From Food to Genes: Transcriptional Regulation of Metabolism by Lipids and Carbohydrates
Abstract
1. Introduction
2. Gene Expression Regulation by Lipids
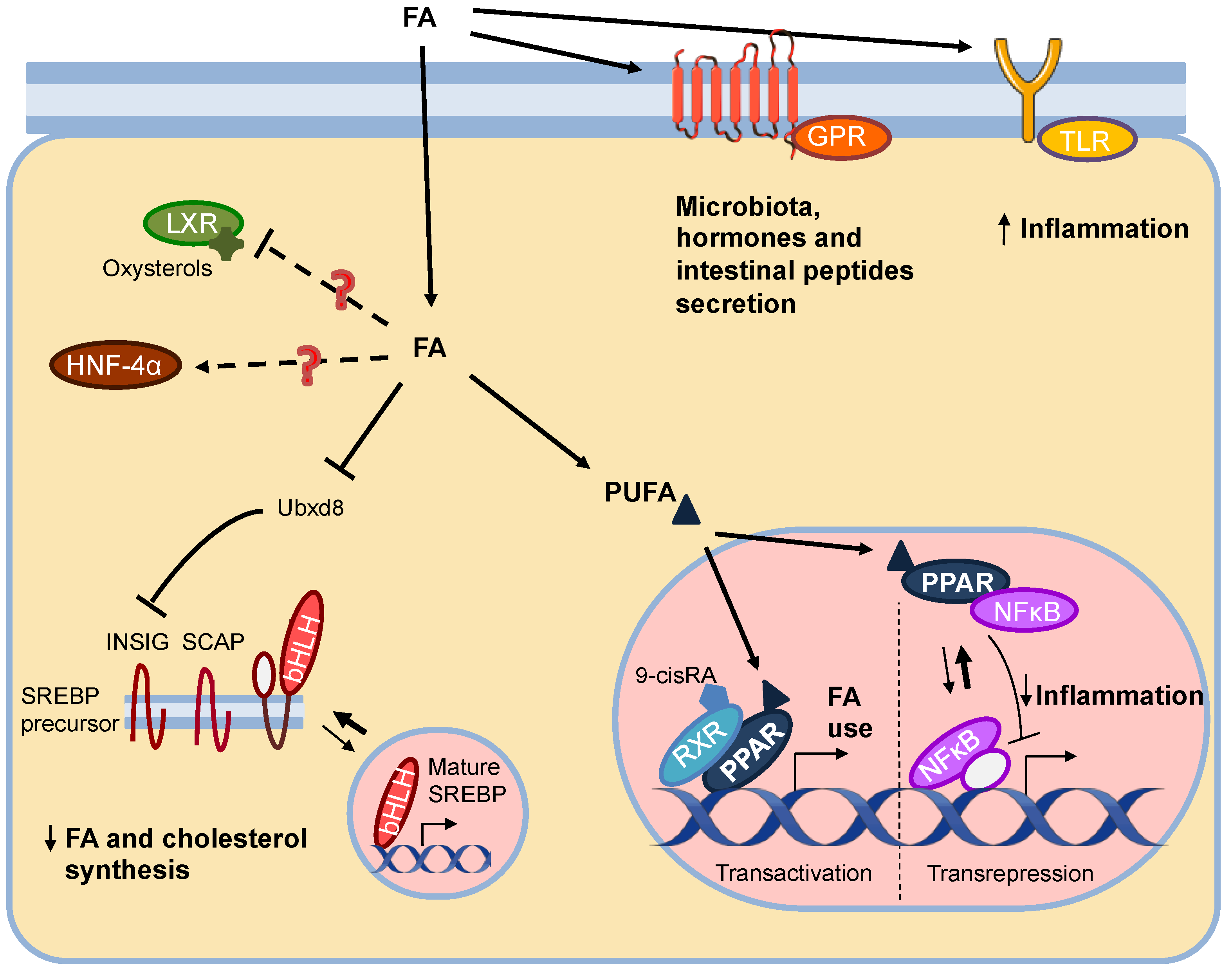
2.1. Fatty Acid Sensors: Nuclear and Membrane Receptors, and Transcription Factors
2.2. PPARs
2.2.1. PPARs and Their Ligands
| Receptor. | Nutrient-Derived Activating Agents | Synthetic Activating Agents |
|---|---|---|
| PPARα | Saturated and unsaturated fatty acids [38,41] Leukotriene B4 [45] 8-hydroxyeicosatetraenoic acid [41,42] | Fibrates [33] |
| PPARβ | Polyunsaturated fatty acids [38,41] 15-hydroxy-eicosatetraenoic acid [43] Prostacyclin [47] | GW501516 |
| PPARγ | Polyunsaturated fatty acids [38] 15-deoxy Δ12,14-prostaglandin J2 [46] 15-hydroxyeicosatetraenoic acid [37,44] 9- and 13-hydroxyoctadecadienoic acid [37,44] | Thiazolidinediones [35] |
| MondoA, ChREBPα/β | Glucose-6-phosphate [49] Xylulose-5-phosphate [50] Fructose-2,6-bisphosphate [51] | ------ |
2.2.2. PPAR Mechanism of Action
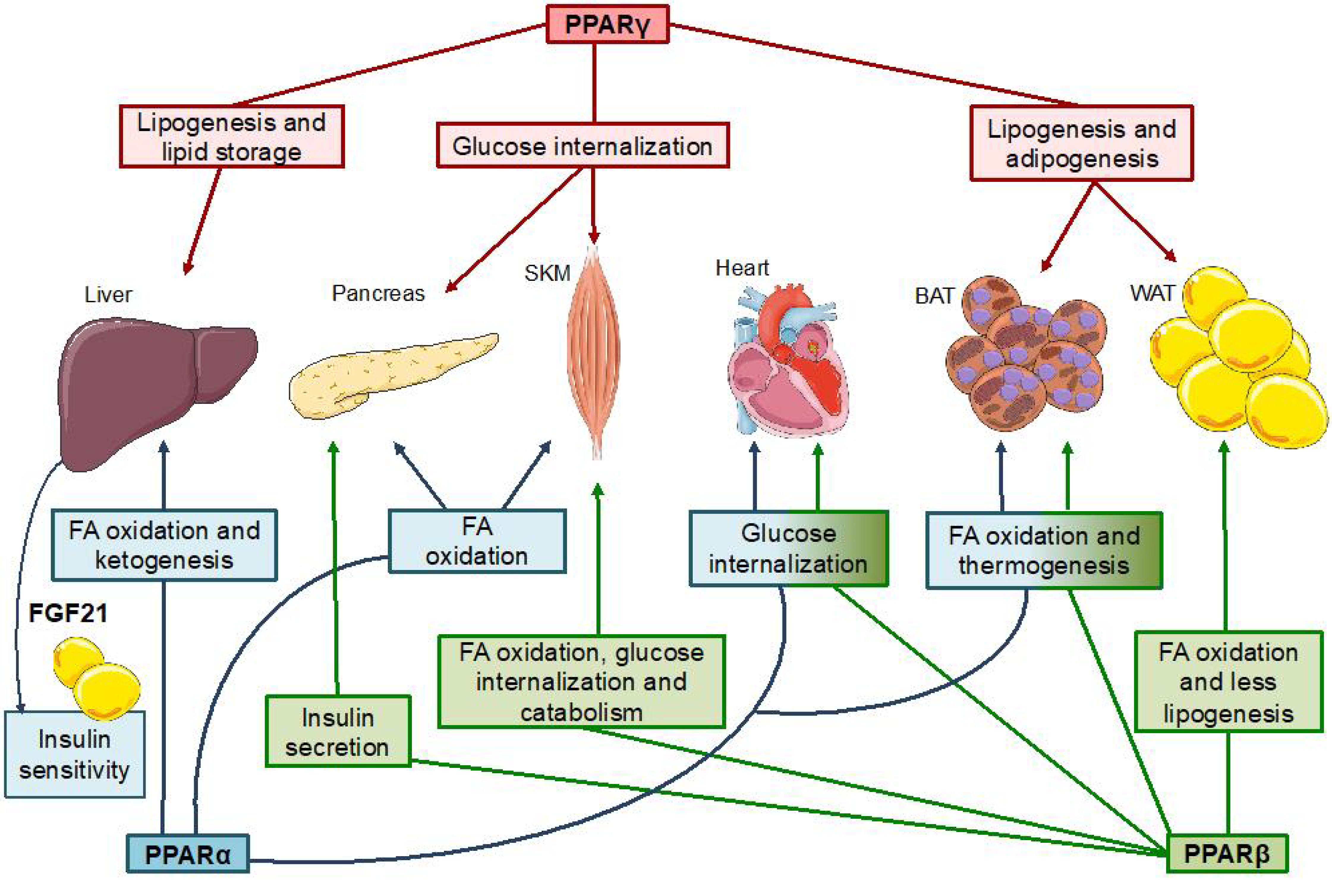
2.2.3. PPAR Metabolic Functions
PPARα
PPARγ
PPARβ
2.2.4. PPAR Anti-Inflammatory Functions
3. Gene Expression Regulation by Carbohydrates
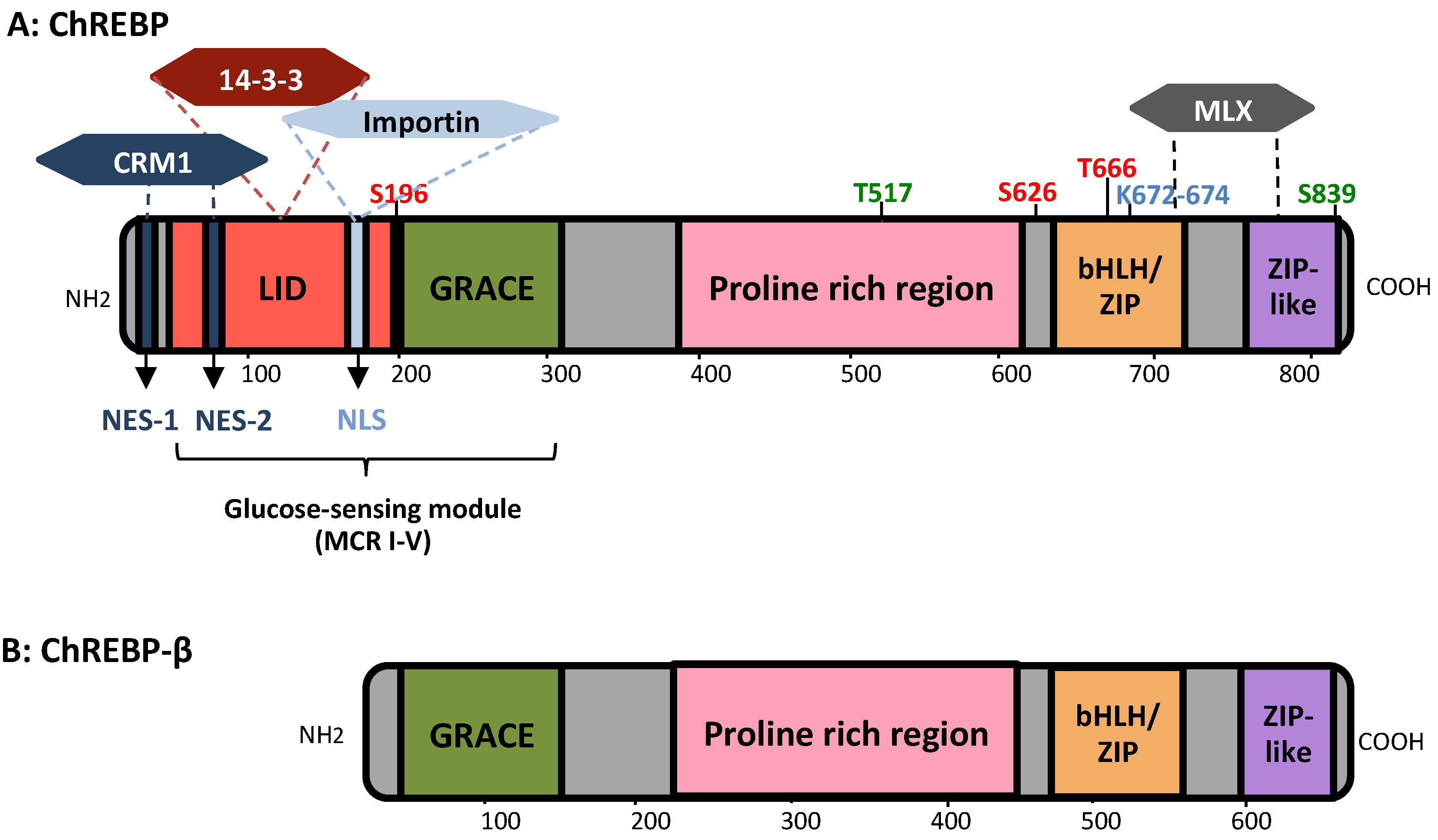
3.1. MondoA/ChREBP Structural Features
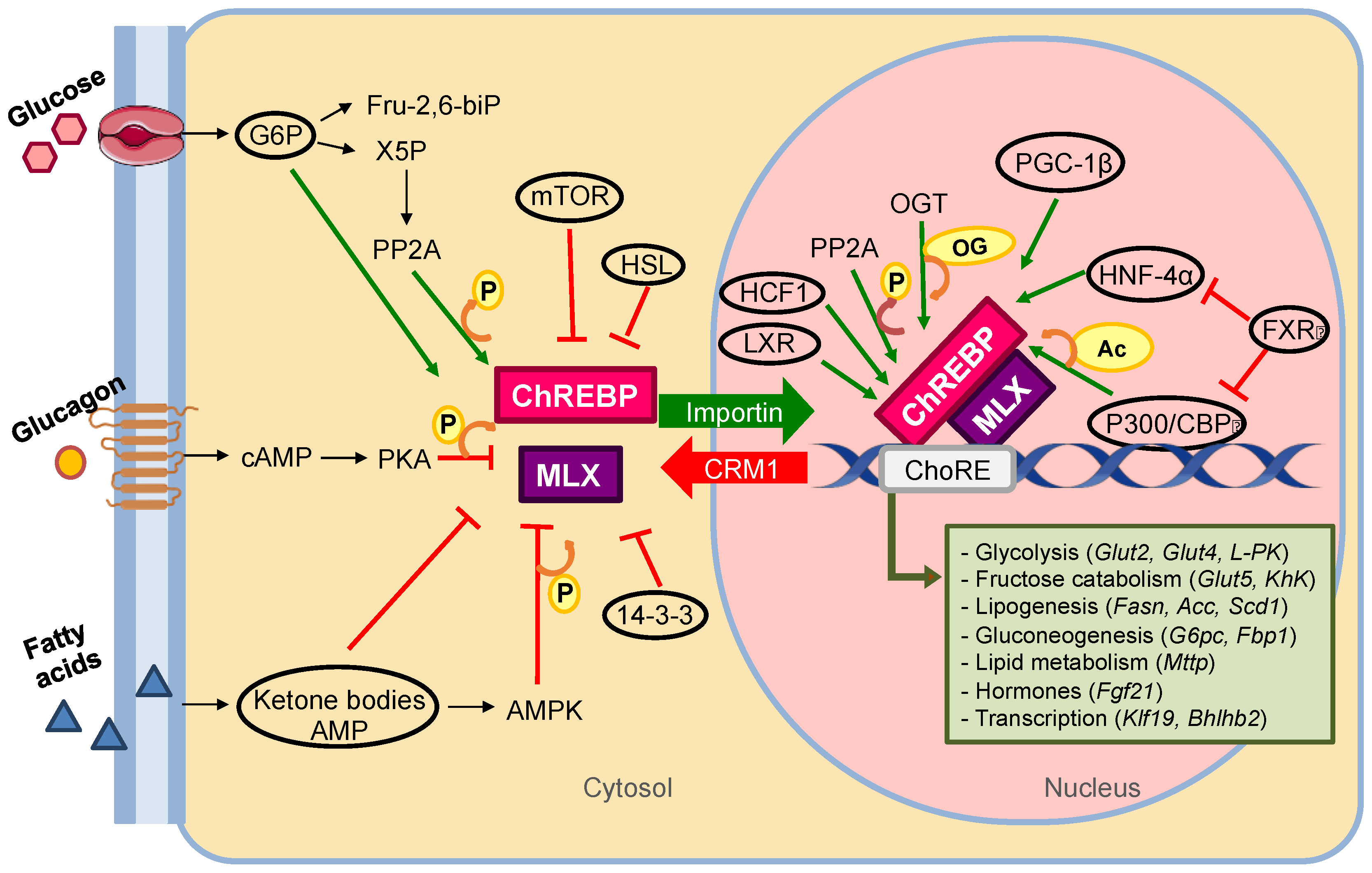
3.2. ChREBP Mechanism of Action
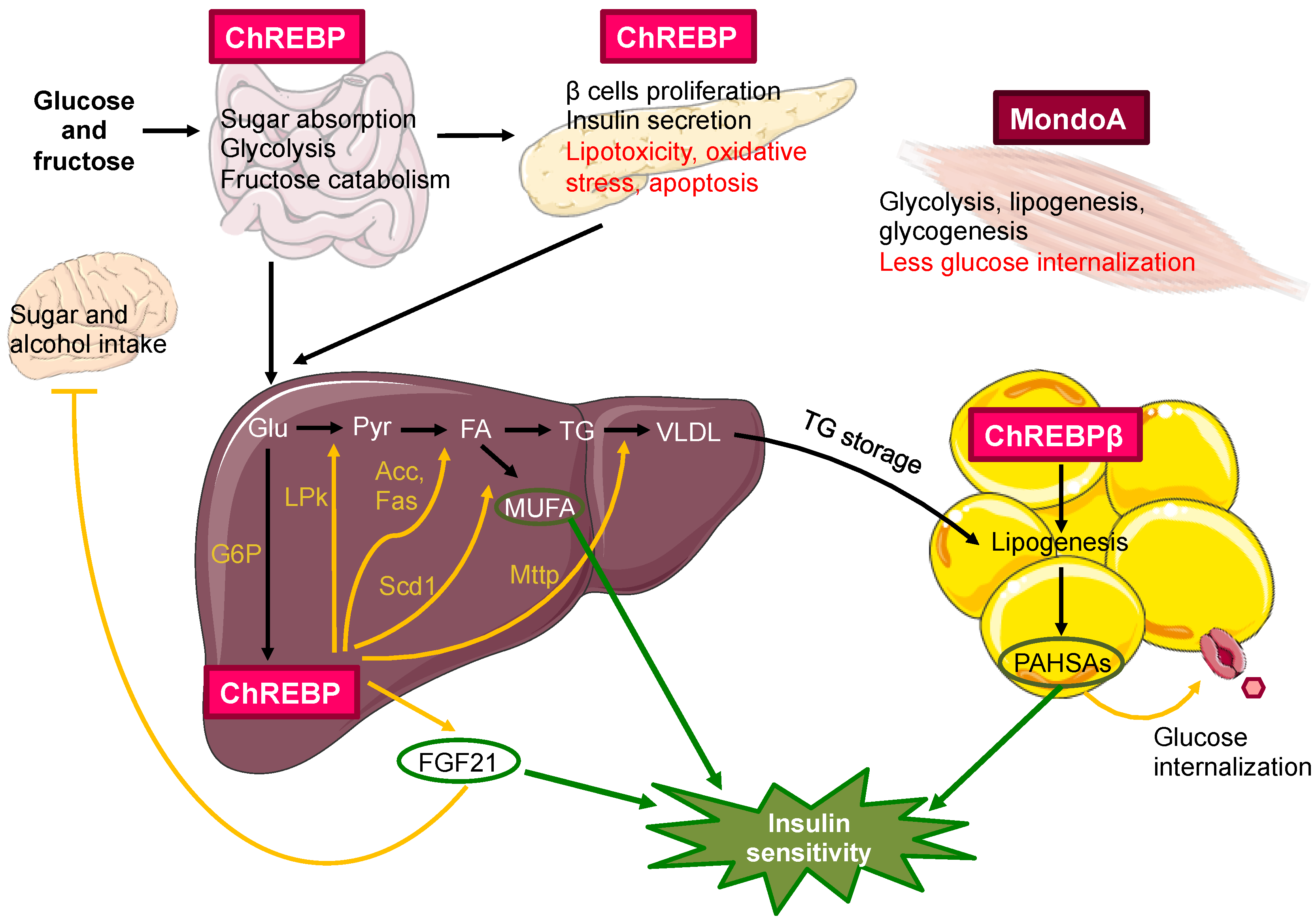
3.3. MondoA/ChREBP Metabolic Functions
4. Conclusions and Outlook
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Jacob, F.; Monod, J. Genetic regulatory mechanisms in the synthesis of proteins. J. Mol. Biol. 1961, 3, 318–356. [Google Scholar] [CrossRef]
- Kadayifci, F.Z.; Zheng, S.; Pan, Y.X. Molecular mechanisms underlying the link between diet and DNA methylation. Int. J. Mol. Sci. 2018, 19, 4055. [Google Scholar] [CrossRef] [PubMed]
- Daliu, P.; Santini, A.; Novellino, E. A decade of nutraceutical patents: Where are we now in 2018? Expert Opin. Ther. Pat. 2018, 28, 875–882. [Google Scholar] [CrossRef]
- Lauritzen, L.; Hansen, H.S.; Jorgensen, M.H.; Michaelsen, K.F. The essentiality of long chain n-3 fatty acids in relation to development and function of the brain and retina. Prog. Lipid Res. 2001, 40, 1–94. [Google Scholar] [CrossRef]
- Serhan, C.N.; Chiang, N. Endogenous pro-resolving and anti-inflammatory lipid mediators: A new pharmacologic genus. Br. J. Pharmacol. 2008, 153, S200–S215. [Google Scholar] [CrossRef] [PubMed]
- Simopoulos, A.P. An increase in the Omega-6/Omega-3 fatty acid ratio increases the risk for obesity. Nutrients 2016, 8, 128. [Google Scholar] [CrossRef]
- Hu, F.B.; Manson, J.A.E.; Willett, W.C. Types of Dietary Fat and Risk of Coronary Heart Disease: A Critical Review. J. Am. Coll. Nutr. 2001, 20, 5–19. [Google Scholar] [CrossRef] [PubMed]
- Galgani, J.E.; Uauy, R.D.; Aguirre, C.A.; Díaz, E.O. Effect of the dietary fat quality on insulin sensitivity. Br. J. Nutr. 2008, 100, 471–479. [Google Scholar] [CrossRef]
- Sellem, L.; Srour, B.; Guéraud, F.; Pierre, F.; Kesse-Guyot, E.; Fiolet, T.; Lavalette, C.; Egnell, M.; Latino-Martel, P.; Fassier, P.; et al. Saturated, mono- and polyunsaturated fatty acid intake and cancer risk: Results from the French prospective cohort NutriNet-Santé. Eur. J. Nutr. 2019, 58, 1515–1527. [Google Scholar] [CrossRef]
- Dreyer, C.; Krey, G.; Keller, H.; Givel, F.; Helftenbein, G.; Wahli, W. Control of the peroxisomal β-oxidation pathway by a novel family of nuclear hormone receptors. Cell 1992, 68, 879–887. [Google Scholar] [CrossRef]
- Aranda, A.; Pascual, A. Nuclear hormone receptors and gene expression. Physiol. Rev. 2001, 81, 1269–1304. [Google Scholar] [CrossRef]
- Georgiadi, A.; Kersten, S. Mechanisms of gene regulation by fatty acids. Adv. Nutr. 2012, 3, 127–134. [Google Scholar] [CrossRef] [PubMed]
- Evans, R.M.; Barish, G.D.; Wang, Y.X. PPARs and the complex journey to obesity. Nat. Med. 2004, 10, 355–361. [Google Scholar] [CrossRef]
- Repa, J.J.; Mangelsdorf, D.J. The role of orphan nuclear receptors in the regulation of cholesterol homeostasis. Annu. Rev. Cell Dev. Biol. 2000, 16, 459–481. [Google Scholar] [CrossRef] [PubMed]
- Lehmann, J.M.; Kliewer, S.A.; Moore, L.B.; Smith-Oliver, T.A.; Oliver, B.B.; Su, J.L.; Sundseth, S.S.; Winegar, D.A.; Blanchard, D.E.; Spencer, T.A.; et al. Activation of the nuclear receptor LXR by oxysterols defines a new hormone response pathway. J. Biol. Chem. 1997, 272, 3137–3140. [Google Scholar] [CrossRef]
- Jump, D.B. Fatty Acid Regulation of Gene Transcription. Crit. Rev. Clin. Lab. Sci. 2004, 41, 41–78. [Google Scholar] [CrossRef] [PubMed]
- Tobin, K.A.R.; Steineger, H.H.; Alberti, S.; Spydevold, Ø.; Auwerx, J.; Gustafsson, J.-Å.; Nebb, H.I. Cross-Talk between Fatty Acid and Cholesterol Metabolism Mediated by Liver X Receptor-α. Mol. Endocrinol. 2000, 14, 741–752. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Hayhurst, G.P.; Lee, Y.-H.; Lambert, G.; Ward, J.M.; Gonzalez, F.J. Hepatocyte Nuclear Factor 4α (Nuclear Receptor 2A1) Is Essential for Maintenance of Hepatic Gene Expression and Lipid Homeostasis. Mol. Cell. Biol. 2001, 21, 1393–1403. [Google Scholar] [CrossRef]
- Hertz, R.; Magenheim, J.; Berman, I.; Bar-Tana, J. Fatty acyl-CoA thioesters are ligands of hepatic nuclear factor-4α. Nature 1998, 392, 512–516. [Google Scholar] [CrossRef]
- Dhe-Paganon, S.; Duda, K.; Iwamoto, M.; Chi, Y.I.; Shoelson, S.E. Crystal structure of the HNF4α ligand binding domain in complex with endogenous fatty acid ligand. J. Biol. Chem. 2002, 277, 37973–37976. [Google Scholar] [CrossRef]
- Yuan, X.; Ta, T.C.; Lin, M.; Evans, J.R.; Dong, Y.; Bolotin, E.; Sherman, M.A.; Forman, B.M.; Sladek, F.M. Identification of an endogenous ligand bound to a native orphan nuclear receptor. PLoS ONE 2009, 4, e5609. [Google Scholar] [CrossRef] [PubMed]
- Lengqvist, J.; Mata de Urquiza, A.; Bergman, A.C.; Willson, T.M.; Sjövall, J.; Perlmann, T.; Griffiths, W.J. Polyunsaturated fatty acids including docosahexaenoic and arachidonic acid bind to the retinoid X receptor α ligand-binding domain. Mol. Cell. Proteomics 2004, 3, 692–703. [Google Scholar] [CrossRef] [PubMed]
- Horton, J.D.; Goldstein, J.L.; Brown, M.S. SREBPs: Activators of the complete program of cholesterol and fatty acid synthesis in the liver. J. Clin. Invest. 2002, 109, 1125–1131. [Google Scholar] [CrossRef] [PubMed]
- Yang, T.; Espenshade, P.J.; Wright, M.E.; Yabe, D.; Gong, Y.; Aebersold, R.; Goldstein, J.L.; Brown, M.S. Crucial step in cholesterol homeostasis: Sterols promote binding of SCAP to INSIG-1, a membrane protein that facilitates retention of SREBPs in ER. Cell 2002, 110, 489–500. [Google Scholar] [CrossRef]
- Lee, J.N.; Kim, H.; Yao, H.; Chen, Y.; Weng, K.; Ye, J. Identification of Ubxd8 protein as a sensor for unsaturated fatty acids and regulator of triglyceride synthesis. Proc. Natl. Acad. Sci. USA 2010, 107, 21424–21429. [Google Scholar] [CrossRef] [PubMed]
- Botolin, D.; Wang, Y.; Christian, B.; Jump, D.B. Docosahexaneoic acid (22:6,n-3) regulates rat hepatocyte SREBP-1 nuclear abundance by Erk- and 26S proteasome-dependent pathways. J. Lipid Res. 2006, 47, 181–192. [Google Scholar] [CrossRef]
- Takeuchi, Y.; Yahagi, N.; Izumida, Y.; Nishi, M.; Kubota, M.; Teraoka, Y.; Yamamoto, T.; Matsuzaka, T.; Nakagawa, Y.; Sekiya, M.; et al. Polyunsaturated fatty acids selectively suppress sterol regulatory element-binding protein-1 through proteolytic processing and autoloop regulatory circuit. J. Biol. Chem. 2010, 285, 11681–11691. [Google Scholar] [CrossRef] [PubMed]
- Ou, J.; Tu, H.; Shan, B.; Luk, A.; DeBose-Boyd, R.A.; Bashmakov, Y.; Goldstein, J.L.; Brown, M.S. Unsaturated fatty acids inhibit transcription of the sterol regulatory element-binding protein-1c (SREBP-1c) gene by antagonizing ligand-dependent activation of the LXR. Proc. Natl. Acad. Sci. USA 2001, 98, 6027–6032. [Google Scholar] [CrossRef]
- Shi, H.; Kokoeva, M.V.; Inouye, K.; Tzameli, I.; Yin, H.; Flier, J.S. TLR4 links innate immunity and fatty acid-induced insulin resistance. J. Clin. Investig. 2006, 116, 3015–3025. [Google Scholar] [CrossRef]
- Covington, D.K.; Briscoe, C.A.; Brown, A.J.; Jayawickreme, C.K. The G-protein-coupled receptor 40 family (GPR40-GPR43) and its role in nutrient sensing. Biochem. Soc. Trans. 2006, 34, 770–773. [Google Scholar] [CrossRef]
- Tomita, T.; Hosoda, K.; Fujikura, J.; Inagaki, N.; Nakao, K. The G-protein-coupled long-chain fatty acid receptor GPR40 and glucose metabolism. Front. Endocrinol. (Lausanne) 2014, 5, 152. [Google Scholar] [CrossRef]
- Suckow, A.T.; Polidori, D.; Yan, W.; Chon, S.; Ma, J.Y.; Leonard, J.; Briscoe, C.P. Alteration of the glucagon axis in GPR120 (FFAR4) knockout mice: A role for gpr120 in glucagon secretion. J. Biol. Chem. 2014, 289, 15751–15763. [Google Scholar] [CrossRef] [PubMed]
- Hess, R.; Stäubli, W.; Riess, W. Nature of the hepatomegalic effect produced by ethyl-chlorophenoxy-isobutyrate in the rat. Nature 1965, 208, 856–858. [Google Scholar] [CrossRef]
- Issemann, I.; Green, S. Activation of a member of the steroid hormone receptor superfamily by peroxisome proliferators. Nature 1990, 347, 645–650. [Google Scholar] [CrossRef]
- Lehmann, J.M.; Moore, L.B.; Smith-Oliver, T.A.; Wilkison, W.O.; Willson, T.M.; Kliewer, S.A. An antidiabetic thiazolidinedione is a high affinity ligand for peroxisome proliferator-activated receptor γ (PPARγ). J. Biol. Chem. 1995, 270, 12953–12956. [Google Scholar] [CrossRef] [PubMed]
- Hong, F.; Pan, S.; Guo, Y.; Xu, P.; Zhai, Y. PPARs as nuclear receptors for nutrient and energy metabolism. Molecules 2019, 24, 2545. [Google Scholar] [CrossRef]
- Varga, T.; Czimmerer, Z.; Nagy, L. PPARs are a unique set of fatty acid regulated transcription factors controlling both lipid metabolism and inflammation. Biochim. Biophys. Acta Mol. Basis Dis. 2011, 1812, 1007–1022. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.E.; Lambert, M.H.; Montana, V.G.; Parks, D.J.; Blanchard, S.G.; Brown, P.J.; Sternbach, D.D.; Lehmann, J.M.; Wisely, G.B.; Willson, T.M.; et al. Molecular recognition of fatty acids by peroxisome proliferator- activated receptors. Mol. Cell 1999, 3, 397–403. [Google Scholar] [CrossRef]
- Coleman, R.A.; Lewin, T.M.; Van Horn, C.G.; Gonzalez-Baró, M.R. Do long-chain acyl-CoA synthetases regulate fatty acid entry into synthetic versus degradative pathways? J. Nutr. 2002, 132, 2123–2126. [Google Scholar] [CrossRef] [PubMed]
- Pawar, A.; Jump, D.B. Unsaturated fatty acid regulation of peroxisome proliferator-activated receptor α activity in rat primary hepatoctes. J. Biol. Chem. 2003, 278, 35931–35939. [Google Scholar] [CrossRef] [PubMed]
- Forman, B.M.; Chen, J.; Evans, R.M. Hypolipidemic drugs, polyunsaturated fatty acids, and eicosanoids are ligands for peroxisome proliferator-activated receptors α and δ. Proc. Natl. Acad. Sci. USA 1997, 94, 4312–4317. [Google Scholar] [CrossRef] [PubMed]
- Krey, G.; Braissant, O.; L’Horset, F.; Kalkhoven, E.; Perroud, M.; Parker, M.G.; Wahli, W. Fatty acids, eicosanoids, and hypolipidemic agents identified as ligands of peroxisome proliferator-activated receptors by coactivator-dependent receptor ligand assay. Mol. Endocrinol. 1997, 11, 779–791. [Google Scholar] [CrossRef] [PubMed]
- Naruhn, S.; Meissner, W.; Adhikary, T.; Kaddatz, K.; Klein, T.; Watzer, B.; Müller-Brüsselbach, S.; Müller, R. 15-Hydroxyeicosatetraenoic acid is a preferential peroxisome proliferator-activated receptor β/δ agonist. Mol. Pharmacol. 2010, 77, 171–184. [Google Scholar] [CrossRef]
- Ringseis, R.; Eder, K. Regulation of genes involved in lipid metabolism by dietary oxidized fat. Mol. Nutr. Food Res. 2011, 55, 109–121. [Google Scholar] [CrossRef] [PubMed]
- Devchand, P.R.; Keller, H.; Peters, J.M.; Vazquez, M.; Gonzalez, F.J.; Wahli, W. The PPARα–leukotriene B 4 pathway to inflammation control. Nature 1996, 384, 39–43. [Google Scholar] [CrossRef]
- Forman, B.M.; Tontonoz, P.; Chen, J.; Brun, R.P.; Spiegelman, B.M.; Evans, R.M. 15-Deoxy-Δ12,14-Prostaglandin J2 is a ligand for the adipocyte determination factor PPARγ. Cell 1995, 83, 803–812. [Google Scholar] [CrossRef]
- Gupta, R.A.; Tan, J.; Krause, W.F.; Geraci, M.W.; Willson, T.M.; Dey, S.K.; DuBois, R.N. Prostacyclin-mediated activation of peroxisome proliferator-activated receptor δ in colorectal cancer. Proc. Natl. Acad. Sci. USA 2000, 97, 13275–13280. [Google Scholar] [CrossRef]
- Rigano, D.; Sirignano, C.; Taglialatela-Scafati, O. The potential of natural products for targeting PPARα. Acta Pharm. Sin. B 2017, 7, 427–438. [Google Scholar] [CrossRef]
- McFerrin, L.G.; Atchley, W.R. A Novel N-Terminal Domain May Dictate the Glucose Response of Mondo Proteins. PLoS ONE 2012, 7, e34803. [Google Scholar] [CrossRef]
- Kabashima, T.; Kawaguchi, T.; Wadzinski, B.E.; Uyeda, K. Xylulose 5-phosphate mediates glucose-induced lipogenesis by xylulose 5-phosphate-activated protein phosphatase in rat liver. Proc. Natl. Acad. Sci. USA 2003, 100, 5107–5112. [Google Scholar] [CrossRef]
- Arden, C.; Tudhope, S.J.; Petrie, J.L.; Al-Oanzi, Z.H.; Cullen, K.S.; Lange, A.J.; Towle, H.C.; Agius, L. Fructose 2, 6-bisphosphate is essential for glucose-regulated gene transcription of glucose-6-phosphatase and other ChREBP target genes in hepatocytes. Biochem. J. 2012, 443, 111–123. [Google Scholar] [CrossRef]
- Kliewer, S.A.; Umesono, K.; Noonan, D.J.; Heyman, R.A.; Evans, R.M. Convergence of 9-cis retinoic acid and peroxisome proliferator signalling pathways through heterodimer formation of their receptors. Nature 1992, 358, 771–774. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, R.; Grøntved, L.; Stunnenberg, H.G.; Mandrup, S. Peroxisome Proliferator-Activated Receptor Subtype- and Cell-Type-Specific Activation of Genomic Target Genes upon Adenoviral Transgene Delivery. Mol. Cell. Biol. 2006, 26, 5698–5714. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Széles, L.; Póliska, S.; Nagy, G.; Szatmari, I.; Szanto, A.; Pap, A.; Lindstedt, M.; Santegoets, S.J.A.M.; Rühl, R.; Dezsö, B.; et al. Research resource: Transcriptome profiling of genes regulated by RXR and its permissive and nonpermissive partners in differentiating monocyte-derived dendritic cells. Mol. Endocrinol. 2010, 24, 2218–2231. [Google Scholar] [CrossRef] [PubMed]
- Haro, D.; Marrero, P.F.; Relat, J. Nutritional regulation of gene expression: Carbohydrate-, fat-and amino acid-dependent modulation of transcriptional activity. Int. J. Mol. Sci. 2019, 20, 1386. [Google Scholar] [CrossRef] [PubMed]
- Sengupta, S.; Peterson, T.R.; Laplante, M.; Oh, S.; Sabatini, D.M. MTORC1 controls fasting-induced ketogenesis and its modulation by ageing. Nature 2010, 468, 1100–1106. [Google Scholar] [CrossRef] [PubMed]
- Dubois, V.; Eeckhoute, J.; Lefebvre, P.; Staels, B. Distinct but complementary contributions of PPAR isotypes to energy homeostasis. J. Clin. Investig. 2017, 127, 1202–1214. [Google Scholar] [CrossRef]
- Goto, T. A review of the studies on food-derived factors which regulate energy metabolism via the modulation of lipid-sensing nuclear receptors. Biosci. Biotechnol. Biochem. 2019, 83, 579–588. [Google Scholar] [CrossRef]
- Rachid, T.L.; Penna-de-Carvalho, A.; Bringhenti, I.; Aguila, M.B.; Mandarim-de-Lacerda, C.A.; Souza-Mello, V. Fenofibrate (PPARalpha agonist) induces beige cell formation in subcutaneous white adipose tissue from diet-induced male obese mice. Mol. Cell. Endocrinol. 2015, 402, 86–94. [Google Scholar] [CrossRef]
- Shah, A.; Rader, D.J.; Millar, J.S. The effect of PPAR-α agonism on apolipoprotein metabolism in humans. Atherosclerosis 2010, 210, 35–40. [Google Scholar] [CrossRef]
- Guerre-Millo, M.; Gervois, P.; Raspé, E.; Madsen, L.; Poulain, P.; Derudas, B.; Herbert, J.M.; Winegar, D.A.; Willson, T.M.; Fruchart, J.C.; et al. Peroxisome proliferator-activated receptor α activators improve insulin sensitivity and reduce adiposity. J. Biol. Chem. 2000, 275, 16638–16642. [Google Scholar] [CrossRef]
- Kersten, S.; Seydoux, J.; Peters, J.M.; Gonzalez, F.J.; Desvergne, B.; Wahli, W. Peroxisome proliferator-activated receptor α mediates the adaptive response to fasting. J. Clin. Investig. 1999, 103, 1489–1498. [Google Scholar] [CrossRef] [PubMed]
- Chakravarthy, M.V.; Pan, Z.; Zhu, Y.; Tordjman, K.; Schneider, J.G.; Coleman, T.; Turk, J.; Semenkovich, C.F. “New” hepatic fat activates PPARα to maintain glucose, lipid, and cholesterol homeostasis. Cell Metab. 2005, 1, 309–322. [Google Scholar] [CrossRef]
- Mukherjee, R.; Jow, L.; Croston, G.E.; Paterniti, J.R. Identification, characterization, and tissue distribution of human peroxisome proliferator-activated receptor (PPAR) isoforms PPARγ2 versus PPARγ1 and activation with retinoid X receptor agonists and antagonists. J. Biol. Chem. 1997, 272, 8071–8076. [Google Scholar] [CrossRef] [PubMed]
- Rosen, E.D.; Sarraf, P.; Troy, A.E.; Bradwin, G.; Moore, K.; Milstone, D.S.; Spiegelman, B.M.; Mortensen, R.M. PPARγ is required for the differentiation of adipose tissue in vivo and in vitro. Mol. Cell 1999, 4, 611–617. [Google Scholar] [CrossRef]
- He, W.; Barak, Y.; Hevener, A.; Olson, P.; Liao, D.; Le, J.; Nelson, M.; Ong, E.; Olefsky, J.M.; Evans, R.M. Adipose-specific peroxisome proliferator-activated receptor γ knockout causes insulin resistance in fat and liver but not in muscle. Proc. Natl. Acad. Sci. USA 2003, 100, 15712–15717. [Google Scholar] [CrossRef]
- Agarwal, A.K.; Garg, A. A Novel Heterozygous Mutation in Peroxisome Proliferator-Activated Receptor-γ Gene in a Patient with Familial Partial Lipodystrophy. J. Clin. Endocrinol. Metab. 2002, 87, 408. [Google Scholar] [CrossRef] [PubMed]
- Myers, M.G.; Burant, C.F. PPAR-γ action: It’s all in your head. Nat. Med. 2011, 17, 544–545. [Google Scholar] [CrossRef]
- Peraldi, P.; Xu, M.; Spiegelman, B.M. Thiazolidinediones block tumor necrosis factor-α-induced inhibition of insulin signaling. J. Clin. Investig. 1997, 100, 1863–1869. [Google Scholar] [CrossRef] [PubMed]
- Steppan, C.M.; Bailey, S.T.; Bhat, S.; Brown, E.J.; Banerjee, R.R.; Wright, C.M.; Patel, H.R.; Ahima, R.S.; Lazar, M.A. The hormone resistin links obesity to diabetes. Nature 2001, 409, 307–312. [Google Scholar] [CrossRef]
- Berg, A.H.; Combs, T.P.; Du, X.; Brownlee, M.; Scherer, P.E. The adipocyte-secreted protein Acrp30 enhances hepatic insulin action. Nat. Med. 2001, 7, 947–953. [Google Scholar] [CrossRef]
- Ribon, V.; Johnson, J.H.; Camp, H.S.; Saltiel, A.R. Thiazolidinediones and insulin resistance: Peroxisome proliferator-activated receptor γ activation stimulates expression of the CAP gene. Proc. Natl. Acad. Sci. USA 1998, 95, 14751–14756. [Google Scholar] [CrossRef] [PubMed]
- Inagaki, T.; Sakai, J.; Kajimura, S. Transcriptional and epigenetic control of brown and beige adipose cell fate and function. Nat. Rev. Mol. Cell Biol. 2016, 17, 480–495. [Google Scholar] [CrossRef]
- Schnuck, J.K.; Sunderland, K.L.; Gannon, N.P.; Kuennen, M.R.; Vaughan, R.A. Leucine stimulates PPARβ/δ-dependent mitochondrial biogenesis and oxidative metabolism with enhanced GLUT4 content and glucose uptake in myotubes. Biochimie 2016, 128, 1–7. [Google Scholar] [CrossRef]
- Vázquez-Carrera, M. Unraveling the Effects of PPARβ/δ on Insulin Resistance and Cardiovascular Disease. Trends Endocrinol. Metab. 2016, 27, 319–334. [Google Scholar] [CrossRef]
- Wang, Y.X. PPARs: Diverse regulators in energy metabolism and metabolic diseases. Cell Res. 2010, 20, 124–137. [Google Scholar] [CrossRef]
- Bojic, L.A.; Telford, D.E.; Fullerton, M.D.; Ford, R.J.; Sutherland, B.G.; Edwards, J.Y.; Sawyez, C.G.; Gros, R.; Kemp, B.E.; Steinberg, G.R.; et al. PPAR—Activation attenuates hepatic steatosis in Ldlr Mice by enhanced fat oxidation, reduced lipogenesis, and improved insulin sensitivity. J. Lipid Res. 2014, 55, 1254–1266. [Google Scholar] [CrossRef]
- Dressel, U.; Allen, T.L.; Pippal, J.B.; Rohde, P.R.; Lau, P.; Muscat, G.E.O. The Peroxisome Proliferator-Activated Receptor β/δ Agonist, GW501516, Regulates the Expression of Genes Involved in Lipid Catabolism and Energy Uncoupling in Skeletal Muscle Cells. Mol. Endocrinol. 2003, 17, 2477–2493. [Google Scholar] [CrossRef]
- Gupta, R.A.; Wang, D.; Katkuri, S.; Wang, H.; Dey, S.K.; DuBois, R.N. Activation of nuclear hormone receptor peroxisome proliferator-activated receptor-δ accelerates intestinal adenoma growth. Nat. Med. 2004, 10, 245–247. [Google Scholar] [CrossRef]
- Korbecki, J.; Bobiński, R.; Dutka, M. Self-regulation of the inflammatory response by peroxisome proliferator-activated receptors. Inflamm. Res. 2019, 68, 443–458. [Google Scholar] [CrossRef]
- Delerive, P.; De Bosscher, K.; Besnard, S.; Vanden Berghe, W.; Peters, J.M.; Gonzalez, F.J.; Fruchart, J.C.; Tedgui, A.; Haegeman, G.; Staels, B. Peroxisome proliferator-activated receptor α negatively regulates the vascular inflammatory gene response by negative cross-talk with transcription factors NF-κB and AP-1. J. Biol. Chem. 1999, 274, 32048–32054. [Google Scholar] [CrossRef]
- Ding, G.; Fu, M.; Qin, Q.; Lewis, W.; Kim, H.W.; Fukai, T.; Bacanamwo, M.; Chen, Y.E.; Schneider, M.D.; Mangelsdorf, D.J.; et al. Cardiac peroxisome proliferator-activated receptor γ is essential in protecting cardiomyocytes from oxidative damage. Cardiovasc. Res. 2007, 76, 269–279. [Google Scholar] [CrossRef] [PubMed]
- Girnun, G.D.; Domann, F.E.; Moore, S.A.; Robbins, M.E.C. Identification of a functional peroxisome proliferator-activated receptor response element in the rat catalase promoter. Mol. Endocrinol. 2002, 16, 2793–2801. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Pascual, G.; Glass, C.K. Peroxisome Proliferator-Activated Receptor γ-Dependent Repression of the Inducible Nitric Oxide Synthase Gene. Mol. Cell. Biol. 2000, 20, 4699–4707. [Google Scholar] [CrossRef]
- Bougarne, N.; Paumelle, R.; Caron, S.; Hennuyer, N.; Mansouri, R.; Gervois, P.; Staels, B.; Haegeman, G.; De Bosscher, K. PPARα blocks glucocorticoid receptor α-mediated transactivation but cooperates with the activated glucocorticoid receptor α for transrepression on NF-κB. Proc. Natl. Acad. Sci. USA 2009, 106, 7397–7402. [Google Scholar] [CrossRef] [PubMed]
- Lo Verme, J.; Fu, J.; Astarita, G.; La Rana, G.; Russo, R.; Calignano, A.; Piomelli, D. The nuclear receptor peroxisome proliferator-activated receptor-α mediates the anti-inflammatory actions of palmitoylethanolamide. Mol. Pharmacol. 2005, 67, 15–19. [Google Scholar] [CrossRef] [PubMed]
- Meade, E.A.; McIntyre, T.M.; Zimmerman, G.A.; Prescott, S.M. Peroxisome proliferators enhance cyclooxygenase-2 expression in epithelial cells. J. Biol. Chem. 1999, 274, 8328–8334. [Google Scholar] [CrossRef]
- Wang, D.; Fu, L.; Ning, W.; Guo, L.; Sun, X.; Dey, S.K.; Chaturvedi, R.; Wilson, K.T.; DuBois, R.N. Peroxisome proliferator-activated receptor ä promotes colonic inflammation and tumor growth. Proc. Natl. Acad. Sci. USA 2014, 111, 7084–7089. [Google Scholar] [CrossRef]
- Yuan, H.; Lu, J.; Xiao, J.; Upadhyay, G.; Umans, R.; Kallakury, B.; Yin, Y.; Fant, M.E.; Kopelovich, L.; Glazer, R.I. PPARδ induces estrogen receptor-positive mammary neoplasia through an inflammatory and metabolic phenotype linked to mTOR activation. Cancer Res. 2013, 73, 4349–4361. [Google Scholar] [CrossRef]
- Bougarne, N.; Weyers, B.; Desmet, S.J.; Deckers, J.; Ray, D.W.; Staels, B.; De Bosscher, K. Molecular actions of PPARα in lipid metabolism and inflammation. Endocr. Rev. 2018, 39, 760–802. [Google Scholar] [CrossRef]
- Cheng, H.S.; Tan, W.R.; Low, Z.S.; Marvalim, C.; Lee, J.Y.H.; Tan, N.S. Exploration and Development of PPAR Modulators in Health and Disease: An Update of Clinical Evidence. Int. J. Mol. Sci. 2019, 20, 5055. [Google Scholar] [CrossRef]
- Girard, J.; Ferré, P.; Foufelle, F. Mechanisms by which carbohydrates regulate expression of genes for glycolytic and lipogenic enzymes. Annu. Rev. Nutr. 1997, 17, 325–352. [Google Scholar] [CrossRef]
- Decaux, J.F.; Antoine, B.; Kahn, A. Regulation of the expression of the L-type pyruvate kinase gene in adult rat hepatocytes in primary culture. J. Biol. Chem. 1989, 264, 11584–11590. [Google Scholar] [CrossRef]
- Prip-Buus, C.; Perdereau, D.; Foufelle, F.; Maury, J.; Ferre, P.; Girard, J. Induction of Fatty-Acid-Synthase Gene Expression by Glucose in Primary Culture of Rat Hepatocytes. Dependency upon Glucokinase Activity. Eur. J. Biochem. 1995, 230, 309–315. [Google Scholar] [CrossRef] [PubMed]
- Thompson, K.; Towle, H. Localization of the carbohydrate response element of the rat L-type pyruvate kinase gene. J. Biol. Chem. 1991, 266, 8679–8682. [Google Scholar] [CrossRef]
- Billin, A.N.; Eilers, A.L.; Coulter, K.L.; Logan, J.S.; Ayer, D.E. MondoA, a Novel Basic Helix-Loop-Helix–Leucine Zipper Transcriptional Activator That Constitutes a Positive Branch of a Max-Like Network. Mol. Cell. Biol. 2000, 20, 8845–8854. [Google Scholar] [CrossRef] [PubMed]
- De Luis, O.; Valero, M.C.; Pérez Jurado, L.A. WBSCR14, a putative transcription factor gene deleted in Williams-Beuren syndrome: Complete characterisation of the human gene and the mouse ortholog. Eur. J. Hum. Genet. 2000, 8, 215–222. [Google Scholar] [CrossRef] [PubMed]
- Yamashita, H.; Takenoshita, M.; Sakurai, M.; Bruick, R.K.; Henzel, W.J.; Shillinglaw, W.; Arnot, D.; Uyeda, K. A glucose-responsive transcription factor that regulates carbohydrate metabolism in the liver. Proc. Natl. Acad. Sci. USA 2001, 98, 9116–9121. [Google Scholar] [CrossRef]
- Herman, M.A.; Peroni, O.D.; Villoria, J.; Schön, M.R.; Abumrad, N.A.; Blüher, M.; Klein, S.; Kahn, B.B. A novel ChREBP isoform in adipose tissue regulates systemic glucose metabolism. Nature 2012, 484, 333–338. [Google Scholar] [CrossRef]
- Amati, B.; Land, H. Myc-Max-Mad: A transcription factor network controlling cell cycle progression, differentiation and death. Curr. Opin. Genet. Dev. 1994, 4, 102–108. [Google Scholar] [CrossRef]
- Billin, A.N.; Eilers, A.L.; Queva, C.; Ayer, D.E. Mlx, a novel Max-like BHLHZip protein that interacts with the max network of transcription factors. J. Biol. Chem. 1999, 274, 36344–36350. [Google Scholar] [CrossRef] [PubMed]
- Shih, H.M.; Liu, Z.; Towle, H.C. Two CACGTG motifs with proper spacing dictate the carbohydrate regulation of hepatic gene transcription. J. Biol. Chem. 1995, 270, 21991–21997. [Google Scholar] [CrossRef] [PubMed]
- Stoeckman, A.K.; Ma, L.; Towle, H.C. Mlx Is the Functional Heteromeric Partner of the Carbohydrate Response Element-binding Protein in Glucose Regulation of Lipogenic Enzyme Genes. J. Biol. Chem. 2004, 279, 15662–15669. [Google Scholar] [CrossRef]
- Ma, L.; Robinson, L.N.; Towle, H.C. ChREBP·Mlx is the principal mediator of glucose-induced gene expression in the liver. J. Biol. Chem. 2006, 281, 28721–28730. [Google Scholar] [CrossRef]
- Fukasawa, M.; Ge, Q.; Wynn, R.M.; Ishii, S.; Uyeda, K. Coordinate regulation/localization of the carbohydrate responsive binding protein (ChREBP) by two nuclear export signal sites: Discovery of a new leucine-rich nuclear export signal site. Biochem. Biophys. Res. Commun. 2010, 391, 1166–1169. [Google Scholar] [CrossRef]
- Sakiyama, H.; Wynn, R.M.; Lee, W.R.; Fukasawa, M.; Mizuguchi, H.; Gardner, K.H.; Repa, J.J.; Uyeda, K. Regulation of nuclear import/export of carbohydrate response element-binding protein (ChREBP): Interaction of anα-helix of ChREBP with the 14-3-3 proteins and regulation by phosphorylation. J. Biol. Chem. 2008, 283, 24899–24908. [Google Scholar] [CrossRef] [PubMed]
- Li, M.V.; Chang, B.; Imamura, M.; Poungvarin, N.; Chan, L. Glucose-dependent transcriptional regulation by an evolutionarily conserved glucose-sensing module. Diabetes 2006, 55, 1179–1189. [Google Scholar] [CrossRef]
- Davies, M.N.; O’Callaghan, B.L.; Towle, H.C. Activation and repression of glucose-stimulated ChREBP requires the concerted action of multiple domains within the MondoA conserved region. Am. J. Physiol. Endocrinol. Metab. 2010, 299, E665. [Google Scholar] [CrossRef][Green Version]
- Jing, G.; Chen, J.; Xu, G.; Shalev, A. Islet ChREBP-β is increased in diabetes and controls ChREBP-α and glucose-induced gene expression via a negative feedback loop. Mol. Metab. 2016, 5, 1208–1215. [Google Scholar] [CrossRef]
- Richards, P.; Ourabah, S.; Montagne, J.; Burnol, A.F.; Postic, C.; Guilmeau, S. MondoA/ChREBP: The usual suspects of transcriptional glucose sensing; Implication in pathophysiology. Metabolism 2017, 70, 133–151. [Google Scholar] [CrossRef]
- Ortega-Prieto, P.; Postic, C. Carbohydrate sensing through the transcription factor ChREBP. Front. Genet. 2019, 10, 472. [Google Scholar] [CrossRef]
- Kawaguchi, T.; Takenoshita, M.; Kabashima, T.; Uyeda, K. Glucose and cAMP regulate the L-type pyruvate kinase gene by phosphorylation/dephosphorylation of the carbohydrate response element binding protein. Proc. Natl. Acad. Sci. USA 2001, 98, 13710–13715. [Google Scholar] [CrossRef]
- Guinez, C.; Filhoulaud, G.; Rayah-Benhamed, F.; Marmier, S.; Dubuquoy, C.; Dentin, R.; Moldes, M.; Burnol, A.F.; Yang, X.; Lefebvre, T.; et al. O-GlcNAcylation increases ChREBP protein content and transcriptional activity in the liver. Diabetes 2011, 60, 1399–1413. [Google Scholar] [CrossRef]
- Li, M.V.; Chen, W.; Harmancey, R.N.; Nuotio-Antar, A.M.; Imamura, M.; Saha, P.; Taegtmeyer, H.; Chan, L. Glucose-6-phosphate mediates activation of the carbohydrate responsive binding protein (ChREBP). Biochem. Biophys. Res. Commun. 2010, 395, 395–400. [Google Scholar] [CrossRef]
- Iizuka, K.; Wu, W.; Horikawa, Y.; Takeda, J. Role of glucose-6-phosphate and xylulose-5-phosphate in the regulation of glucose-stimulated gene expression in the pancreatic β cell line, INS-1E. Endocr. J. 2013, 60, 473–482. [Google Scholar] [CrossRef]
- Dentin, R.; Tomas-Cobos, L.; Foufelle, F.; Leopold, J.; Girard, J.; Postic, C.; Ferré, P. Glucose 6-phosphate, rather than xylulose 5-phosphate, is required for the activation of ChREBP in response to glucose in the liver. J. Hepatol. 2012, 56, 199–209. [Google Scholar] [CrossRef]
- Tsatsos, N.G.; Towle, H.C. Glucose activation of ChREBP in hepatocytes occurs via a two-step mechanism. Biochem. Biophys. Res. Commun. 2006, 340, 449–456. [Google Scholar] [CrossRef]
- Bricambert, J.; Miranda, J.; Benhamed, F.; Girard, J.; Postic, C.; Dentin, R. Salt-inducible kinase 2 links transcriptional coactivator p300 phosphorylation to the prevention of ChREBP-dependent hepatic steatosis in mice. J. Clin. Investig. 2010, 120, 4316–4331. [Google Scholar] [CrossRef]
- Poupeau, A.; Postic, C. Cross-regulation of hepatic glucose metabolism via ChREBP and nuclear receptors. Biochim. Biophys. Acta Mol. Basis Dis. 2011, 1812, 995–1006. [Google Scholar] [CrossRef]
- Adamson, A.W.; Suchankova, G.; Rufo, C.; Nakamura, M.T.; Teran-Garcia, M.; Clarke, S.D.; Gettys, T.W. Hepatocyte nuclear factor-4α contributes to carbohydrate-induced transcriptional activation of hepatic fatty acid synthase. Biochem. J. 2006, 399, 285–295. [Google Scholar] [CrossRef]
- Burke, S.J.; Collier, J.J.; Scott, D.K. cAMP opposes the glucose-mediated induction of the L-PK gene by preventing the recruitment of a complex containing ChREBP, HNF4α, and CBP. FASEB J. 2009, 23, 2855–2865. [Google Scholar] [CrossRef]
- Caron, S.; Huaman Samanez, C.; Dehondt, H.; Ploton, M.; Briand, O.; Lien, F.; Dorchies, E.; Dumont, J.; Postic, C.; Cariou, B.; et al. Farnesoid X Receptor Inhibits the Transcriptional Activity of Carbohydrate Response Element Binding Protein in Human Hepatocytes. Mol. Cell. Biol. 2013, 33, 2202–2211. [Google Scholar] [CrossRef]
- Chambers, K.T.; Chen, Z.; Lai, L.; Leone, T.C.; Towle, H.C.; Kralli, A.; Crawford, P.A.; Finck, B.N. PGC-1β and ChREBP partner to cooperatively regulate hepatic lipogenesis in a glucose concentration-dependent manner. Mol. Metab. 2013, 2, 194–204. [Google Scholar] [CrossRef]
- Fan, Q.; Nørgaard, R.C.; Grytten, I.; Ness, C.M.; Lucas, C.; Vekterud, K.; Soedling, H.; Matthews, J.; Lemma, R.B.; Gabrielsen, O.S.; et al. LXR Regulates ChREBP Transactivity in a Target Gene-Specific Manner through an Agonist-Modulated LBD-LID InteractionLBD-LID Interaction. Cells 2020, 9, 1214. [Google Scholar] [CrossRef]
- Lane, E.A.; Choi, D.W.; Garcia-Haro, L.; Levine, Z.G.; Tedoldi, M.; Walker, S.; Danial, N.N. HCF-1 Regulates De Novo Lipogenesis through a Nutrient-Sensitive Complex with ChREBP. Mol. Cell 2019, 75, 357–371.e7. [Google Scholar] [CrossRef]
- Anthonisen, E.H.; Berven, L.; Holm, S.; Nygård, M.; Nebb, H.I.; Grønning-Wang, L.M. Nuclear receptor liver X receptor is O-GlcNAc-modified in response to glucose. J. Biol. Chem. 2010, 285, 1607–1615. [Google Scholar] [CrossRef]
- Nakagawa, T.; Ge, Q.; Pawlosky, R.; Wynn, R.M.; Veech, R.L.; Uyeda, K. Metabolite regulation of nucleo-cytosolic trafficking of carbohydrate response element-binding protein (ChREBP): Role of ketone bodies. J. Biol. Chem. 2013, 288, 28358–28367. [Google Scholar] [CrossRef]
- Sato, S.; Jung, H.; Nakagawa, T.; Pawlosky, R.; Takeshima, T.; Lee, W.R.; Sakiyama, H.; Laxman, S.; Wynn, R.M.; Tu, B.P.; et al. Metabolite regulation of nuclear localization of carbohydrate-response element-binding protein (ChREBP): Role of amp as an allosteric inhibitor. J. Biol. Chem. 2016, 291, 10515–10527. [Google Scholar] [CrossRef]
- Ido-Kitamura, Y.; Sasaki, T.; Kobayashi, M.; Kim, H.-J.; Lee, Y.-S.; Kikuchi, O.; Yokota-Hashimoto, H.; Iizuka, K.; Accili, D.; Kitamura, T. Hepatic FoxO1 Integrates Glucose Utilization and Lipid Synthesis through Regulation of Chrebp O-Glycosylation. PLoS ONE 2012, 7, e47231. [Google Scholar] [CrossRef]
- Chau, G.C.; Im, D.U.; Kang, T.M.; Bae, J.M.; Kim, W.; Pyo, S.; Moon, E.Y.; Um, S.H. mTOR controls ChREBP transcriptional activity and pancreatic β cell survival under diabetic stress. J. Cell Biol. 2017, 216, 2091–2105. [Google Scholar] [CrossRef]
- Morigny, P.; Houssier, M.; Mairal, A.; Ghilain, C.; Mouisel, E.; Benhamed, F.; Masri, B.; Recazens, E.; Denechaud, P.D.; Tavernier, G.; et al. Interaction between hormone-sensitive lipase and ChREBP in fat cells controls insulin sensitivity. Nat. Metab. 2019, 1, 133–146. [Google Scholar] [CrossRef]
- Cha, J.Y.; Repa, J.J. The Liver X Receptor (LXR) and hepatic lipogenesis: The carbohydrate-response element-binding protein is a target gene of LXR. J. Biol. Chem. 2007, 282, 743–751. [Google Scholar] [CrossRef]
- Meng, J.; Feng, M.; Dong, W.; Zhu, Y.; Li, Y.; Zhang, P.; Wu, L.; Li, M.; Lu, Y.; Chen, H.; et al. Identification of HNF-4α as a key transcription factor to promote ChREBP expression in response to glucose. Sci. Rep. 2016, 6, 23944. [Google Scholar] [CrossRef]
- Iizuka, K.; Bruick, R.K.; Liang, G.; Horton, J.D.; Uyeda, K. Deficiency of carbohydrate response element-binding protein (ChREBP) reduces lipogenesis as well as glycolysis. Proc. Natl. Acad. Sci. USA 2004, 101, 7281–7286. [Google Scholar] [CrossRef]
- Benhamed, F.; Denechaud, P.D.; Lemoine, M.; Robichon, C.; Moldes, M.; Bertrand-Michel, J.; Ratziu, V.; Serfaty, L.; Housset, C.; Capeau, J.; et al. The lipogenic transcription factor ChREBP dissociates hepatic steatosis from insulin resistance in mice and humans. J. Clin. Investig. 2012, 122, 2176–2194. [Google Scholar] [CrossRef]
- Song, Z.; Yang, H.; Zhou, L.; Yang, F. Glucose-sensing transcription factor mondoa/chrebp as targets for type 2 diabetes: Opportunities and challenges. Int. J. Mol. Sci. 2019, 20, 5132. [Google Scholar] [CrossRef]
- Ahn, B.; Wan, S.; Jaiswal, N.; Vega, R.B.; Ayer, D.E.; Titchenell, P.M.; Han, X.; Won, K.J.; Kelly, D.P. MondoA drives muscle lipid accumulation and insulin resistance. JCI Insight 2019, 4, e129119. [Google Scholar] [CrossRef]
- Ahn, B.; Soundarapandian, M.M.; Sessions, H.; Peddibhotla, S.; Roth, G.P.; Li, J.L.; Sugarman, E.; Koo, A.; Malany, S.; Wang, M.; et al. MondoA coordinately regulates skeletal myocyte lipid homeostasis and insulin signaling. J. Clin. Investig. 2016, 126, 3567–3579. [Google Scholar] [CrossRef]
- Linden, A.G.; Li, S.; Choi, H.Y.; Fang, F.; Fukasawa, M.; Uyeda, K.; Hammer, R.E.; Horton, J.D.; Engelking, L.J.; Liang, G. Interplay between ChREBP and SREBP-1c coordinates postprandial glycolysis and lipogenesis in livers of mice. J. Lipid Res. 2018, 59, 475–487. [Google Scholar] [CrossRef]
- Iizuka, K.; Miller, B.; Uyeda, K. Deficiency of carbohydrate-activated transcription factor ChREBP prevents obesity and improves plasma glucose control in leptin-deficient (ob/ob) mice. Am. J. Physiol. Endocrinol. Metab. 2006, 291, E358–E364. [Google Scholar] [CrossRef]
- Niwa, H.; Iizuka, K.; Kato, T.; Wu, W.; Tsuchida, H.; Takao, K.; Horikawa, Y.; Takeda, J. ChREBP rather than SHP regulates hepatic VLDL secretion. Nutrients 2018, 10, 321. [Google Scholar] [CrossRef]
- Stamatikos, A.D.; Da Silva, R.P.; Lewis, J.T.; Douglas, D.N.; Kneteman, N.M.; Jacobs, R.L.; Paton, C.M. Tissue Specific Effects of Dietary Carbohydrates and Obesity on ChREBPα and ChREBPβ Expression. Lipids 2016, 51, 95–104. [Google Scholar] [CrossRef]
- Zhang, X.; Yeung, D.C.Y.; Karpisek, M.; Stejskal, D.; Zhou, Z.G.; Liu, F.; Wong, R.L.C.; Chow, W.S.; Tso, A.W.K.; Lam, K.S.L.; et al. Serum FGF21 levels are increased in obesity and are independently associated with the metabolic syndrome in humans. Diabetes 2008, 57, 1246–1253. [Google Scholar] [CrossRef]
- Inagaki, T.; Dutchak, P.; Zhao, G.; Ding, X.; Gautron, L.; Parameswara, V.; Li, Y.; Goetz, R.; Mohammadi, M.; Esser, V.; et al. Endocrine Regulation of the Fasting Response by PPARα-Mediated Induction of Fibroblast Growth Factor 21. Cell Metab. 2007, 5, 415–425. [Google Scholar] [CrossRef] [PubMed]
- Iizuka, K.; Takeda, J.; Horikawa, Y. Glucose induces FGF21 mRNA expression through ChREBP activation in rat hepatocytes. FEBS Lett. 2009, 583, 2882–2886. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Garza, Ú.; Torres-Oteros, D.; Yarritu-Gallego, A.; Marrero, P.F.; Haro, D.; Relat, J. Fibroblast growth factor 21 and the adaptive response to nutritional challenges. Int. J. Mol. Sci. 2019, 20, 4692. [Google Scholar] [CrossRef] [PubMed]
- Talukdar, S.; Owen, B.M.; Song, P.; Hernandez, G.; Zhang, Y.; Zhou, Y.; Scott, W.T.; Paratala, B.; Turner, T.; Smith, A.; et al. FGF21 regulates sweet and alcohol preference. Cell Metab. 2016, 23, 344–349. [Google Scholar] [CrossRef] [PubMed]
- Fisher, F.M.; Kim, M.S.; Doridot, L.; Cunniff, J.C.; Parker, T.S.; Levine, D.M.; Hellerstein, M.K.; Hudgins, L.C.; Maratos-Flier, E.; Herman, M.A. A critical role for ChREBP-mediated FGF21 secretion in hepatic fructose metabolism. Mol. Metab. 2017, 6, 14–21. [Google Scholar] [CrossRef]
- Kato, T.; Iizuka, K.; Takao, K.; Horikawa, Y.; Kitamura, T.; Takeda, J. ChREBP-knockout mice show sucrose intolerance and fructose malabsorption. Nutrients 2018, 10, 340. [Google Scholar] [CrossRef]
- Schalkwijk, C.G.; Stehouwer, C.D.A.; van Hinsbergh, V.W.M. Fructose-mediated non-enzymatic glycation: Sweet coupling or bad modification. Diabetes. Metab. Res. Rev. 2004, 20, 369–382. [Google Scholar] [CrossRef]
- Iizuka, K. The role of carbohydrate response element binding protein in intestinal and hepatic fructose metabolism. Nutrients 2017, 9, 181. [Google Scholar] [CrossRef]
- Pedersen, K.B.; Zhang, P.; Doumen, C.; Charbonnet, M.; Lu, D.; Newgard, C.B.; Haycock, J.W.; Lange, A.J.; Scott, D.K. The promoter for the gene encoding the catalytic subunit of rat glucose-6-phosphatase contains two distinct glucose-responsive regions. Am. J. Physiol. Endocrinol. Metab. 2007, 292, E788–E801. [Google Scholar] [CrossRef] [PubMed]
- Iizuka, K.; Tomita, R.; Takeda, J.; Horikawa, Y. Rat glucagon receptor mRNA is directly regulated by glucose through transactivation of the carbohydrate response element binding protein. Biochem. Biophys. Res. Commun. 2012, 417, 1107–1112. [Google Scholar] [CrossRef] [PubMed]
- Douard, V.; Ferraris, R.P. The role of fructose transporters in diseases linked to excessive fructose intake. J. Physiol. 2013, 591, 401–414. [Google Scholar] [CrossRef] [PubMed]
- Ebert, K.; Witt, H. Fructose malabsorption. Mol. Cell. Pediatr. 2016, 3, 10. [Google Scholar] [CrossRef]
- Zhang, D.; Tong, X.; Van Dommelen, K.; Gupta, N.; Stamper, K.; Brady, G.F.; Meng, Z.; Lin, J.; Rui, L.; Omary, M.B.; et al. Lipogenic transcription factor ChREBP mediates fructose-induced metabolic adaptations to prevent hepatotoxicity. J. Clin. Investig. 2017, 127, 2855–2867. [Google Scholar] [CrossRef]
- Vijayakumar, A.; Aryal, P.; Wen, J.; Syed, I.; Vazirani, R.P.; Moraes-Vieira, P.M.; Camporez, J.P.; Gallop, M.R.; Perry, R.J.; Peroni, O.D.; et al. Absence of Carbohydrate Response Element Binding Protein in Adipocytes Causes Systemic Insulin Resistance and Impairs Glucose Transport. Cell Rep. 2017, 21, 1021–1035. [Google Scholar] [CrossRef]
- Yore, M.M.; Syed, I.; Moraes-Vieira, P.M.; Zhang, T.; Herman, M.A.; Homan, E.A.; Patel, R.T.; Lee, J.; Chen, S.; Peroni, O.D.; et al. Discovery of a class of endogenous mammalian lipids with anti-diabetic and anti-inflammatory effects. Cell 2014, 159, 318–332. [Google Scholar] [CrossRef]
- Eissing, L.; Scherer, T.; Tödter, K.; Knippschild, U.; Greve, J.W.; Buurman, W.A.; Pinnschmidt, H.O.; Rensen, S.S.; Wolf, A.M.; Bartelt, A.; et al. De novo lipogenesis in human fat and liver is linked to ChREBP-β and metabolic health. Nat. Commun. 2013, 4, 1528. [Google Scholar] [CrossRef]
- Sanchez-Gurmaches, J.; Tang, Y.; Jespersen, N.Z.; Wallace, M.; Martinez Calejman, C.; Gujja, S.; Li, H.; Edwards, Y.J.K.; Wolfrum, C.; Metallo, C.M.; et al. Brown Fat AKT2 Is a Cold-Induced Kinase that Stimulates ChREBP-Mediated De Novo Lipogenesis to Optimize Fuel Storage and Thermogenesis. Cell Metab. 2018, 27, 195–209.e6. [Google Scholar] [CrossRef]
- Metukuri, M.R.; Zhang, P.; Basantani, M.K.; Chin, C.; Stamateris, R.E.; Alonso, L.C.; Takane, K.K.; Gramignoli, R.; Strom, S.C.; O’Doherty, R.M.; et al. ChREBP mediates glucose-stimulated pancreatic β-cell proliferation. Diabetes 2012, 61, 2004–2015. [Google Scholar] [CrossRef] [PubMed]
- Poungvarin, N.; Lee, J.K.; Yechoor, V.K.; Li, M.V.; Assavapokee, T.; Suksaranjit, P.; Thepsongwajja, J.J.; Saha, P.K.; Oka, K.; Chan, L. Carbohydrate response element-binding protein (ChREBP) plays a pivotal role in beta cell glucotoxicity. Diabetologia 2012, 55, 1783–1796. [Google Scholar] [CrossRef]
- Iizuka, K.; Horikawa, Y. Regulation of lipogenesis via BHLHB2/DEC1 and ChREBP feedback looping. Biochem. Biophys. Res. Commun. 2008, 374, 95–100. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bravo-Ruiz, I.; Medina, M.Á.; Martínez-Poveda, B. From Food to Genes: Transcriptional Regulation of Metabolism by Lipids and Carbohydrates. Nutrients 2021, 13, 1513. https://doi.org/10.3390/nu13051513
Bravo-Ruiz I, Medina MÁ, Martínez-Poveda B. From Food to Genes: Transcriptional Regulation of Metabolism by Lipids and Carbohydrates. Nutrients. 2021; 13(5):1513. https://doi.org/10.3390/nu13051513
Chicago/Turabian StyleBravo-Ruiz, Inés, Miguel Ángel Medina, and Beatriz Martínez-Poveda. 2021. "From Food to Genes: Transcriptional Regulation of Metabolism by Lipids and Carbohydrates" Nutrients 13, no. 5: 1513. https://doi.org/10.3390/nu13051513
APA StyleBravo-Ruiz, I., Medina, M. Á., & Martínez-Poveda, B. (2021). From Food to Genes: Transcriptional Regulation of Metabolism by Lipids and Carbohydrates. Nutrients, 13(5), 1513. https://doi.org/10.3390/nu13051513








