Effects of Three-Month Administration of High-Saturated Fat Diet and High-Polyunsaturated Fat Diets with Different Linoleic Acid (LA, C18:2n–6) to α-Linolenic Acid (ALA, C18:3n–3) Ratio on the Mouse Liver Proteome
Abstract
:1. Introduction
2. Materials and Methods
2.1. Animals, Diets and Sampling
2.2. Plasma Biochemical Parameters
2.3. Two-Dimensional Electrophoresis
2.3.1. Homogenization
2.3.2. First Dimension—Isoelectrofocusing (IEF)
2.3.3. Second Dimension—SDS-PAGE
2.3.4. Image Staining and Analysis
2.4. Matrix-Assisted Laser Desorption Ionization–Time of Flight Mass Spectrometry (MALDI-TOF MS)
2.5. Protein Expression Analysis by Western Blotting
2.6. RNA Extraction and Quantitative RT-PCR
2.6.1. RNA Isolation
2.6.2. Reverse Transcription (RT)
2.6.3. Primers Design
2.6.4. Amplified Products Verification and Real-Time PCR Analysis
2.7. Statistical Analysis
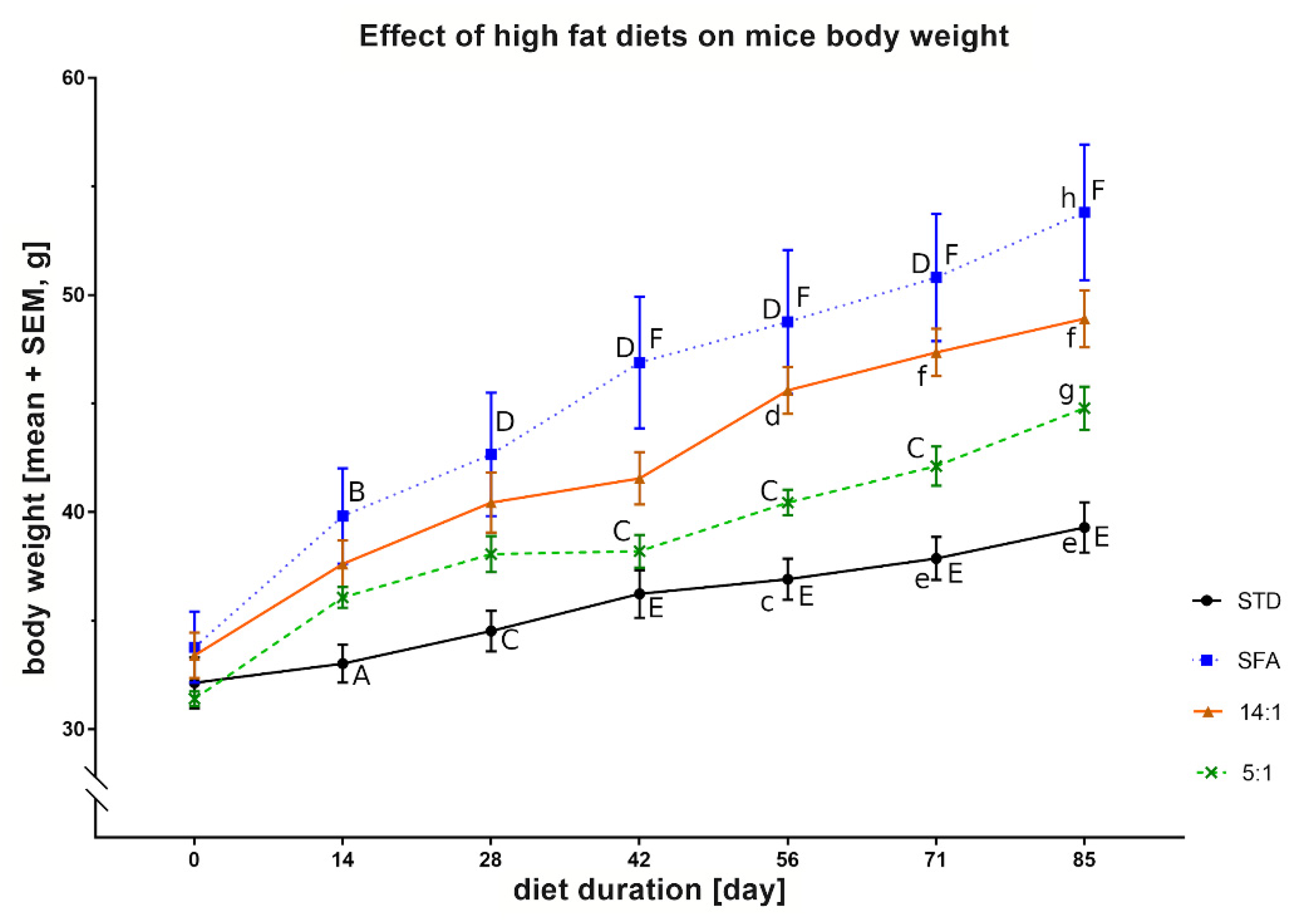
3. Results
3.1. Total Fat Content in Diets
3.2. Biochemical Blood Parameters
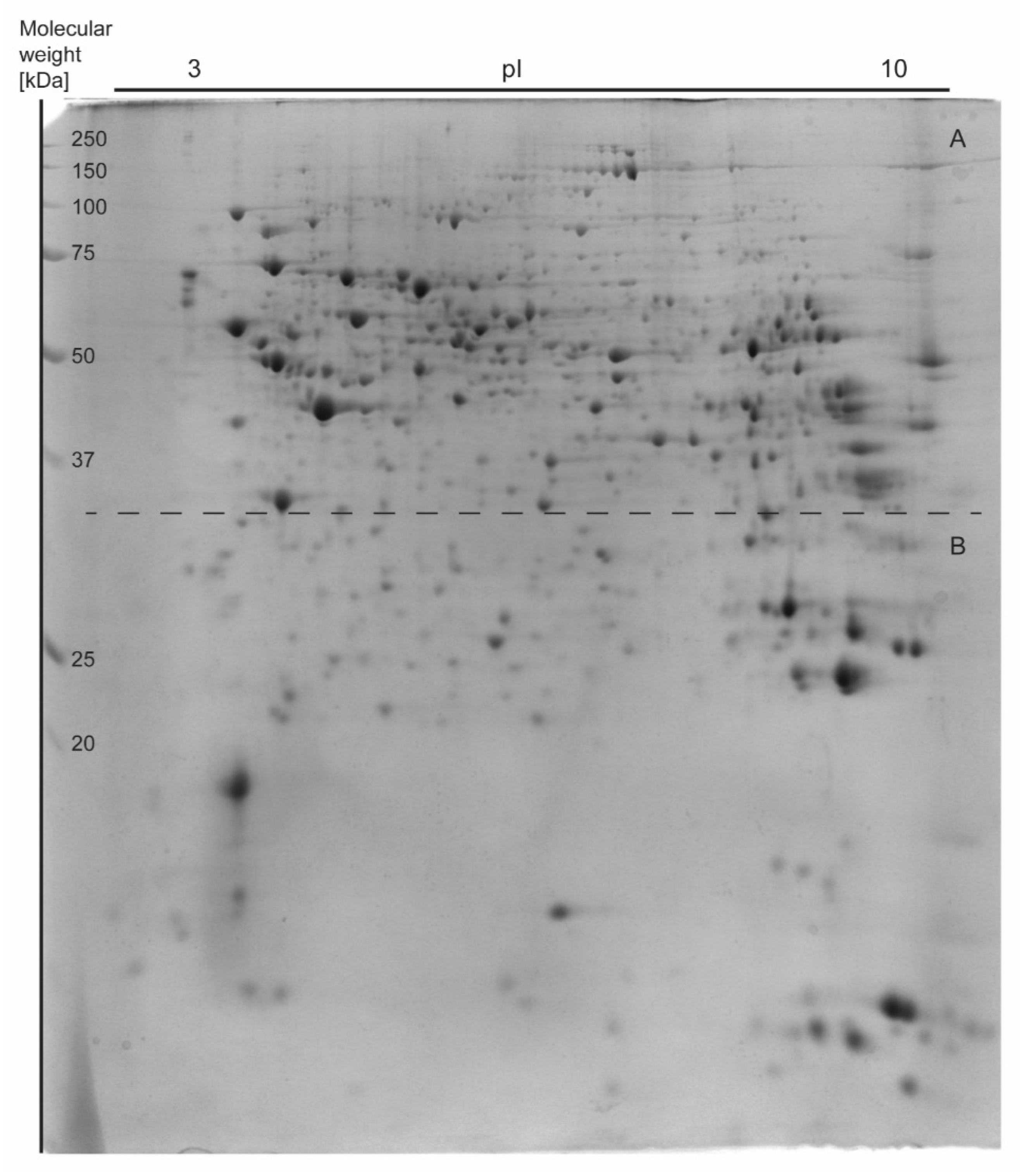
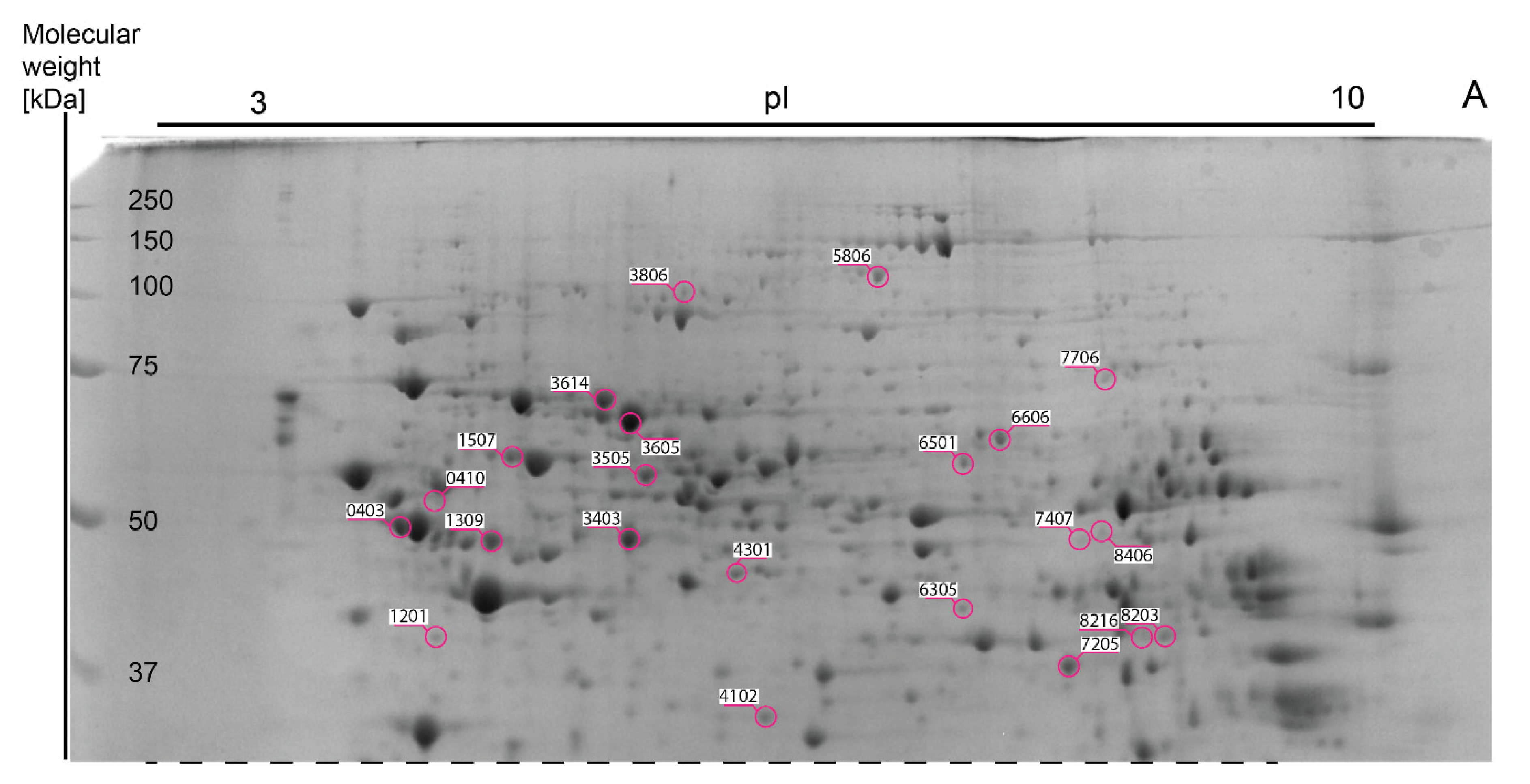
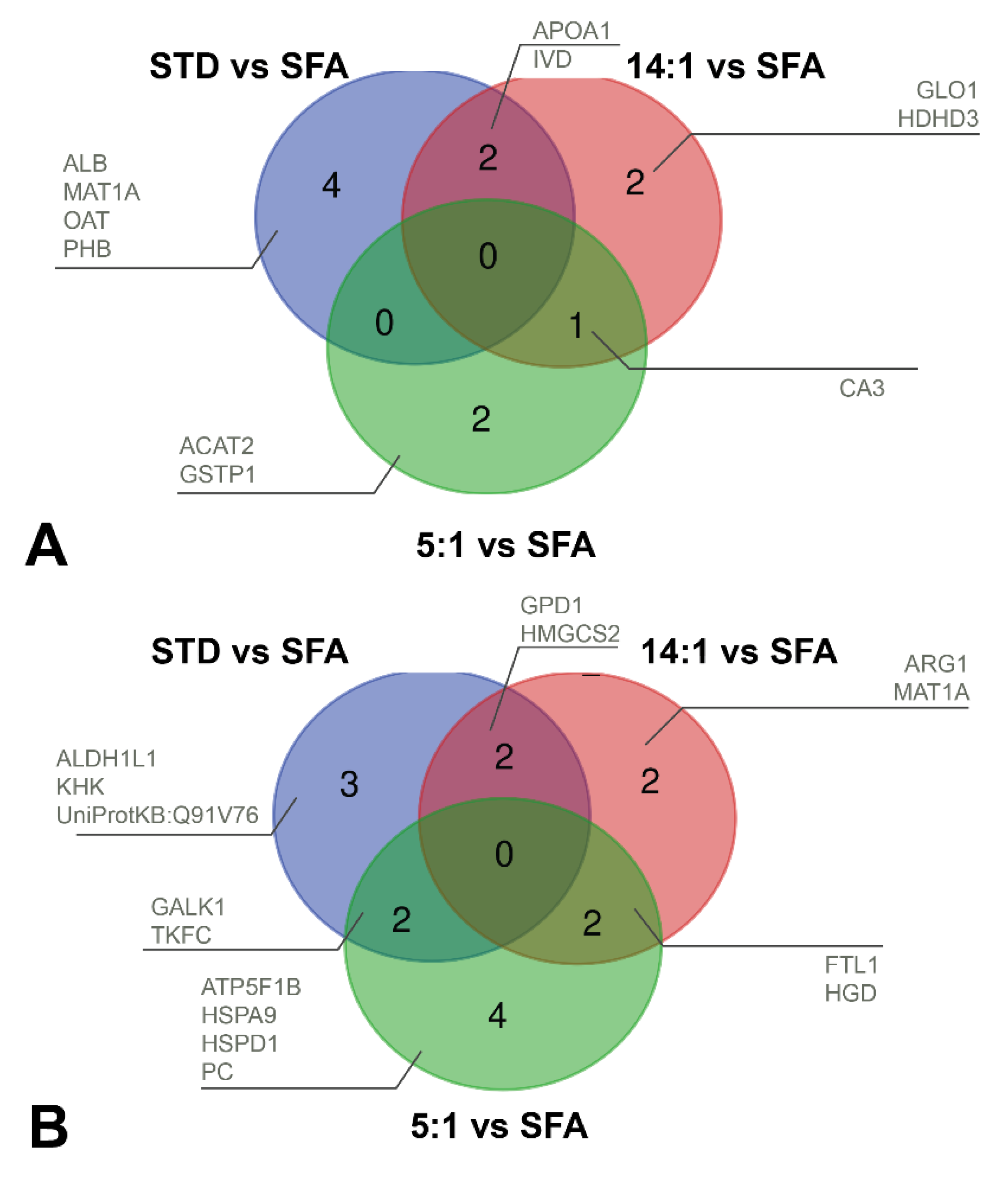
3.3. Analysis of Liver Proteome Differences
3.4. Controlling mRNA and Protein Expression Levels by High-Fat Diets
4. Discussion
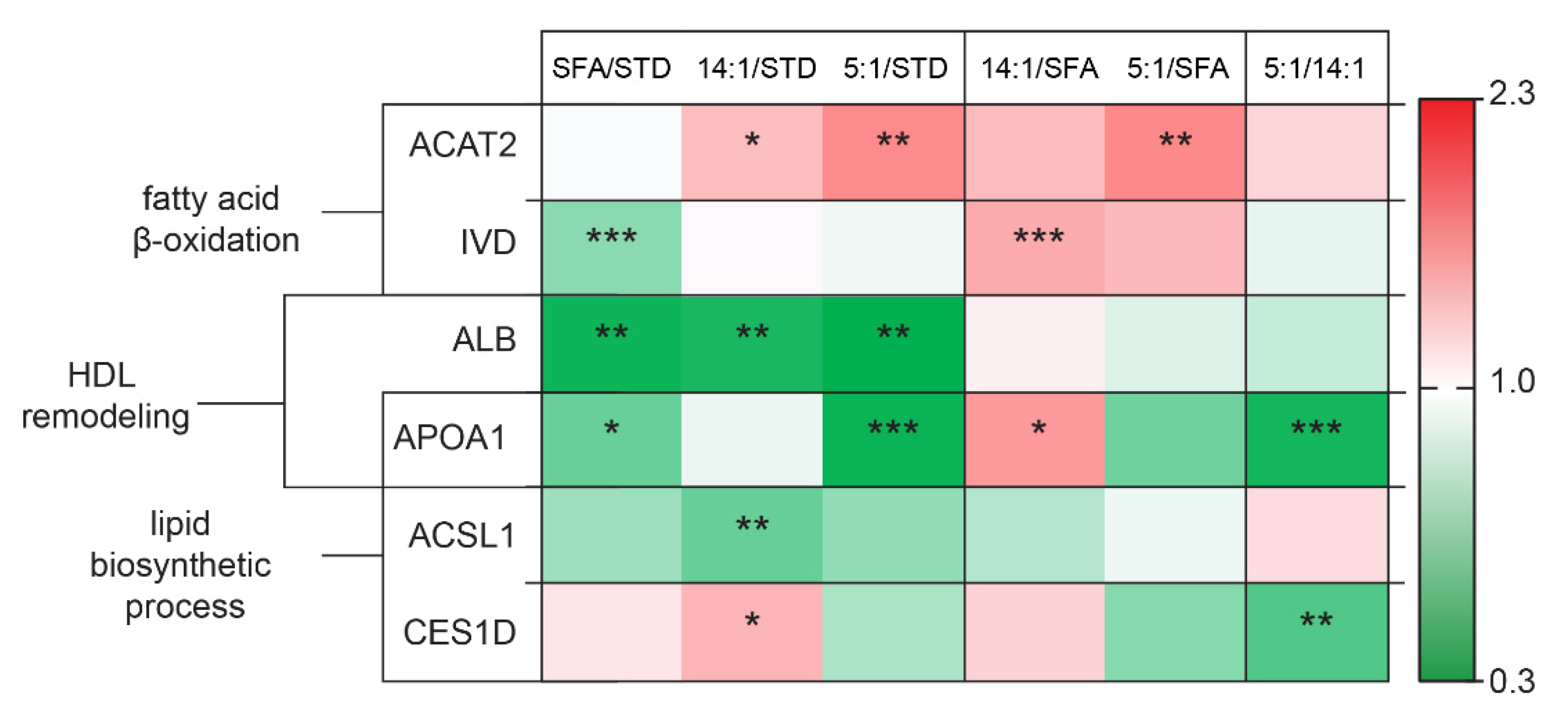
4.1. Differences in the Expression of Proteins Related to Lipid Metabolism
4.2. Changes in the Expression of Proteins Involved in Carbohydrate Metabolism
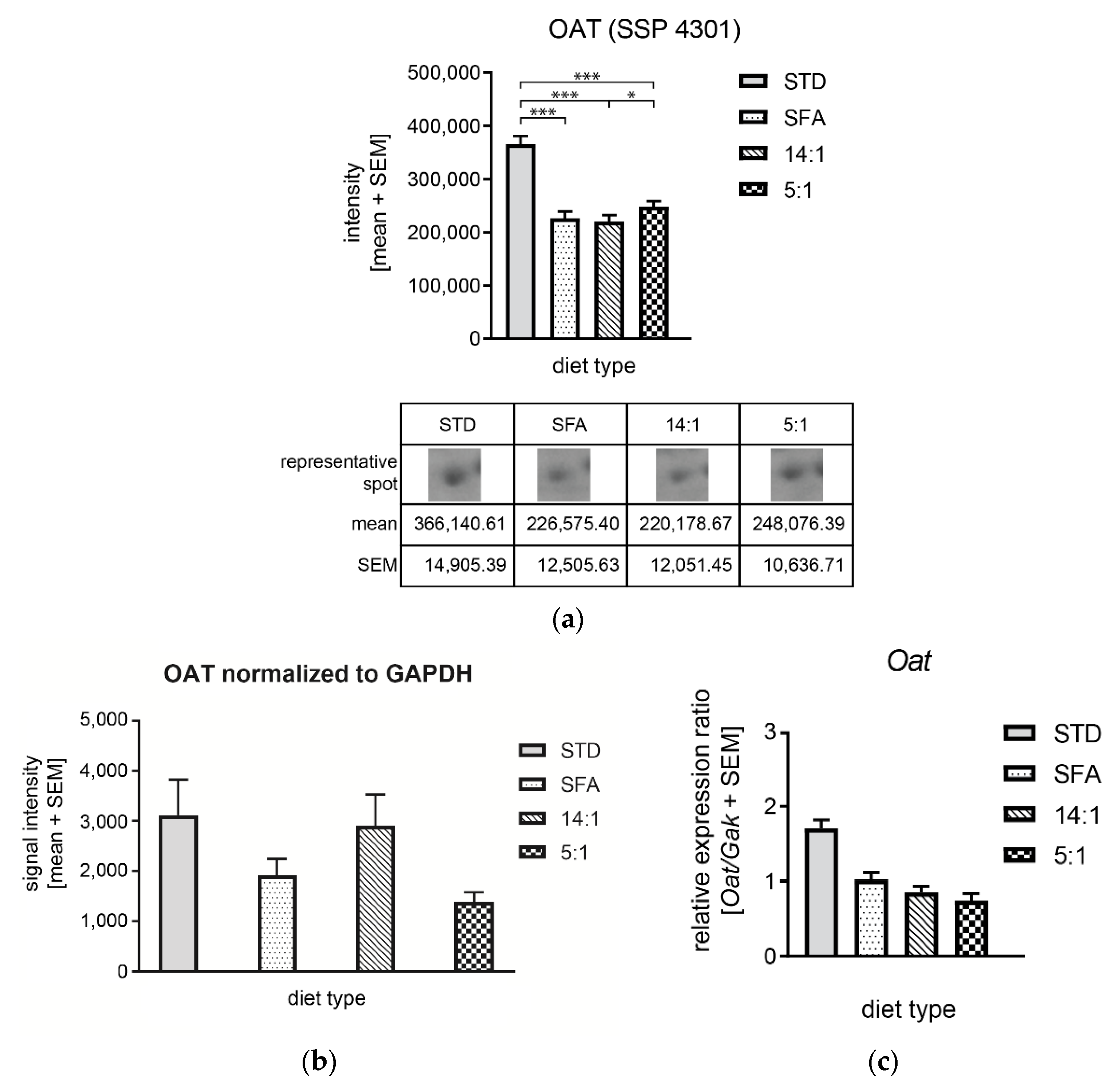
4.3. Changes in the Expression of Proteins Associated with Amino Acid Metabolism.
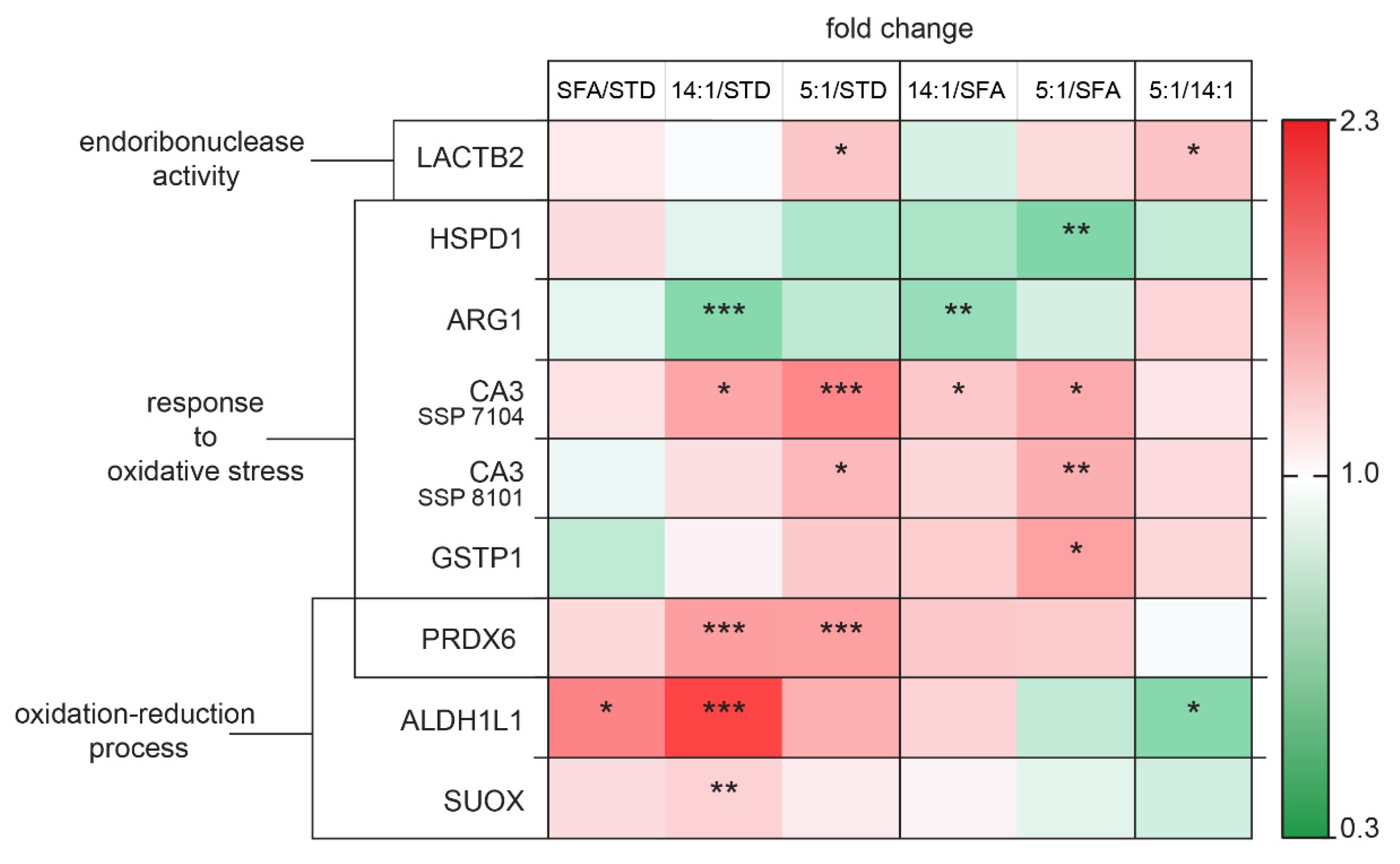
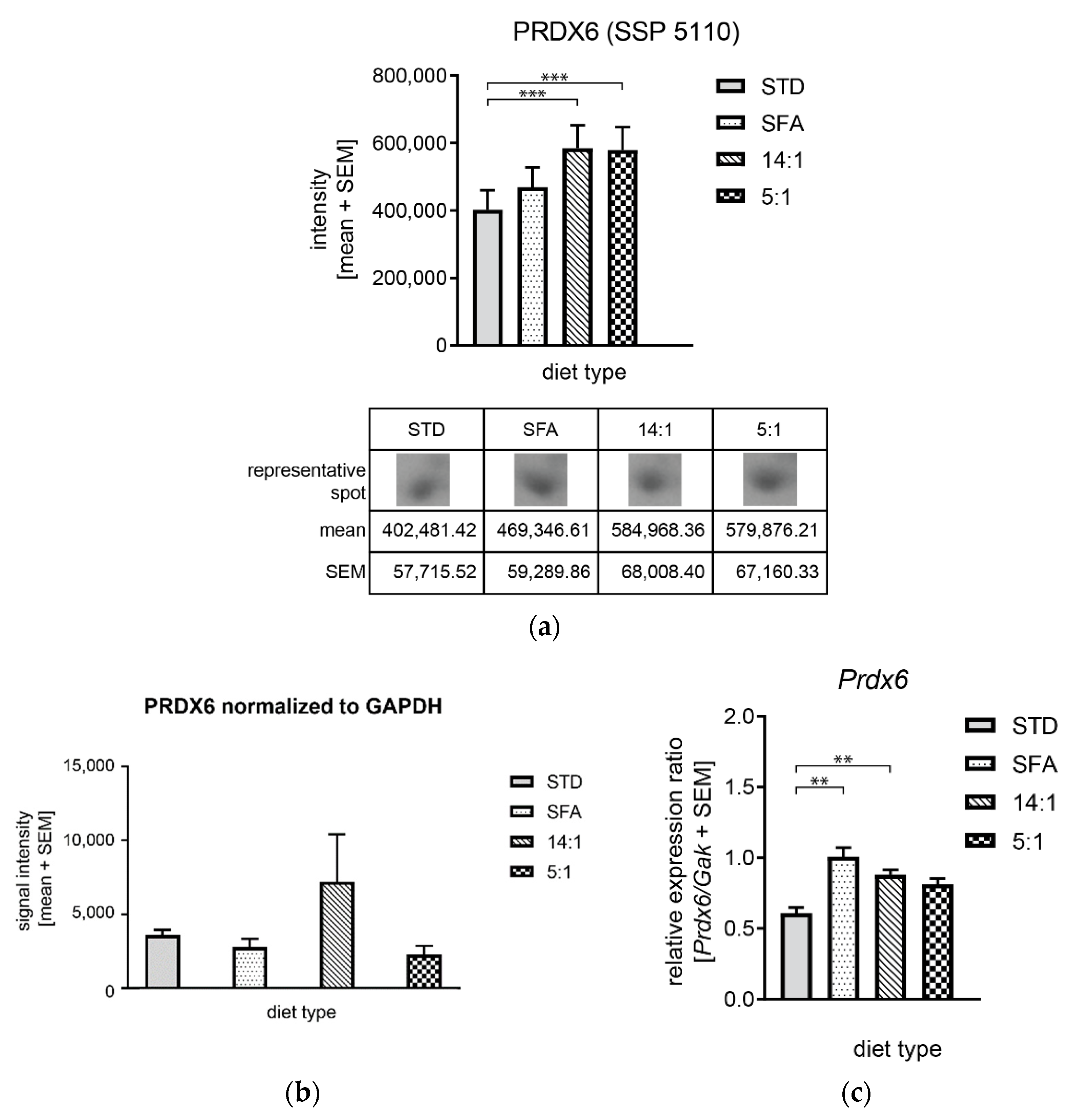
4.4. Changes in the Expression of Proteins Involved in Oxidative Stress Regulation
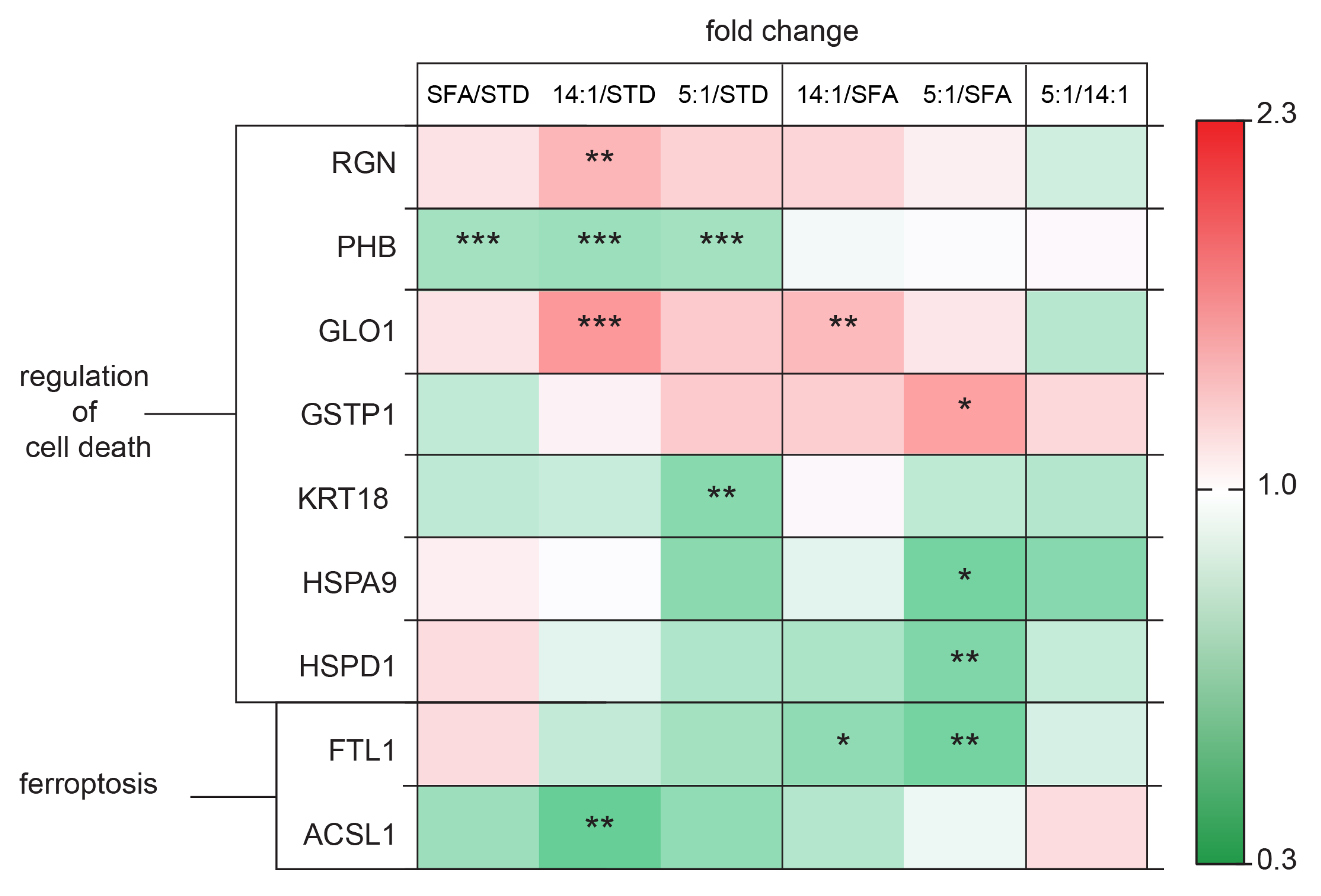
4.5. Changes in the Expression of Proteins Involved in Cell Death Processes
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Diao, P.; Wang, X.; Jia, F.; Kimura, T.; Hu, X.; Shirotori, S.; Nakamura, I.; Sato, Y.; Nakayama, J.; Moriya, K.; et al. A saturated fatty acid-rich diet enhances hepatic lipogenesis and tumorigenesis in HCV core gene transgenic mice. J. Nutr. Biochem. 2020, 85, 108460. [Google Scholar] [CrossRef]
- Lappano, R.; Sebastiani, A.; Cirillo, F.; Rigiracciolo, D.C.; Galli, G.R.; Curcio, R.; Malaguarnera, R.; Belfiore, A.; Cappello, A.R.; Maggiolini, M. The lauric acid-activated signaling prompts apoptosis in cancer cells. Cell Death Discov. 2017, 3, 17063. [Google Scholar] [CrossRef]
- Lima, R.D.S.; Block, J.M. Coconut oil: What do we really know about it so far? Food Qual. Saf. 2019, 3, 61–72. [Google Scholar] [CrossRef]
- Rothwell, N.J.; Stock, M.J. Stimulation of thermogenesis and brown fat activity in rats fed medium chain triglyceride. Metabolism 1987, 36, 128–130. [Google Scholar] [CrossRef]
- Van Wymelbeke, V.; Louis-Sylvestre, J.; Fantino, M. Substrate oxidation and control of food intake in men after a fat-substitute meal compared with meals supplemented with an isoenergetic load of carbohydrate, long-chain triacylglycerols, or medium-chain triacylglycerols. Am. J. Clin. Nutr. 2001, 74, 620–630. [Google Scholar] [CrossRef] [Green Version]
- Clegg, M.E. They say coconut oil can aid weight loss, but can it really? Eur. J. Clin. Nutr. 2017, 71, 1139–1143. [Google Scholar] [CrossRef]
- Department of Health and Human Services and U.S. Department of Agriculture (HHS) and (USDA). Dietary Guidelines for Americans, 8th ed.; US Department of Health and Human Services: Washington, DC, USA, 2015. Available online: http://health.gov/dietaryguidelines/2015/guidelines/ (accessed on 13 May 2021).
- Eckel, R.H.; Jakicic, J.M.; Ard, J.D.; de Jesus, J.M.; Houston Miller, N.; Hubbard, V.S.; Lee, I.M.; Lichtenstein, A.H.; Loria, C.M.; Millen, B.E.; et al. 2013 AHA/ACC guideline on lifestyle management to reduce cardiovascular risk: A report of the American College of Cardiology/American Heart Association Task Force on Practice Guidelines. Circulation 2014, 129, S76–S99. [Google Scholar] [CrossRef] [Green Version]
- Kromhout, D.; Menotti, A.; Bloemberg, B.; Aravanis, C.; Blackburn, H.; Buzina, R.; Dontas, A.S.; Fidanza, F.; Giampaoli, S.; Jansen, A.; et al. Dietary saturated and trans fatty acids and cholesterol and 25-year mortality from coronary heart disease: The Seven Countries Study. Prev. Med. 1995, 24, 308–315. [Google Scholar] [CrossRef]
- Billingsley, H.E.; Carbone, S.; Lavie, C.J. Dietary Fats and Chronic Noncommunicable Diseases. Nutrients 2018, 10, 1385. [Google Scholar] [CrossRef] [Green Version]
- Wang, D.D.; Li, Y.; Chiuve, S.E.; Stampfer, M.J.; Manson, J.E.; Rimm, E.B.; Willett, W.C.; Hu, F.B. Association of Specific Dietary Fats With Total and Cause-Specific Mortality. JAMA Intern. Med. 2016, 176, 1134–1145. [Google Scholar] [CrossRef] [PubMed]
- Gouaref, I.; Bouazza, A.; Abderrhmane, S.A.; Koceir, E.-A. Lipid Profile Modulates Cardiometabolic Risk Biomarkers Including Hypertension in People with Type-2 Diabetes: A Focus on Unbalanced Ratio of Plasma Polyunsaturated/Saturated Fatty Acids. Molecules 2020, 25, 4315. [Google Scholar] [CrossRef]
- Brennan, S.F.; Woodside, J.V.; Lunny, P.M.; Cardwell, C.R.; Cantwell, M.M. Dietary fat and breast cancer mortality: A systematic review and meta-analysis. Crit. Rev. Food Sci. Nutr. 2017, 57, 1999–2008. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Billingsley, H.E.; Hummel, S.L.; Carbone, S. The role of diet and nutrition in heart failure: A state-of-the-art narrative review. Prog. Cardiovasc. Dis. 2020, 63, 538–551. [Google Scholar] [CrossRef]
- Siri-Tarino, P.W.; Sun, Q.; Hu, F.B.; Krauss, R.M. Meta-analysis of prospective cohort studies evaluating the association of saturated fat with cardiovascular disease. Am. J. Clin. Nutr. 2010, 91, 535–546. [Google Scholar] [CrossRef] [Green Version]
- Unger, A.L.; Torres-Gonzalez, M.; Kraft, J. Dairy Fat Consumption and the Risk of Metabolic Syndrome: An Examination of the Saturated Fatty Acids in Dairy. Nutrients 2019, 11, 2200. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lepczyński, A.; Ożgo, M.; Michałek, K.; Dratwa-Chałupnik, A.; Grabowska, M.; Herosimczyk, A.; Liput, K.P.; Poławska, E.; Kram, A.; Pierzchała, M. Effects of Three-Month Feeding High Fat Diets with Different Fatty Acid Composition on Myocardial Proteome in Mice. Nutrients 2021, 13, 330. [Google Scholar] [CrossRef]
- Pink, M.; Verma, N.; Rettenmeier, A.W.; Schmitz-Spanke, S. CBB staining protocol with higher sensitivity and mass spectrometric compatibility. Electrophoresis 2010, 31, 593–598. [Google Scholar] [CrossRef]
- Dratwa-Chałupnik, A.; Lepczyński, A.; Ożgo, M.; Herosimczyk, A.; Michałek, K.; Niemcewicz, M.; Pałyszka, A.; Skrzypczak, W. Analysis of the efficiency of post-electrophoretic protein staining using Colloidal Coomassie Blue G-250. Acta Sci. Pol. Zootechnica 2015, 14, 67–76. [Google Scholar]
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P.; et al. STRING v11: Protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019, 47, D607–D613. [Google Scholar] [CrossRef] [Green Version]
- UniProt, C. UniProt: A worldwide hub of protein knowledge. Nucleic Acids Res. 2018, 47, D506–D515. [Google Scholar]
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T.; et al. Gene Ontology: Tool for the unification of biology. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carbon, S.; Douglass, E.; Dunn, N.; Good, B.; Harris, N.L.; Lewis, S.E.; Mungall, C.J.; Basu, S.; Chisholm, R.L.; Dodson, R.J.; et al. The Gene Ontology Resource: 20 years and still GOing strong. Nucleic Acids Res. 2018, 47, D330–D338. [Google Scholar]
- Cheng, X.; Xiao, X.; Chou, K.-C. pLoc-mEuk: Predict subcellular localization of multi-label eukaryotic proteins by extracting the key GO information into general PseAAC. Genomics 2018, 110, 50–58. [Google Scholar] [CrossRef]
- Gong, H.; Sun, L.; Chen, B.; Han, Y.; Pang, J.; Wu, W.; Qi, R.; Zhang, T.-M. Evaluation of candidate reference genes for RT-qPCR studies in three metabolism related tissues of mice after caloric restriction. Sci. Rep. 2016, 6, 38513. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hruz, T.; Wyss, M.; Docquier, M.; Pfaffl, M.W.; Masanetz, S.; Borghi, L.; Verbrugghe, P.; Kalaydjieva, L.; Bleuler, S.; Laule, O.; et al. RefGenes: Identification of reliable and condition specific reference genes for RT-qPCR data normalization. BMC Genom. 2011, 12, 156. [Google Scholar] [CrossRef] [Green Version]
- Andersen, C.L.; Jensen, J.L.; Ørntoft, T.F. Normalization of Real-Time Quantitative Reverse Transcription-PCR Data: A Model-Based Variance Estimation Approach to Identify Genes Suited for Normalization, Applied to Bladder and Colon Cancer Data Sets. Cancer Res. 2004, 64, 5245. [Google Scholar] [CrossRef] [Green Version]
- Ruijter, J.M.; Ramakers, C.; Hoogaars, W.M.H.; Karlen, Y.; Bakker, O.; van den Hoff, M.J.B.; Moorman, A.F.M. Amplification efficiency: Linking baseline and bias in the analysis of quantitative PCR data. Nucleic Acids Res. 2009, 37, e45. [Google Scholar] [CrossRef] [Green Version]
- Pfaffl, M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001, 29, e45. [Google Scholar] [CrossRef]
- Tyburski, J.; Studzińska, A.; Daca, P.; Tretyn, A. PCR w czasie rzeczywistym. Metody analizy danych. Biotechnologia 2008, 1, 86–96. [Google Scholar]
- Timmers, S.; de Vogel-van den Bosch, J.; de Wit, N.; Schaart, G.; van Beurden, D.; Hesselink, M.; van der Meer, R.; Schrauwen, P. Differential effects of saturated versus unsaturated dietary fatty acids on weight gain and myocellular lipid profiles in mice. Nutr. Diabetes 2011, 1, e11. [Google Scholar] [CrossRef] [Green Version]
- Beulen, Y.; Martínez-González, M.A.; van de Rest, O.; Salas-Salvadó, J.; Sorlí, J.V.; Gómez-Gracia, E.; Fiol, M.; Estruch, R.; Santos-Lozano, J.M.; Schröder, H.; et al. Quality of Dietary Fat Intake and Body Weight and Obesity in a Mediterranean Population: Secondary Analyses within the PREDIMED Trial. Nutrients 2018, 10, 2011. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rosqvist, F.; Iggman, D.; Kullberg, J.; Cedernaes, J.; Johansson, H.E.; Larsson, A.; Johansson, L.; Ahlström, H.; Arner, P.; Dahlman, I.; et al. Overfeeding polyunsaturated and saturated fat causes distinct effects on liver and visceral fat accumulation in humans. Diabetes 2014, 63, 2356–2368. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- He, Y.; Yang, T.; Du, Y.; Qin, L.; Ma, F.; Wu, Z.; Ling, H.; Yang, L.; Wang, Z.; Zhou, Q.; et al. High fat diet significantly changed the global gene expression profile involved in hepatic drug metabolism and pharmacokinetic system in mice. Nutr. Metab. 2020, 17, 37. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Li, J.; Cao, P.; Liu, Y. Sinapine-enriched rapeseed oils reduced fatty liver formation in high-fat diet-fed C57BL/6J mice. RSC Adv. 2020, 10, 21248–21258. [Google Scholar] [CrossRef]
- Kralova Lesna, I.; Suchanek, P.; Kovar, J.; Stavek, P.; Poledne, R. Replacement of dietary saturated FAs by PUFAs in diet and reverse cholesterol transport. J. Lipid Res. 2008, 49, 2414–2418. [Google Scholar] [CrossRef] [Green Version]
- Gaundal, L.; Myhrstad, M.C.W.; Leder, L.; Byfuglien, M.G.; Gjøvaag, T.; Rud, I.; Retterstøl, K.; Holven, K.B.; Ulven, S.M.; Telle-Hansen, V.H. Beneficial effect on serum cholesterol levels, but not glycaemic regulation, after replacing SFA with PUFA for 3 d: A randomised crossover trial. Br. J. Nutr. 2021, 125, 915–925. [Google Scholar] [CrossRef]
- Diniz, Y.S.A.; Cicogna, A.C.; Padovani, C.R.; Santana, L.S.; Faine, L.A.; Novelli, E.L.B. Diets rich in saturated and polyunsaturated fatty acids: Metabolic shifting and cardiac health. Nutrition 2004, 20, 230–234. [Google Scholar] [CrossRef]
- Soltis, A.R.; Kennedy, N.J.; Xin, X.; Zhou, F.; Ficarro, S.B.; Yap, Y.S.; Matthews, B.J.; Lauffenburger, D.A.; White, F.M.; Marto, J.A.; et al. Hepatic Dysfunction Caused by Consumption of a High-Fat Diet. Cell Rep. 2017, 21, 3317–3328. [Google Scholar] [CrossRef] [Green Version]
- Yoshimura, S.; Nakashima, S.; Tomiga, Y.; Kawakami, S.; Uehara, Y.; Higaki, Y. Short- and long-term effects of high-fat diet feeding and voluntary exercise on hepatic lipid metabolism in mice. Biochem. Biophys. Res. Commun. 2018, 507, 291–296. [Google Scholar] [CrossRef]
- Wang, Y.-J.; Bian, Y.; Luo, J.; Lu, M.; Xiong, Y.; Guo, S.-Y.; Yin, H.-Y.; Lin, X.; Li, Q.; Chang, C.C.Y.; et al. Cholesterol and fatty acids regulate cysteine ubiquitylation of ACAT2 through competitive oxidation. Nat. Cell Biol. 2017, 19, 808–819. [Google Scholar] [CrossRef] [Green Version]
- Temel, R.E.; Hou, L.; Rudel, L.L.; Shelness, G.S. ACAT2 stimulates cholesteryl ester secretion in apoB-containing lipoproteins. J. Lipid Res. 2007, 48, 1618–1627. [Google Scholar] [CrossRef] [Green Version]
- Bell, T.A.; Kelley, K.; Wilson, M.D.; Sawyer, J.K.; Rudel, L.L. Dietary Fat–Induced Alterations in Atherosclerosis Are Abolished by ACAT2-Deficiency in ApoB100 Only, LDLr−/− Mice. Arterioscler. Thromb. Vasc. Biol. 2007, 27, 1396–1402. [Google Scholar] [CrossRef] [Green Version]
- Guo, Y.; Darshi, M.; Ma, Y.; Perkins, G.A.; Shen, Z.; Haushalter, K.J.; Saito, R.; Chen, A.; Lee, Y.S.; Patel, H.H.; et al. Quantitative proteomic and functional analysis of liver mitochondria from high fat diet (HFD) diabetic mice. Mol. Cell Proteom. 2013, 12, 3744–3758. [Google Scholar] [CrossRef] [Green Version]
- Schlune, A.; Riederer, A.; Mayatepek, E.; Ensenauer, R. Aspects of Newborn Screening in Isovaleric Acidemia. Int. J. Neonatal Screen. 2018, 4, 7. [Google Scholar] [CrossRef] [Green Version]
- Solano, A.F.; Leipnitz, G.; De Bortoli, G.M.; Seminotti, B.; Amaral, A.U.; Fernandes, C.G.; Latini, A.S.; Dutra-Filho, C.S.; Wajner, M. Induction of oxidative stress by the metabolites accumulating in isovaleric acidemia in brain cortex of young rats. Free Radic. Res. 2008, 42, 707–715. [Google Scholar] [CrossRef]
- Botham, K.M.; Avella, M.; Cantafora, A.; Bravo, E. The lipolysis of chylomicrons derived from different dietary fats by lipoprotein lipase in vitro. Biochim. Biophys. Acta Lipids Lipid Metab. 1997, 1349, 257–263. [Google Scholar] [CrossRef]
- Ruan, X.; Li, Z.; Zhang, Y.; Yang, L.; Pan, Y.; Wang, Z.; Feng, G.-S.; Chen, Y. Apolipoprotein A-I possesses an anti-obesity effect associated with increase of energy expenditure and up-regulation of UCP1 in brown fat. J. Cell Mol. Med. 2011, 15, 763–772. [Google Scholar] [CrossRef] [Green Version]
- Wei, H.; Averill, M.M.; McMillen, T.S.; Dastvan, F.; Mitra, P.; Subramanian, S.; Tang, C.; Chait, A.; LeBoeuf, R.C. Modulation of adipose tissue lipolysis and body weight by high-density lipoproteins in mice. Nutr. Diabetes 2014, 4, e108. [Google Scholar] [CrossRef] [Green Version]
- Han, R.; Lai, R.; Ding, Q.; Wang, Z.; Luo, X.; Zhang, Y.; Cui, G.; He, J.; Liu, W.; Chen, Y. Apolipoprotein A-I stimulates AMP-activated protein kinase and improves glucose metabolism. Diabetologia 2007, 50, 1960–1968. [Google Scholar] [CrossRef] [Green Version]
- Lian, J.; Wei, E.; Groenendyk, J.; Das, S.K.; Hermansson, M.; Li, L.; Watts, R.; Thiesen, A.; Oudit, G.Y.; Michalak, M.; et al. Ces3/TGH Deficiency Attenuates Steatohepatitis. Sci. Rep. 2016, 6, 25747. [Google Scholar] [CrossRef] [Green Version]
- Lian, J.; Wei, E.; Wang, S.P.; Quiroga, A.D.; Li, L.; Di Pardo, A.; van der Veen, J.; Sipione, S.; Mitchell, G.A.; Lehner, R. Liver specific inactivation of carboxylesterase 3/triacylglycerol hydrolase decreases blood lipids without causing severe steatosis in mice. Hepatology 2012, 56, 2154–2162. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez, J.T.; Betts, J.A. Dietary sugars, exercise and hepatic carbohydrate metabolism. Proc. Nutr. Soc. 2018, 78, 246–256. [Google Scholar] [CrossRef]
- Softic, S.; Gupta, M.K.; Wang, G.-X.; Fujisaka, S.; O’Neill, B.T.; Rao, T.N.; Willoughby, J.; Harbison, C.; Fitzgerald, K.; Ilkayeva, O.; et al. Divergent effects of glucose and fructose on hepatic lipogenesis and insulin signaling. J. Clin. Investig. 2017, 127, 4059–4074. [Google Scholar] [CrossRef] [Green Version]
- Kumashiro, N.; Beddow, S.A.; Vatner, D.F.; Majumdar, S.K.; Cantley, J.L.; Guebre-Egziabher, F.; Fat, I.; Guigni, B.; Jurczak, M.J.; Birkenfeld, A.L.; et al. Targeting pyruvate carboxylase reduces gluconeogenesis and adiposity and improves insulin resistance. Diabetes 2013, 62, 2183–2194. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cai, D.; Yuan, M.; Liu, H.; Han, Z.; Pan, S.; Yang, Y.; Zhao, R. Epigenetic and SP1-mediated regulation is involved in the repression of galactokinase 1 gene in the liver of neonatal piglets born to betaine-supplemented sows. Eur. J. Nutr. 2017, 56, 1899–1909. [Google Scholar] [CrossRef]
- Coelho, A.I.; Berry, G.T.; Rubio-Gozalbo, M.E. Galactose metabolism and health. Curr. Opin. Clin. Nutr. Metab. Care 2015, 18, 422–427. [Google Scholar] [CrossRef]
- Ramani, P.; Arya, K. Galactokinase Deficiency. StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing. Available online: https://www.ncbi.nlm.nih.gov/books/NBK560683/ (accessed on 13 May 2021).
- Bashir, S.; Sharma, Y.; Jairajpuri, D.; Rashid, F.; Nematullah, M.; Khan, F. Alteration of adipose tissue immune cell milieu towards the suppression of inflammation in high fat diet fed mice by flaxseed oil supplementation. PLoS ONE 2019, 14, e0223070. [Google Scholar] [CrossRef]
- Ito, T.; Kubo, M.; Nagaoka, K.; Funakubo, N.; Setiawan, H.; Takemoto, K.; Eguchi, E.; Fujikura, Y.; Ogino, K. Early obesity leads to increases in hepatic arginase I and related systemic changes in nitric oxide and l-arginine metabolism in mice. J. Physiol. Biochem. 2018, 74, 9–16. [Google Scholar] [CrossRef]
- Johnson, F.K.; Peyton, K.J.; Liu, X.-M.; Azam, M.A.; Shebib, A.R.; Johnson, R.A.; Durante, W. Arginase promotes endothelial dysfunction and hypertension in obese rats. Obesity 2015, 23, 383–390. [Google Scholar] [CrossRef] [Green Version]
- Erdely, A.; Kepka-Lenhart, D.; Salmen-Muniz, R.; Chapman, R.; Hulderman, T.; Kashon, M.; Simeonova, P.P.; Morris, S.M., Jr. Arginase activities and global arginine bioavailability in wild-type and ApoE-deficient mice: Responses to high fat and high cholesterol diets. PLoS ONE 2010, 5, e15253. [Google Scholar] [CrossRef] [Green Version]
- Romero, M.J.; Platt, D.H.; Tawfik, H.E.; Labazi, M.; El-Remessy, A.B.; Bartoli, M.; Caldwell, R.B.; Caldwell, R.W. Diabetes-induced coronary vascular dysfunction involves increased arginase activity. Circ. Res. 2008, 102, 95–102. [Google Scholar] [CrossRef]
- Chung, J.H.; Moon, J.; Lee, Y.S.; Chung, H.-K.; Lee, S.-M.; Shin, M.-J. Arginase inhibition restores endothelial function in diet-induced obesity. Biochem. Biophys. Res. Commun. 2014, 451, 179–183. [Google Scholar] [CrossRef]
- Moon, J.; Do, H.J.; Cho, Y.; Shin, M.-J. Arginase inhibition ameliorates hepatic metabolic abnormalities in obese mice. PLoS ONE 2014, 9, e103048. [Google Scholar] [CrossRef] [Green Version]
- Ahmed, A.A.; Balogun, K.A.; Bykova, N.V.; Cheema, S.K. Novel regulatory roles of omega-3 fatty acids in metabolic pathways: A proteomics approach. Nutr. Metab. 2014, 11, 6. [Google Scholar] [CrossRef] [Green Version]
- Luo, M.; Mengos, A.E.; Stubblefield, T.M.; Mandarino, L.J. High fat diet-induced changes in hepatic protein abundance in mice. J. Proteom. Bioinform. 2012, 5, 60–66. [Google Scholar] [CrossRef] [Green Version]
- Ventura, G.; De Bandt, J.-P.; Segaud, F.; Perret, C.; Robic, D.; Levillain, O.; Le Plenier, S.; Godard, C.; Cynober, L.; Moinard, C. Overexpression of ornithine aminotransferase: Consequences on amino acid homeostasis. Br. J. Nutr. 2008, 101, 843–851. [Google Scholar] [CrossRef] [Green Version]
- Rakhshandehroo, M.; Knoch, B.; Müller, M.; Kersten, S. Peroxisome Proliferator-Activated Receptor Alpha Target Genes. PPAR Res. 2010, 2010, 612089. [Google Scholar] [CrossRef] [Green Version]
- Zúñiga, J.; Cancino, M.; Medina, F.; Varela, P.; Vargas, R.; Tapia, G.; Videla, L.A.; Fernández, V. N-3 PUFA supplementation triggers PPAR-α activation and PPAR-α/NF-κB interaction: Anti-inflammatory implications in liver ischemia-reperfusion injury. PLoS ONE 2011, 6, e28502. [Google Scholar] [CrossRef] [Green Version]
- Zigmond, E.; Ben Ya’acov, A.; Lee, H.; Lichtenstein, Y.; Shalev, Z.; Smith, Y.; Zolotarov, L.; Ziv, E.; Kalman, R.; Le, H.V.; et al. Suppression of Hepatocellular Carcinoma by Inhibition of Overexpressed Ornithine Aminotransferase. ACS Med. Chem. Lett. 2015, 6, 840–844. [Google Scholar] [CrossRef] [Green Version]
- Méndez, L.; Ciordia, S.; Fernández, M.S.; Juárez, S.; Ramos, A.; Pazos, M.; Gallardo, J.M.; Torres, J.L.; Nogués, M.R.; Medina, I. Changes in liver proteins of rats fed standard and high-fat and sucrose diets induced by fish omega-3 PUFAs and their combination with grape polyphenols according to quantitative proteomics. J. Nutr. Biochem. 2017, 41, 84–97. [Google Scholar] [CrossRef] [Green Version]
- Ciapaite, J.; van den Broek, N.M.; te Brinke, H.; Nicolay, K.; Jeneson, J.A.; Houten, S.M.; Prompers, J.J. Differential effects of short- and long-term high-fat diet feeding on hepatic fatty acid metabolism in rats. Biochim. Biophys. Acta Mol. Cell Biol. Lipids. 2011, 1811, 441–451. [Google Scholar] [CrossRef]
- Su, L.-J.; Zhang, J.-H.; Gomez, H.; Murugan, R.; Hong, X.; Xu, D.; Jiang, F.; Peng, Z.-Y. Reactive Oxygen Species-Induced Lipid Peroxidation in Apoptosis, Autophagy, and Ferroptosis. Oxid. Med. Cell Longev. 2019, 2019, 5080843. [Google Scholar] [CrossRef] [Green Version]
- Fisher, A.B. Peroxiredoxin 6: A bifunctional enzyme with glutathione peroxidase and phospholipase A₂ activities. Antioxid. Redox Signal. 2011, 15, 831–844. [Google Scholar] [CrossRef] [Green Version]
- Arriga, R.; Pacifici, F.; Capuani, B.; Coppola, A.; Orlandi, A.; Scioli, M.G.; Pastore, D.; Andreadi, A.; Sbraccia, P.; Tesauro, M.; et al. Peroxiredoxin 6 Is a Key Antioxidant Enzyme in Modulating the Link between Glycemic and Lipogenic Metabolism. Oxid. Med. Cell Longev. 2019, 2019, 9685607. [Google Scholar] [CrossRef] [PubMed]
- Tu, Q.; Xiong, Y.; Fan, L.; Qiao, B.; Xia, Z.; Hu, L.; Wang, Y.; Peng, G.; Ye, Q. Peroxiredoxin 6 attenuates ischemia- and hypoxia-induced liver damage of brain-dead donors. Mol. Med. Rep. 2016, 13, 753–761. [Google Scholar] [CrossRef] [Green Version]
- Lee, D.H.; Jung, Y.Y.; Park, M.H.; Jo, M.R.; Han, S.B.; Yoon, D.Y.; Roh, Y.S.; Hong, J.T. Peroxiredoxin 6 Confers Protection Against Nonalcoholic Fatty Liver Disease Through Maintaining Mitochondrial Function. Antioxid. Redox Signal. 2019, 31, 387–402. [Google Scholar] [CrossRef]
- Voos, W.; Röttgers, K. Molecular chaperones as essential mediators of mitochondrial biogenesis. Biochim. Biophys. Acta Mol. Cell Res. 2002, 1592, 51–62. [Google Scholar] [CrossRef] [Green Version]
- Renner, S.W.; Walker, L.M.; Forsberg, L.J.; Sexton, J.Z.; Brenman, J.E. Carbonic anhydrase III (Car3) is not required for fatty acid synthesis and does not protect against high-fat diet induced obesity in mice. PLoS ONE 2017, 12, e0176502. [Google Scholar] [CrossRef] [Green Version]
- Zimmerman, U.-J.P.; Wang, P.; Zhang, X.; Bogdanovich, S.; Forster, R.E. Anti-Oxidative Response of Carbonic Anhydrase III in Skeletal Muscle. IUBMB Life 2004, 56, 343–347. [Google Scholar]
- Malcolm, T.; Aroor, A.; Bivin, W.; Sowers, J.; Woodman, R.; Lockette, W. Carbonic anhydrase mediates hepatic steatosis and vascular stiffness induced by a Western diet. FASEB J. Off. Publ. Fed. of Am. Soc. Exp. Biol. 2016, 30, 956.958. [Google Scholar]
- Xia, L.; Liu, Y.; Zhang, S.; Yang, Y.; Zhou, Z.; Tu, J. Can Prohibitin 1 be a Safeguard against liver disease? Ann. Hepatol. 2019, 18, 790–795. [Google Scholar] [CrossRef]
- Peng, Y.-T.; Chen, P.; Ouyang, R.-Y.; Song, L. Multifaceted role of prohibitin in cell survival and apoptosis. Apoptosis 2015, 20, 1135–1149. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ko, K.S.; Tomasi, M.L.; Iglesias-Ara, A.; French, B.A.; French, S.W.; Ramani, K.; Lozano, J.J.; Oh, P.; He, L.; Stiles, B.L.; et al. Liver-specific deletion of prohibitin 1 results in spontaneous liver injury, fibrosis, and hepatocellular carcinoma in mice. Hepatology 2010, 52, 2096–2108. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yamaguchi, M. The role of regucalcin in nuclear regulation of regenerating liver. Biochem. Biophys. Res. Commun. 2000, 276, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi, M. Regucalcin, a novel regulatory protein implicated in obesity and diabetes. Integr. Mol. Med. 2014, 1, 7–10. [Google Scholar] [CrossRef]
- Yamaguchi, M.; Tsurusaki, Y.; Misawa, H.; Inagaki, S.; Ma, Z.J.; Takahashi, H. Potential role of regucalcin as a specific biochemical marker of chronic liver injury with carbon tetrachloride administration in rats. Mol. Cell Biochem. 2002, 241, 61–67. [Google Scholar] [CrossRef]
- Kucukoglu, O.; Guldiken, N.; Chen, Y.; Usachov, V.; El-Heliebi, A.; Haybaeck, J.; Denk, H.; Trautwein, C.; Strnad, P. High-fat diet triggers Mallory-Denk body formation through misfolding and crosslinking of excess keratin 8. Hepatology 2014, 60, 169–178. [Google Scholar] [CrossRef] [PubMed]
- Singla, A.; Moons, D.S.; Snider, N.T.; Wagenmaker, E.R.; Jayasundera, V.B.; Omary, M.B. Oxidative stress, Nrf2 and keratin up-regulation associate with Mallory-Denk body formation in mouse erythropoietic protoporphyria. Hepatology 2012, 56, 322–331. [Google Scholar] [CrossRef] [Green Version]
- Guldiken, N.; Usachov, V.; Levada, K.; Trautwein, C.; Ziol, M.; Nahon, P.; Strnad, P. Keratins 8 and 18 are type II acute-phase responsive genes overexpressed in human liver disease. Liver Int. 2015, 35, 1203–1212. [Google Scholar] [CrossRef]
- Harada, M.; Strnad, P.; Resurreccion, E.Z.; Ku, N.-O.; Omary, M.B. Keratin 18 overexpression but not phosphorylation or filament organization blocks mouse Mallory body formation. Hepatology 2007, 45, 88–96. [Google Scholar] [CrossRef]
- Bettermann, K.; Mehta, A.K.; Hofer, E.M.; Wohlrab, C.; Golob-Schwarzl, N.; Svendova, V.; Schimek, M.G.; Stumptner, C.; Thüringer, A.; Speicher, M.R.; et al. Keratin 18-deficiency results in steatohepatitis and liver tumors in old mice: A model of steatohepatitis-associated liver carcinogenesis. Oncotarget 2016, 7, 73309–73322. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wolf, J.H.; Bhatti, T.R.; Fouraschen, S.; Chakravorty, S.; Wang, L.; Kurian, S.; Salomon, D.; Olthoff, K.M.; Hancock, W.W.; Levine, M.H. Heat shock protein 70 is required for optimal liver regeneration after partial hepatectomy in mice. Liver Transpl. 2014, 20, 376–385. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bondia-Pons, I.; Boqué, N.; Paternain, L.; Santamaría, E.; Fernández, J.; Campión, J.; Milagro, F.; Corrales, F.; Martínez, J.A. Liver proteome changes induced by a short-term high-fat sucrose diet in wistar rats. J. Nutrigenet Nutrigenomics 2011, 4, 344–353. [Google Scholar] [CrossRef]
- Maio, N.; Rouault, T.A. Outlining the Complex Pathway of Mammalian Fe-S Cluster Biogenesis. Trends Biochem. Sci. 2020, 45, 411–426. [Google Scholar] [CrossRef] [PubMed]
- Kang, Q.; Zou, H.; Yang, X.; Cai, J.-B.; Liu, L.-X.; Xie, N.; Wang, L.-M.; Li, Y.-H.; Zhang, X.-W. Characterization and prognostic significance of mortalin, Bcl-2 and Bax in intrahepatic cholangiocarcinoma. Oncol. Lett. 2018, 15, 2161–2168. [Google Scholar] [CrossRef]
- Yi, X.; Luk, J.M.; Lee, N.P.; Peng, J.; Leng, X.; Guan, X.-Y.; Lau, G.K.; Beretta, L.; Fan, S.-T. Association of Mortalin (HSPA9) with Liver Cancer Metastasis and Prediction for Early Tumor Recurrence. Mol. Cell Proteomics 2008, 7, 315–325. [Google Scholar] [CrossRef] [Green Version]
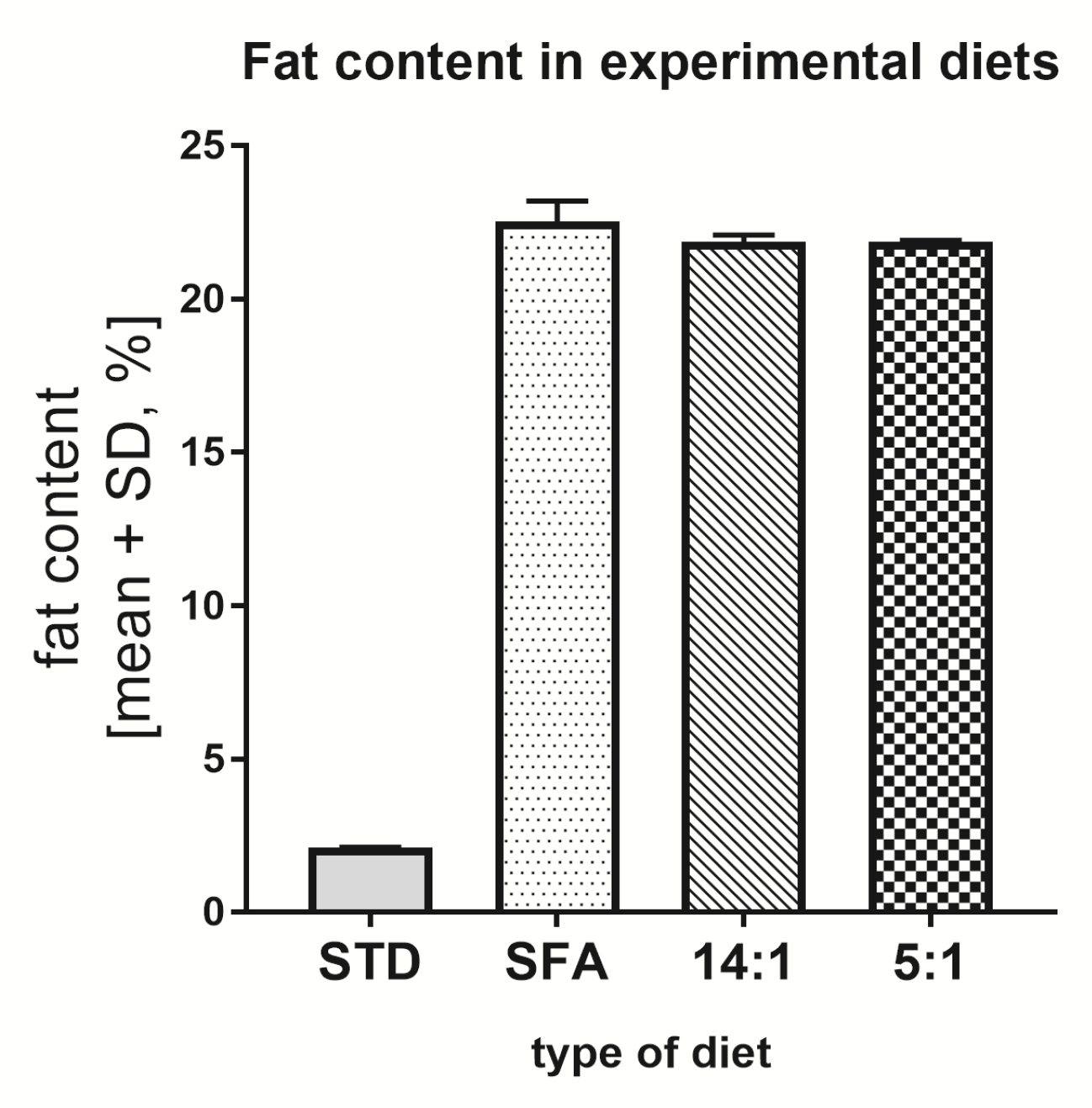



| Group | Components | [g] | LA/ALA | % SFA | % PUFA | % MUFA |
|---|---|---|---|---|---|---|
| SFA | Labofeed H | 790 | 1.41 | 76.87 | 11.04 | 12.09 |
| virgin coconut oil | 200 | |||||
| pumpkin seed oil | 10 | |||||
| 14:1 | Labofeed H | 790 | 13.76 | 1.68 | 82.21 | 16.10 |
| pumpkin seed oil | 210 | |||||
| 5:1 | Labofeed H | 790 | 5.00 | 9.91 | 79.69 | 10.40 |
| sunflower seed oil | 80 | |||||
| pumpkin seed oil | 65 | |||||
| avocado oil | 20 | |||||
| virgin coconut oil | 20 | |||||
| hemp seed oil | 15 | |||||
| corn oil | 10 |
| Parameter | STD | SFA | 14:1 | 5:1 |
|---|---|---|---|---|
| albumin [g/L] | 33.34 (1.51) | 32.39 (1.74) | 34.01 (1.91) | 31.54 (1.36) |
| total protein [g/L] | 53.61 (4.19) | 57.09 (7.38) | 59.01 (8.48) | 49.80 (2.21) |
| total bilirubin [μmol/L] | 1.58 (0.65) | 1.65 (0.34) | 1.09 (0.54) | 1.49 (0.59) |
| alanine aminotransferase [U/L] | 41.00 (17.78) | 36.25 (10.31) | 38.25 (16.00) | 32.75 (6.02) |
| aspartate aminotransferase [U/L] | 138.13 (62.66) | 93.00 (21.84) | 106.50 (47.88) | 109.13 (65.12) |
| cholinesterase [U/L] | 8219.75 (2215.14) | 6931.88 (981.76) | 6659.50 (1134.28) | 6563.75 (488.00) |
| lipase [U/L] | 13.66 (1.15) | 21.04 (2.95) | 23.39 (6.48) | 18.49 (2.97) |
| iron [μmol/L] | 19.99 (4.52) | 26.33 (5.22) | 26.14 (4.50) | 25.84 (4.42) |
| glucose [mmol/L] | 5.29 C,E (1.66) | 12.01 D (1.32) | 10.60 D (1.68) | 11.40 F (1.44) |
| total cholesterol [mmol/L] | 2.83 A,C (0.31) | 3.98 B (0.48) | 4.44 D (0.63) | 3.60 (0.58) |
| high-density lipoprotein-cholesterol [mmol/L] | 2.79 (0.33) | 3.81 (0.64) | 4.12 (0.86) | 3.53 (0.58) |
| low-density lipoprotein-cholesterol [mmol/L] | 0.12 (0.13) | 0.39 (0.18) | 0.37 (0.27) | 0.31 (0.20) |
| triacylglycerols [mmol/L] | 0.91 (0.28) | 1.39 (0.38) | 1.13 (0.17) | 1.17 (0.31) |
| Spot Number (SSP) | Protein Name | Accession Number UniProtKB/NCBI | Gene Symbol | MASCOT Score | E-Value | Sequence Coverage | Peptides Matched /Peptides Searched | Theoretical pI/MW (Da) | Fold Change | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SFA/STD | 14:1/STD | 5:1/STD | 14:1/SFA | 5:1/SFA | 5:1/14:1 | |||||||||
| 0106 | Regucalcin | Q64374/NP_033086.1 | Rgn | 188 | 1.1e-14 | 61% | 16/28 | 5.15/ 33,899 | 1.13 | 1.34 (**) | 1.20 | 1.19 | 1.07 | 0.90 |
| 0403 | ATP synthase subunit beta, mitochondrial | P56480/NP_058054.2 | Atp5f1b | 346 | 1.7e-30 | 70% | 32/49 | 5.19/ 56,265 | 1.39 | 1.29 | 0.78 | 0.92 | 0.56 (**) | 0.61 (*) |
| 0410 | ATP synthase subunit beta, mitochondrial | P56480/NP_058054.2 | Atp5f1b | 298 | 1.1e-25 | 59% | 28/37 | 5.19/ 56,265 | 0.79 | 0.72 | 1.58 | 0.91 | 2.00 | 2.21 (*) |
| 1001 | Lactoylglutathione lyase | Q9CPU0/NP_001107032.1 | Glo1 | 72 | 3.90e-03 | 30% | 5/13 | 5.24/ 20,967 | 1.12 | 1.48 (***) | 1.24 | 1.32 (**) | 1.11 | 0.84 |
| 1002 | Apolipoprotein A-I | Q00623/NP_033822.2 | Apoa1 | 168 | 1.1e-12 | 48% | 18/46 | 5.51/ 30,597 | 0.65 (*) | 0.96 | 0.44 (***) | 1.47 (*) | 0.67 | 0.46 (***) |
| 1201 | Galactokinase | Q9R0N0/NP_058601.2 | Galk1 | 95 | 2.1e-05 | 33% | 11/40 | 5.17/ 42,668 | 1.36 (**) | 1.36 (**) | 1.05 | 1.00 | 0.78 (*) | 0.77 (*) |
| 1309 | Keratin, type I cytoskeletal 18 | P05784/NP_034794.2 | Krt18 | 256 | 1.7e-21 | 66% | 27/61 | 5.22/ 47,509 | 0.86 | 0.88 | 0.73 (**) | 1.03 | 0.86 | 0.83 |
| 1507 | 60 kDa heat shock protein, mitochondrial | P63038/NP_034607.3 | Hspd1 | 213 | 3.4e-17 | 42% | 22/38 | 5.91/ 61,088 | 1.15 | 0.94 | 0.82 | 0.82 | 0.71 (**) | 0.87 |
| 2102 | Prohibitin | P67778/NP_032857.1 | Phb | 184 | 2.7e-14 | 65% | 13/26 | 5.57/ 29,859 | 0.80 (***) | 0.78 (***) | 0.79 (***) | 0.98 | 1.00 | 1.02 |
| 2104 | NADH dehydrogenase [ubiquinone] iron-sulfur protein 3, mitochondrial | Q9DCT2/NP_080964.1 | Ndufs3 | 155 | 2.1e-11 | 47% | 11/20 | 6.67/ 30,302 | 0.89 | 0.85 (*) | 0.81 (**) | 0.95 | 0.91 | 0.96 |
| 3010 | Ferritin light chain 1 | P29391/NP_034370.2 | Ftl1 | 108 | 1.1e-06 | 53% | 8/23 | 5.66/ 20,847 | 1.16 | 0.87 | 0.79 | 0.75 (*) | 0.69 (**) | 0.92 |
| 3103 | Ketohexokinase | P97328/NP_032465.2 | Khk | 61 | 4.90e-02 | 26% | 5/14 | 5.59/ 33,300 | 1.44 (**) | 1.29 | 1.32 | 0.89 | 0.91 | 1.02 |
| 3403 | S-adenosylmethionine synthase isoform type-1 | Q91X83/NP_598414.1 | Mat1a | 179 | 8.5e-14 | 50% | 20/43 | 5.51/ 44,051 | 0.66 (*) | 0.53 (***) | 0.62 (***) | 0.80 (*) | 0.93 | 1.16 |
| 3505 | Sulfite oxidase, mitochondrial | Q8R086/NP_776094.2 | Suox | 215 | 2.1e-17 | 45% | 16/26 | 6.07/ 61,231 | 1.15 | 1.20 (**) | 1.09 | 1.04 | 0.94 | 0.90 |
| 3605 | Albumin | P07724/NP_033784.2 | Alb | 288 | 1.1e-24 | 54% | 29/55 | 5.75/ 70,700 | 0.45 (**) | 0.47 (**) | 0.41 (**) | 1.06 | 0.93 | 0.87 |
| 3614 | Stress-70 protein, mitochondrial | P38647/NP_034611.2 | Hspa9 | 245 | 2.1e-20 | 48% | 29/46 | 5.81/ 73,701 | 1.07 | 1.01 | 0.73 | 0.94 | 0.69 (*) | 0.73 |
| 3806 | Cytosolic 10-formyltetrahydrofolate dehydrogenase | Q8R0Y6/NP_081682.1 | Aldh1l1 | 114 | 2.7e-07 | 21% | 13/23 | 5.64/ 99,502 | 1.57 (*) | 1.88 (***) | 1.37 | 1.19 | 0.87 | 0.73 (*) |
| 4101 | Indolethylamine N-methyltransferase | P40936/NP_033375.1 | Inmt | 113 | 3.4e-07 | 51% | 10/23 | 5.75/ 30,068 | 0.88 | 0.87 | 0.82 (*) | 0.98 | 0.93 | 0.94 |
| 4102 | ester hydrolase C11orf54 homolog | Q91V76/NP_001186413.1 | N/A | 120 | 6.7e-08 | 46% | 13/40 | 5.86/ 35,430 | 1.30 (**) | 1.35 (**) | 1.51 (***) | 1.04 | 1.16 | 1.12 |
| 4301 | Ornithine aminotransferase, mitochondrial | P29758/NP_058674.1 | Oat | 205 | 2.1e-16 | 53% | 20/36 | 6.19/ 48,723 | 0.62 (***) | 0.60 (***) | 0.68 (***) | 0.97 | 1.09 | 1.13 (*) |
| 5101 | Endoribonuclease LACTB2 | Q99KR3/NP_663356.1 | Lactb2 | 86 | 1.80e-04 | 26% | 8/17 | 5.89/ 33,019 | 1.09 | 1.00 | 1.26 (*) | 0.92 | 1.16 | 1.27 (*) |
| 5110 | Peroxiredoxin-6 | O08709/NP_031479.1 | Prdx6 | 189 | 8.5e-15 | 72% | 15/33 | 5.71/ 24,969 | 1.17 | 1.45 (***) | 1.44 (***) | 1.25 | 1.24 | 0.99 |
| 5806 | Pyruvate carboxylase, mitochondrial | Q05920/NP_001156418.1 | Pc | 101 | 5.3e-06 | 15% | 13/22 | 6.25/ 130,344 | 1.27 | 1.19 | 0.91 | 0.94 | 0.71 (**) | 0.76 |
| 6003 | Haloacid dehalogenase-like hydrolase domain–containing protein 3 | Q9CYW4/NP_077219.1 | Hdhd3 | 117 | 1.3e-07 | 35% | 10/24 | 6.31/ 28,237 | 1.01 | 1.21 (*) | 1.11 | 1.20 (*) | 1.10 | 0.92 |
| 6305 | Isovaleryl-CoA dehydrogenase, mitochondrial | Q9JHI5/NP_062800.1 | Ivd | 95 | 2.2e-05 | 34% | 12/38 | 8.53/ 46,695 | 0.74 (***) | 1.02 | 0.97 | 1.39 (***) | 1.32 | 0.95 |
| 6501 | Carboxylesterase 1D | Q8VCT4/NP_444430.2 | Ces1d | 124 | 2.7e-08 | 33% | 15/31 | 6.17/ 62,034 | 1.12 | 1.34 (*) | 0.81 | 1.20 | 0.73 | 0.60 (**) |
| 6606 | Triokinase/FMN cyclase | Q8VC30/NP_663471.1 | Tkfc | 249 | 8.5e-21 | 55% | 27/40 | 6.44/ 59,938 | 1.45 (***) | 1.28 | 1.15 | 0.88 | 0.79 (*) | 0.90 |
| 7104 | Carbonic anhydrase 3 | P16015/NP_031632.2 | Ca3 | 231 | 5.3e-19 | 81% | 16/34 | 6.89/ 29,633 | 1.13 | 1.41 (*) | 1.56 (***) | 1.25 (*) | 1.39 (*) | 1.11 |
| 7205 | Arginase-1 | Q61176/NP_031508.1 | Arg1 | 225 | 2.1e-18 | 72% | 19/52 | 6.51/ 34,957 | 0.95 | 0.73 (***) | 0.86 | 0.77 (**) | 0.91 | 1.18 |
| 7407 | Hydroxymethylglutaryl-CoA synthase, mitochondrial | P54869/NP_032282.2 | Hmgcs2 | 97 | 1.4e-05 | 25% | 13/32 | 8.65/ 57,300 | 1.39 (**) | 0.92 | 1.02 | 0.66 (*) | 0.73 | 1.10 |
| 7706 | long-chain–fatty-acid--CoA ligase 1 | P41216/NP_032007.2 | Acsl1 | 116 | 1.7e-07 | 24% | 14/31 | 6.81/ 78,928 | 0.78 | 0.65 (**) | 0.75 | 0.83 | 0.97 | 1.16 |
| 8011 | Glutathione S-transferase P 1 | P19157/NP_038569.1 | Gstp1 | 86 | 1.90e-04 | 48% | 13/53 | 7.68/ 23,765 | 0.86 | 1.05 | 1.24 | 1.22 | 1.44 (*) | 1.18 |
| 8101 | Carbonic anhydrase 3 | P16015/NP_031632.2 | Ca3 | 237 | 1.3e-19 | 81% | 18/81 | 6.89/ 29,633 | 0.97 | 1.14 | 1.33 (*) | 1.18 | 1.37 (**) | 1.16 |
| 8106 | Glycerol-3-phosphate dehydrogenase [NAD(+)], cytoplasmic | P13707/NP_034401.1 | Gpd1 | 119 | 8.5e-08 | 40% | 12/32 | 6.75/ 38,176 | 1.42 (**) | 1.06 | 1.42 (**) | 0.75 (*) | 1.00 | 1.33 (*) |
| 8203 | Acetyl-CoA acetyltransferase, cytosolic | Q8CAY6/NP_033364.2 | Acat2 | 83 | 3.40e-04 | 27% | 8/30 | 7.16/ 41,727 | 1.00 | 1.30 (*) | 1.55 (**) | 1.31 | 1.55 (**) | 1.19 |
| 8216 | Serine--pyruvate aminotransferase, mitochondrial | O35423/NP_057911.2 | Agxt | 92 | 4.1e-05 | 33% | 11/44 | 8.37/ 46,282 | 1.20 | 1.24 | 1.93 (*) | 1.03 | 1.61 | 1.56 |
| 8406 | Homogentisate 1,2-dioxygenase | O09173/NP_038575.2 | Hgd | 103 | 3.4e-06 | 28% | 11/28 | 6.86/ 50,726 | 1.18 | 0.62 (*) | 0.77 | 0.52 (***) | 0.65 (**) | 1.25 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liput, K.P.; Lepczyński, A.; Nawrocka, A.; Poławska, E.; Ogłuszka, M.; Jończy, A.; Grzybek, W.; Liput, M.; Szostak, A.; Urbański, P.; et al. Effects of Three-Month Administration of High-Saturated Fat Diet and High-Polyunsaturated Fat Diets with Different Linoleic Acid (LA, C18:2n–6) to α-Linolenic Acid (ALA, C18:3n–3) Ratio on the Mouse Liver Proteome. Nutrients 2021, 13, 1678. https://doi.org/10.3390/nu13051678
Liput KP, Lepczyński A, Nawrocka A, Poławska E, Ogłuszka M, Jończy A, Grzybek W, Liput M, Szostak A, Urbański P, et al. Effects of Three-Month Administration of High-Saturated Fat Diet and High-Polyunsaturated Fat Diets with Different Linoleic Acid (LA, C18:2n–6) to α-Linolenic Acid (ALA, C18:3n–3) Ratio on the Mouse Liver Proteome. Nutrients. 2021; 13(5):1678. https://doi.org/10.3390/nu13051678
Chicago/Turabian StyleLiput, Kamila P., Adam Lepczyński, Agata Nawrocka, Ewa Poławska, Magdalena Ogłuszka, Aneta Jończy, Weronika Grzybek, Michał Liput, Agnieszka Szostak, Paweł Urbański, and et al. 2021. "Effects of Three-Month Administration of High-Saturated Fat Diet and High-Polyunsaturated Fat Diets with Different Linoleic Acid (LA, C18:2n–6) to α-Linolenic Acid (ALA, C18:3n–3) Ratio on the Mouse Liver Proteome" Nutrients 13, no. 5: 1678. https://doi.org/10.3390/nu13051678
APA StyleLiput, K. P., Lepczyński, A., Nawrocka, A., Poławska, E., Ogłuszka, M., Jończy, A., Grzybek, W., Liput, M., Szostak, A., Urbański, P., Roszczyk, A., Pareek, C. S., & Pierzchała, M. (2021). Effects of Three-Month Administration of High-Saturated Fat Diet and High-Polyunsaturated Fat Diets with Different Linoleic Acid (LA, C18:2n–6) to α-Linolenic Acid (ALA, C18:3n–3) Ratio on the Mouse Liver Proteome. Nutrients, 13(5), 1678. https://doi.org/10.3390/nu13051678






