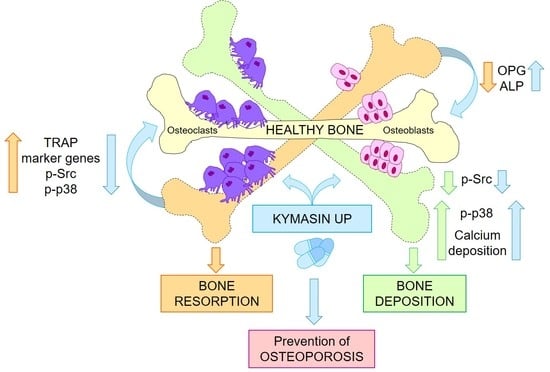
KYMASIN UP Natural Product Inhibits Osteoclastogenesis and Improves Osteoblast Activity by Modulating Src and p38 MAPK
Abstract
:1. Introduction
2. Results and Discussion
2.1. KYMASIN UP Suppresses the Gene Expression of Osteoclastic Markers
2.2. KYMASIN UP Inhibits RANKL-Induced Osteoclastogenesis
2.3. KYMASIN UP Restore the RANKL/OPG Ratio in Osteoporotic Conditions
2.4. W. somnifera Is the Main Responsible for the Effects of KYMASIN UP on Osteoclasts
2.5. KYMASIN UP Does Not Affect BMP2-Dependent C2C12 Osteoblast Transdifferentiation
2.6. KYMASIN UP Improves C2C12-Derived Osteoblast Mineralization Activity
2.7. Effects of KYMASIN UP on Human Osteoblast Activity
2.8. KYMASIN UP Affects Src and p38 MAPK Activation
3. Materials and Methods
3.1. KYMASIN UP
3.2. Cell Cultures
3.3. MTT 3-(4,5-Dimethylthiazol-2-yl)-2,5-Diphenyltetrazolium Bromide) Assay
3.4. TRAP Staining and Activity
3.5. F-Actin Ring-Formation Assay
3.6. Real-Time PCR
3.7. Western Blotting
3.8. Immunoflorescence (IF) for Desmin Expression
3.9. ALP Activity and Staining
3.10. Mineralization Assay
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Raisz, L.G. Pathogenesis of osteoporosis: Concepts, conflicts, and prospects. J. Clin. Investig. 2005, 115, 3318–3325. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Choi, M.H.; Yang, J.H.; Seo, J.S.; Kim, Y.-J.; Kang, S.-W. Prevalence and diagnosis experience of osteoporosis in postmenopausal women over 50: Focusing on socioeconomic factors. PLoS ONE 2021, 16, e0248020. [Google Scholar] [CrossRef] [PubMed]
- Song, S.; Guo, Y.; Yang, Y.; Fu, D. Advances in pathogenesis and therapeutic strategies for osteoporosis. Pharmacol. Ther. 2022, 237, 108168. [Google Scholar] [CrossRef] [PubMed]
- Corrado, A.; Cici, D.; Rotondo, C.; Maruotti, N.; Cantatore, F.P. Molecular Basis of Bone Aging. Int. J. Mol. Sci. 2020, 21, 3679. [Google Scholar] [CrossRef] [PubMed]
- Noh, J.-Y.; Yang, Y.; Jung, H. Molecular Mechanisms and Emerging Therapeutics for Osteoporosis. Int. J. Mol. Sci. 2020, 21, 7623. [Google Scholar] [CrossRef]
- Gao, Y.; Patil, S.; Jia, J. The Development of Molecular Biology of Osteoporosis. Int. J. Mol. Sci. 2021, 22, 8182. [Google Scholar] [CrossRef]
- An, J.; Yang, H.; Zhang, Q.; Liu, C.; Zhao, J.; Zhang, L.; Chen, B. Natural products for treatment of osteoporosis: The effects and mechanisms on promoting osteoblast-mediated bone formation. Life Sci. 2016, 147, 46–58. [Google Scholar] [CrossRef] [PubMed]
- Salvadori, L.; Mandrone, M.; Manenti, T.; Ercolani, C.; Cornioli, L.; Lianza, M.; Tomasi, P.; Chiappalupi, S.; Di Filippo, E.S.; Fulle, S.; et al. Identification of Withania somnifera-Silybum marianum-Trigonella foenum-graecum Formulation as a Nutritional Supplement to Contrast Muscle Atrophy and Sarcopenia. Nutrients 2020, 13, 49. [Google Scholar] [CrossRef]
- Rathi, A.; Ishaq, M.; Najmi, A.K.; Akhtar, M. Trigonelline Demonstrated Ameliorative Effects in Dexamethasone Induced Osteoporotic Rats. Drug Res. 2020, 70, 257–264. [Google Scholar] [CrossRef]
- Anjaneyulu, K.; Bhat, K.M.; Srinivasa, S.R.; Devkar, R.A.; Henry, T. Beneficial Role of Hydro-alcoholic Seed Extract of Trigonella foenum graecum on Bone Structure and Strength in Menopause Induced Osteopenia. Ethiop. J. Health Sci. 1970, 28, 787–794. [Google Scholar] [CrossRef]
- Khedgikar, V.; Kushwaha, P.; Gautam, J.; Verma, A.; Changkija, B.; Kumar, A.; Sharma, S.; Nagar, G.K.; Singh, D.; Trivedi, P.K.; et al. Withaferin A: A proteasomal inhibitor promotes healing after injury and exerts anabolic effect on osteoporotic bone. Cell Death Dis. 2013, 4, e778. [Google Scholar] [CrossRef] [Green Version]
- Kim, J.-L.; Kang, S.-W.; Kang, M.-K.; Gong, J.-H.; Lee, E.-S.; Han, S.J.; Kang, Y.-H. Osteoblastogenesis and osteoprotection enhanced by flavonolignan silibinin in osteoblasts and osteoclasts. J. Cell. Biochem. 2011, 113, 247–259. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.-L.; Park, S.-H.; Jeong, D.; Nam, J.-S.; Kang, Y.-H. Osteogenic activity of silymarin through enhancement of alkaline phosphatase and osteocalcin in osteoblasts and tibia-fractured mice. Exp. Biol. Med. 2012, 237, 417–428. [Google Scholar] [CrossRef]
- Kim, S.; Kang, S.-S.; Choi, S.-I.; Kim, G.-H.; Imm, J.-Y. Ecklonia cava Extract Containing Dieckol Suppresses RANKL-Induced Osteoclastogenesis via MAP Kinase/NF-κB Pathway Inhibition and Heme Oxygenase-1 Induction. J. Microbiol. Biotechnol. 2019, 29, 11–20. [Google Scholar] [CrossRef] [Green Version]
- Orecchini, E.; Mondanelli, G.; Orabona, C.; Volpi, C.; Adorisio, S.; Calvitti, M.; Thuy, T.T.; Delfino, D.V.; Belladonna, M.L. Artocarpus tonkinensis Extract Inhibits LPS-Triggered Inflammation Markers and Suppresses RANKL-Induced Osteoclastogenesis in RAW264.7. Front. Pharmacol. 2021, 11, 593829. [Google Scholar] [CrossRef]
- Katagiri, T.; Yamaguchi, A.; Komaki, M.; Abe, E.; Takahashi, N.; Ikeda, T.; Rosen, V.; Wozney, J.M.; Fujisawa-Sehara, A.; Suda, T. Bone morphogenetic protein-2 converts the differentiation pathway of C2C12 myoblasts into the osteoblast lineage. J. Cell Biol. 1994, 127, 1755–1766. [Google Scholar] [CrossRef] [Green Version]
- Hidaka, Y.; Chiba-Ohkuma, R.; Karakida, T.; Onuma, K.; Yamamoto, R.; Fujii-Abe, K.; Saito, M.M.; Yamakoshi, Y.; Kawahara, H. Combined Effect of Midazolam and Bone Morphogenetic Protein-2 for Differentiation Induction from C2C12 Myoblast Cells to Osteoblasts. Pharmaceutics 2020, 12, 218. [Google Scholar] [CrossRef] [Green Version]
- Hwang, J.-S.; Lien, A.S.-Y.; Jiang, Y.-D. Commentary on the effects of receptor activator of nuclear factor—B ligand inhibition on bone mass and muscle strength. J. Diabetes Investig. 2019, 11, 287–289. [Google Scholar] [CrossRef]
- Roodman, G.D. Advances in bone biology: The osteoclast. Endocr. Rev. 1996, 17, 308–332. [Google Scholar] [CrossRef]
- Georges, S.; Velasco, C.R.; Trichet, V.; Fortun, Y.; Heymann, D.; Padrines, M. Proteases and bone remodelling. Cytokine Growth Factor Rev. 2009, 20, 29–41. [Google Scholar] [CrossRef]
- Kim, J.-L.; Kim, Y.-H.; Kang, M.-K.; Gong, J.-H.; Han, S.-J.; Kang, Y.-H. Antiosteoclastic activity of milk thistle extract after ovariectomy to suppress estrogen deficiency-induced osteoporosis. BioMed Res. Int. 2013, 2013, 919374. [Google Scholar] [CrossRef] [Green Version]
- Kavitha, C.V.; Deep, G.; Gangar, S.C.; Jain, A.K.; Agarwal, C.; Agarwal, R. Silibinin inhibits prostate cancer cells- and RANKL-induced osteoclastogenesis by targeting NFATc1, NF-κB, and AP-1 activation in RAW264.7 cells. Mol. Carcinog. 2012, 53, 169–180. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, H.; Wang, J.; Sun, Q.; Chen, G.; Sun, S.; Ma, X.; Qiu, H.; Liu, X.; Xu, L.; Liu, M. Jatrorrhizine hydrochloride suppresses RANKL-induced osteoclastogenesis and protects against wear particle-induced osteolysis. Int. J. Mol. Sci. 2018, 19, 3698. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Chen, G.; Zhang, Q.; Zhao, F.; Yu, X.; Ma, X.; Liu, M. Phillyrin Attenuates Osteoclast Formation and Function and Prevents LPS-Induced Osteolysis in Mice. Front. Pharmacol. 2019, 10, 1188. [Google Scholar] [CrossRef] [Green Version]
- Shishodia, S.; Aggarwal, B.B. Diosgenin inhibits osteoclastogenesis, invasion, and proliferation through the downregulation of Akt, IκB kinase activation and NF-κB-regulated gene expression. Oncogene 2005, 25, 1463–1473. [Google Scholar] [CrossRef]
- Kim, J.H.; Kim, K.; Jin, H.M.; Song, I.; Youn, B.U.; Lee, J.; Kim, N. Silibinin inhibits osteoclast differentiation mediated by TNF family members. Mol. Cells 2009, 28, 201–207. [Google Scholar] [CrossRef] [PubMed]
- Srivastava, R.K.; Azam, Z.; Pandey, V.; Gupta, N.; Sapra, L.; Dar, H.Y.; Shokeen, N.; Bhardwaj, A.; Ahmad, A.; Soni, V. Phytoconstituents as novel osteo-protective agents Implications in bone health. Front. Biosci. 2020, 25, 1259–1296. [Google Scholar] [CrossRef] [PubMed]
- Nagareddy, P.R.; Lakshmana, M. Withania somnifera improves bone calcification in calcium-deficient ovariectomized rats. J. Pharm. Pharmacol. 2006, 58, 513–519. [Google Scholar] [CrossRef] [PubMed]
- Boyce, B.F.; Xing, L. Functions of RANKL/RANK/OPG in bone modeling and remodeling. Arch. Biochem. Biophys. 2008, 473, 139–146. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Azizieh, F.Y.; Shehab, D.; Al Jarallah, K.; Gupta, R.; Raghupathy, R. Circulatory Levels of RANKL, OPG, and Oxidative Stress Markers in Postmenopausal Women With Normal or Low Bone Mineral Density. Biomark. Insights 2019, 14, 1177271919843825. [Google Scholar] [CrossRef]
- Zinnia, M.A.; Islam, A.B.M.M.K. Fenugreek steroidal saponins hinder osteoclastogenic bone resorption by targeting CSF-1R which diminishes the RANKL/OPG ratio. Int. J. Biol. Macromol. 2021, 186, 351–364. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Song, C.; Fu, X.; Liu, M.; Li, Y.; Pan, J.; Liu, H.; Wang, S.; Xiang, L.; Xiao, G.G.; et al. High-Dose Diosgenin Reduces Bone Loss in Ovariectomized Rats via Attenuation of the RANKL/OPG Ratio. Int. J. Mol. Sci. 2014, 15, 7130. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ichikawa, H.; Takada, Y.; Shishodia, S.; Jayaprakasam, B.; Nair, M.G.; Aggarwal, B.B. Withanolides potentiate apoptosis, inhibit invasion, and abolish osteoclastogenesis through suppression of nuclear factor-κB (NF-κB) activation and NF-κB–regulated gene expression. Mol. Cancer Ther. 2006, 5, 1434–1445. [Google Scholar] [CrossRef] [Green Version]
- Halloran, D.; Durbano, H.W.; Nohe, A. Bone Morphogenetic Protein-2 in Development and Bone Homeostasis. J. Dev. Biol. 2020, 8, 19. [Google Scholar] [CrossRef]
- Li, H.; Choudhary, S.K.; Milner, D.J.; I Munir, M.; Kuisk, I.R.; Capetanaki, Y. Inhibition of desmin expression blocks myoblast fusion and interferes with the myogenic regulators MyoD and myogenin. J. Cell Biol. 1994, 124, 827–841. [Google Scholar] [CrossRef] [Green Version]
- Schiaffino, S.; Reggiani, C. Myosin isoforms in mammalian skeletal muscle. J. Appl. Physiol. 1994, 77, 493–501. [Google Scholar] [CrossRef]
- Amarasekara, D.S.; Kim, S.; Rho, J. Regulation of Osteoblast Differentiation by Cytokine Networks. Int. J. Mol. Sci. 2021, 22, 2851. [Google Scholar] [CrossRef]
- Folwarczna, J.; Zych, M.; Nowińska, B.; Pytlik, M. Effects of fenugreek (Trigonella foenum-graecum L.) seed on bone mechanical properties in rats. Eur. Rev. Med. Pharmacol. Sci. 2014, 18, 1937–1947. [Google Scholar]
- Folwarczna, J.; Zych, M.; Nowińska, B.; Pytlik, M.; Janas, A. Unfavorable effect of trigonelline, an alkaloid present in coffee and fenugreek, on bone mechanical properties in estrogen-deficient rats. Mol. Nutr. Food Res. 2014, 58, 1457–1464. [Google Scholar] [CrossRef] [PubMed]
- Bosetti, M.; Boccafoschi, F.; Leigheb, M.; Cannas, M.F. Effect of different growth factors on human osteoblasts activities: A possible application in bone regeneration for tissue engineering. Biomol. Eng. 2007, 24, 613–618. [Google Scholar] [CrossRef]
- Ji, L.; Li, X.; He, S.; Chen, S. Regulation of osteoclast-mediated bone resorption by microRNA. Cell. Mol. Life Sci. 2022, 79, 287. [Google Scholar] [CrossRef] [PubMed]
- Wu, M.; Chen, G.; Li, Y.-P. TGF-β and BMP signaling in osteoblast, skeletal development, and bone formation, homeostasis and disease. Bone Res. 2016, 4, 16009. [Google Scholar] [CrossRef] [PubMed]
- Zhi, X.; Fang, C.; Gu, Y.; Chen, H.; Chen, X.; Cui, J.; Hu, Y.; Weng, W.; Zhou, Q.; Wang, Y.; et al. Guaiacol suppresses osteoclastogenesis by blocking interactions of RANK with TRAF6 and C-Src and inhibiting NF-κB, MAPK and AKT pathways. J. Cell. Mol. Med. 2020, 24, 5122–5134. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Destaing, O.; Sanjay, A.; Itzstein, C.; Horne, W.C.; Toomre, D.; De Camilli, P.; Baron, R. The Tyrosine Kinase Activity of c-Src Regulates Actin Dynamics and Organization of Podosomes in Osteoclasts. Mol. Biol. Cell 2008, 19, 394–404. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marzia, M.; Sims, N.; Voit, S.; Migliaccio, S.; Taranta, A.; Bernardini, S.; Faraggiana, T.; Yoneda, T.; Mundy, G.R.; Boyce, B.F.; et al. Decreased C-Src Expression Enhances Osteoblast Differentiation and Bone Formation. J. Cell Biol. 2000, 151, 311–320. [Google Scholar] [CrossRef] [Green Version]
- Cong, Q.; Jia, H.; Li, P.; Qiu, S.; Yeh, J.; Wang, Y.; Zhang, Z.-L.; Ao, J.; Li, B.; Liu, H. p38α MAPK regulates proliferation and differentiation of osteoclast progenitors and bone remodeling in an aging-dependent manner. Sci. Rep. 2017, 7, 45964. [Google Scholar] [CrossRef] [Green Version]
- Bosetti, M.; Fusaro, L.; Nicoli, E.; Borrone, A.; Aprile, S.; Cannas, M. Poly-L-lactide acid-modified scaffolds for osteoinduction and osteoconduction. J. Biomed. Mater. Res. Part A 2013, 102, 3531–3539. [Google Scholar] [CrossRef]
- Chiappalupi, S.; Sorci, G.; Vukasinovic, A.; Salvadori, L.; Sagheddu, R.; Coletti, D.; Renga, G.; Romani, L.; Donato, R.; Riuzzi, F. Targeting RAGE prevents muscle wasting and prolongs survival in cancer cachexia. J. Cachexia Sarcopenia Muscle 2020, 11, 929–946. [Google Scholar] [CrossRef] [Green Version]
- Bosetti, M.; Borrone, A.; Leigheb, M.; Shastri, V.P.; Cannas, M. Injectable Graft Substitute Active on Bone Tissue Regeneration. Tissue Eng. Part A 2017, 23, 1413–1422. [Google Scholar] [CrossRef]
- Kirk, B.; Zanker, J.; Duque, G. Osteosarcopenia: Epidemiology, diagnosis, and treatment—facts and numbers. J. Cachexia Sarcopenia Muscle 2020, 11, 609–618. [Google Scholar] [CrossRef] [Green Version]
- He, C.; He, W.; Hou, J.; Chen, K.; Huang, M.; Yang, M.; Luo, X.; Li, C. Bone and Muscle Crosstalk in Aging. Front. Cell Dev. Biol. 2020, 8, 585644. [Google Scholar] [CrossRef] [PubMed]
- de Sire, A.; Invernizzi, M.; Baricich, A.; Lippi, L.; Ammendolia, A.; Grassi, F.A.; Leigheb, M. Optimization of transdisciplinary management of elderly with femur proximal extremity fracture: A patient-tailored plan from orthopaedics to rehabilitation. World J. Orthop. 2021, 12, 456–466. [Google Scholar] [CrossRef] [PubMed]
- Jilka, R.L. The Relevance of Mouse Models for Investigating Age-Related Bone Loss in Humans. J. Gerontol. Ser. A 2013, 68, 1209–1217. [Google Scholar] [CrossRef] [PubMed]
- Gigliotti, C.L.; Boggio, E.; Clemente, N.; Shivakumar, Y.; Toth, E.; Sblattero, D.; D’Amelio, P.; Isaia, G.C.; Dianzani, C.; Yagi, J.; et al. ICOS-Ligand Triggering Impairs Osteoclast Differentiation and Function In Vitro and In Vivo. J. Immunol. 2016, 197, 3905–3916. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tomimori, Y.; Mori, K.; Koide, M.; Nakamichi, Y.; Ninomiya, T.; Udagawa, N.; Yasuda, H. Evaluation of pharmaceuticals with a novel 50-hour animal model of bone loss. J. Bone Miner. Res. 2009, 24, 1194–1205. [Google Scholar] [CrossRef] [PubMed]







| Gene | Forward Primer 5′-3′ | Reverse Primer 5′-3′ |
|---|---|---|
| Gapdh | GCCTTCCGTGTTCCTACCC | CAGTGGGCCCTCAGATGC |
| Acp5 | CTGCCTTGTCAAGAACTTGC | ACCTTTCGTTGATGTCGCAC |
| Mmp9 | TAGCACAACAGCTGACTACG | ATCCTGGTCATAGTTGGCTG |
| CalcR | TCATCATCCACCTGGTTGAG | CACAGCCATGACAATCAGAG |
| Ctsk | AGAAGACTCACCAGAAGCAG | CAGGTTATGGGCAGAGATTTG |
| Osx | CCCCTTGTCGTCATGGTTACAG | AGAGAAAGCCTTTGCCCACCTA |
| Col1a1 | AGCACGTCTGGTTTGGAGAG | GCTGTAGGTGAAGCGACTGT |
| Bglap | AAGCAGGAGGGCAATAAGGT | TTTGTAGGCGGTCTTCAAGC |
| Runx2 | GCCGGGAATGATGAGAACTA | GGACCGTCCACTGTCACTTT |
| Primary Antibody | Molecular | Dilution | Source |
|---|---|---|---|
| Weight (kDa) | |||
| Mouse monoclonal anti-MyHC-II (MF20) | 220 | 1:10,000 | eBiosciences, San Diego, CA, USA |
| Mouse monoclonal anti-Myogenin (F5D) | 34 | 1:1000 | Santa Cruz Biotechnol., Dallas, TX, USA |
| Mouse monoclonal anti-α-Tubulin (DM1A) | 55 | 1:2000 | Santa Cruz Biotechnol., Dallas, TX, USA |
| Rabbit polyclonal anti-p38 MAPK | 40 | 1:1000 | Cell Signaling Technol., Danvers, MA, USA |
| Rabbit monoclonal anti-phospho-p38 MAPK | 43 | 1.2 | Cell Signaling Technol., Danvers, MA, USA |
| (Thr180/Tyr182) (D3F9) XP | |||
| Rabbit polyclonal anti-Phospho-Src Family (Tyr416) #2101 | 60 | 1:1000 | Cell Signaling Technol., Danvers, MA, USA |
| Rabbit monoclonal anti-Src (36D10) | 60 | 1:5000 | Cell Signaling Technol., Danvers, MA, USA |
| Rabbit polyclonal anti-phospho-p44/42 MAPK (Erk1/2) (Thr202/Tyr204) | 42/44 | 1:2000 | Cell Signaling Technol., Danvers, MA, USA |
| Rabbit polyclonal anti-MAP Kinase (ERK-1, ERK-2) | 42/44 | 1:10,000 | Cell Signaling Technol., Danvers, MA, USA |
| Rabbit monoclonal anti-phospho-Akt (Ser473) (D9E) | 60 | 1:2000 | Cell Signaling Technol., Danvers, MA, USA |
| Rabbit polyclonal anti-Akt (Thr308) (D25E6) | 60 | 1:2000 | Cell Signaling Technol., Danvers, MA, USA |
| Mouse monoclonal anti-GAPDH (6C5) | 37 | 1:5000 | Santa Cruz Biotechnol., Dallas, TX, USA |
| Goat anti-rabbit IgG/IgM-HRP conjugated | Sigma-Aldrich, St. Louis, MO, USA | ||
| Goat anti-mouse IgG/IgM-HRP conjugated | Sigma-Aldrich, St. Louis, MO, USA |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Salvadori, L.; Belladonna, M.L.; Castiglioni, B.; Paiella, M.; Panfili, E.; Manenti, T.; Ercolani, C.; Cornioli, L.; Chiappalupi, S.; Gentili, G.; et al. KYMASIN UP Natural Product Inhibits Osteoclastogenesis and Improves Osteoblast Activity by Modulating Src and p38 MAPK. Nutrients 2022, 14, 3053. https://doi.org/10.3390/nu14153053
Salvadori L, Belladonna ML, Castiglioni B, Paiella M, Panfili E, Manenti T, Ercolani C, Cornioli L, Chiappalupi S, Gentili G, et al. KYMASIN UP Natural Product Inhibits Osteoclastogenesis and Improves Osteoblast Activity by Modulating Src and p38 MAPK. Nutrients. 2022; 14(15):3053. https://doi.org/10.3390/nu14153053
Chicago/Turabian StyleSalvadori, Laura, Maria Laura Belladonna, Beatrice Castiglioni, Martina Paiella, Eleonora Panfili, Tommaso Manenti, Catia Ercolani, Luca Cornioli, Sara Chiappalupi, Giulia Gentili, and et al. 2022. "KYMASIN UP Natural Product Inhibits Osteoclastogenesis and Improves Osteoblast Activity by Modulating Src and p38 MAPK" Nutrients 14, no. 15: 3053. https://doi.org/10.3390/nu14153053
APA StyleSalvadori, L., Belladonna, M. L., Castiglioni, B., Paiella, M., Panfili, E., Manenti, T., Ercolani, C., Cornioli, L., Chiappalupi, S., Gentili, G., Leigheb, M., Sorci, G., Bosetti, M., Filigheddu, N., & Riuzzi, F. (2022). KYMASIN UP Natural Product Inhibits Osteoclastogenesis and Improves Osteoblast Activity by Modulating Src and p38 MAPK. Nutrients, 14(15), 3053. https://doi.org/10.3390/nu14153053