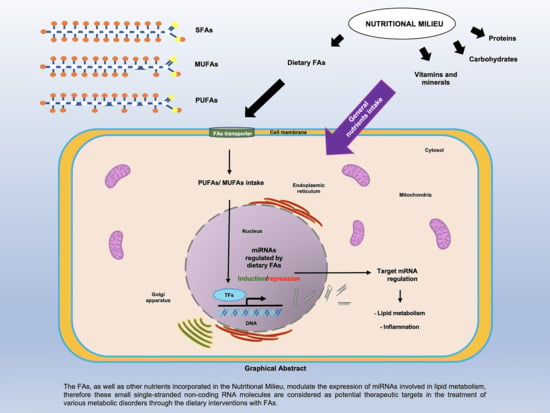
Effect of Dietary Fatty Acids on MicroRNA Expression Related to Metabolic Disorders and Inflammation in Human and Animal Trials
Abstract
1. Introduction
2. Materials and Methods
3. Results
3.1. DFAs Alter miRNA Expression Associated with Metabolic Disorders
3.2. DFAs Alter miRNA Expression Associated with Inflammation
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Scorletti, E.; Byrne, C.D. Omega-3 Fatty Acids, Hepatic Lipid Metabolism, and Nonalcoholic Fatty Liver Disease. Annu. Rev. Nutr. 2013, 33, 231–248. [Google Scholar] [CrossRef]
- Cardiovascular Diseases (CVDs). Available online: http://www.who.int/news-room/fact-sheets/detail/cardiovascular-diseases-(cvds) (accessed on 22 September 2018).
- García-Segura, L.; Pérez-Andrade, M.; Miranda-Ríos, J. The Emerging Role of MicroRNAs in the Regulation of Gene Expression by Nutrients. J. Nutr. Nutr. 2013, 6, 16–31. [Google Scholar] [CrossRef]
- Temple, N.J. Fat, sugar, whole grains and heart disease: 50 years of confusion. Nutrients 2018, 10, 39. [Google Scholar] [CrossRef]
- Gershuni, V.M. Saturated Fat: Part of a Healthy Diet. Curr. Nutr. Rep. 2018, 7, 85–96. [Google Scholar] [CrossRef] [PubMed]
- DiNicolantonio, J.J.; Lucan, S.C.; O’Keefe, J.H. The Evidence for Saturated Fat and for Sugar Related to Coronary Heart Disease. Prog. Cardiovasc. Dis. 2016, 58, 464–472. [Google Scholar] [CrossRef] [PubMed]
- Kuipers, R.S.; de Graaf, D.J.; Luxwolda, M.F.; Muskiet, M.H.A.; Dijck-Brouwer, D.A.J.; Muskiet, F.A.J. Saturated fat, carbohydrates and cardiovascular disease. Neth. J. Med. 2011, 69, 372–378. [Google Scholar] [PubMed]
- Astrup, A.; Magkos, F.; Bier, D.M.; Brenna, J.T.; de Oliveira Otto, M.C.; Hill, J.O.; King, J.C.; Mente, A.; Ordovas, J.M.; Volek, J.S.; et al. Saturated Fats and Health: A Reassessment and Proposal for Food-Based Recommendations: JACC State-of-the-Art Review. J. Am. Coll. Cardiol. 2020, 76, 844–857. [Google Scholar] [CrossRef]
- Siri-Tarino, P.W.; Sun, Q.; Hu, F.B.; Krauss, R.M. Meta-analysis of prospective cohort studies evaluating the association of saturated fat with cardiovascular disease. Am. J. Clin. Nutr. 2010, 91, 535–546. [Google Scholar] [CrossRef] [PubMed]
- Zock, P.L.; Blom, W.A.M.; Nettleton, J.A.; Hornstra, G. Progressing Insights into the Role of Dietary Fats in the Prevention of Cardiovascular Disease. Curr. Cardiol. Rep. 2016, 18, 111. [Google Scholar] [CrossRef]
- Vannice, G.; Rasmussen, H. Position of the academy of nutrition and dietetics: Dietary fatty acids for healthy adults. J. Acad. Nutr. Diet. 2014, 114, 136–153. [Google Scholar] [CrossRef]
- Briggs, M.A.; Petersen, K.S.; Kris-Etherton, P.M. Saturated Fatty Acids and Cardiovascular Disease: Replacements for Saturated Fat to Reduce Cardiovascular Risk. Healthcare 2017, 5, 29. [Google Scholar] [CrossRef] [PubMed]
- Desgagné, V.; Guay, S.-P.; Guérin, R.; Corbin, F.; Couture, P.; Lamarche, B.; Bouchard, L. Variations in HDL-carried miR-223 and miR-135a concentrations after consumption of dietary trans fat are associated with changes in blood lipid and inflammatory markers in healthy men—An exploratory study. Epigenetics 2016, 11, 438–448. [Google Scholar] [CrossRef] [PubMed]
- White, B. Dietary fatty acids. Am. Fam. Physician 2009, 80, 345–350. [Google Scholar] [PubMed]
- Tvrzicka, E.; Kremmyda, L.-S.; Stankova, B.; Zak, A. Fatty acids as biocompounds: Their role in human metabolism, health and disease—A review. Part 1: Classification, dietary sources and biological functions. Biomed. Pap. Med. Fac. Univ. Palacky. Olomouc. Czech. Repub. 2011, 155, 117–130. [Google Scholar] [CrossRef]
- Desgagné, V.; Guérin, R.; Guay, S.P.; Corbin, F.; Couture, P.; Lamarche, B.; Bouchard, L. Changes in high-density lipoprotein-carried miRNA contribution to the plasmatic pool after consumption of dietary trans fat in healthy men. Epigenomics 2017, 9, 669–688. [Google Scholar] [CrossRef] [PubMed]
- Calder, P.C. Functional Roles of Fatty Acids and Their Effects on Human Health. J. Parenter. Enter. Nutr. 2015, 39, 18S–32S. [Google Scholar] [CrossRef]
- Boigues, J.F.H.; Mach, N. Efecto de los ácidos grasos poliinsaturados en la prevención de la obesidad a través de modificaciones epigenéticas. Endocrinología y Nutrición 2015, 62, 338–349. [Google Scholar] [CrossRef] [PubMed]
- Takamura, T.; Honda, M.; Sakai, Y.; Ando, H.; Shimizu, A.; Ota, T.; Sakurai, M.; Misu, H.; Kurita, S.; Matsuzawa-Nagata, N.; et al. Gene expression profiles in peripheral blood mononuclear cells reflect the pathophysiology of type 2 diabetes. Biochem. Biophys. Res. Commun. 2007, 361, 379–384. [Google Scholar] [CrossRef]
- Sheikh, M.S.A. Role of Plasma Soluble Lectin-Like Oxidized Low-Density Lipoprotein Receptor-1 and microRNA-98 in Severity and Risk of Coronary Artery Disease. Balk. Med. J. 2021, 38, 13–22. [Google Scholar] [CrossRef]
- Xu, Y.; Miao, C.; Cui, J.; Bian, X. Mir-92a-3p promotes ox-ldl induced-apoptosis in huvecs via targeting sirt6 and activating mapk signaling pathway. Braz. J. Med. Biol. Res. 2021, 54, 1–8. [Google Scholar] [CrossRef]
- Deiuliis, J.A. MicroRNAs as regulators of metabolic disease: Pathophysiologic significance and emerging role as biomarkers and therapeutics. Int. J. Obes. 2016, 40, 88–101. [Google Scholar] [CrossRef]
- Treiber, T.; Treiber, N.; Meister, G. Regulation of microRNA biogenesis and its crosstalk with other cellular pathways. Nat. Rev. Mol. Cell Biol. 2019, 20, 5–20. [Google Scholar] [CrossRef]
- O’Brien, J.; Hayder, H.; Zayed, Y.; Peng, C. Overview of microRNA biogenesis, mechanisms of actions, and circulation. Front. Endocrinol. 2018, 9, 402. [Google Scholar] [CrossRef]
- Yang, Z.; Cappello, T.; Wang, L. Emerging role of microRNAs in lipid metabolism. Acta Pharm. Sin. B 2015, 5, 145–150. [Google Scholar] [CrossRef] [PubMed]
- Ameres, S.L.; Zamore, P.D. Diversifying microRNA sequence and function. Nat. Rev. Mol. Cell Biol. 2013, 14, 475–488. [Google Scholar] [CrossRef]
- Desvignes, T.; Batzel, P.; Berezikov, E.; Eilbeck, K.; Eppig, J.T.; McAndrews, M.S.; Singer, A.; Postlethwait, J.H. MiRNA Nomenclature: A View Incorporating Genetic Origins, Biosynthetic Pathways, and Sequence Variants. Trends Genet. 2015, 31, 613–626. [Google Scholar] [CrossRef]
- Gulyaeva, L.F.; Kushlinskiy, N.E. Regulatory mechanisms of microRNA expression. J. Transl. Med. 2016, 14, 143. [Google Scholar] [CrossRef]
- Ross, S.A.; Davis, C.D. The Emerging Role of microRNAs and Nutrition in Modulating Health and Disease. Annu. Rev. Nutr. 2014, 34, 305–336. [Google Scholar] [CrossRef]
- Carotenuto, F.; Albertini, M.C.; Coletti, D.; Vilmercati, A.; Campanella, L.; Darzynkiewicz, Z.; Teodori, L. How Diet Intervention via Modulation of DNA Damage Response through MicroRNAs May Have an Effect on Cancer Prevention and Aging, an in Silico Study. Int. J. Mol. Sci. 2016, 17, 752. [Google Scholar] [CrossRef] [PubMed]
- Baselga-Escudero, L.; Arola-Arnal, A.; Pascual-Serrano, A.; Ribas-Latre, A.; Casanova, E.; Salvadó, M.-J.; Arola, L.; Blade, C. Chronic Administration of Proanthocyanidins or Docosahexaenoic Acid Reversess the Increase of miR-33a and miR-122 in Dyslipidemic Obese Rats. PLoS ONE 2013, 8, e69817. [Google Scholar] [CrossRef]
- Kim, J.; Okla, M.; Erickson, A.; Carr, T.; Natarajan, S.K.; Chung, S. Eicosapentaenoic acid potentiates brown thermogenesis through FFAR4-dependent up-regulation of miR-30b and miR-378. J. Biol. Chem. 2016, 291, 20551–20562. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Shao, Y.; Yuan, F.; Feng, H.; Li, N.; Zhang, H.; Wu, C.; Liu, Z. Fish oil feeding modulates the expression of hepatic MicroRNAs in a western-style diet-induced nonalcoholic fatty liver disease rat model. Biomed. Res. Int. 2017, 2017, 1–11. [Google Scholar] [CrossRef]
- Chartoumpekis, D.V.; Zaravinos, A.; Ziros, P.G.; Iskrenova, R.P.; Psyrogiannis, A.I.; Kyriazopoulou, V.E.; Habeos, I.G. Differential Expression of MicroRNAs in Adipose Tissue after Long-Term High-Fat Diet-Induced Obesity in Mice. PLoS ONE 2012, 7, e34872. [Google Scholar] [CrossRef] [PubMed]
- Nesca, V.; Guay, C.; Jacovetti, C.; Menoud, V.; Peyot, M.-L.; Laybutt, D.R.; Prentki, M.; Regazzi, R. Identification of particular groups of microRNAs that positively or negatively impact on beta cell function in obese models of type 2 diabetes. Diabetologia 2013, 56, 2203–2212. [Google Scholar] [CrossRef] [PubMed]
- Vinciguerra, M.; Sgroi, A.; Veyrat-Durebex, C.; Rubbia-Brandt, L.; Buhler, L.H.; Foti, M. Unsaturated fatty acids inhibit the expression of tumor suppressor phosphatase and tensin homolog (PTEN) via microRNA-21 up-regulation in hepatocytes. Hepatology 2009, 49, 1176–1184. [Google Scholar] [CrossRef]
- Ahn, J.; Lee, H.; Chung, C.H.; Ha, T. High fat diet induced downregulation of microRNA-467b increased lipoprotein lipase in hepatic steatosis. Biochem. Biophys. Res. Commun. 2011, 414, 664–669. [Google Scholar] [CrossRef]
- Cirera, S.; Birck, M.; Busk, P.K.; Fredholm, M. Expression profiles of miRNA-122 and its target CAT1 in minipigs (Sus scrofa) fed a high-cholesterol diet. Comp. Med. 2010, 60, 136–141. [Google Scholar]
- Gil-Zamorano, J.; Martin, R.; Daimiel, L.; Richardson, K.; Giordano, E.; Nicod, N.; García-Carrasco, B.; Soares, S.M.A.; Iglesias-Gutiérrez, E.; Lasunción, M.A.; et al. Docosahexaenoic Acid Modulates the Enterocyte Caco-2 Cell Expression of MicroRNAs Involved in Lipid Metabolism. J. Nutr. 2014, 144, 575–585. [Google Scholar] [CrossRef]
- Daimiel-Ruiz, L.; Klett-Mingo, M.; Konstantinidou, V.; Micó, V.; Aranda, J.F.; García, B.; Martínez-Botas, J.; Dávalos, A.; Fernández-Hernando, C.; Ordovás, J.M. Dietary lipids modulate the expression of miR-107, an miRNA that regulates the circadian system. Mol. Nutr. Food Res. 2015, 59, 552–565. [Google Scholar] [CrossRef]
- Parra, P.; Serra, F.; Palou, A. Expression of Adipose MicroRNAs Is Sensitive to Dietary Conjugated Linoleic Acid Treatment in Mice. PLoS ONE 2010, 5, e13005. [Google Scholar] [CrossRef]
- Ortega, F.J.; Cardona-Alvarado, M.I.; Mercader, J.M.; Moreno-Navarrete, J.M.; Moreno, M.; Sabater, M.; Fuentes-Batllevell, N.; Ramírez-Chávez, E.; Ricart, W.; Molina-Torres, J.; et al. Circulating profiling reveals the effect of a polyunsaturated fatty acid-enriched diet on common microRNAs. J. Nutr. Biochem. 2015, 26, 1095–1101. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Z.; Ge, Y.; Zhang, J.; Xue, M.; Li, Q.; Lin, D.; Ma, W. PUFA diets alter the microRNA expression profiles in an inflammation rat model. Mol. Med. Rep. 2015, 11, 4149–4157. [Google Scholar] [CrossRef] [PubMed]
- D’Amore, S.; Vacca, M.; Cariello, M.; Graziano, G.; D’Orazio, A.; Salvia, R.; Sasso, R.C.; Sabbà, C.; Palasciano, G.; Moschetta, A. Genes and miRNA expression signatures in peripheral blood mononuclear cells in healthy subjects and patients with metabolic syndrome after acute intake of extra virgin olive oil. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2016, 1861, 1671–1680. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.M.; Min, K.H.; Lee, W. Induction of mir-96 by dietary saturated fatty acids exacerbates hepatic insulin resistance through the suppression of insr and irs-1. PLoS ONE 2016, 11, e0169039. [Google Scholar] [CrossRef]
- Ali, O.; Darwish, H.A.; Eldeib, K.M.; Abdel Azim, S.A. miR-26a potentially contributes to the regulation of fatty acid and sterol metabolism in vitro human HepG2 cell model of nonalcoholic fatty liver disease. Oxid. Med. Cell. Longev. 2018, 2018, 8515343. [Google Scholar] [CrossRef]
- De Gottardi, A.; Vinciguerra, M.; Sgroi, A.; Moukil, M.; Ravier-Dall’Antonia, F.; Pazienza, V.; Pugnale, P.; Foti, M.; Hadengue, A. Microarray analyses and molecular profiling of steatosis induction in immortalized human hepatocytes. Lab. Investig. 2007, 87, 792–806. [Google Scholar] [CrossRef]
- Georgiadi, A.; Kersten, S. Mechanisms of gene regulation by fatty acids. Adv. Nutr. 2012, 3, 127–134. [Google Scholar] [CrossRef]
- Jimenez-Lopez, C.; Carpena, M.; Lourenço-Lopes, C.; Gallardo-Gomez, M.; Lorenzo, J.M.; Barba, F.J.; Prieto, M.A.; Simal-Gandara, J. Bioactive compounds and quality of extra virgin olive oil. Foods 2020, 9, 1014. [Google Scholar] [CrossRef]
- Betz, M.J.; Enerbäck, S. Targeting thermogenesis in brown fat and muscle to treat obesity and metabolic disease. Nat. Rev. Endocrinol. 2018, 14, 77–87. [Google Scholar] [CrossRef]
- Motard-Bélanger, A.; Charest, A.; Grenier, G.; Paquin, P.; Chouinard, Y.; Lemieux, S.; Couture, P.; Lamarche, B. Study of the effect of trans fatty acids from ruminants on blood lipids and other risk factors for cardiovascular disease. Am. J. Clin. Nutr. 2008, 87, 593–599. [Google Scholar] [CrossRef]
- Forsythe, C.E.; Phinney, S.D.; Fernandez, M.L.; Quann, E.E.; Wood, R.J.; Bibus, D.M.; Kraemer, W.J.; Feinman, R.D.; Volek, J.S. Comparison of low fat and low carbohydrate diets on circulating fatty acid composition and markers of inflammation. Lipids 2008, 43, 65–77. [Google Scholar] [CrossRef]
- Fritsche, K.L. The Science of Fatty Acids and Inflammation. Adv. Nutr. 2015, 6, 293S–301S. [Google Scholar] [CrossRef]
- Rietschel, E.T.; Kirikae, T.; Schade, F.U.; Mamat, U.; Schmidt, G.; Loppnow, H.; Ulmer, A.J.; Zähringer, U.; Seydel, U.; Di Padova, F.; et al. Bacterial endotoxin: Molecular relationships of structure to activity and function. FASEB J. 1994, 8, 217–225. [Google Scholar] [CrossRef]
- Cani, P.D.; Amar, J.; Iglesias, M.A.; Poggi, M.; Knauf, C.; Bastelica, D.; Neyrinck, A.M.; Fava, F.; Tuohy, K.M.; Chabo, C.; et al. Metabolic endotoxemia initiates obesity and insulin resistance. Diabetes 2007, 56, 1761–1772. [Google Scholar] [CrossRef]
- Erridge, C. The capacity of foodstuffs to induce innate immune activation of human monocytes in vitro is dependent on food content of stimulants of Toll-like receptors 2 and 4. Br. J. Nutr. 2011, 105, 15–23. [Google Scholar] [CrossRef]
- Hoshino, K.; Takeuchi, O.; Kawai, T.; Sanjo, H.; Ogawa, T.; Takeda, Y.; Takeda, K.; Akira, S. Cutting edge: Toll-like receptor 4 (TLR4)-deficient mice are hyporesponsive to lipopolysaccharide: Evidence for TLR4 as the Lps gene product. J. Immunol. 1999, 162, 3749–3752. [Google Scholar]
- Takeda, K.; Kaisho, T.; Akira, S. Toll-like receptors. Annu. Rev. Immunol. 2003, 21, 335–376. [Google Scholar] [CrossRef]
- Laugerette, F.; Vors, C.; Géloën, A.; Chauvin, M.A.; Soulage, C.; Lambert-Porcheron, S.; Peretti, N.; Alligier, M.; Burcelin, R.; Laville, M.; et al. Emulsified lipids increase endotoxemia: Possible role in early postprandial low-grade inflammation. J. Nutr. Biochem. 2011, 22, 53–59. [Google Scholar] [CrossRef]
- Bailey, M.A.; Holscher, H.D. Microbiome-mediated effects of the Mediterranean diet on inflammation. Adv. Nutr. 2018, 9, 93–206. [Google Scholar] [CrossRef] [PubMed]


| Type of DFAs | miRNAs | Type of Regulation | Organism/Cell/Tissue Type, Sample Size | Dose/Duration | Gene Expression | Ref |
|---|---|---|---|---|---|---|
| DHA | miR-33a, miR-122 | repressed | Liver and PBMCs. Dyslipidemic cafeteria diet-fed male Wistar 150 g rats supplemented with DHA and/or proanthocyanidins. Five groups (n = 7) | Standard chow diet and cafeteria diet as a high-fat model. DHA: 515 mg PUFAs/kg (of body weight) dissolved in arabic gum, with sunflower lecithin and rosemary extract (flavoring) × 3 wks | qRT-PCR | [31] |
| EPA | miR-30b, miR193b, miR-365, miR-196a, miR-378 | induced | Murine brown preadipocytes. iBAT from mice. C57BL/six male mice, (iBAT) adipocyte precursor cells, four diet groups (n = 8) | Murine primary brown adipogenic precursor cells from iBAT treated with 100 μM PA, OO, or EPA. Mice fed iso-caloric HF diet (50% cal from fat) with palm oil (PO), fish oil (FO), or olive oil (OO) × 12 wks. | qPCR | [32] |
| Fish oil, DHA, EPA | miR-345-5p, miR-34a-5p, miR-3556b, miR-3558-3p, miR-3590-5p, miR-362-3p, miR-374-3p, miR-374-5p, miR-455-5p, miR-466c-5p, miR-490-3p, miR-497-5p, miR-499-5p, miR-503-5p, miR-505-5p, miR-511-3p, miR-511-5p, miR-547-3p, miR-664-3p, miR-871-3p, miR-872-5p, miR-99a-3p, miR-9a-5p, let-7f-1-3p, miR-101a-5p, miR-106b-5p, miR-126b, miR-130a-3p, miR-142-3p, miR-142-5p, miR-144-3p, miR-144-5p, miR-146a-3p, miR-15b-3p, miR-17-5p, miR-18a-5p, miR-190a-5p, miR-193-3p, miR-19a-3p, miR-19b-3p, miR-22-5p, miR-223-3p, miR-23a-5p, miR-29b-3p, miR-29c-3p, miR-301a-3p, miR-32-5p, miR-33-5p, miR-330-5p, miR-331-3p, miR-339-5p | repressed | Nine-week-old male Sprague–Dawley rats, liver tissue | Lard-rich western diet (45 kcal% fat, 2% cholesterol) or Fish oil rich diet (45 kcal% fat and 2% cholesterol, 10% fish oil) | Illumina sequencing, RT-qPCR | [33] |
| miR-100-5p, miR-10a-5p, miR-1249, miR-139-3p, miR-140-3p, miR-143-3p, miR-146b-5p, miR-148b-3p, miR-151-3p, miR-151-5p, miR-152-5p, miR-182, miR-203b-3p, miR-219-1-3p, miR-27b-5p, miR-28-5p, miR-293-5p, miR-30d-5p, miR-3102, miR-328a-3p, miR-3586-3p, miR-370-3p, miR-375-3p, miR-425-5p, miR-598-3p, miR-92a-3p, miR-99a-5p, miR-99b-5p | induced | |||||
| SFA (lard, cholesterol) | miR-342-3p, miR-146a miR-146b miR-222 miR-221 miR-142-3p miR-142-5p miR-21 miR-335-5p miR-146a miR-146b miR-647 miR-379 | induced | White adipose tissue excised from the epididymal fat pad, C57BL6J wild type male mice eight weeks old fed HFD | Mice were fed HDF (60% fat, lard, and cholesterol) × five months. | microarray, qPCR | [34] |
| MiR-141, miR-200a, miR-200a miR-200b miR-200c miR-122 miR-204 miR-133b miR-1 miR-30a miR-192 miR-193a-3p miR-203 miR-378 miR-30e | repressed | |||||
| HFD (cholesterol and lard) | miR-132, miR-199a-5p | induced | Dissociated islet cells from mice. Five wk old male C57BL/six mice and cell lines B-cells | Mice were fed an HFD (60% energy from fat) × 8 wks | Microarray | [35] |
| miR-199a-3p, miR-184 miR-203 miR-210 miR-383 | repressed | |||||
| Oleic acid | miR-21 | induced | HepG2 cells, human primary hepatocytes, Wistar rat liver tissues | Rats HFD: 19.5% fat (36.6% SFAs, 41.9% UFAs, 33.9% PUFAs), 50 μM on HepG2 cells and HPHs. | RT-qPCR | [36] |
| Stearic acid | miR-467b | repressed | Liver tissues, and hepatocytes, five wk old male C57BL/6J mice in two groups (n = 10), murine Hepa 1–6 hepatocytes | Mice fed HFD (20%lard) × 8 wks. In cells, saturated fatty acid-induced steatosis with 50 microM stearic acid SA × 24 h. | RT-qPCR. | [37] |
| Cholesterol, monounsaturated fatty acids (MUFAs), polyunsaturated fatty acids (PUFAs) | miR-122 | repressed | Liver tissues, hepatocytes, Mini Pig Sus scrofa Standard diet (n = 5), High cholesterol diet (n = 7) | High cholesterol diet × 11 wks (22.77% crude fat, 19.3 MJ/kg, 2% cholesterol 0.55% MUFAs, 6.86% PUFAs) | qPCR | [38] |
| DHA | miR-141-3p, miR-221-3p, miR-30c, miR-192, miR-1283 | induced | Caco-2, HepG2 hepatocytes | Fatty acids delivered to cells as lipid micelles with phosphatidylcholine × 24 h with 200 μM/L oleic and palmitic acid × 24 h. | Microarray, qRT-PCR | [39] |
| miR-30a | repressed | |||||
| DHA + Palmitic fatty acid | miR-1 | induced | ||||
| Palmitic fatty acid | miR-106b | induced | ||||
| DHA + Oleic Acid | let-7f, miR-181a-5p | induced | ||||
| Cholesterol | miR-151b | induced | Caco-2 cells, murine brain, cerebellum and kidney tissues. Mice and cell lines | In cells: cholesterol, CLA, and DHA delivered to cells as micelles with lysophosphatidylcholine and sodium taurocholate. In animals: 8wk-old C57BL6/J mice fed a normal chow diet or an HFD containing 1.25% of cholesterol × 16 wks after mice sacrificed and tissue samples collected. | RT-qPCR | [40] |
| miR-215 | repressed | |||||
| CLA (conjugated linoleic acid) | miR-224, miR-106b miR-16 miR-122 miR-151-3p miR-107 miR-151b | induced | ||||
| miR-192, miR-215 miR-3141 miR-4739 miR-4534 | repressed | |||||
| DHA | miR-23a, miR-1260b let-7i miR-30d miR-183 miR-92b miR-107 miR-320e miR-151b | induced | ||||
| miR-192, miR-215 miR-4454 miR-4787-5p miR-3960 miR-4739 miR-3665 miR-3141 miR-3940-5p miR-4687-3p | repressed | |||||
| CLA 1 | miR-143 | induced | Mice, retroperitoneal adipose tissue | CLA 1 = Standard diet (SD)+ conjugated linoleic acids (CLA) 3 mg × 37 days, CLA 2 = SD + CLA 10 mg, CLA 3= HFD + CLA 6 mg, CLA 4= HFD + CLA 20 mg | qPCR | [41] |
| CLA 2, 4 | repressed | |||||
| CLA 3 | no change | |||||
| CLA 1, 2, 3 | miR-103 | induced | ||||
| CLA 4 | repressed | |||||
| CLA 1, 2, 4 | miR-107 | repressed | ||||
| CLA 3 | induced | |||||
| CLA 1, 2, 4 | miR-221 | induced | ||||
| CLA 3 | no change | |||||
| CLA 1, 2, 3, 4 | miR-222 | induced | ||||
| CLA 2, 4 CLA 3 | miR-328, miR-330-3p, miR-221, miR-125a-5p | repressed | Blood plasma. In the first study, 20 miRNAs were differentially expressed n = 10 healthy women, BMI 30–35. Validation study n = 20 (8 m, 12 w) | 30 g almonds and walnuts × 8 wks (2.02 g n-3, 11.1 n-6) in a normocaloric diet. | Microarray, RT-qPCR | [42] |
| miR-192, miR-486-5p, miR-19b, miR-106a, miR-18a, miR-130b | no change | |||||
| CLA 1, 2, 3 | miR-103 | induced | Serum, PBMCs, adipocytes, hepatocytes | AIN-93G rat chow mixed with lard and corn oil, plus EPA and DHA or omega-6 1(0 μL/100 g/day) × 16 wks | Microarray, RT-qPCR | [43] |
| CLA 4 CLA 1, 2, 4 | miR-1286, miR-619-3p, miR-302c-5p, miR-519b-3p, miR-614, miR-23b-3p | repressed | Healthy women, PBMCs | 50 mL (44 g) single dose 8 a.m., samples were taken after 4. | Microarray, RT-qPCR | [44] |
| miR-107 | repressed | |||||
| CLA 3 | miR-96 | induced | Mice. The liver and gastrocnemius skeletal muscle, Hep2 cells | HFD, 60% calories from fat × 14 weeks, cells treated with palmitate (0.5 mM) or oleate (0 ± 0.5 mM) for 18 h | RT-qPCR | [45] |
| CLA 1, 2, 4 | miR-221 | induced | HepG2 cell line | Palmitic, oleic acid (1:2) long-chain mixture different concentrations for 24 h. | RT-qPCR | [46] |
| CLA 3 | miR-223, miR-135a | no change | Men, purified HDL from plasma | high in iTFA (10.2 g/2500 kcal, 3.7% daily energy), high in rTFA (10.2 g/2500 kcal, 3.7% daily energy), control diet low in TFAs (2.2 g/2500 kcal, 0.8% daily energy (each for four weeks, >3 week wash-out period) | RT-qPCR | [13] |
| CLA 1, 2, 3, 4 | miR-222 | induced | Men, purified HDL from plasma | High in iTFA (10.2 g/2500 kcal, 3.7% daily energy), high in rTFA (10.2 g/2500 kcal, 3.7% daily energy), control diet low in TFAs (2.2 g/2500 kcal, 0.8% daily energy (each for four weeks, >3 week wash-out period) | microarrays, RT-qPCR | [16] |
| iTFA (vaccenic acid 28 g/100 g) vs. Control | miR-199a-5p, miR-30a-5p | induced | ||||
| rTFA, iTFA vs. control | miR-328-3p, miR-423-3p, miR-124-3p, miR-150-5p, miR-31-5p, miR-375 | repressed | ||||
| iTFA vs. rTFA | miR-133a-3p | repressed |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
MacDonald-Ramos, K.; Martínez-Ibarra, A.; Monroy, A.; Miranda-Ríos, J.; Cerbón, M. Effect of Dietary Fatty Acids on MicroRNA Expression Related to Metabolic Disorders and Inflammation in Human and Animal Trials. Nutrients 2021, 13, 1830. https://doi.org/10.3390/nu13061830
MacDonald-Ramos K, Martínez-Ibarra A, Monroy A, Miranda-Ríos J, Cerbón M. Effect of Dietary Fatty Acids on MicroRNA Expression Related to Metabolic Disorders and Inflammation in Human and Animal Trials. Nutrients. 2021; 13(6):1830. https://doi.org/10.3390/nu13061830
Chicago/Turabian StyleMacDonald-Ramos, Karla, Alejandra Martínez-Ibarra, Adriana Monroy, Juan Miranda-Ríos, and Marco Cerbón. 2021. "Effect of Dietary Fatty Acids on MicroRNA Expression Related to Metabolic Disorders and Inflammation in Human and Animal Trials" Nutrients 13, no. 6: 1830. https://doi.org/10.3390/nu13061830
APA StyleMacDonald-Ramos, K., Martínez-Ibarra, A., Monroy, A., Miranda-Ríos, J., & Cerbón, M. (2021). Effect of Dietary Fatty Acids on MicroRNA Expression Related to Metabolic Disorders and Inflammation in Human and Animal Trials. Nutrients, 13(6), 1830. https://doi.org/10.3390/nu13061830