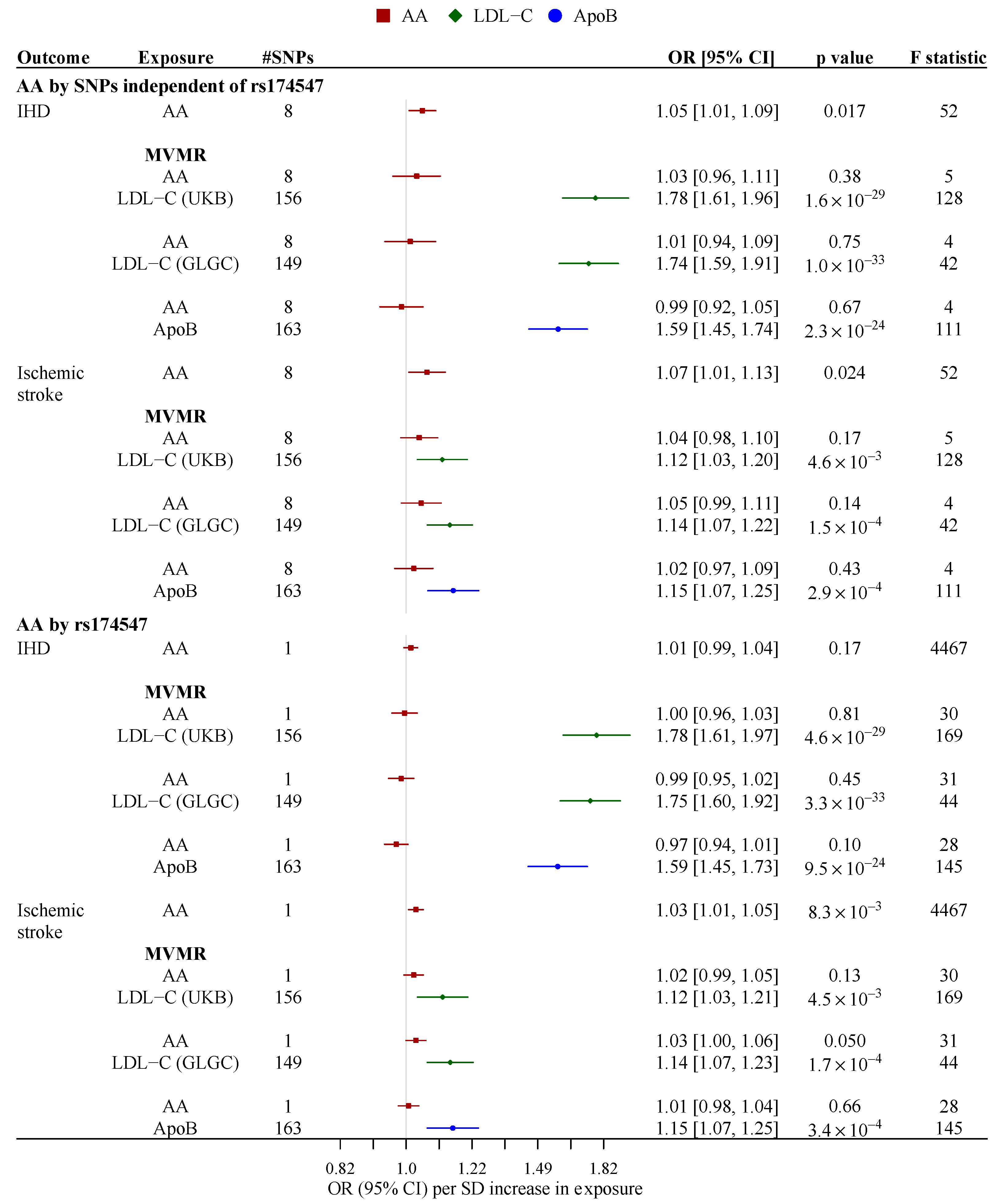
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
List of Abbreviations
| Abbreviation | Full Name |
| AA | arachidonic acid |
| ApoA-I | apolipoprotein A-I |
| ApoB | apolipoprotein B |
| APTT | activated partial thromboplastin time |
| BBJ | the Biobank Japan |
| BMI | body mass index |
| CHARGE | Cohorts for Heart and Aging Research in Genomic Epidemiology |
| CI | confidence interval |
| CIWG | the Cohorts for Heart and Aging Research in Genomic Epidemiology Inflammation Working Group |
| CRP | C-reactive protein |
| CVD | cardiovascular disease |
| GWAS | genome-wide association study |
| DBP | diastolic blood pressure |
| DHA | docosahexaenoic acid |
| EPA | eicosapentaenoic acid |
| GIANT | the Genetic Investigation of ANthropometric Traits Consortium |
| GLGC | the Global Lipids Genetics Consortium |
| HDL-C | high-density lipoprotein cholesterol |
| IHD | ischemic heart disease |
| IL-1β | interleukin-1 β |
| IL-1Ra | interleukin-1 receptor antagonist |
| IL-6 | interleukin-6 |
| IL-6R | IL-6 receptor |
| IVW | inverse variance-weighted |
| LDL-C | low-density lipoprotein cholesterol |
| MR | Mendelian randomization |
| OR | odds ratio |
| PT | prothrombin time |
| PUFA | polyunsaturated fatty acid |
| RCT | randomized controlled trial |
| SBP | systolic blood pressure |
| SD | standard deviation |
| SNP | single-nucleotide polymorphisms |
| TG | triglycerides |
| TNF-α | tumor necrosis factor-α |
| UKB | UK Biobank |
References
- Keys, A. Diet and the epidemiology of coronary heart disease. J. Am. Med. Assoc. 1957, 164, 1912–1919. [Google Scholar] [CrossRef]
- US Senate Select Committee on Nutrition and Human Needs. Dietary Goals for the United States, 1st ed.; US Government Printing Office: Washington, DC, USA, 1977.
- US Department of Health and Human Services and U.S. 2015–2020 Dietary Guidelines for Americans, 8th ed. Available online: https://health.gov/dietaryguidelines/2015/guidelines/ (accessed on 10 January 2021).
- Bhatt, D.L.; Steg, P.G.; Miller, M.; Brinton, E.A.; Jacobson, T.A.; Ketchum, S.B.; Doyle, R.T., Jr.; Juliano, R.A.; Jiao, L.; Granowitz, C.; et al. Cardiovascular Risk Reduction with Icosapent Ethyl for Hypertriglyceridemia. N. Engl. J. Med. 2019, 380, 11–22. [Google Scholar] [CrossRef]
- Aung, T.; Halsey, J.; Kromhout, D.; Gerstein, H.C.; Marchioli, R.; Tavazzi, L.; Geleijnse, J.M.; Rauch, B.; Ness, A.; Galan, P.; et al. Associations of Omega-3 Fatty Acid Supplement Use With Cardiovascular Disease Risks: Meta-analysis of 10 Trials Involving 77917 Individuals. JAMA Cardiol. 2018, 3, 225–234. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kawashima, H. Intake of arachidonic acid-containing lipids in adult humans: Dietary surveys and clinical trials. Lipids Health Dis. 2019, 18, 101. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Piper, K.; Garelnabi, M. Eicosanoids: Atherosclerosis and cardiometabolic health. J. Clin. Transl. Endocrinol. 2020, 19, 100216. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Zhao, J.V.; Schooling, C.M. The associations of plasma phospholipid arachidonic acid with cardiovascular diseases: A Mendelian randomization study. EBioMedicine 2021, 63, 103189. [Google Scholar] [CrossRef] [PubMed]
- Yuan, S.; Back, M.; Bruzelius, M.; Mason, A.M.; Burgess, S.; Larsson, S. Plasma Phospholipid Fatty Acids, FADS1 and Risk of 15 Cardiovascular Diseases: A Mendelian Randomisation Study. Nutrients 2019, 11, 3001. [Google Scholar] [CrossRef] [Green Version]
- Yuan, T.; Si, S.; Li, Y.; Li, W.; Chen, X.; Liu, C.; Li, J.; Wang, B.; Hou, L.; Liu, Y.; et al. Roles for circulating polyunsaturated fatty acids in ischemic stroke and modifiable factors: A Mendelian randomization study. Nutr. J. 2020, 19, 70. [Google Scholar] [CrossRef] [PubMed]
- Motoyama, K.R.; Curb, J.D.; Kadowaki, T.; El-Saed, A.; Abbott, R.D.; Okamura, T.; Evans, R.W.; Nakamura, Y.; Sutton-Tyrrell, K.; Rodriquez, B.L.; et al. Association of serum n-6 and n-3 polyunsaturated fatty acids with lipids in 3 populations of middle-aged men. Am. J. Clin. Nutr. 2009, 90, 49–55. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ferrucci, L.; Cherubini, A.; Bandinelli, S.; Bartali, B.; Corsi, A.; Lauretani, F.; Martin, A.; Andres-Lacueva, C.; Senin, U.; Guralnik, J.M. Relationship of plasma polyunsaturated fatty acids to circulating inflammatory markers. J. Clin. Endocrinol. Metab. 2006, 91, 439–446. [Google Scholar] [CrossRef]
- Markworth, J.F.; Mitchell, C.J.; D’Souza, R.F.; Aasen, K.M.M.; Durainayagam, B.R.; Mitchell, S.M.; Chan, A.H.C.; Sinclair, A.J.; Garg, M.; Cameron-Smith, D. Arachidonic acid supplementation modulates blood and skeletal muscle lipid profile with no effect on basal inflammation in resistance exercise trained men. Prostaglandinsleukotrienesand Essent. Fat. Acids 2018, 128, 74–86. [Google Scholar] [CrossRef]
- Kusumoto, A.; Ishikura, Y.; Kawashima, H.; Kiso, Y.; Takai, S.; Miyazaki, M. Effects of arachidonate-enriched triacylglycerol supplementation on serum fatty acids and platelet aggregation in healthy male subjects with a fish diet. Br. J. Nutr. 2007, 98, 626–635. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nelson, G.J.; Schmidt, P.C.; Bartolini, G.; Kelley, D.S.; Phinney, S.D.; Kyle, D.; Silbermann, S.; Schaefer, E.J. The effect of dietary arachidonic acid on plasma lipoprotein distributions, apoproteins, blood lipid levels, and tissue fatty acid composition in humans. Lipids 1997, 32, 427–433. [Google Scholar] [CrossRef]
- Kakutani, S.; Ishikura, Y.; Tateishi, N.; Horikawa, C.; Tokuda, H.; Kontani, M.; Kawashima, H.; Sakakibara, Y.; Kiso, Y.; Shibata, H.; et al. Supplementation of arachidonic acid-enriched oil increases arachidonic acid contents in plasma phospholipids, but does not increase their metabolites and clinical parameters in Japanese healthy elderly individuals: A randomized controlled study. Lipids Health Dis. 2011, 10, 241. [Google Scholar] [CrossRef] [Green Version]
- Roberts, M.D.; Iosia, M.; Kerksick, C.M.; Taylor, L.W.; Campbell, B.; Wilborn, C.D.; Harvey, T.; Cooke, M.; Rasmussen, C.; Greenwood, M.; et al. Effects of arachidonic acid supplementation on training adaptations in resistance-trained males. J. Int. Soc. Sports Nutr. 2007, 4, 21. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Seyberth, H.W.; Oelz, O.; Kennedy, T.; Sweetman, B.J.; Danon, A.; Frölich, J.C.; Heimberg, M.; Oates, J.A. Increased arachidonate in lipids after administration to man: Effects on prostaglandin biosynthesis. Clin. Pharm. 1975, 18, 521–529. [Google Scholar] [CrossRef]
- Pantaleo, P.; Marra, F.; Vizzutti, F.; Spadoni, S.; Ciabattoni, G.; Galli, C.; La Villa, G.; Gentilini, P.; Laffi, G. Effects of dietary supplementation with arachidonic acid on platelet and renal function in patients with cirrhosis. Clin. Sci. 2004, 106, 27–34. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nelson, G.J.; Schmidt, P.C.; Bartolini, G.; Kelley, D.S.; Kyle, D. The effect of dietary arachidonic acid on platelet function, platelet fatty acid composition, and blood coagulation in humans. Lipids 1997, 32, 421–425. [Google Scholar] [CrossRef]
- Tucci, S.; Vohr, S.H.; McCoy, R.C.; Vernot, B.; Robinson, M.R.; Barbieri, C.; Nelson, B.J.; Fu, W.; Purnomo, G.A.; Sudoyo, H.; et al. Evolutionary history and adaptation of a human pygmy population of Flores Island, Indonesia. Science 2018, 361, 511–516. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Richardson, T.G.; Sanderson, E.; Palmer, T.M.; Ala-Korpela, M.; Ference, B.A.; Davey Smith, G.; Holmes, M.V. Evaluating the relationship between circulating lipoprotein lipids and apolipoproteins with risk of coronary heart disease: A multivariable Mendelian randomisation analysis. PLoS Med. 2020, 17, e1003062. [Google Scholar] [CrossRef] [Green Version]
- Yuan, S.; Tang, B.; Zheng, J.; Larsson, S.C. Circulating Lipoprotein Lipids, Apolipoproteins and Ischemic Stroke. Ann. Neurol. 2020, 88, 1229–1236. [Google Scholar] [CrossRef]
- Guan, W.; Steffen, B.T.; Lemaitre, R.N.; Wu, J.H.Y.; Tanaka, T.; Manichaikul, A.; Foy, M.; Rich, S.S.; Wang, L.; Nettleton, J.A.; et al. Genome-wide association study of plasma N6 polyunsaturated fatty acids within the cohorts for heart and aging research in genomic epidemiology consortium. Circ. Cardiovasc. Genet. 2014, 7, 321–331. [Google Scholar] [CrossRef] [Green Version]
- Bycroft, C.; Freeman, C.; Petkova, D.; Band, G.; Elliott, L.T.; Sharp, K.; Motyer, A.; Vukcevic, D.; Delaneau, O.; O’Connell, J.; et al. The UK Biobank resource with deep phenotyping and genomic data. Nature 2018, 562, 203–209. [Google Scholar] [CrossRef] [Green Version]
- Willer, C.J.; Schmidt, E.M.; Sengupta, S.; Peloso, G.M.; Gustafsson, S.; Kanoni, S.; Ganna, A.; Chen, J.; Buchkovich, M.L.; Mora, S.; et al. Discovery and refinement of loci associated with lipid levels. Nat. Genet. 2013, 45, 1274–1283. [Google Scholar] [CrossRef] [Green Version]
- Yengo, L.; Sidorenko, J.; Kemper, K.E.; Zheng, Z.; Wood, A.R.; Weedon, M.N.; Frayling, T.M.; Hirschhorn, J.; Yang, J.; Visscher, P.M. Meta-analysis of genome-wide association studies for height and body mass index in ∼700000 individuals of European ancestry. Hum. Mol. Genet. 2018, 27, 3641–3649. [Google Scholar] [CrossRef]
- Ahola-Olli, A.V.; Würtz, P.; Havulinna, A.S.; Aalto, K.; Pitkänen, N.; Lehtimäki, T.; Kähönen, M.; Lyytikäinen, L.P.; Raitoharju, E.; Seppälä, I.; et al. Genome-wide Association Study Identifies 27 Loci Influencing Concentrations of Circulating Cytokines and Growth Factors. Am. J. Hum. Genet. 2017, 100, 40–50. [Google Scholar] [CrossRef] [Green Version]
- Kalaoja, M.; Corbin, L.J.; Tan, V.Y.; Ahola-Olli, A.V.; Havulinna, A.S.; Santalahti, K.; Pitkänen, N.; Lehtimäki, T.; Lyytikäinen, L.P.; Raitoharju, E.; et al. The Role of Inflammatory Cytokines as Intermediates in the Pathway from Increased Adiposity to Disease. Obesity (Silver Spring) 2021, 29, 428–437. [Google Scholar] [CrossRef]
- Sun, B.B.; Maranville, J.C.; Peters, J.E.; Stacey, D.; Staley, J.R.; Blackshaw, J.; Burgess, S.; Jiang, T.; Paige, E.; Surendran, P.; et al. Genomic atlas of the human plasma proteome. Nature 2018, 558, 73–79. [Google Scholar] [CrossRef]
- Ligthart, S.; Vaez, A.; Võsa, U.; Stathopoulou, M.G.; de Vries, P.S.; Prins, B.P.; Van der Most, P.J.; Tanaka, T.; Naderi, E.; Rose, L.M.; et al. Genome Analyses of >200,000 Individuals Identify 58 Loci for Chronic Inflammation and Highlight Pathways that Link Inflammation and Complex Disorders. Am. J. Hum. Genet. 2018, 103, 691–706. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kanai, M.; Akiyama, M.; Takahashi, A.; Matoba, N.; Momozawa, Y.; Ikeda, M.; Iwata, N.; Ikegawa, S.; Hirata, M.; Matsuda, K.; et al. Genetic analysis of quantitative traits in the Japanese population links cell types to complex human diseases. Nat. Genet. 2018, 50, 390–400. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Evangelou, E.; Warren, H.R.; Mosen-Ansorena, D.; Mifsud, B.; Pazoki, R.; Gao, H.; Ntritsos, G.; Dimou, N.; Cabrera, C.P.; Karaman, I.; et al. Genetic analysis of over 1 million people identifies 535 new loci associated with blood pressure traits. Nat. Genet. 2018, 50, 1412–1425. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Locke, A.E.; Kahali, B.; Berndt, S.I.; Justice, A.E.; Pers, T.H.; Day, F.R.; Powell, C.; Vedantam, S.; Buchkovich, M.L.; Yang, J.; et al. Genetic studies of body mass index yield new insights for obesity biology. Nature 2015, 518, 197–206. [Google Scholar] [CrossRef] [Green Version]
- Sliz, E.; Kalaoja, M.; Ahola-Olli, A.; Raitakari, O.; Perola, M.; Salomaa, V.; Lehtimäki, T.; Karhu, T.; Viinamäki, H.; Salmi, M.; et al. Genome-wide association study identifies seven novel loci associating with circulating cytokines and cell adhesion molecules in Finns. J. Med. Genet. 2019, 56, 607–616. [Google Scholar] [CrossRef] [Green Version]
- Herder, C.; Nuotio, M.L.; Shah, S.; Blankenberg, S.; Brunner, E.J.; Carstensen, M.; Gieger, C.; Grallert, H.; Jula, A.; Kähönen, M.; et al. Genetic determinants of circulating interleukin-1 receptor antagonist levels and their association with glycemic traits. Diabetes 2014, 63, 4343–4359. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Swerdlow, D.I.; Holmes, M.V.; Kuchenbaecker, K.B.; Engmann, J.E.; Shah, T.; Sofat, R.; Guo, Y.; Chung, C.; Peasey, A.; Pfister, R.; et al. The interleukin-6 receptor as a target for prevention of coronary heart disease: A mendelian randomisation analysis. Lancet 2012, 379, 1214–1224. [Google Scholar] [CrossRef] [Green Version]
- Sarwar, N.; Butterworth, A.S.; Freitag, D.F.; Gregson, J.; Willeit, P.; Gorman, D.N.; Gao, P.; Saleheen, D.; Rendon, A.; Nelson, C.P.; et al. Interleukin-6 receptor pathways in coronary heart disease: A collaborative meta-analysis of 82 studies. Lancet 2012, 379, 1205–1213. [Google Scholar] [CrossRef] [Green Version]
- Tang, W.; Schwienbacher, C.; Lopez, L.M.; Ben-Shlomo, Y.; Oudot-Mellakh, T.; Johnson, A.D.; Samani, N.J.; Basu, S.; Gögele, M.; Davies, G.; et al. Genetic associations for activated partial thromboplastin time and prothrombin time, their gene expression profiles, and risk of coronary artery disease. Am. J. Hum. Genet. 2012, 91, 152–162. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- de Vries, P.S.; Chasman, D.I.; Sabater-Lleal, M.; Chen, M.H.; Huffman, J.E.; Steri, M.; Tang, W.; Teumer, A.; Marioni, R.E.; Grossmann, V.; et al. A meta-analysis of 120 246 individuals identifies 18 new loci for fibrinogen concentration. Hum. Mol. Genet. 2016, 25, 358–370. [Google Scholar] [CrossRef]
- Nikpay, M.; Goel, A.; Won, H.H.; Hall, L.M.; Willenborg, C.; Kanoni, S.; Saleheen, D.; Kyriakou, T.; Nelson, C.P.; Hopewell, J.C.; et al. A comprehensive 1,000 Genomes-based genome-wide association meta-analysis of coronary artery disease. Nat. Genet. 2015, 47, 1121–1130. [Google Scholar] [CrossRef] [Green Version]
- Malik, R.; Chauhan, G.; Traylor, M.; Sargurupremraj, M.; Okada, Y.; Mishra, A.; Rutten-Jacobs, L.; Giese, A.K.; van der Laan, S.W.; Gretarsdottir, S.; et al. Multiancestry genome-wide association study of 520,000 subjects identifies 32 loci associated with stroke and stroke subtypes. Nat. Genet. 2018, 50, 524–537. [Google Scholar] [CrossRef] [Green Version]
- Bowden, J.; Del Greco, M.F.; Minelli, C.; Davey Smith, G.; Sheehan, N.A.; Thompson, J.R. Assessing the suitability of summary data for two-sample Mendelian randomization analyses using MR-Egger regression: The role of the I2 statistic. Int. J. Epidemiol. 2016, 45, 1961–1974. [Google Scholar] [CrossRef] [Green Version]
- Burgess, S.; Butterworth, A.; Thompson, S.G. Mendelian randomization analysis with multiple genetic variants using summarized data. Genet. Epidemiol. 2013, 37, 658–665. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Burgess, S.; Thompson, S.G. Interpreting findings from Mendelian randomization using the MR-Egger method. Eur. J. Epidemiol. 2017, 32, 377–389. [Google Scholar] [CrossRef] [Green Version]
- Yamagishi, K.; Nettleton, J.A.; Folsom, A.R. Plasma fatty acid composition and incident heart failure in middle-aged adults: The Atherosclerosis Risk in Communities (ARIC) Study. Am. Heart J. 2008, 156, 965–974. [Google Scholar] [CrossRef] [Green Version]
- Sanderson, E.; Davey Smith, G.; Windmeijer, F.; Bowden, J. An examination of multivariable Mendelian randomization in the single-sample and two-sample summary data settings. Int. J. Epidemiol. 2019, 48, 713–727. [Google Scholar] [CrossRef] [Green Version]
- Rees, J.M.B.; Wood, A.M.; Burgess, S. Extending the MR-Egger method for multivariable Mendelian randomization to correct for both measured and unmeasured pleiotropy. Stat. Med. 2017, 36, 4705–4718. [Google Scholar] [CrossRef] [Green Version]
- Burgess, S. Sample size and power calculations in Mendelian randomization with a single instrumental variable and a binary outcome. Int. J. Epidemiol. 2014, 43, 922–929. [Google Scholar] [CrossRef]
- Schwartz, G.G.; Olsson, A.G.; Abt, M.; Ballantyne, C.M.; Barter, P.J.; Brumm, J.; Chaitman, B.R.; Holme, I.M.; Kallend, D.; Leiter, L.A.; et al. Effects of dalcetrapib in patients with a recent acute coronary syndrome. N. Engl. J. Med. 2012, 367, 2089–2099. [Google Scholar] [CrossRef] [Green Version]
- Karjalainen, M.K.; Holmes, M.V.; Wang, Q.; Anufrieva, O.; Kähönen, M.; Lehtimäki, T.; Havulinna, A.S.; Kristiansson, K.; Salomaa, V.; Perola, M.; et al. Apolipoprotein A-I concentrations and risk of coronary artery disease: A Mendelian randomization study. Atherosclerosis 2020, 299, 56–63. [Google Scholar] [CrossRef]
- Choo, J.; Ueshima, H.; Curb, J.D.; Shin, C.; Evans, R.W.; El-Saed, A.; Kadowaki, T.; Okamura, T.; Nakata, K.; Otake, T.; et al. Serum n-6 fatty acids and lipoprotein subclasses in middle-aged men: The population-based cross-sectional ERA-JUMP study. Am. J. Clin. Nutr. 2010, 91, 1195–1203. [Google Scholar] [CrossRef] [Green Version]
- Leiviskä, J.; Sundvall, J.; Alfthan, G.; Jauhiainen, M.; Salomaa, V. Apolipoprotein A-I, apolipoprotein B, and apolipoprotein B/apolipoprotein A-I ratio: Reference intervals compared with values in different pathophysiological conditions from the FINRISK 2007 study. Clin. Chim. Acta 2011, 412, 1146–1150. [Google Scholar] [CrossRef]
- Arrol, S.; Mackness, M.I.; Durrington, P.N. The effects of fatty acids on apolipoprotein B secretion by human hepatoma cells (HEP G2). Atherosclerosis 2000, 150, 255–264. [Google Scholar] [CrossRef]
- Thies, F.; Miles, E.A.; Nebe-von-Caron, G.; Powell, J.R.; Hurst, T.L.; Newsholme, E.A.; Calder, P.C. Influence of dietary supplementation with long-chain n-3 or n-6 polyunsaturated fatty acids on blood inflammatory cell populations and functions and on plasma soluble adhesion molecules in healthy adults. Lipids 2001, 36, 1183–1193. [Google Scholar] [CrossRef]
- Yuan, S.; Larsson, S.C. Association of genetic variants related to plasma fatty acids with type 2 diabetes mellitus and glycaemic traits: A Mendelian randomisation study. Diabetologia 2020, 63, 116–123. [Google Scholar] [CrossRef] [Green Version]
- Lopez, P.M.; Subramanian, S.V.; Schooling, C.M. Effect measure modification conceptualized using selection diagrams as mediation by mechanisms of varying population-level relevance. J. Clin. Epidemiol. 2019, 113, 123–128. [Google Scholar] [CrossRef]
- Schooling, C.M.; Lopez, P.M.; Yang, Z.; Zhao, J.V.; Au Yeung, S.L.; Huang, J. Use of multivariable Mendelian randomization to address biases due to competing risk before recruitment. Front. Genet. 2020. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, T.; Au Yeung, S.-L.; Schooling, C.M. Associations of Arachidonic Acid Synthesis with Cardiovascular Risk Factors and Relation to Ischemic Heart Disease and Stroke: A Univariable and Multivariable Mendelian Randomization Study. Nutrients 2021, 13, 1489. https://doi.org/10.3390/nu13051489
Zhang T, Au Yeung S-L, Schooling CM. Associations of Arachidonic Acid Synthesis with Cardiovascular Risk Factors and Relation to Ischemic Heart Disease and Stroke: A Univariable and Multivariable Mendelian Randomization Study. Nutrients. 2021; 13(5):1489. https://doi.org/10.3390/nu13051489
Chicago/Turabian StyleZhang, Ting, Shiu-Lun Au Yeung, and C. Mary Schooling. 2021. "Associations of Arachidonic Acid Synthesis with Cardiovascular Risk Factors and Relation to Ischemic Heart Disease and Stroke: A Univariable and Multivariable Mendelian Randomization Study" Nutrients 13, no. 5: 1489. https://doi.org/10.3390/nu13051489





