The Effects of Ionizing Radiation on Gut Microbiota, a Systematic Review
Abstract
:1. Introduction
2. Materials and Methods
2.1. Search Strategy and Selection Criteria
2.2. Inclusion and Exclusion Criteria
2.3. Study Selection and Data Extraction
2.4. Risk of Bias in Individual Studies
3. Results
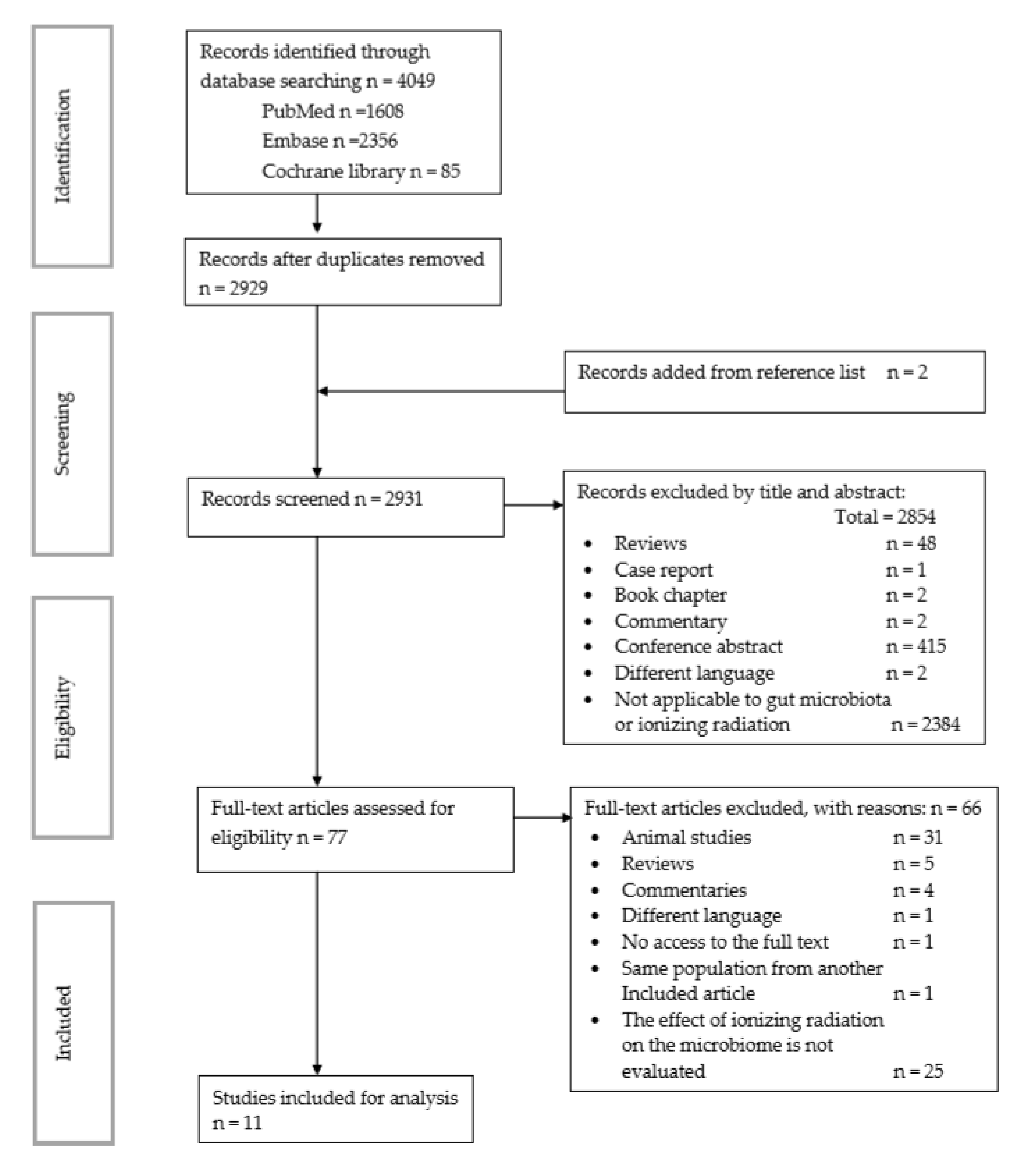
3.1. Search Results
3.2. Study Characteristics
3.3. Sampling and Microbiota Analysis
3.4. Findings
3.4.1. Diversity and Richness Analysis
3.4.2. Gut Microbial Composition
4. Discussion
4.1. Limitations of the Studies
4.2. Limitations of the Review
5. Conclusions
Future Directions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Acknowledgments
Conflicts of Interest
References
- Kumagai, T.; Rahman, F.; Smith, A.M. The Microbiome and Radiation Induced-Bowel Injury: Evidence for Potential Mechanistic Role in Disease Pathogenesis. Nutrients 2018, 10, 1405. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lynch, S.V.; Pedersen, O. The Human Intestinal Microbiome in Health and Disease. N. Engl. J. Med. 2016, 375, 2369–2379. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhernakova, A.; Kurilshikov, A.; Bonder, M.J.; Tigchelaar, E.F.; Schirmer, M.; Vatanen, T.; Mujagic, Z.; Vila, A.V.; Falony, G.; Vieira-Silva, S.; et al. Population-based metagenomics analysis reveals markers for gut microbiome composition and diversity. Science 2016, 352, 565–569. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marchesi, J.R.; Adams, D.H.; Fava, F.; Hermes, G.D.; Hirschfield, G.M.; Hold, G.; Quraishi, M.N.; Kinross, J.; Smidt, H.; Tuohy, K.M.; et al. The gut microbiota and host health: A new clinical frontier. Gut 2016, 65, 330–339. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nam, Y.D.; Kim, H.J.; Seo, J.G.; Kang, S.W.; Bae, J.-W. Impact of Pelvic Radiotherapy on Gut Microbiota of Gynecological Cancer Patients Revealed by Massive Pyrosequencing. PLoS ONE 2013, 8, e82659. [Google Scholar] [CrossRef]
- Crawford, P.A.; Gordon, J.I. Microbial regulation of intestinal radiosensitivity. Proc. Natl. Acad. Sci. USA 2005, 102, 13254–13259. [Google Scholar] [CrossRef] [Green Version]
- Wang, A.; Ling, Z.; Yang, Z.; Kiela, P.R.; Wang, T.; Wang, C.; Cao, L.; Geng, F.; Shen, M.; Ran, X.; et al. Gut Microbial Dysbiosis May Predict Diarrhea and Fatigue in Patients Undergoing Pelvic Cancer Radiotherapy: A Pilot Study. PLoS ONE 2015, 10, e0126312. [Google Scholar] [CrossRef] [Green Version]
- Desouky, O.; Ding, N.; Zhou, G. Targeted and non-targeted effects of ionizing radiation. J. Radiat. Res. Appl. Sci. 2015, 8, 247–254. [Google Scholar] [CrossRef] [Green Version]
- Mollà, M.; Panes, J. Radiation-induced intestinal inflammation. World J. Gastroenterol. 2007, 13, 3043–3046. [Google Scholar] [CrossRef] [PubMed]
- Leibowitz, B.J.; Wei, L.; Zhang, L.; Ping, X.; Epperly, M.; Greenberger, J.; Cheng, T.; Yu, J. Ionizing irradiation induces acute haematopoietic syndrome and gastrointestinal syndrome independently in mice. Nat. Commun. 2014, 5, 3494. [Google Scholar] [CrossRef] [Green Version]
- Booth, C.; Tudor, G.; Tudor, J.; Katz, B.P.; MacVittie, T.J. Acute Gastrointestinal Syndrome in High-Dose Irradiated Mice. Health Phys. 2012, 103, 383–399. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Villa, J.K.; Han, R.; Tsai, C.-H.; Chen, A.; Sweet, P.; Franco, G.; Vaezian, R.; Tkavc, R.; Daly, M.J.; Contreras, L.M. A small RNA regulates pprM, a modulator of pleiotropic proteins promoting DNA repair, in Deinococcus radiodurans under ionizing radiation. Sci. Rep. 2021, 11, 12949. [Google Scholar] [CrossRef] [PubMed]
- Šiková, M.; Janoušková, M.; Ramaniuk, O.; Páleníková, P.; Pospisil, J.; Bartl, P.; Suder, A.; Pajer, P.; Kubičková, P.; Pavliš, O.; et al. Ms1 RNA increases the amount of RNA polymerase inMycobacterium smegmatis. Mol. Microbiol. 2019, 111, 354–372. [Google Scholar] [CrossRef] [PubMed]
- Andreyev, J. Gastrointestinal complications of pelvic radiotherapy: Are they of any importance? Gut 2005, 54, 1051–1054. [Google Scholar] [CrossRef] [Green Version]
- Hauer-Jensen, M.; Wang, J.; Boerma, M.; Fu, Q.; Denham, J.W. Radiation damage to the gastrointestinal tract: Mechanisms, diagnosis, and management. Curr. Opin. Support. Palliat. Care 2007, 1, 23–29. [Google Scholar] [CrossRef]
- Shi, W.; Shen, L.; Zou, W.; Wang, J.; Yang, J.; Wang, Y.; Liu, B.; Xie, L.; Zhu, J.; Zhang, Z. The Gut Microbiome Is Associated With Therapeutic Responses and Toxicities of Neoadjuvant Chemoradiotherapy in Rectal Cancer Patients—A Pilot Study. Front. Cell. Infect. Microbiol. 2020, 10, 562463. [Google Scholar] [CrossRef]
- Yi, Y.; Shen, L.; Shi, W.; Xia, F.; Zhang, H.; Wang, Y.; Zhang, J.; Wang, Y.; Sun, X.; Zhang, Z.; et al. Gut Microbiome Components Predict Response to Neoadjuvant Chemoradiotherapy in Patients with Locally Advanced Rectal Cancer: A Prospective, Longitudinal Study. Off. J. Am. Assoc. Cancer Res. 2021, 27, 1329–1340. [Google Scholar] [CrossRef]
- Mitra, A.; Grossman Biegert, G.W.; Delgado, A.Y.; Karpinets, T.V.; Solley, T.N.; Mezzari, M.P.; Yoshida-Court, K.; Petrosino, J.F.; Mikkelson, M.D.; Lin, L.; et al. Microbial Diversity and Composition Is Associated with Patient-Reported Toxicity during Chemoradiation Therapy for Cervical Cancer. Int. J. Radiat. Oncol. 2020, 107, 163–171. [Google Scholar] [CrossRef]
- El Alam, M.B.; Sims, T.T.; Kouzy, R.; Biegert, G.W.G.; Jaoude, J.; Karpinets, T.V.; Yoshida-Court, K.; Wu, X.; Delgado-Medrano, A.Y.; Mezzari, M.P.; et al. A prospective study of the adaptive changes in the gut microbiome during standard-of-care chemoradiotherapy for gynecologic cancers. PLoS ONE 2021, 16, e0247905. [Google Scholar] [CrossRef]
- Ulluwishewa, D.; Anderson, R.C.; McNabb, W.C.; Moughan, P.J.; Wells, J.M.; Roy, N.C. Regulation of Tight Junction Permeability by Intestinal Bacteria and Dietary Components. J. Nutr. 2011, 141, 769–776. [Google Scholar] [CrossRef] [Green Version]
- Stringer, A.; Gibson, R.; Yeoh, A.; Bowen, J.; Keefe, D. Chemotherapy-induced diarrhoea is associated with a modified intestinal microbiome and intestinal inflammation. Support. Care Cancer 2011, 19, S153. [Google Scholar]
- Stringer, A.M. Interaction between Host Cells and Microbes in Chemotherapy-Induced Mucositis. Nutrients 2013, 5, 1488–1499. [Google Scholar] [CrossRef] [Green Version]
- Cuzzolin, L.; Zambreri, D.; Donini, M.; Griso, C.; Benoni, G. Influence of Radiotherapy on Intestinal Microflora in Cancer Patients. J. Chemother. 1992, 4, 176–179. [Google Scholar] [CrossRef]
- Sheikh Sajjadieh, M.R.; Kuznetsova, L.V.; Bojenko, V.B. Dysbiosis in Ukrainian Children with Irritable Bowel Syndrome Affected by Natural Radiation. Iran. J. Pediatr. 2012, 22, 364–368. [Google Scholar]
- García-Peris, P.; Velasco, C.; Lozano, M.A.; Moreno, Y.; Paron, L.; de la Cuerda, C.; Bretón, I.; Camblor, M.; García-Hernández, J.; Guarner, F.; et al. Effect of a mixture of inulin and fructo-oligosaccharide on Lactobacillus and Bifidobacterium intestinal microbiota of patients receiving radiotherapy: A randomised, double-blind, placebo-controlled trial. Nutr. Hosp. 2012, 27, 1908–1915. [Google Scholar] [CrossRef]
- Wang, Z.; Wang, Q.; Wang, X.; Zhu, L.; Chen, J.; Zhang, B.; Chen, Y.; Yuan, Z. Gut microbial dysbiosis is associated with development and progression of radiation enteritis during pelvic radiotherapy. J. Cell. Mol. Med. 2019, 23, 3747–3756. [Google Scholar] [CrossRef] [PubMed]
- Sahly, N.; Moustafa, A.; Zaghloul, M.; Salem, T.Z. Effect of radiotherapy on the gut microbiome in pediatric cancer patients: A pilot study. PeerJ 2019, 7, e7683. [Google Scholar] [CrossRef] [PubMed]
- Higgins, J.P.T.; Thomas, J.; Chandler, J.; Cumpston, M.; Li, T.; Page, M.J.; Welch, V.A. (Eds.) Cochrane Handbook for Systematic Reviews of Interventions Version 6.2 (Updated February 2021). Cochrane. 2021. Available online: www.training.cochrane.org/handbook (accessed on 5 November 2020).
- Wells, G.A.; Shea, B.; O’Connell, D.; Peterson, J.; Welch, V.; Losos, M.; Tugwell, P. The Newcastle-Ottawa Scale (NOS) for Assessing the Quality of Nonrandomised Studies in Meta-Analyses. Available online: http://www.ohri.ca/programs/clinical_epidemiology/oxford.asp#.YPyh9YR9Gg0.google (accessed on 11 April 2021).
- Manichanh, C.; Varela, E.; Martinez, C.; Antolin, M.; Llopis, M.; Doré, J.; Giralt, J.; Guarner, F.; Malagelada, J.-R. The Gut Microbiota Predispose to the Pathophysiology of Acute Postradiotherapy Diarrhea. Am. J. Gastroenterol. 2008, 103, 1754–1761. [Google Scholar] [CrossRef] [PubMed]
- Ritchie, L.E.; Taddeo, S.S.; Weeks, B.R.; Lima, F.; Bloomfield, S.A.; Azcarate-Peril, M.A.; Zwart, S.R.; Smith, S.M.; Turner, N.D. Space Environmental Factor Impacts upon Murine Colon Microbiota and Mucosal Homeostasis. PLoS ONE 2015, 10, e0125792. [Google Scholar] [CrossRef] [PubMed]
- Gupta, N.; Kainthola, A.; Tiwari, M.; Agrawala, P.K. Gut microbiota response to ionizing radiation and its modulation by HDAC inhibitor TSA. Int. J. Radiat. Biol. 2020, 96, 1560–1570. [Google Scholar] [CrossRef]
- Thomsen, M.; Clarke, S.; Vitetta, L. The role of adjuvant probiotics to attenuate intestinal inflammatory responses due to cancer treatments. Benef. Microbes 2018, 9, 899–916. [Google Scholar] [CrossRef]
- Cromer, W.E.; Zawieja, D.C. Acute exposure to space flight results in evidence of reduced lymph Transport, tissue fluid Shifts, and immune alterations in the rat gastrointestinal system. Life Sci. Space Res. 2018, 17, 74–82. [Google Scholar] [CrossRef] [PubMed]
- Kondrashova, V.G.; Vdovenko, V.Y.; Kolpakov, I.E.; Popova, A.S.; Mishchenko, L.P.; Gritsenko, T.V.; Stepanova, E.I. Gut microbiota among children living in areas contaminated by radiation and having the cardiac connective tissue dysplasia syndrome. Probl. Radiatsiinoi Medytsyny Radiobiolohii 2014, 19, 277–286. [Google Scholar]
- Claesson, M.J.; Clooney, A.G.; O’Toole, P.W. A clinician’s guide to microbiome analysis. Nat. Rev. Gastroenterol. Hepatol. 2017, 14, 585–595. [Google Scholar] [CrossRef] [PubMed]
- Kumar, R.; Eipers, P.; Little, R.B.; Crowley, M.; Crossman, D.K.; Lefkowitz, E.J.; Morrow, C.D. Getting Started with Microbiome Analysis: Sample Acquisition to Bioinformatics. Curr. Protoc. Hum. Genet. 2014, 82, 18.8.1–18.8.29. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jost, L. Partitioning diversity into independent alpha and beta components. Ecology 2007, 88, 2427–2439. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sankar, S.A.; Lagier, J.C.; Pontarotti, P.; Raoult, D.; Fournier, P.E. The human gut microbiome, a taxonomic conundrum. Syst. Appl. Microbiol. 2015, 38, 276–286. [Google Scholar] [CrossRef]
- Mosca, A.; Leclerc, M.; Hugot, J.-P. Gut Microbiota Diversity and Human Diseases: Should We Reintroduce Key Predators in Our Ecosystem? Front. Microbiol. 2016, 7, 455. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eckburg, P.B.; Bik, E.M.; Bernstein, C.N.; Purdom, E.; Dethlefsen, L.; Sargent, M.; Gill, S.R.; Nelson, K.E.; Relman, D.A. Diversity of the Human Intestinal Microbial Flora. Science 2005, 308, 1635–1638. [Google Scholar] [CrossRef] [Green Version]
- Yatsunenko, T.; Rey, F.E.; Manary, M.J.; Trehan, I.; Dominguez-Bello, M.G.; Contreras, M.; Magris, M.; Hidalgo, G.; Baldassano, R.N.; Anokhin, A.P.; et al. Human gut microbiome viewed across age and geography. Nature 2012, 486, 222–227. [Google Scholar] [CrossRef]
- Zhao, Z.; Cheng, W.; Qu, W.; Shao, G.; Liu, S. Antibiotic Alleviates Radiation-Induced Intestinal Injury by Remodeling Microbiota, Reducing Inflammation, and Inhibiting Fibrosis. ACS Omega 2020, 5, 2967–2977. [Google Scholar] [CrossRef]
- Wang, M.; Dong, Y.; Wu, J.; Li, H.; Zhang, Y.; Fan, S.; Li, D. Baicalein ameliorates ionizing radiation-induced injuries by rebalancing gut microbiota and inhibiting apoptosis. Life Sci. 2020, 261, 118463. [Google Scholar] [CrossRef]
- Suau, A.; Rochet, V.; Sghir, A.; Gramet, G.; Brewaeys, S.; Sutren, M.; Rigottier-Gois, L.; Doré, J. Fusobacterium prausnitzii and Related Species Represent a Dominant Group Within the Human Fecal Flora. Syst. Appl. Microbiol. 2001, 24, 139–145. [Google Scholar] [CrossRef] [PubMed]
- Sokol, H.; Seksik, P.; Furet, J.P.; Firmesse, O.; Nion-Larmurier, I.; Beaugerie, L.; Cosnes, J.; Corthier, G.; Marteau, P.; Doré, J. Low counts of Faecalibacterium prausnitzii in colitis microbiota. Inflamm. Bowel Dis. 2009, 15, 1183–1189. [Google Scholar] [CrossRef] [PubMed]
- Bennett, K.W.; Eley, A. Fusobacteria: New taxonomy and related diseases. J. Med. Microbiol. 1993, 39, 246–254. [Google Scholar] [CrossRef] [PubMed]
- Magne, F.; Gotteland, M.; Gauthier, L.; Zazueta, A.; Pesoa, S.; Navarrete, P.; Balamurugan, R. The Firmicutes/Bacteroidetes Ratio: A Relevant Marker of Gut Dysbiosis in Obese Patients? Nutrients 2020, 12(5), 1474. [Google Scholar] [CrossRef] [PubMed]
- Tseng, C.H.; Wu, C.Y. The gut microbiome in obesity. J. Formos. Med. Assoc. 2019, 118, S3–S9. [Google Scholar] [CrossRef]
- Yamanouchi, K.; Tsujiguchi, T.; Sakamoto, Y.; Ito, K. Short-term follow-up of intestinal flora in radiation-exposed mice. J. Radiat. Res. 2019, 60, 328–332. [Google Scholar] [CrossRef] [Green Version]
- Goudarzi, M.; Mak, T.D.; Jacobs, J.P.; Moon, B.H.; Strawn, S.J.; Braun, J.; Brenner, D.J.; Fornace, A.J., Jr.; Li, H.H. An Integrated Multi-Omic Approach to Assess Radiation Injury on the Host-Microbiome Axis. Radiat. Res. 2016, 186, 219–234. [Google Scholar] [CrossRef] [Green Version]
- Salyers, A.A. Bacteroides of the Human Lower Intestinal Tract. Annu. Rev. Microbiol. 1984, 38, 293–313. [Google Scholar] [CrossRef]
- Wexler, H.M. Bacteroides: The Good, the Bad, and the Nitty-Gritty. Clin. Microbiol. Rev. 2007, 20, 593–621. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huttenhower, C.; Gevers, D.; Knight, R.; Abubucker, S.; Badger, J.H.; Chinwalla, A.T.; Creasy, H.H.; Earl, A.M.; FitzGerald, M.G.; Fulton, R.S.; et al. Structure, function and diversity of the healthy human microbiome. Nature 2012, 486, 207–214. [Google Scholar] [CrossRef] [Green Version]
- Iizumi, T.; Battaglia, T.; Ruiz, V.; Perez Perez, G.I. Gut Microbiome and Antibiotics. Arch. Med. Res. 2017, 48, 727–734. [Google Scholar] [CrossRef] [PubMed]
- Villéger, R.; Lopès, A.; Carrier, G.; Veziant, J.; Billard, E.; Barnich, N.; Gagnière, J.; Vazeille, E.; Bonnet, M. Intestinal Microbiota: A Novel Target to Improve Anti-Tumor Treatment? Int. J. Mol. Sci. 2019, 20, 4584. [Google Scholar] [CrossRef] [PubMed] [Green Version]
| Search Number | Search Terms |
|---|---|
| Search #1 | “microbiota” OR “gastrointestinal microbiome” OR “microbiome” OR “16s rRNA” |
| Search #2 | “radiation” OR “radiotherapy” |
| Search #3 | Search #1 AND Search #2 |
| Search Number | Search Terms |
|---|---|
| Search #1 | “microbiota” OR “gastrointestinal microbiome” OR “microbiome” OR “16s rRNA” OR “microflora” |
| Search #2 | “radiation” OR “radiotherapy” |
| Search #3 | English OR Spanish OR Portuguese |
| Search #4 | Search #1 AND Search #2 AND Search #3 |
| Search Number | Search Terms |
|---|---|
| Search #1 | “microbiota” OR “gastrointestinal microbiome” OR “microbiome” OR “16s rRNA” OR “microflora” |
| Search #2 | “radiation” OR “radiotherapy” |
| Search #3 | Search #1 AND Search #2 |
| Author, Year / DOI | Study Design | Participant Demographics: N; Sex (M:F); Age; Type of Cancer/Other | Type of Radiation | Microbiome Assessment Method / Software for Sequencing and For Identification | Type of Sample / Number of Samples | Main Findings | Antibiotic Used as Exclusion? / Comments |
|---|---|---|---|---|---|---|---|
| Cuzzolin et al., 1992 [23] / 10.1080/1120009x.1992.11739160 | Prospective cohort | N = 15 (0:15) / 45–79 years / Gynecological cancer | Pelvic RT / 4000 cGy in 4 to 5 weeks overall 175–200 cGy daily 5 days per week | Culture counts / Agar-based methods | Fecal / 5 samples: - 1 before - 4 after irradiation fractions |
| Yes / Other exclusion criteria: cytotoxic chemotherapy |
| Sajjadieh et al., 2012 [24] / PMID: 23400266; PMCID: PMC3564093 | Prospective cohort | N = 75 Control group n = 20 / 4–18 years / Living in a contaminated area near Chernobyl | Ambient radiation / Internal whole-body radioactivity Cs-137 measured by γ-ray spectrometry | Bacterial culture; colony-forming units / CPLX agar Bifidobacterium; LBS agar Lactobacillus; COBA agar Enterococcus; DHL agar Enterobacter | Fecal / 1 sample |
| Yes / - |
| García-Peris et al., 2012 [25] / 10.3305/nh.2012.27.6.5992 | RCT | N = 31 (0:31) / 36–77 years (median 59) / Gynecological cancer | Pelvic RT / 52.2 Gy 1.8 Gy/day 5 times a week 29 sessions | Culture counts / Fluorescent in situ hybridization, genus-specific probes (Bifidobacterium: Bif164 and Lactobacillus: LAC158) | Fecal / 4 samples: - 7 days before RT - 15 days after RT - At the end of the treatment - 3 weeks after RT |
| Yes / Other exclusion criteria: previous RT; previous or adjuvant QT; immunosuppressive |
| Nam et al., 2013 [5] / 10.1371/journal.pone.0082659 | Prospective cohort | N = 9 (0:9) Control N = 6 / 35–63 years / Gynecologic cancer (cervix and endometrium) | Pelvic RT / 50.4 Gy 5 times a week 5 week period 25 fractions | 16S rRNA V1/V2 / QIIME MOTHUR UPARSE / Ribosomal and SILVA databases | Fecal / 4 samples:- 1 week before - After the first RT - At the end of the fifth RT - 1–3 months after final RT |
| Yes / QT (two individuals did not take QT during radiotherapy) |
| Wang A et al., 2015 [7] / 10.1371/journal.pone.0126312 | Prospective cohort | N = 11 (2:9) Control: N = 4 / 41–65 years (median 51) / Cervical, anal, and colorectal cancer | Pelvic RT / 44–50 Gy 1.8–2.0 Gy/day 5 times a week 5 week period 25 fractions | 16s rRNA V3 region / SILVA ribosomal RNA database/MOTHUR | Fecal 2 samples: - Immediately before - Just after RT |
| Yes / Other exclusion criteria: chemotherapy, steroid, immunosuppressor 1 month before / Comparison between patients that developed diarrhea and those who did not |
| Yi et al., 2021 [17] / 10.1158/1078-0432.CCR-20-3445 | Prospective cohort | N = 84 (58:26) Control N = 31 / Nonresponder group 56.46 ± 9.47 years Responder 56.64 ± 10.43 / Locally advanced rectal cancer | Pelvic RT / 45–50 Gy daily fraction 1.8–2 Gy | 16S rRNA gene V3–V4 region / Ribosomal Database Project classifier /Illumina Miseq / VSearch; USearch STAMP | Fecal / 2 samples :- Initial day (n = 84) - Within three days upon completion of (n = 83) nCRT treatment |
| No / Exclusion: exposure to prebiotics, probiotics, steroids, or immunosuppressants / QTconcurrent |
| Wang Z et al., 2019 [26] / 10.1111/jcmm.14289 | Prospective cohort | N = 18 (0:18) / 30–67 years (median 57) / Cervical cancer | Pelvic RT / 50.4 Gy 180cGy/fraction | 16s rRNA / Illumina Hiseq / QIIME / UPARSE / Greengene database | Fecal / 2 samples: - One day before - First day after the treatment |
| Yes / Other exclusion criteria:recent use of probiotics; proton pump inhibitors; other morbidities such as enteritis or autoimmune condition |
| Sahly et al., 2019 [27] / 10.7717/peerj.7683 | Prospective cohort | N = 3 (3:0) Control N = 2 / 3.5–7 years / Rhabdomyosarcoma near pelvic region | Pelvic RT / 50.4 Gy 180 cGy / fraction 28 fractions | 16s rRNA V3–V5 / Illumina Miseq / QIIME 2 / SILVA database | Fecal / 3 samples: - Before radiotherapy - 12–16 days after - 26–28 days after |
| No / QT weeks before RT |
| Shi et al., 2020 [16] / 10.3389/fcimb.2020.562463 | Prospective cohort | N = 22 (16:6) / 45–72 years (median 61) / Rectal cancer | Pelvic RT / 50Gy 2Gy daily fractions | 16s rRNA V3–4 region / MOTHUR / SILVA database / Ribosomal Database project | Fecal samples / 2 samples: - At treatment initiation - Just after nCRT |
| Yes / concurrent chemotherapy / Exclusion criteria: steroids and immunosuppressants within the previous 6 months |
| Mitra et al., 2020[18] / 10.1016/j.ijrobp.2019.12.040 | Prospective cohort | N = 35 (0:35) / 35–72 years (median 47) / Cervical cancer | Pelvic RT / No information found about doses | 16s rRNA V4 region / Illumina MiSeq / SILVA database / UPARSE | Fecal / 4 samples: - Before RT - During radiation therapy (weeks 1, 3, and 5) |
| No / QT Weekly cisplatin |
| El Alam et al., 2021[19] / 10.1371/journal.pone.0247905 | Prospective cohort | N = 58 (50:8) / Mean 49.36 ± 10.52 years / Gynecologic cancer patients (55 cervical, 2 vulvar, and 1 with vaginal cancer) | 45 Gy (minimum radiation dose) 5 weeks 25 fractions / Either 2 or 5 pulsed dose brachytherapy | 16S rRNA V4 region / Alkek Center for Metagenomics and Microbiome Research at Baylor College of Medicine using a methodology from the Human Microbiome Project | Rectal swabs / 5 samples: - Immediately before treatment - 1, 3, 5, and 12 weeks after treatment initiation |
| No / QTcisplatin and brachytherapy / 53 patients did not provide samples at all time points |
| Cuzzolin et al., 1992 [23] / 10.1080/1120009x.1992.11739160 | Prospective cohort | N = 15 (0:15) / 45–79 years / Gynecological cancer | Pelvic RT / 4000 cGy in 4 to 5 weeks overall 175–200 c Gy daily 5 days per week | Culture counts / Agar-based methods | Fecal / 5 samples: - 1 before - 4 after irradiation fractions |
| Yes / Other exclusion criteria: cytotoxic chemotherapy |
| Sajjadieh et al., 2012 [24] / PMID: 23400266; PMCID: PMC3564093 | Prospective cohort | N = 75 Control group n = 20 / 4–18 years / Living in a contaminated area near Chernobyl | Ambient radiation / Internal whole-body radioactivity Cs-137 measured by γ-ray spectrometry | Bacterial culture; colony-forming units / CPLX agar Bifidobacterium; LBS agar Lactobacillus; COBA agar Enterococcus; DHL agar Enterobacter | Fecal / 1 sample |
| Yes / - |
| García-Peris et al., 2012 [25] / 10.3305/nh.2012.27.6.5992 | RCT | N = 31 (0:31) / 36–77 years (median 59) / Gynecological cancer | Pelvic RT / 52.2 Gy 1.8 Gy/day 5 times a week 29 sessions | Culture counts / Fluorescent in situ hybridization, genus-specific probes (Bifidobacterium: Bif164 and Lactobacillus: LAC158) | Fecal / 4 samples: - 7 days before RT - 15 days after RT - At the end of the treatment - 3 weeks after RT |
| Yes / Other exclusion criteria: previous RT; previous or adjuvant QT; immunosuppressive |
| Nam et al., 2013 [5] / 10.1371/journal.pone.0082659 | Prospective cohort | N = 9 (0:9) Control N = 6 / 35–63 years / Gynecologic cancer (cervix and endometrium) | Pelvic RT / 50.4 Gy 5 times a week 5 week period 25 fractions | 16S rRNA V1/V2 / QIIME MOTHUR UPARSE / Ribosomal and SILVA databases | Fecal / 4 samples: - 1 week before - After the first RT - At the end of the fifth RT - 1–3 months after final RT |
| Yes / QT (two individuals did not take QT during radiotherapy) |
| Wang A et al., 2015 [7] / 10.1371/journal.pone.0126312 | Prospective cohort | N = 11 (2:9) Control: N = 4 / 41–65 years (median 51) / Cervical, anal, and colorectal cancer | Pelvic RT / 44–50 Gy 1.8–2.0 Gy/day 5 times a week 5 week period 25 fractions | 16s rRNA V3 region / SILVA ribosomal RNA database / MOTHUR | Fecal 2 samples: - Immediately before - Just after RT |
| Yes / Other exclusion criteria: chemotherapy, steroid, immunosuppressor 1 month before / Comparison between patients that developed diarrhea and those who did not |
| Yi et al., 2021 [17] / 10.1158/1078-0432.CCR-20-3445 | Prospective cohort | N = 84 (58:26) Control N = 31 / Nonresponder group 56.46 ± 9.47 years Responder 56.64 ± 10.43 / Locally advanced rectal cancer | Pelvic RT / 45–50 Gy daily fraction 1.8–2 Gy | 16S rRNA gene V3–V4 region / Ribosomal Database Project classifier / Illumina Miseq / VSearch; USearch STAMP | Fecal / 2 samples: - Initial day (n = 84) - Within three days upon completion of (n = 83) nCRT treatment |
| No / Exclusion: exposure to prebiotics, probiotics, steroids, or immunosuppressants /QT concurrent |
| Wang Z et al., 2019 [26] / 10.1111/jcmm.14289 | Prospective cohort | N = 18 (0:18) / 30–67 years (median 57) / Cervical cancer | Pelvic RT / 50.4 Gy 180cGy/fraction | 16s rRNA / Illumina Hiseq / QIIME / UPARSE / Greengene database | Fecal / 2 samples:- One day before - First day after the treatment |
| Yes / Other exclusion criteria: recent use of probiotics; proton pump inhibitors; other morbidities such as enteritis or autoimmune condition |
| Sahly et al., 2019 [27] / 10.7717/peerj.7683 | Prospective cohort | N = 3 (3:0) Control N = 2 /3.5–7 years / Rhabdomyosarcoma near pelvic region | Pelvic RT / 50.4 Gy 180 cGy/fraction 28 fractions | 16s rRNA V3–V5 / Illumina Miseq / QIIME 2 / SILVA database | Fecal / 3 samples: - Before radiotherapy - 12–16 days after - 26–28 days after |
| No / QT weeks before RT |
| Shi et al., 2020 [16] / 10.3389/fcimb.2020.562463 | Prospective cohort | N = 22 (16:6) / 45–72 years (median 61) / Rectal cancer | Pelvic RT / 50Gy 2Gy daily fractions | 16s rRNA V3–4 region / MOTHUR / SILVA database / Ribosomal Database project | Fecal samples / 2 samples: - At treatment initiation - Just after nCRT |
| Yes / concurrent chemotherapy / Exclusion criteria: steroids and immunosuppressants within the previous 6 months |
| Mitra et al., 2020[18] / 10.1016/j.ijrobp.2019.12.040 | Prospective cohort | N = 35 (0:35) / 35–72 years (median 47) / Cervical cancer | Pelvic RT / No information found about doses | 16s rRNA V4 region / Illumina MiSeq / SILVA database / UPARSE | Fecal / 4 samples: - Before RT - During radiation therapy (weeks 1, 3, and 5) |
| No / QT Weekly cisplatin |
| El Alam et al., 2021[19] / 10.1371/journal.pone.0247905 | Prospective cohort | N = 58 (50:8) / Mean 49.36 ± 10.52 years / Gynecologic cancer patients (55 cervical, 2 vulvar, and 1 with vaginal cancer) | 45 Gy (minimum radiation dose) 5 weeks 25 fractions / Either 2 or 5 pulsed dose brachytherapy | 16S rRNA V4 region / Alkek Center for Metagenomics and Microbiome Research at Baylor College of Medicine using a methodology from the Human Microbiome Project | Rectal swabs / 5 samples: - Immediately before treatment - 1, 3, 5, and 12 weeks after treatment initiation |
| No / QT cisplatin and brachytherapy / 53 patients did not provide samples at all time points |
| Author, Year | Selection | Comparability | Outcome | Score |
|---|---|---|---|---|
| Cuzzolin et al., 1992 [23] | *0** | *0 | *0* | 5/9 |
| Sajjadieh et al., 2012 [24] | *00* | *0 | *0* | 5/9 |
| Nam et al., 2013 [5] | **** | *0 | *** | 8/9 |
| Wang A et al., 2015 [7] | *0** | *0 | *0* | 6/9 |
| Yi et al., 2021 [17] | **** | *0 | *** | 8/9 |
| Wang Z et al., 2019 [26] | **** | *0 | *0* | 7/9 |
| Sahly et al., 2019 [27] | **** | *0 | *0* | 7/9 |
| Shi et al., 2020 [16] | **** | ** | *0* | 8/9 |
| Mitra et al., 2020 [18] | **** | ** | *0* | 8/9 |
| El Alam et al., 2021 [19] | **** | *0 | **0 | 7/9 |
| Domain | Risk of Bias | Comments |
|---|---|---|
| Sequence generation | High | No information regarding the sequence generation. “patients were randomised to receive...” |
| Allocation concealment | Low | “coded sachets” |
| Blinding of participants, personnel, and outcome assessors | Low | Outcome assessors and participants blinded |
| Incomplete outcome data | Low | “Nine patients were excluded from the study: four because they were prescribed antibiotics, three for personal reasons, and two due to lack of adherence” |
| Selection outcome reporting | Low | Study protocol available and all of study’s pre-specified outcomes have been reported |
| Other sources of bias | Low | Study appears to be free of other sources of bias |
| Dysbiosis | |
|---|---|
| |
| Diversity | |
| Alpha diversity | |
| Alpha diversity |
|
| Shannon index |
|
| Simpson index | |
| Beta diversity | |
| |
| Richness | |
| Richness index |
|
| Chao1 index |
|
| OTUs | |
| Composition | |
| Phylum level | |
| Firmicutes/Bacteroidetes ratio |
|
| Unclassified bacteria | |
| Actinobacteria | |
| Bacteroidetes | |
| Firmicutes | |
| Fusobacteria |
|
| Proteobacteria | |
| Class level | |
| Gammaproteobacteria |
|
| Bacilli |
|
| Clostridia |
|
| Order level | |
| Clostridiales | |
| Lactobacillales |
|
| Fusobacteriales |
|
| Pasteurellales | |
| Family level | |
| Defluviitaleaceae |
|
| Eubacteriaceae |
|
| Fusobacteriaceae |
|
| Lachnospiracea |
|
| Streptococcaceae |
|
| Veillonellaceae |
|
| Enterococcaceae |
|
| Pasteurellaceae |
|
| Ruminococcaceae |
|
| Genus level | |
| Bacteroides | |
| Bifidobacterium | |
| Citrobacter |
|
| Clostridium_XIVa |
|
| Clostridium XI and XVIII and unclassified (others) |
|
| Coprococcus |
|
| Dorea |
|
| Enterobacter |
|
| Enterococcus |
|
| Escherichia–Shigella |
|
| Ezakiella |
|
| Fusobacterium |
|
| Faecalibacterium | |
| Haemophilus |
|
| Lactobacillus | |
| Megamonas |
|
| Oscillibacter | |
| Parvimonas |
|
| Peptostreptococcus |
|
| Porphyromonas |
|
| Roseburia | |
| Ruminococcus |
|
| Serratia |
|
| Streptococcus | |
| Subdoligranulum |
|
| Sutterella |
|
| Veilonella |
|
| Prevotella_2 |
|
| Prevotella_9 |
|
| Species level | |
| Actinomyces odontolyticus |
|
| Adlercreutzia equolifaciens |
|
| Aeromonas hydrophila |
|
| Amphibacillus sp. YIM-kkny6 |
|
| Bacteroides sp. CCUG 39913 |
|
| Butyrate-producingbacterium T1–815 |
|
| Butyrate-producing bacterium |
|
| Butyrate-producingbacterium SS2/1 |
|
| Candidatus Bacilloplasma |
|
| Coriobacterium sp. CCUG 33918 |
|
| Clostridium methylpentosum |
|
| Clostridiales bacterium DJF CP67 |
|
| Clostridium leptum |
|
| Clostridiales bacterium A2–162 |
|
| Clostridium sp. BGC36 |
|
| Clostridium spp. (Cl. histolyticum, Cl. bifermentans, Cl. sporogenes) |
|
| Dialister sp. E2 20 |
|
| Escherichia coli |
|
| Eubacterium eligens |
|
| Eubacterium hallii |
|
| Enterococcus faecium 1 |
|
| Enterobacter sp. mcp11b |
|
| Fusobacterium nucleatum |
|
| Faecalibacterium Prausnitzii |
|
| Faecalibacterium sp. DJF VR20 |
|
| Human intestinal firmicute CB47 |
|
| Klebsiella pneumonia |
|
| Lactobacillus murinus |
|
| Lachnospiraceae bacterium DJF RP14 |
|
| Lachnospira pectinoschiza |
|
| Lactobacillales bacterium |
|
| Lactobacilli aerobi spp. |
|
| Lactobacilli anaerobi spp. |
|
| Oscillospira sp. BA04013493 |
|
| Prevotella stercorea |
|
| Prevotella copri |
|
| Peptococcus and Peptostreptococcus spp. |
|
| Roseburia inulinivorans |
|
| Ruminococcus sp. DJF VR52 |
|
| Ruminococcus sp. CO28 |
|
| Roseburia sp. DJFVR77 |
|
| Ruminococcus sp. CO41 |
|
| Ruminococcus callidus |
|
| Ruminococcus sp. CO28 |
|
| Ruminococcus sp. CS1 |
|
| Swine fecal bacterium FPC110 |
|
| Weissella confuse |
|
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fernandes, A.; Oliveira, A.; Soares, R.; Barata, P. The Effects of Ionizing Radiation on Gut Microbiota, a Systematic Review. Nutrients 2021, 13, 3025. https://doi.org/10.3390/nu13093025
Fernandes A, Oliveira A, Soares R, Barata P. The Effects of Ionizing Radiation on Gut Microbiota, a Systematic Review. Nutrients. 2021; 13(9):3025. https://doi.org/10.3390/nu13093025
Chicago/Turabian StyleFernandes, Ana, Ana Oliveira, Raquel Soares, and Pedro Barata. 2021. "The Effects of Ionizing Radiation on Gut Microbiota, a Systematic Review" Nutrients 13, no. 9: 3025. https://doi.org/10.3390/nu13093025







