The Influence of the Differentiation of Genes Encoding Peroxisome Proliferator-Activated Receptors and Their Coactivators on Nutrient and Energy Metabolism
Abstract
:1. Introduction
2. The PPAR Family

3. The PPARG Gene

4. The PPARD Gene

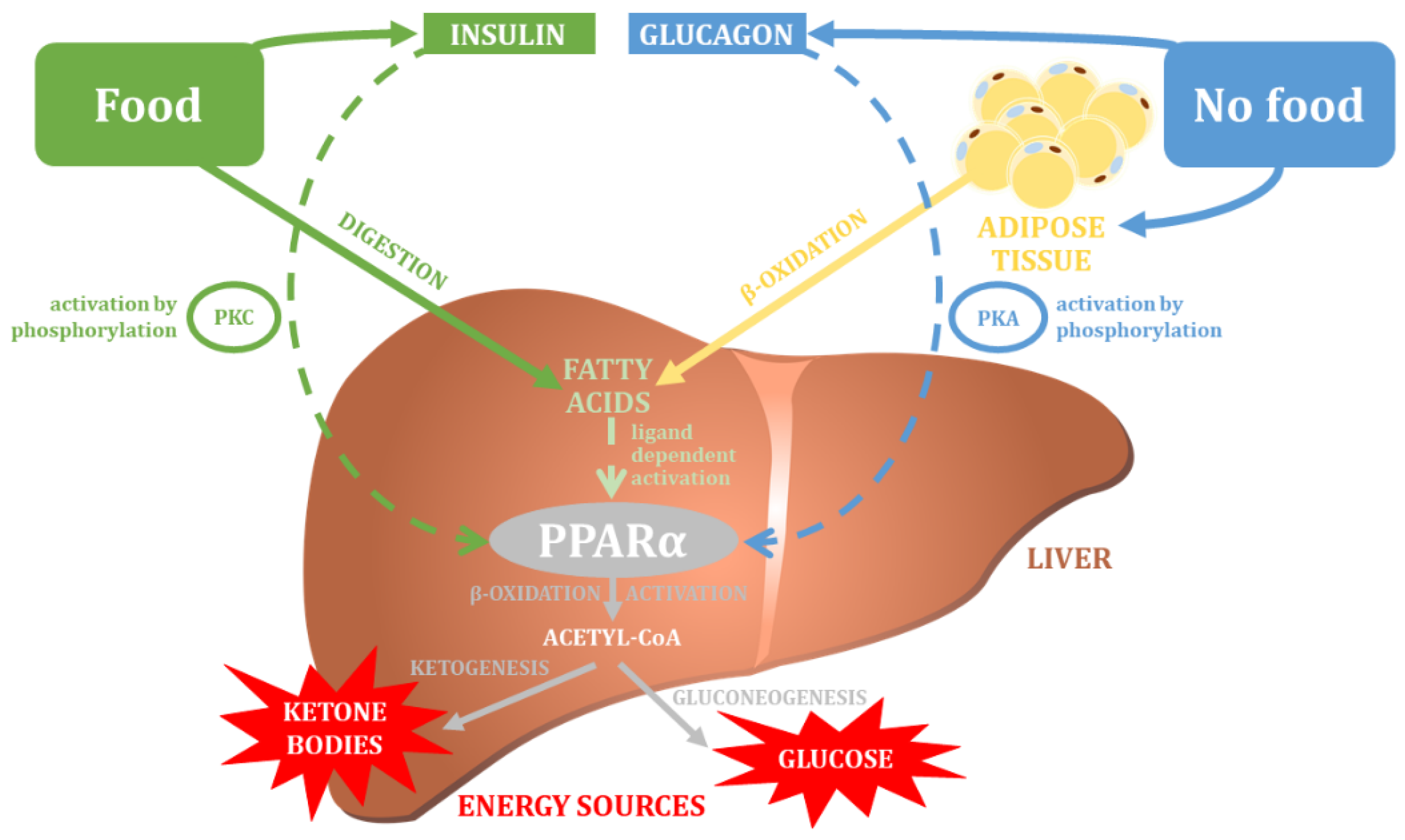
5. The PPARA Gene
6. The PPARGC1A Gene
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Maggio, C.A.; Pi-Sunyer, F.X. Obesity and type 2 diabetes. Endocrinol. Metab. Clin. N. Am. 2003, 32, 805–822. [Google Scholar] [CrossRef] [PubMed]
- Després, J.P.; Lemieux, I. Abdominal obesity and metabolic syndrome. Nature 2006, 444, 881–887. [Google Scholar] [CrossRef] [PubMed]
- Litwin, M.; Kułaga, Z. Obesity, metabolic syndrome, and primary hypertension. Pediatr. Nephrol. 2021, 36, 825–837. [Google Scholar] [CrossRef] [PubMed]
- Avgerinos, K.I.; Spyrou, N.; Mantzoros, C.S.; Dalamaga, M. Obesity and cancer risk: Emerging biological mechanisms and perspectives. Metabolism 2019, 92, 121–135. [Google Scholar] [CrossRef]
- Kopp, W. How Western Diet and Lifestyle Drive the Pandemic of Obesity and Civilization Diseases. Diabetes Metab. Syndr. Obes. 2019, 12, 2221–2236. [Google Scholar] [CrossRef] [Green Version]
- Freire, R. Scientific evidence of diets for weight loss: Different macronutrient composition, intermittent fasting, and popular diets. Nutrition 2020, 69, 110549. [Google Scholar] [CrossRef]
- Thom, G.; Lean, M. Is There an Optimal Diet for Weight Management and Metabolic Health? Gastroenterology 2017, 152, 1739–1751. [Google Scholar] [CrossRef] [Green Version]
- Reed, D.R.; Bachmanov, A.A.; Beauchamp, G.K.; Tordoff, M.G.; Price, R.A. Heritable variation in food preferences and their contribution to obesity. Behav. Genet. 1997, 27, 373–387. [Google Scholar] [CrossRef]
- Loos, R.J.F.; Bouchard, C. Obesity—Is it a genetic disorder? J. Intern. Med. 2003, 254, 401–425. [Google Scholar] [CrossRef]
- Heianza, Y.; Qi, L. Gene-Diet Interaction and Precision Nutrition in Obesity. Int. J. Mol. Sci. 2017, 18, 787. [Google Scholar] [CrossRef]
- Bron, R.; Furness, J.B. Rhythm of digestion: Keeping time in the gastrointestinal tract. Clin. Exp. Pharmacol. Physiol. 2009, 36, 1041–1048. [Google Scholar] [CrossRef] [PubMed]
- Das, U.N. Obesity: Genes, brain, gut, and environment. Nutrition 2010, 26, 459–473. [Google Scholar] [CrossRef]
- Faust, K.; Croes, D.; van Helden, J. Prediction of metabolic pathways from genome-scale metabolic networks. Biosystems 2011, 105, 109–121. [Google Scholar] [CrossRef] [PubMed]
- Mazzoccoli, G.; Pazienza, V.; Vinciguerra, M. Clock genes and clock-controlled genes in the regulation of metabolic rhythms. Chronobiol. Int. 2012, 29, 227–251. [Google Scholar] [CrossRef] [PubMed]
- Kaji, T.; Nonogaki, K. Role of homeobox genes in the hypothalamic development and energy balance. Front. Biosci. 2013, 18, 740–747. [Google Scholar] [CrossRef] [Green Version]
- Cheng, C.F.; Ku, H.C.; Lin, H. PGC-1α as a Pivotal Factor in Lipid and Metabolic Regulation. Int. J. Mol. Sci. 2018, 19, 3447. [Google Scholar] [CrossRef] [Green Version]
- Vesnina, A.; Prosekov, A.; Kozlova, O.; Atuchin, V. Genes and Eating Preferences, Their Roles in Personalized Nutrition. Genes 2020, 11, 357. [Google Scholar] [CrossRef] [Green Version]
- Mullins, V.A.; Bresette, W.; Johnstone, L.; Hallmark, B.; Chilton, F.H. Genomics in Personalized Nutrition: Can You “Eat for Your Genes”? Nutrients 2020, 12, 3118. [Google Scholar] [CrossRef]
- Qi, L. Gene-diet interaction and weight loss. Curr. Opin. Lipidol. 2014, 25, 27–34. [Google Scholar] [CrossRef] [Green Version]
- Crovesy, L.; Rosado, E.L. Interaction between genes involved in energy intake regulation and diet in obesity. Nutrition 2019, 67–68, 110547. [Google Scholar] [CrossRef]
- Rankinen, T.; Bouchard, C. Gene-physical activity interactions: Overview of human studies. Obesity 2008, 16 (Suppl. S3), S47–S50. [Google Scholar] [CrossRef] [PubMed]
- Adamo, K.B.; Tesson, F. Genotype-specific weight loss treatment advice: How close are we? Appl. Physiol. Nutr. Metab. 2007, 32, 351–366. [Google Scholar] [CrossRef] [PubMed]
- Martínez, J.A.; Parra, M.D.; Santos, J.L.; Moreno-Aliaga, M.J.; Marti, A.; Martínez-González, M.A. Genotype-dependent response to energy-restricted diets in obese subjects: Towards personalized nutrition. Asia Pac. J. Clin. Nutr. 2008, 17, 119–122. [Google Scholar] [PubMed]
- Moreno-Aliaga, M.J.; Santos, J.L.; Marti, A.; Martínez, J.A. Does weight loss prognosis depend on genetic make-up? Obes. Rev. 2005, 6, 155–168. [Google Scholar] [CrossRef] [PubMed]
- Joffe, Y.T.; Houghton, C.A. A Novel Approach to the Nutrigenetics and Nutrigenomics of Obesity and Weight Management. Curr. Oncol. Rep. 2016, 18, 43. [Google Scholar] [CrossRef]
- Bayer, S.; Winkler, V.; Hauner, H.; Holzapfel, C. Associations between Genotype-Diet Interactions and Weight Loss-A Systematic Review. Nutrients 2020, 12, 2891. [Google Scholar] [CrossRef]
- Heid, I.M.; Jackson, A.U.; Randall, J.C.; Winkler, T.W.; Qi, L.; Ssteinthorsdottir, V.; Tthorleifsson, G.; Zillikens, C.; Sspeliotes, E.K.; Mägi, R.; et al. Meta-analysis identifies 13 new loci associated with waist-hip ratio and reveals sexual dimorphism in the genetic basis of fat distribution. Nat. Genet. 2010, 42, 949–960. [Google Scholar] [CrossRef] [Green Version]
- Hebebrand, J.; Hinney, A.; Knoll, N.; Volckmar, A.L.; Scherag, A. Molecular genetic aspects of weight regulation. Dtsch. Arztebl. Int. 2013, 110, 338–344. [Google Scholar] [CrossRef] [Green Version]
- Zhang, X.; Qi, Q.; Zhang, C.; Hu, F.B.; Sacks, F.M.; Qi, L. FTO genotype and 2-year change in body composition and fat distribution in response to weight-loss diets: The POUNDS LOST Trial. Diabetes 2012, 61, 3005–3011. [Google Scholar] [CrossRef] [Green Version]
- Pan, Q.; Delahanty, L.M.; Jablonski, K.A.; Knowler, W.C.; Kahn, S.E.; Florez, J.C.; Franks, P.W. Variation at the melanocortin 4 receptor gene and response to weight-loss interventions in the diabetes prevention program. Obesity 2013, 21, E520–E526. [Google Scholar] [CrossRef]
- Garaulet, M.; Smith, C.E.; Hernández-González, T.; Lee, Y.C.; Ordovás, J.M. PPARγ Pro12Ala interacts with fat intake for obesity and weight loss in a behavioural treatment based on the Mediterranean diet. Mol. Nutr. Food Res. 2011, 55, 1771–1779. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bray, M.S.; Hagberg, J.M.; Pérusse, L.; Rankinen, T.; Roth, S.M.; Wolfarth, B.; Bouchard, C. The human gene map for performance and health-related fitness phenotypes: The 2006–2007 update. Med. Sci. Sports Exerc. 2009, 41, 34–72. [Google Scholar] [CrossRef] [PubMed]
- Viecelli, C.; Ewald, C.Y. The non-modifiable factors age, gender, and genetics influence resistance exercise. Front. Aging 2022, 3, 1005848. [Google Scholar] [CrossRef] [PubMed]
- Venckunas, T.; Degens, H. Genetic polymorphisms of muscular fitness in young healthy men. PLoS ONE 2022, 17, e0275179. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Emmerich, A.; Pillon, N.J.; Moore, T.; Hemerich, D.; Cornelis, M.C.; Mazzaferro, E.; Broos, S.; Ahluwalia, T.S.; Bartz, T.M.; et al. Genome-wide association analyses of physical activity and sedentary behavior provide insights into underlying mechanisms and roles in disease prevention. Nat. Genet. 2022, 54, 1332–1344. [Google Scholar] [CrossRef]
- Liberati, A.; Altman, D.G.; Tetzlaff, J.; Mulrow, C.; Gøtzsche, P.C.; Ioannidis, J.P.A.; Clarke, M.; Devereaux, P.J.; Kleijnen, J.; Moher, D. The PRISMA statement for reporting systematic reviews and meta-analyses of studies that evaluate health care interventions: Explanation and elaboration. J. Clin. Epidemiol. 2009, 62, e1–e34. [Google Scholar] [CrossRef] [Green Version]
- Maltais, L.J.; Blake, J.A.; Chu, T.; Lutz, C.M.; Eppig, J.T.; Jackson, I. Rules and guidelines for mouse gene, allele, and mutation nomenclature: A condensed version. Genomics 2002, 79, 471–474. [Google Scholar] [CrossRef]
- Bruford, E.A.; Braschi, B.; Denny, P.; Jones, T.E.M.; Seal, R.L.; Tweedie, S. Guidelines for human gene nomenclature. Nat. Genet. 2020, 52, 754–758. [Google Scholar] [CrossRef]
- Smith, R.L.; Soeters, M.R.; Wüst, R.C.I.; Houtkooper, R.H. Metabolic Flexibility as an Adaptation to Energy Resources and Requirements in Health and Disease. Endocr. Rev. 2018, 39, 489–517. [Google Scholar] [CrossRef] [Green Version]
- Berger, J.; Moller, D.E. The Mechanisms of Action of PPARs. Annu. Rev. Med. 2003, 53, 409–435. [Google Scholar] [CrossRef]
- Rogue, A.; Spire, C.; Brun, M.; Claude, N.; Guillouzo, A. Gene Expression Changes Induced by PPAR Gamma Agonists in Animal and Human Liver. PPAR Res. 2010, 2010, 325183. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Desvergne, B.; Wahli, W. Peroxisome proliferator-activated receptors: Nuclear control of metabolism. Endocr. Rev. 1999, 20, 649–688. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hong, F.; Pan, S.; Guo, Y.; Xu, P.; Zhai, Y. PPARs as Nuclear Receptors for Nutrient and Energy Metabolism. Molecules 2019, 24, 2545. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Woller, A.; Duez, H.; Staels, B.; Lefranc, M. A Mathematical Model of the Liver Circadian Clock Linking Feeding and Fasting Cycles to Clock Function. Cell Rep. 2016, 17, 1087–1097. [Google Scholar] [CrossRef] [Green Version]
- Evans, R.M.; Barish, G.D.; Wang, Y.X. PPARs and the complex journey to obesity. Nat. Med. 2004, 10, 355–361. [Google Scholar] [CrossRef]
- Brunmeir, R.; Xu, F. Functional Regulation of PPARs through Post-Translational Modifications. Int. J. Mol. Sci. 2018, 19, 1738. [Google Scholar] [CrossRef] [Green Version]
- Lehrke, M.; Lazar, M.A. The Many Faces of PPARγ. Cell 2005, 123, 993–999. [Google Scholar] [CrossRef] [Green Version]
- Grygiel-Górniak, B. Peroxisome proliferator-activated receptors and their ligands: Nutritional and clinical implications—A review. Nutr. J. 2014, 13, 17. [Google Scholar] [CrossRef] [Green Version]
- Marion-Letellier, R.; Déchelotte, P.; Lacucci, M.; Ghosh, S. Dietary modulation of peroxisome proliferator-activated receptor gamma. Gut 2009, 58, 586–593. [Google Scholar] [CrossRef]
- Marion-Letellier, R.; Savoye, G.; Ghosh, S. Fatty acids, eicosanoids and PPAR gamma. Eur. J. Pharmacol. 2016, 785, 44–49. [Google Scholar] [CrossRef]
- Tontonoz, P.; Spiegelman, B.M. Fat and beyond: The diverse biology of PPARgamma. Annu. Rev. Biochem. 2008, 77, 289–312. [Google Scholar] [CrossRef] [PubMed]
- Randle, P. Regulatory interactions between lipids and carbohydrates: The glucose fatty acid cycle after 35 years. Diabetes Metab. Rev. 1998, 14, 263–283. [Google Scholar] [CrossRef]
- Maciejewska-Karlowska, A.; Sawczuk, M.; Cieszczyk, P.; Zarebska, A.; Sawczyn, S. Association between the Pro12Ala polymorphism of the peroxisome proliferator-activated receptor gamma gene and strength athlete status. PLoS ONE 2013, 8, e67172. [Google Scholar] [CrossRef]
- Greene, M.E.; Blumberg, B.; McBride, O.W.; Yi, H.F.; Kronquist, K.; Kwan, K.; Hsieh, L.; Greene, G.; Nimer, S.D. Isolation of the human peroxisome proliferator activated receptor gamma cDNA: Expression in hematopoietic cells and chromosomal mapping. Gene Expr. 1995, 4, 281–299. [Google Scholar] [PubMed]
- Zhu, Y.; Qi, C.; Korenberg, J.R.; Chen, X.N.; Noya, D.; Sambasiva Rao, M.; Reddy, J.K. Structural organization of mouse peroxisome proliferator-activated receptor gamma (mPPAR gamma) gene: Alternative promoter use and different splicing yield two mPPAR gamma isoforms. Proc. Natl. Acad. Sci. USA 1995, 92, 7921–7925. [Google Scholar] [CrossRef] [Green Version]
- Omi, T.; Brenig, B.; Špilar Kramer, Š.; Iwamoto, S.; Stranzinger, G.; Neuenschwander, S. Identification and characterization of novel peroxisome proliferator-activated receptor-gamma (PPAR-gamma) transcriptional variants in pig and human. J. Anim. Breed. Genet. 2005, 122 (Suppl. S1), S45–S53. [Google Scholar] [CrossRef]
- Zhou, J.; Wilson, K.M.; Medh, J.D. Genetic analysis of four novel peroxisome proliferator activated receptor-gamma splice variants in monkey macrophages. Biochem. Biophys. Res. Commun. 2002, 293, 274–283. [Google Scholar] [CrossRef] [Green Version]
- Adamo, K.B.; Dent, R.; Langefeld, C.D.; Cox, M.; Williams, K.; Carrick, K.M.; Stuart, J.S.; Sundseth, S.S.; Harper, M.E.; McPherson, R.; et al. Peroxisome proliferator-activated receptor gamma 2 and acyl-CoA synthetase 5 polymorphisms influence diet response. Obesity 2007, 15, 1068–1075. [Google Scholar] [CrossRef]
- Almeida, S.M.; Furtado, J.M.; Mascarenhas, P.; Ferraz, M.E.; Ferreira, J.C.; Monteiro, M.P.; Vilanova, M.; Ferraz, F.P. Association between LEPR, FTO, MC4R, and PPARG-2 polymorphisms with obesity traits and metabolic phenotypes in school-aged children. Endocrine 2018, 60, 466–478. [Google Scholar] [CrossRef] [Green Version]
- Cecil, J.E.; Watt, P.; Palmer, C.N.; Hetherington, M. Energy balance and food intake: The role of PPARgamma gene polymorphisms. Physiol. Behav. 2006, 88, 227–233. [Google Scholar] [CrossRef]
- Matsuo, T.; Nakata, Y.; Katayama, Y.; Iemitsu, M.; Maeda, S.; Okura, T.; Kim, M.K.; Ohkubo, H.; Hotta, K.; Tanaka, K. PPARG genotype accounts for part of individual variation in body weight reduction in response to calorie restriction. Obesity 2009, 17, 1924–1931. [Google Scholar] [CrossRef] [PubMed]
- Lagou, V.; Scott, R.A.; Manios, Y.; Chen, T.L.J.; Wang, G.; Grammatikaki, E.; Kortsalioudaki, C.; Liarigkovinos, T.; Moschonis, G.; Roma-Giannikou, E.; et al. Impact of peroxisome proliferator-activated receptors gamma and delta on adiposity in toddlers and preschoolers in the GENESIS Study. Obesity 2008, 16, 913–918. [Google Scholar] [CrossRef] [PubMed]
- Valeeva, F.V.; Medvedeva, M.S.; Khasanova, K.B.; Valeeva, E.V.; Kiseleva, T.A.; Egorova, E.S.; Pickering, C.; Ahmetov, I.I. Association of gene polymorphisms with body weight changes in prediabetic patients. Mol. Biol. Rep. 2022, 49, 4217–4224. [Google Scholar] [CrossRef] [PubMed]
- Abaj, F.; Rafiee, M.; Koohdani, F. A personalised diet approach study: Interaction between PPAR-γ Pro12Ala and dietary insulin indices on metabolic markers in diabetic patients. J. Hum. Nutr. Diet. 2022, 35, 663–674. [Google Scholar] [CrossRef] [PubMed]
- Liao, W.; Nguyen, M.T.A.; Yoshizaki, T.; Favelyukis, S.; Patsouris, D.; Imamura, T.; Verma, I.M.; Olefsky, J.M. Suppression of PPAR-gamma attenuates insulin-stimulated glucose uptake by affecting both GLUT1 and GLUT4 in 3T3-L1 adipocytes. Am. J. Physiol. Endocrinol. Metab. 2007, 293, E219–E227. [Google Scholar] [CrossRef] [Green Version]
- Yap, K.H.; Yee, G.S.; Candasamy, M.; Tan, S.C.; Md, S.; Majeed, A.B.A.; Bhattamisra, S.K. Catalpol Ameliorates Insulin Sensitivity and Mitochondrial Respiration in Skeletal Muscle of Type-2 Diabetic Mice Through Insulin Signaling Pathway and AMPK/SIRT1/PGC-1α/PPAR-γ Activation. Biomolecules 2020, 10, 1360. [Google Scholar] [CrossRef] [PubMed]
- Kintscher, U.; Law, R.E. PPARgamma-mediated insulin sensitization: The importance of fat versus muscle. Am. J. Physiol. Endocrinol. Metab. 2005, 288, E287–E291. [Google Scholar] [CrossRef] [Green Version]
- Verma, N.K.; Singh, J.; Dey, C.S. PPAR-gamma expression modulates insulin sensitivity in C2C12 skeletal muscle cells. Br. J. Pharmacol. 2004, 143, 1006–1013. [Google Scholar] [CrossRef] [Green Version]
- Saraf, N.; Sharma, P.K.; Mondal, S.C.; Garg, V.K.; Singh, A.K. Role of PPARg2 transcription factor in thiazolidinedione-induced insulin sensitization. J. Pharm. Pharmacol. 2012, 64, 161–171. [Google Scholar] [CrossRef]
- Hevener, A.L.; Olefsky, J.M.; Reichart, D.; Nguyen, M.T.A.; Bandyopadyhay, G.; Leung, H.Y.; Watt, M.J.; Benner, C.; Febbraio, M.A.; Nguyen, A.K.; et al. Macrophage PPAR gamma is required for normal skeletal muscle and hepatic insulin sensitivity and full antidiabetic effects of thiazolidinediones. J. Clin. Investig. 2007, 117, 1658–1669. [Google Scholar] [CrossRef]
- Ortmeyer, H.K.; Sajan, M.P.; Miura, A.; Kanoh, Y.; Rivas, J.; Li, Y.; Standaert, M.L.; Ryan, A.S.; Bodkin, N.L.; Farese, R.V.; et al. Insulin signaling and insulin sensitizing in muscle and liver of obese monkeys: Peroxisome proliferator-activated receptor gamma agonist improves defective activation of atypical protein kinase C. Antioxid. Redox Signal. 2011, 14, 207–219. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yamauchi, T.; Kamon, J.; Waki, H.; Murakami, K.; Motojima, K.; Komeda, K.; Ide, T.; Kubota, N.; Terauchi, Y.; Tobe, K.; et al. The mechanisms by which both heterozygous peroxisome proliferator-activated receptor gamma (PPARgamma) deficiency and PPARgamma agonist improve insulin resistance. J. Biol. Chem. 2001, 276, 41245–41254. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stumvoll, M.; Häring, H. The peroxisome proliferator-activated receptor-gamma2 Pro12Ala polymorphism. Diabetes 2002, 51, 2341–2347. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Broekema, M.F.; Savage, D.B.; Monajemi, H.; Kalkhoven, E. Gene-gene and gene-environment interactions in lipodystrophy: Lessons learned from natural PPARγ mutants. Biochim. Biophys. Acta. Mol. Cell Biol. Lipids 2019, 1864, 715–732. [Google Scholar] [CrossRef]
- Hollenberg, A.N.; Susulic, V.S.; Madura, J.P.; Zhang, B.; Moller, D.E.; Tontonoz, P.; Sarraf, P.; Spiegelman, B.M.; Lowell, B.B. Functional antagonism between CCAAT/Enhancer binding protein-alpha and peroxisome proliferator-activated receptor-gamma on the leptin promoter. J. Biol. Chem. 1997, 272, 5283–5290. [Google Scholar] [CrossRef] [Green Version]
- Sarhangi, N.; Sharifi, F.; Hashemian, L.; Hassani Doabsari, M.; Heshmatzad, K.; Rahbaran, M.; Jamaldini, S.H.; Aghaei Meybodi, H.R.; Hasanzad, M. PPARG (Pro12Ala) genetic variant and risk of T2DM: A systematic review and meta-analysis. Sci. Rep. 2020, 10, 12764. [Google Scholar] [CrossRef]
- Semple, R.K.; Chatterjee, V.K.K.; O’Rahilly, S. PPAR gamma and human metabolic disease. J. Clin. Investig. 2006, 116, 581–589. [Google Scholar] [CrossRef] [Green Version]
- Skat-Rørdam, J.; Højland Ipsen, D.; Lykkesfeldt, J.; Tveden-Nyborg, P. A role of peroxisome proliferator-activated receptor γ in non-alcoholic fatty liver disease. Basic Clin. Pharmacol. Toxicol. 2019, 124, 528–537. [Google Scholar] [CrossRef]
- Yen, C.-J.; Beamer, B.A.; Negri, C.; Silver, K.; Brown, K.A.; Yarnall, D.P.; Burns, D.K.; Roth, J.; Shuldiner, A.R. Molecular scanning of the human peroxisome proliferator activated receptor gamma (hPPAR gamma) gene in diabetic Caucasians: Identification of a Pro12Ala PPAR gamma 2 missense mutation. Biochem. Biophys. Res. Commun. 1997, 241, 270–274. [Google Scholar] [CrossRef]
- Deeb, S.S.; Fajas, L.; Nemoto, M.; Pihlajamäki, J.; Mykkänen, L.; Kuusisto, J.; Laakso, M.; Fujimoto, W.; Auwerx, J. A Pro12Ala substitution in PPARgamma2 associated with decreased receptor activity, lower body mass index and improved insulin sensitivity. Nat. Genet. 1998, 20, 284–287. [Google Scholar] [CrossRef]
- Masugi, J.; Tamori, Y.; Mori, H.; Koike, T.; Kasuga, M. Inhibitory effect of a proline-to-alanine substitution at codon 12 of peroxisome proliferator-activated receptor-gamma 2 on thiazolidinedione-induced adipogenesis. Biochem. Biophys. Res. Commun. 2000, 268, 178–182. [Google Scholar] [CrossRef] [PubMed]
- Werman, A.; Hollenberg, A.; Solanes, G.; Bjørbæk, C.; Vidal-Puig, A.J.; Flier, J.S. Ligand-independent activation domain in the N terminus of peroxisome proliferator-activated receptor gamma (PPARgamma). Differential activity of PPARgamma1 and -2 isoforms and influence of insulin. J. Biol. Chem. 1997, 272, 20230–20235. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Adams, M.; Reginato, M.J.; Shao, D.; Lazar, M.A.; Chatterjee, V.K. Transcriptional activation by peroxisome proliferator-activated receptor gamma is inhibited by phosphorylation at a consensus mitogen-activated protein kinase site. J. Biol. Chem. 1997, 272, 5128–5132. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Geiss-Friedlander, R.; Melchior, F. Concepts in sumoylation: A decade on. Nat. Rev. Mol. Cell Biol. 2007, 8, 947–956. [Google Scholar] [CrossRef]
- Shao, D.; Rangwala, S.M.; Bailey, S.T.; Krakow, S.L.; Reginato, M.J.; Lazar, M.A. Interdomain communication regulating ligand binding by PPAR-gamma. Nature 1998, 396, 377–380. [Google Scholar] [CrossRef]
- Ek, J.; Urhammer, S.A.; Sørensen, T.I.A.; Andersen, T.; Auwerx, J.; Pedersen, O. Homozygosity of the Pro12Ala variant of the peroxisome proliferation-activated receptor-gamma2 (PPAR-gamma2): Divergent modulating effects on body mass index in obese and lean Caucasian men. Diabetologia 1999, 42, 892–895. [Google Scholar] [CrossRef] [Green Version]
- da Silva, R.S.B.; Persuhn, D.C.; Barbosa, F.K.L.; de Souza, M.F.; Sena, K. de F.; Costa, M. da S.; Franca, G.A.M.; de Assis, C.S.; Cardoso, G.A.; Silva, A.S. Relationship of the Pro12Ala Polymorphism on the PPARy2 Gene with the Body Composition of Practitioners of Cyclic Exercises. Front. Physiol. 2021, 11, 633721. [Google Scholar] [CrossRef]
- Danawati, C.W.; Nagata, M.; Moriyama, H.; Hara, K.; Yasuda, H.; Nakayama, M.; Kotani, R.; Yamada, K.; Sakata, M.; Kurohara, M.; et al. A possible association of Pro12Ala polymorphism in peroxisome proliferator-activated receptor gamma2 gene with obesity in native Javanese in Indonesia. Diabetes. Metab. Res. Rev. 2005, 21, 465–469. [Google Scholar] [CrossRef]
- Bhatt, S.P.; Misra, A.; Sharma, M.; Luthra, K.; Guleria, R.; Pandey, R.M.; Vikram, N.K. Ala/Ala genotype of Pro12Ala polymorphism in the peroxisome proliferator-activated receptor-γ2 gene is associated with obesity and insulin resistance in Asian Indians. Diabetes Technol. Ther. 2012, 14, 828–834. [Google Scholar] [CrossRef] [Green Version]
- Yao, Y.S.; Li, J.; Jin, Y.L.; Chen, Y.; He, L.P. Association between PPAR-γ2 Pro12Ala polymorphism and obesity: A meta-analysis. Mol. Biol. Rep. 2015, 42, 1029–1038. [Google Scholar] [CrossRef]
- Galbete, C.; Toledo, E.; Martínez-González, M.A.; Martínez, J.A.; Guillén-Grima, F.; Marti, A. Pro12Ala variant of the PPARG2 gene increases body mass index: An updated meta-analysis encompassing 49,092 subjects. Obesity 2013, 21, 1486–1495. [Google Scholar] [CrossRef]
- Lapice, E.; Vaccaro, O. Interaction between Pro12Ala polymorphism of PPARγ2 and diet on adiposity phenotypes. Curr. Atheroscler. Rep. 2014, 16, 462. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mansoori, A.; Amini, M.; Kolahdooz, F.; Seyedrezazadeh, E. Obesity and Pro12Ala Polymorphism of Peroxisome Proliferator-Activated Receptor-Gamma Gene in Healthy Adults: A Systematic Review and Meta-Analysis. Ann. Nutr. Metab. 2015, 67, 104–118. [Google Scholar] [CrossRef] [PubMed]
- Beamer, B.A.; Yen, C.J.; Andersen, R.E.; Muller, D.; Elahi, D.; Cheskin, L.J.; Andres, R.; Roth, J.; Shuldiner, A.R. Association of the Pro12Ala variant in the peroxisome proliferator-activated receptor-gamma2 gene with obesity in two Caucasian populations. Diabetes 1998, 47, 1806–1808. [Google Scholar] [CrossRef] [PubMed]
- Masud, S.; Ye, S. Effect of the peroxisome proliferator activated receptor-gamma gene Pro12Ala variant on body mass index: A meta-analysis. J. Med. Genet. 2003, 40, 773–780. [Google Scholar] [CrossRef] [Green Version]
- Luan, J.; Browne, P.O.; Harding, A.H.; Halsall, D.J.; O’Rahilly, S.; Chatterjee, V.K.K.; Wareham, N.J. Evidence for gene-nutrient interaction at the PPARgamma locus. Diabetes 2001, 50, 686–689. [Google Scholar] [CrossRef] [Green Version]
- Franks, P.W.; Luan, J.; Browne, P.O.; Harding, A.H.; O’Rahilly, S.; Chatterjee, V.K.K.; Wareham, N.J. Does peroxisome proliferator-activated receptor γ genotype (Pro12ala) modify the association of physical activity and dietary fat with fasting insulin level? Metabolism 2004, 53, 11–16. [Google Scholar] [CrossRef]
- Zarebska, A.; Jastrzebski, Z.; Cieszczyk, P.; Leonska-Duniec, A.; Kotarska, K.; Kaczmarczyk, M.; Sawczuk, M.; Maciejewska-Karlowska, A. The Pro12Ala polymorphism of the peroxisome proliferator-activated receptor gamma gene modifies the association of physical activity and body mass changes in Polish women. PPAR Res. 2014, 2014, 373782. [Google Scholar] [CrossRef] [Green Version]
- Ek, J.; Andersen, G.; Urhammer, S.A.; Hansen, L.; Carstensen, B.; Borch-Johnsen, K.; Drivsholm, T.; Berglund, L.; Hansen, T.; Lithell, H.; et al. Studies of the Pro12Ala polymorphism of the peroxisome proliferator-activated receptor-gamma2 (PPAR-gamma2) gene in relation to insulin sensitivity among glucose tolerant caucasians. Diabetologia 2001, 44, 1170–1176. [Google Scholar] [CrossRef] [Green Version]
- Frederiksen, L.; Brødbæk, K.; Fenger, M.; Jørgensen, T.; Borch-Johnsen, K.; Madsbad, S.; Urhammer, S.A. Comment: Studies of the Pro12Ala polymorphism of the PPAR-gamma gene in the Danish MONICA cohort: Homozygosity of the Ala allele confers a decreased risk of the insulin resistance syndrome. J. Clin. Endocrinol. Metab. 2002, 87, 3989–3992. [Google Scholar] [CrossRef]
- Vidal-Puig, A.J.; Considine, R.V.; Jimenez-Liñan, M.; Werman, A.; Pories, W.J.; Caro, J.F.; Flier, J.S. Peroxisome proliferator-activated receptor gene expression in human tissues. Effects of obesity, weight loss, and regulation by insulin and glucocorticoids. J. Clin. Investig. 1997, 99, 2416–2422. [Google Scholar] [CrossRef]
- Altshuler, D.; Hirschhorn, J.N.; Klannemark, M.; Lindgren, C.M.; Vohl, M.C.; Nemesh, J.; Lane, C.R.; Schaffner, S.F.; Bolk, S.; Brewer, C.; et al. The common PPARgamma Pro12Ala polymorphism is associated with decreased risk of type 2 diabetes. Nat. Genet. 2000, 26, 76–80. [Google Scholar] [CrossRef] [PubMed]
- Vidal-Puig, A.; Jimenez-Liñan, M.; Lowell, B.B.; Hamann, A.; Hu, E.; Spiegelman, B.; Flier, J.S.; Moller, D.E. Regulation of PPAR gamma gene expression by nutrition and obesity in rodents. J. Clin. Investig. 1996, 97, 2553–2561. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Polimanti, R.; Yang, B.Z.; Zhao, H.; Gelernter, J. Evidence of Polygenic Adaptation in the Systems Genetics of Anthropometric Traits. PLoS ONE 2016, 11, e0160654. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barak, Y.; Liao, D.; He, W.; Ong, E.S.; Nelson, M.C.; Olefsky, J.M.; Boland, R.; Evans, R.M. Effects of peroxisome proliferator-activated receptor delta on placentation, adiposity, and colorectal cancer. Proc. Natl. Acad. Sci. USA 2002, 99, 303–308. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peters, J.M.; Lee, S.S.T.; Li, W.; Ward, J.M.; Gavrilova, O.; Everett, C.; Reitman, M.L.; Hudson, L.D.; Gonzalez, F.J. Growth, adipose, brain, and skin alterations resulting from targeted disruption of the mouse peroxisome proliferator-activated receptor beta(delta). Mol. Cell. Biol. 2000, 20, 5119–5128. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schmidt, A.; Endo, N.; Rutledge, S.J.; Vogel, R.; Shinar, D.; Rodan, G.A. Identification of a new member of the steroid hormone receptor superfamily that is activated by a peroxisome proliferator and fatty acids. Mol. Endocrinol. 1992, 6, 1634–1641. [Google Scholar] [CrossRef] [Green Version]
- Yoshikawa, T.; Brkanac, Z.; Dupont, B.R.; Xing, G.Q.; Leach, R.J.; Detera-Wadleigh, S.D. Assignment of the human nuclear hormone receptor, NUC1 (PPARD), to chromosome 6p21.1-p21.2. Genomics 1996, 35, 637–638. [Google Scholar] [CrossRef] [PubMed]
- Lundell, K.; Thulin, P.; Hamsten, A.; Ehrenborg, E. Alternative splicing of human peroxisome proliferator-activated receptor delta (PPAR delta): Effects on translation efficiency and trans-activation ability. BMC Mol. Biol. 2007, 8, 70. [Google Scholar] [CrossRef] [Green Version]
- Skogsberg, J.; Kannisto, K.; Roshani, L.; Gagne, E.; Hamsten, A.; Larsson, C.; Ehrenborg, E. Characterization of the human peroxisome proliferator activated receptor delta gene and its expression. Int. J. Mol. Med. 2000, 6, 73–81. [Google Scholar] [CrossRef]
- Holst, D.; Luquet, S.; Nogueira, V.; Kristiansen, K.; Leverve, X.; Grimaldi, P.A. Nutritional regulation and role of peroxisome proliferator-activated receptor δ in fatty acid catabolism in skeletal muscle. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2003, 1633, 43–50. [Google Scholar] [CrossRef] [PubMed]
- Oliver, W.R.; Shenk, J.L.; Snaith, M.R.; Russell, C.S.; Plunket, K.D.; Bodkin, N.L.; Lewis, M.C.; Winegar, D.A.; Sznaidman, M.L.; Lambert, M.H.; et al. A selective peroxisome proliferator-activated receptor delta agonist promotes reverse cholesterol transport. Proc. Natl. Acad. Sci. USA 2001, 98, 5306–5311. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Y.X.; Lee, C.H.; Tiep, S.; Yu, R.T.; Ham, J.; Kang, H.; Evans, R.M. Peroxisome-proliferator-activated receptor δ activates fat metabolism to prevent obesity. Cell 2003, 113, 159–170. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Luquet, S.; Lopez-Soriano, J.; Holst, D.; Fredenrich, A.; Melki, J.; Rassoulzadegan, M.; Grimaldi, P.A. Peroxisome proliferator-activated receptor delta controls muscle development and oxidative capability. FASEB J. 2003, 17, 2299–2301. [Google Scholar] [CrossRef] [PubMed]
- Gupta, R.A.; Wang, D.; Katkuri, S.; Wang, H.; Dey, S.K.; DuBois, R.N. Activation of nuclear hormone receptor peroxisome proliferator-activated receptor-delta accelerates intestinal adenoma growth. Nat. Med. 2004, 10, 245–247. [Google Scholar] [CrossRef] [PubMed]
- Karpe, F.; Ehrenborg, E.E. PPARdelta in humans: Genetic and pharmacological evidence for a significant metabolic function. Curr. Opin. Lipidol. 2009, 20, 333–336. [Google Scholar] [CrossRef] [PubMed]
- Song, J.; Li, N.; Hu, R.; Yu, Y.; Xu, K.; Ling, H.; Lu, Q.; Yang, T.; Wang, T.; Yin, X. Effects of PPARD gene variants on the therapeutic responses to exenatide in chinese patients with type 2 diabetes mellitus. Front. Endocrinol. 2022, 13, 949990. [Google Scholar] [CrossRef]
- Maciejewska-Skrendo, A.; Pawlik, A.; Sawczuk, M.; Rać, M.; Kusak, A.; Safranow, K.; Dziedziejko, V. PPARA, PPARD and PPARG gene polymorphisms in patients with unstable angina. Gene 2019, 711, 143947. [Google Scholar] [CrossRef]
- Andrulionyte, L.; Peltola, P.; Chiasson, J.L.; Laakso, M. Single Nucleotide Polymorphisms of PPARD in Combination with the Gly482Ser Substitution of PGC-1A and the Pro12Ala Substitution of PPARG2 Predict the Conversion from Impaired Glucose Tolerance to Type 2 Diabetes: The STOP-NIDDM Trial. Diabetes 2006, 55, 2148–2152. [Google Scholar] [CrossRef] [Green Version]
- Ticha, I.; Gnosa, S.; Lindblom, A.; Liu, T.; Sun, X.F. Variants of the PPARD gene and their clinicopathological significance in colorectal cancer. PLoS ONE 2013, 8, e83952. [Google Scholar] [CrossRef]
- Uemura, H.; Hiyoshi, M.; Arisawa, K.; Yamaguchi, M.; Naito, M.; Kawai, S.; Hamajima, N.; Matsuo, K.; Taguchi, N.; Takashima, N.; et al. Gene variants in PPARD and PPARGC1A are associated with timing of natural menopause in the general Japanese population. Maturitas 2012, 71, 369–375. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.; Kim, M.; Yoo, H.J.; Shon, J.; Lee, J.H. Associations between hypertension and the peroxisome proliferator-activated receptor-δ (PPARD) gene rs7770619 C>T polymorphism in a Korean population. Hum. Genom. 2018, 12, 28. [Google Scholar] [CrossRef] [PubMed]
- Maculewicz, E.; Mastalerz, A.; Maciejewska-Skrendo, A.; Cieszczyk, P.; Cywinska, A.; Borecka, A.; Garbacz, A.; Szarska, E.; Dziuda, L.; Lorenz, K.; et al. Association between peroxisome proliferator-activated receptor-alpha, -delta and -gamma gene (PPARA, PPARD, PPARG) polymorphisms and overweight parameters in physically active men. Biol. Sport 2021, 38, 767–776. [Google Scholar] [CrossRef]
- Ehrenborg, E.; Skogsberg, J. Peroxisome proliferator-activated receptor delta and cardiovascular disease. Atherosclerosis 2013, 231, 95–106. [Google Scholar] [CrossRef]
- Zhang, Y.; Gao, T.; Hu, S.; Lin, B.; Yan, D.; Xu, Z.; Zhang, Z.; Mao, Y.; Mao, H.; Wang, L.; et al. The Functional SNPs in the 5′ Regulatory Region of the Porcine PPARD Gene Have Significant Association with Fat Deposition Traits. PLoS ONE 2015, 10, e0143734. [Google Scholar] [CrossRef] [Green Version]
- Kim, M.; Kim, M.; Yoo, H.J.; Sun, Y.; Lee, S.H.; Lee, J.H. PPARD rs7770619 polymorphism in a Korean population: Association with plasma malondialdehyde and impaired fasting glucose or newly diagnosed type 2 diabetes. Diabetes Vasc. Dis. Res. 2018, 15, 360–363. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maciejewska-Karlowska, A.; Hanson, E.D.; Sawczuk, M.; Cieszczyk, P.; Eynon, N. Genomic haplotype within the Peroxisome Proliferator-Activated Receptor Delta (PPARD) gene is associated with elite athletic status. Scand. J. Med. Sci. Sports 2014, 24, e148–e155. [Google Scholar] [CrossRef]
- Skogsberg, J.; Kannisto, K.; Cassel, T.N.; Hamsten, A.; Eriksson, P.; Ehrenborg, E. Evidence that peroxisome proliferator-activated receptor delta influences cholesterol metabolism in men. Arterioscler. Thromb. Vasc. Biol. 2003, 23, 637–643. [Google Scholar] [CrossRef] [Green Version]
- Skogsberg, J.; McMahon, A.D.; Karpe, F.; Hamsten, A.; Packard, C.J.; Ehrenborg, E. Peroxisome proliferator activated receptor delta genotype in relation to cardiovascular risk factors and risk of coronary heart disease in hypercholesterolaemic men. J. Intern. Med. 2003, 254, 597–604. [Google Scholar] [CrossRef]
- Chen, S.; Tsybouleva, N.; Ballantyne, C.M.; Gotto, A.M.; Marian, A.J. Effects of PPARalpha, gamma and delta haplotypes on plasma levels of lipids, severity and progression of coronary atherosclerosis and response to statin therapy in the lipoprotein coronary atherosclerosis study. Pharmacogenetics 2004, 14, 61–71. [Google Scholar] [CrossRef]
- Aberle, J.; Hopfer, I.; Beil, F.U.; Seedorf, U. Association of the T+294C polymorphism in PPAR delta with low HDL cholesterol and coronary heart disease risk in women. Int. J. Med. Sci. 2006, 3, 108–111. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vänttinen, M.; Nuutila, P.; Kuulasmaa, T.; Pihlajamäki, J.; Hällsten, K.; Virtanen, K.A.; Lautamäki, R.; Peltoniemi, P.; Takala, T.; Viljanen, A.P.M.; et al. Single nucleotide polymorphisms in the peroxisome proliferator-activated receptor delta gene are associated with skeletal muscle glucose uptake. Diabetes 2005, 54, 3587–3591. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reilly, S.M.; Lee, C.H. PPAR delta as a therapeutic target in metabolic disease. FEBS Lett. 2008, 582, 26–31. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leońska-Duniec, A.; Cieszczyk, P.; Jastrzębski, Z.; Jażdżewska, A.; Lulińska-Kuklik, E.; Moska, W.; Ficek, K.; Niewczas, M.; Maciejewska-Skrendo, A. The polymorphisms of the PPARD gene modify post-training body mass and biochemical parameter changes in women. PLoS ONE 2018, 13, e0202557. [Google Scholar] [CrossRef] [Green Version]
- Thamer, C.; Machann, J.; Stefan, N.; Schäfer, S.A.; Machicao, F.; Staiger, H.; Laakso, M.; Böttcher, M.; Claussen, C.; Schick, F.; et al. Variations in PPARD determine the change in body composition during lifestyle intervention: A whole-body magnetic resonance study. J. Clin. Endocrinol. Metab. 2008, 93, 1497–1500. [Google Scholar] [CrossRef] [Green Version]
- Stefan, N.; Thamer, C.; Staiger, H.; Machicao, F.; Machann, J.; Schick, F.; Venter, C.; Niess, A.; Laakso, M.; Fritsche, A.; et al. Genetic variations in PPARD and PPARGC1A determine mitochondrial function and change in aerobic physical fitness and insulin sensitivity during lifestyle intervention. J. Clin. Endocrinol. Metab. 2007, 92, 1827–1833. [Google Scholar] [CrossRef] [Green Version]
- Robitaille, J.; Gaudet, D.; Pérusse, L.; Vohl, M.C. Features of the metabolic syndrome are modulated by an interaction between the peroxisome proliferator-activated receptor-delta-87T>C polymorphism and dietary fat in French-Canadians. Int. J. Obes. 2007, 31, 411–417. [Google Scholar] [CrossRef] [Green Version]
- Shin, H.D.; Park, B.L.; Kim, L.H.; Jung, H.S.; Cho, Y.M.; Moon, M.K.; Park, Y.J.; Lee, H.K.; Park, K.S. Genetic polymorphisms in peroxisome proliferator-activated receptor delta associated with obesity. Diabetes 2004, 53, 847–851. [Google Scholar] [CrossRef] [Green Version]
- Sáez, M.E.; Grilo, A.; Morón, F.J.; Manzano, L.; Martínez-Larrad, M.T.; González-Pérez, A.; Serrano-Hernando, J.; Ruiz, A.; Ramírez-Lorca, R.; Serrano-Ríos, M. Interaction between Calpain 5, Peroxisome proliferator-activated receptor-gamma and Peroxisome proliferator-activated receptor-delta genes: A polygenic approach to obesity. Cardiovasc. Diabetol. 2008, 7, 23. [Google Scholar] [CrossRef] [Green Version]
- Grarup, N.; Albrechtsen, A.; Ek, J.; Borch-Johnsen, K.; Jørgensen, T.; Schmitz, O.; Hansen, T.; Pedersen, O. Variation in the peroxisome proliferator-activated receptor delta gene in relation to common metabolic traits in 7495 middle-aged white people. Diabetologia 2007, 50, 1201–1208. [Google Scholar] [CrossRef]
- Corrales, P.; Vidal-Puig, A.; Medina-Gómez, G. PPARs and Metabolic Disorders Associated with Challenged Adipose Tissue Plasticity. Int. J. Mol. Sci. 2018, 19, 2124. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gross, B.; Pawlak, M.; Lefebvre, P.; Staels, B. PPARs in obesity-induced T2DM, dyslipidaemia and NAFLD. Nat. Rev. Endocrinol. 2016, 13, 36–49. [Google Scholar] [CrossRef] [PubMed]
- Peeters, A.; Baes, M. Role of PPARα in Hepatic Carbohydrate Metabolism. PPAR Res. 2010, 2010, 572405. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vu-Dac, N.; Gervois, P.; Jakel, H.; Nowak, M.; Baugé, E.; Lè Ne Dehondt, H.; Staels, B.; Pennacchio, L.A.; Rubin, E.M.; Fruchart-Najib, J.; et al. Apolipoprotein A5, a Crucial Determinant of Plasma Triglyceride Levels, Is Highly Responsive to Peroxisome Proliferator-activated Receptor α Activators. J. Biol. Chem. 2003, 278, 17982–17985. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schoonjans, K.; Peinado-Onsurbe, J.; Lefebvre, A.M.; Heyman, R.A.; Briggs, M.; Deeb, S.; Staels, B.; Auwerx, J. PPARalpha and PPARgamma activators direct a distinct tissue-specific transcriptional response via a PPRE in the lipoprotein lipase gene. EMBO J. 1996, 15, 5336–5348. [Google Scholar] [CrossRef] [PubMed]
- Hiukka, A.; Leinonen, E.; Jauhiainen, M.; Sundvall, J.; Ehnholm, C.; Keech, A.C.; Taskinen, M.R. Long-term effects of fenofibrate on VLDL and HDL subspecies in participants with type 2 diabetes mellitus. Diabetologia 2007, 50, 2067–2075. [Google Scholar] [CrossRef] [Green Version]
- Vu-Dac, N.; Schoonjans, K.; Kosykh, V.; Dallongeville, J.; Fruchart, J.C.; Staels, B.; Auwerx, J. Fibrates increase human apolipoprotein A-II expression through activation of the peroxisome proliferator-activated receptor. J. Clin. Investig. 1995, 96, 741–750. [Google Scholar] [CrossRef] [PubMed]
- Colin, S.; Briand, O.; Touche, V.; Wouters, K.; Baron, M.; Pattou, F.; Hanf, R.; Tailleux, A.; Chinetti, G.; Staels, B.; et al. Activation of intestinal peroxisome proliferator-activated receptor-α increases high-density lipoprotein production. Eur. Heart J. 2013, 34, 2566–2574. [Google Scholar] [CrossRef]
- Dubois, V.; Eeckhoute, J.; Lefebvre, P.; Staels, B. Distinct but complementary contributions of PPAR isotypes to energy homeostasis. J. Clin. Investig. 2017, 127, 1202–1214. [Google Scholar] [CrossRef] [Green Version]
- Pawlak, M.; Lefebvre, P.; Staels, B. Molecular mechanism of PPARα action and its impact on lipid metabolism, inflammation and fibrosis in non-alcoholic fatty liver disease. J. Hepatol. 2015, 62, 720–733. [Google Scholar] [CrossRef]
- Goto, T.; Lee, J.Y.; Teraminami, A.; Kim, Y.I.; Hirai, S.; Uemura, T.; Inoue, H.; Takahashi, N.; Kawada, T. Activation of peroxisome proliferator-activated receptor-alpha stimulates both differentiation and fatty acid oxidation in adipocytes. J. Lipid Res. 2011, 52, 873–884. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ginsberg, H.; Elam, M.; Lovato, L.; Crouse, J.; Leiter, L.; Linz, P.; Friedewald, W.; Buse, J.; Gerstein, H.; Probstfield, J.; et al. Effects of combination lipid therapy in type 2 diabetes mellitus. N. Engl. J. Med. 2010, 362, 1563–1574. [Google Scholar] [CrossRef] [PubMed]
- Keech, A.; Simes, R.; Barter, P.; Best, J.; Scott, R.; Taskinen, M.; Forder, P.; Pillai, A.; Davis, T.; Glasziou, P.; et al. Effects of long-term fenofibrate therapy on cardiovascular events in 9795 people with type 2 diabetes mellitus (the FIELD study): Randomised controlled trial. Lancet 2005, 366, 1849–1861. [Google Scholar] [CrossRef] [PubMed]
- Chakravarthy, M.V.; Zhu, Y.; López, M.; Yin, L.; Wozniak, D.F.; Coleman, T.; Hu, Z.; Wolfgang, M.; Vidal-Puig, A.; Lane, M.D.; et al. Brain fatty acid synthase activates PPARalpha to maintain energy homeostasis. J. Clin. Investig. 2007, 117, 2539–2552. [Google Scholar] [CrossRef]
- Fu, J.; Gaetani, S.; Oveisi, F.; Lo Verme, J.; Serrano, A.; De Fonseca, F.R.; Rosengarth, A.; Luecke, H.; Di Giacomo, B.; Tarzia, G.; et al. Oleylethanolamide regulates feeding and body weight through activation of the nuclear receptor PPAR-alpha. Nature 2003, 425, 90–93. [Google Scholar] [CrossRef]
- Sher, T.; Gonzalez, F.J.; Yi, H.F.; McBride, O.W. cDNA cloning, chromosomal mapping, and functional characterization of the human peroxisome proliferator activated receptor. Biochemistry 1993, 32, 5598–5604. [Google Scholar] [CrossRef]
- Ruscica, M.; Busnelli, M.; Runfola, E.; Corsini, A.; Sirtori, C.R. Impact of PPAR-Alpha Polymorphisms-The Case of Metabolic Disorders and Atherosclerosis. Int. J. Mol. Sci. 2019, 20, 4378. [Google Scholar] [CrossRef] [Green Version]
- Vohl, M.C.; Lepage, P.; Gaudet, D.; Brewer, C.G.; Bétard, C.; Perron, P.; Houde, G.; Cellier, C.; Faith, J.M.; Després, J.P.; et al. Molecular scanning of the human PPARα gene: Association of the L162V mutation with hyperapobetalipoproteinemia. J. Lipid Res. 2000, 41, 945–952. [Google Scholar] [CrossRef]
- Flavell, D.M.; Pineda Torra, I.; Jamshidi, Y.; Evans, D.; Diamond, J.R.; Elkeles, R.S.; Bujac, S.R.; Miller, G.; Talmud, P.J.; Staels, B.; et al. Variation in the PPARalpha gene is associated with altered function in vitro and plasma lipid concentrations in Type II diabetic subjects. Diabetologia 2000, 43, 673–680. [Google Scholar] [CrossRef]
- Hsu, M.H.; Palmer, C.N.A.; Song, W.; Griffin, K.J.; Johnson, E.F. A carboxyl-terminal extension of the zinc finger domain contributes to the specificity and polarity of peroxisome proliferator-activated receptor DNA binding. J. Biol. Chem. 1998, 273, 27988–27997. [Google Scholar] [CrossRef]
- Sapone, A.; Peters, J.M.; Sakai, S.; Tomita, S.; Papiha, S.S.; Dai, R.; Friedman, F.K.; Gonzalez, F.J. The human peroxisome proliferator-activated receptor alpha gene: Identification and functional characterization of two natural allelic variants. Pharmacogenetics 2000, 10, 321–333. [Google Scholar] [CrossRef] [PubMed]
- Uthurralt, J.; Gordish-Dressman, H.; Bradbury, M.; Tesi-Rocha, C.; Devaney, J.; Harmon, B.; Reeves, E.K.; Brandoli, C.; Hansen, B.C.; Seip, R.L.; et al. PPARalpha L162V underlies variation in serum triglycerides and subcutaneous fat volume in young males. BMC Med. Genet. 2007, 8, 55. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tai, E.S.; Demissie, S.; Cupples, L.A.; Corella, D.; Wilson, P.W.; Schaefer, E.J.; Ordovas, J.M. Association between the PPARA L162V polymorphism and plasma lipid levels: The Framingham Offspring Study. Arterioscler. Thromb. Vasc. Biol. 2002, 22, 805–810. [Google Scholar] [CrossRef] [PubMed]
- Lacquemant, C.; Lepretre, F.; Pineda Torra, I.; Manraj, M.; Charpentier, G.; Ruiz, J.; Staels, B.; Froguel, P.H. Mutation screening of the PPARalpha gene in type 2 diabetes associated with coronary heart disease. Diabetes Metab. 2000, 26, 393–401. [Google Scholar] [PubMed]
- Sparsø, T.; Hussain, M.S.; Andersen, G.; Hainerova, I.; Borch-Johnsen, K.; Jørgensen, T.; Hansen, T.; Pedersen, O. Relationships between the functional PPARalpha Leu162Val polymorphism and obesity, type 2 diabetes, dyslipidaemia, and related quantitative traits in studies of 5799 middle-aged white people. Mol. Genet. Metab. 2007, 90, 205–209. [Google Scholar] [CrossRef] [PubMed]
- Silbernagel, G.; Stefan, N.; Hoffmann, M.M.; Machicao-Arano, F.; Machann, J.; Schick, F.; Winkelmann, B.R.; Boehm, B.O.; Häring, H.U.; Fritsche, A.; et al. The L162V polymorphism of the peroxisome proliferator activated receptor alpha gene (PPARA) is not associated with type 2 diabetes, BMI or body fat composition. Exp. Clin. Endocrinol. Diabetes 2009, 117, 113–118. [Google Scholar] [CrossRef]
- Tai, E.S.; Corella, D.; Demissie, S.; Cupples, L.A.; Coltell, O.; Schaefer, E.J.; Tucker, K.L.; Ordovas, J.M. Polyunsaturated fatty acids interact with the PPARA-L162V polymorphism to affect plasma triglyceride and apolipoprotein C-III concentrations in the Framingham Heart Study. J. Nutr. 2005, 135, 397–403. [Google Scholar] [CrossRef] [Green Version]
- Paradis, A.M.; Fontaine-Bisson, B.; Bossé, Y.; Robitaille, J.; Lemieux, S.; Jacques, H.; Lamarche, B.; Tchernof, A.; Couture, P.; Vohl, M.C. The peroxisome proliferator-activated receptor alpha Leu162Val polymorphism influences the metabolic response to a dietary intervention altering fatty acid proportions in healthy men. Am. J. Clin. Nutr. 2005, 81, 523–530. [Google Scholar] [CrossRef] [Green Version]
- Chan, E.; Tan, C.S.; Deurenberg-Yap, M.; Chia, K.S.; Chew, S.K.; Tai, E.S. The V227A polymorphism at the PPARA locus is associated with serum lipid concentrations and modulates the association between dietary polyunsaturated fatty acid intake and serum high density lipoprotein concentrations in Chinese women. Atherosclerosis 2006, 187, 309–315. [Google Scholar] [CrossRef]
- Yamakawa-Kobayashi, K.; Ishiguro, H.; Arinami, T.; Miyazaki, R.; Hamaguchi, H. A Val227Ala polymorphism in the peroxisome proliferator activated receptor alpha (PPARalpha) gene is associated with variations in serum lipid levels. J. Med. Genet. 2002, 39, 189–191. [Google Scholar] [CrossRef]
- Mei, H.L.; Li, J.; Shen, P.; Husna, B.; Tai, E.S.; Yong, E.L. A natural polymorphism in peroxisome proliferator-activated receptor-alpha hinge region attenuates transcription due to defective release of nuclear receptor corepressor from chromatin. Mol. Endocrinol. 2008, 22, 1078–1092. [Google Scholar] [CrossRef] [Green Version]
- Dongiovanni, P.; Valenti, L. Peroxisome proliferator-activated receptor genetic polymorphisms and nonalcoholic Fatty liver disease: Any role in disease susceptibility? PPAR Res. 2013, 2013. [Google Scholar] [CrossRef] [PubMed]
- Naito, H.; Yamanoshita, O.; Kamijima, M.; Katoh, T.; Matsunaga, T.; Lee, C.H.; Kim, H.; Aoyama, T.; Gonzalez, F.J.; Nakajima, T. Association of V227A PPARalpha polymorphism with altered serum biochemistry and alcohol drinking in Japanese men. Pharmacogenet. Genom. 2006, 16, 569–577. [Google Scholar] [CrossRef] [PubMed]
- Jamshidi, Y.; Montgomery, H.E.; Hense, H.W.; Myerson, S.G.; Pineda Torra, I.; Staels, B.; World, M.J.; Doering, A.; Erdmann, J.; Hengstenberg, C.; et al. Peroxisome proliferator—Activated receptor alpha gene regulates left ventricular growth in response to exercise and hypertension. Circulation 2002, 105, 950–955. [Google Scholar] [CrossRef] [Green Version]
- Flavell, D.M.; Jamshidi, Y.; Hawe, E.; Pineda Torra, I.; Taskinen, M.R.; Frick, M.H.; Nieminen, M.S.; Kesäniemi, Y.A.; Pasternack, A.; Staels, B.; et al. Peroxisome proliferator-activated receptor alpha gene variants influence progression of coronary atherosclerosis and risk of coronary artery disease. Circulation 2002, 105, 1440–1445. [Google Scholar] [CrossRef] [Green Version]
- Ahmetov, I.I.; Mozhayskaya, I.A.; Flavell, D.M.; Astratenkova, I.V.; Komkova, A.I.; Lyubaeva, E.V.; Tarakin, P.P.; Shenkman, B.S.; Vdovina, A.B.; Netreba, A.I.; et al. PPARalpha gene variation and physical performance in Russian athletes. Eur. J. Appl. Physiol. 2006, 97, 103–108. [Google Scholar] [CrossRef]
- Sack, M.N.; Rader, T.A.; Park, S.; Bastin, J.; McCune, S.A.; Kelly, D.P. Fatty acid oxidation enzyme gene expression is downregulated in the failing heart. Circulation 1996, 94, 2837–2842. [Google Scholar] [CrossRef]
- Doney, A.S.F.; Fischer, B.; Lee, S.; Morris, A.D.; Leese, G.P.; Palmer, C.N.A. Association of common variation in the PPARA gene with incident myocardial infarction in individuals with type 2 diabetes: A Go-DARTS study. Nucl. Recept. 2005, 3, 4. [Google Scholar] [CrossRef] [Green Version]
- Purushothaman, S.; Ajitkumar, V.K.; Renuka Nair, R. Association of PPARα Intron 7 Polymorphism with Coronary Artery Disease: A Cross-Sectional Study. ISRN Cardiol. 2011, 2011, 816025. [Google Scholar] [CrossRef] [Green Version]
- Chen, E.S.; Mazzotti, D.R.; Furuya, T.K.; Cendoroglo, M.S.; Ramos, L.R.; Araujo, L.Q.; Burbano, R.R.; Smith, M.D.A.C. Association of PPARalpha gene polymorphisms and lipid serum levels in a Brazilian elderly population. Exp. Mol. Pathol. 2010, 88, 197–201. [Google Scholar] [CrossRef]
- Pan, W.; Liu, C.; Zhang, J.; Gao, X.; Yu, S.; Tan, H.; Yu, J.; Qian, D.; Li, J.; Bian, S.; et al. Association Between Single Nucleotide Polymorphisms in PPARA and EPAS1 Genes and High-Altitude Appetite Loss in Chinese Young Men. Front. Physiol. 2019, 10, 59. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Horscroft, J.A.; Kotwica, A.O.; Laner, V.; West, J.A.; Hennis, P.J.; Levett, D.Z.H.; Howard, D.J.; Fernandez, B.O.; Burgess, S.L.; Ament, Z.; et al. Metabolic basis to Sherpa altitude adaptation. Proc. Natl. Acad. Sci. USA 2017, 114, 6382–6387. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Duraisamy, A.J.; Bayen, S.; Saini, S.; Sharma, A.K.; Vats, P.; Singh, S.B. Changes in ghrelin, CCK, GLP-1, and peroxisome proliferator-activated receptors in a hypoxia-induced anorexia rat model. Endokrynol. Pol. 2015, 66, 334–341. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Volcik, K.A.; Nettleton, J.A.; Ballantyne, C.M.; Boerwinkle, E. Peroxisome proliferator-activated receptor [alpha] genetic variation interacts with n-6 and long-chain n-3 fatty acid intake to affect total cholesterol and LDL-cholesterol concentrations in the Atherosclerosis Risk in Communities Study. Am. J. Clin. Nutr. 2008, 87, 1926–1931. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Esterbauer, H.; Oberkofler, H.; Krempler, F.; Patsch, W. Human peroxisome proliferator activated receptor gamma coactivator 1 (PPARGC1) gene: cDNA sequence, genomic organization, chromosomal localization, and tissue expression. Genomics 1999, 62, 98–102. [Google Scholar] [CrossRef]
- Puigserver, P.; Wu, Z.; Park, C.W.; Graves, R.; Wright, M.; Spiegelman, B.M. A cold-inducible coactivator of nuclear receptors linked to adaptive thermogenesis. Cell 1998, 92, 829–839. [Google Scholar] [CrossRef] [Green Version]
- Liang, H.; Ward, W.F. PGC-1α: A key regulator of energy metabolism. Am. J. Physiol. Adv. Physiol. Educ. 2006, 30, 145–151. [Google Scholar] [CrossRef]
- Kelly, D.P.; Scarpulla, R.C. Transcriptional regulatory circuits controlling mitochondrial biogenesis and function. Genes Dev. 2004, 18, 357–368. [Google Scholar] [CrossRef] [Green Version]
- Chinsomboon, J.; Ruas, J.; Gupta, R.K.; Thom, R.; Shoag, J.; Rowe, G.C.; Sawada, N.; Raghuram, S.; Arany, Z. The transcriptional coactivator PGC-1alpha mediates exercise-induced angiogenesis in skeletal muscle. Proc. Natl. Acad. Sci. USA 2009, 106, 21401–21406. [Google Scholar] [CrossRef] [Green Version]
- Yan, Z.; Okutsu, M.; Akhtar, Y.N.; Lira, V.A. Regulation of exercise-induced fiber type transformation, mitochondrial biogenesis, and angiogenesis in skeletal muscle. J. Appl. Physiol. 2011, 110, 264–274. [Google Scholar] [CrossRef]
- Westerterp, K.R. Physical activity and physical activity induced energy expenditure in humans: Measurement, determinants, and effects. Front. Physiol. 2013, 4, 90. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sacks, H.S.; Fain, J.N.; Holman, B.; Cheema, P.; Chary, A.; Parks, F.; Karas, J.; Optican, R.; Bahouth, S.W.; Garrett, E.; et al. Uncoupling protein-1 and related messenger ribonucleic acids in human epicardial and other adipose tissues: Epicardial fat functioning as brown fat. J. Clin. Endocrinol. Metab. 2009, 94, 3611–3615. [Google Scholar] [CrossRef] [PubMed]
- Boström, P.; Wu, J.; Jedrychowski, M.P.; Korde, A.; Ye, L.; Lo, J.C.; Rasbach, K.A.; Boström, E.A.; Choi, J.H.; Long, J.Z.; et al. A PGC1-α-dependent myokine that drives brown-fat-like development of white fat and thermogenesis. Nature 2012, 481, 463–468. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wenz, T.; Rossi, S.G.; Rotundo, R.L.; Spiegelman, B.M.; Moraes, C.T. Increased muscle PGC-1alpha expression protects from sarcopenia and metabolic disease during aging. Proc. Natl. Acad. Sci. USA 2009, 106, 20405–20410. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ek, J.; Andersen, G.; Urhammer, S.A.; Gæde, P.H.; Drivsholm, T.; Borch-Johnsen, K.; Hansen, T.; Pedersen, O. Mutation analysis of peroxisome proliferator-activated receptor-γ coactivator-1 (PGC-1) and relationships of identified amino acid polymorphisms to Type II diabetes mellitus. Diabetologia 2001, 44, 2220–2226. [Google Scholar] [CrossRef] [Green Version]
- Ling, C.; Del Guerra, S.; Lupi, R.; Rönn, T.; Granhall, C.; Luthman, H.; Masiello, P.; Marchetti, P.; Groop, L.; Del Prato, S. Epigenetic regulation of PPARGC1A in human type 2 diabetic islets and effect on insulin secretion. Diabetologia 2008, 51, 615–622. [Google Scholar] [CrossRef] [Green Version]
- Ling, C.; Poulsen, P.; Carlsson, E.; Ridderstråle, M.; Almgren, P.; Wojtaszewski, J.; Beck-Nielsen, H.; Groop, L.; Vaag, A. Multiple environmental and genetic factors influence skeletal muscle PGC-1alpha and PGC-1beta gene expression in twins. J. Clin. Investig. 2004, 114, 1518–1526. [Google Scholar] [CrossRef] [Green Version]
- Patti, M.E.; Butte, A.J.; Crunkhorn, S.; Cusi, K.; Berria, R.; Kashyap, S.; Miyazaki, Y.; Kohane, I.; Costello, M.; Saccone, R.; et al. Coordinated reduction of genes of oxidative metabolism in humans with insulin resistance and diabetes: Potential role of PGC1 and NRF1. Proc. Natl. Acad. Sci. USA 2003, 100, 8466–8471. [Google Scholar] [CrossRef] [Green Version]
- ZHANG, S.; LU, W.; YAN, L.; WU, M.; XU, M.; CHEN, L.; CHENG, H. Association between peroxisome proliferator-activated receptor-γ coactivator-1α gene polymorphisms and type 2 diabetes in southern Chinese population: Role of altered interaction with myocyte enhancer factor 2C. Chin. Med. J. 2007, 120, 1878–1885. [Google Scholar] [CrossRef]
- Michael, L.F.; Wu, Z.; Cheatham, R.B.; Puigserver, P.; Adelmant, G.; Lehman, J.J.; Kelly, D.P.; Spiegelman, B.M. Restoration of insulin-sensitive glucose transporter (GLUT4) gene expression in muscle cells by the transcriptional coactivator PGC-1. Proc. Natl. Acad. Sci. USA 2001, 98, 3820–3825. [Google Scholar] [CrossRef]
- Handschin, C.; Rhee, J.; Lin, J.; Tarr, P.T.; Spiegelman, B.M. An autoregulatory loop controls peroxisome proliferator-activated receptor gamma coactivator 1alpha expression in muscle. Proc. Natl. Acad. Sci. USA 2003, 100, 7111–7116. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- James, D.E.; Brown, R.; Navarro, J.; Pilch, P.F. Insulin-regulatable tissues express a unique insulin-sensitive glucose transport protein. Nature 1988, 333, 183–185. [Google Scholar] [CrossRef] [PubMed]
- Fukumoto, H.; Kayano, T.; Buse, J.B.; Edwards, Y.; Pilch, P.F.; Bell, G.I.; Seino, S. Cloning and characterization of the major insulin-responsive glucose transporter expressed in human skeletal muscle and other insulin-responsive tissues. J. Biol. Chem. 1989, 264, 7776–7779. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.L.; Gibbs, E.M.; Mccoid, S.C.; Milici, A.J.; Stukenbrok, H.A.; Mcpherson, R.K.; Treadway, J.L.; Pessin, J.E. Transgenic mice expressing the human GLUT4/muscle-fat facilitative glucose transporter protein exhibit efficient glycemic control. Proc. Natl. Acad. Sci. USA 1993, 90, 11346–11350. [Google Scholar] [CrossRef] [Green Version]
- Esterbauer, H.; Oberkofler, H.; Linnemayr, V.; Iglseder, B.; Hedegger, M.; Wolfsgruber, P.; Paulweber, B.; Fastner, G.; Krempler, F.; Patsch, W. Peroxisome proliferator-activated receptor-γ coactivator-1 gene locus: Associations with obesity indices in middle-aged women. Diabetes 2002, 51, 1281–1286. [Google Scholar] [CrossRef] [Green Version]
- Muller, Y.L.; Bogardus, C.; Pedersen, O.; Baier, L. A Gly482Ser missense mutation in the peroxisome proliferator-activated receptor gamma coactivator-1 is associated with altered lipid oxidation and early insulin secretion in Pima Indians. Diabetes 2003, 52, 895–898. [Google Scholar] [CrossRef] [Green Version]
- Kunej, T.; Globočnik Petrovic, M.; Dovč, P.; Peterlin, B.; Petrovič, D. A Gly482Ser polymorphism of the peroxisome proliferator-activated receptor-gamma coactivator-1 (PGC-1) gene is associated with type 2 diabetes in Caucasians. Folia Biol. 2004, 50, 157–158. [Google Scholar]
- Barroso, I.; Luan, J.; Sandhu, M.S.; Franks, P.W.; Crowley, V.; Schafer, A.J.; O’Rahilly, S.; Wareham, N.J. Meta-analysis of the Gly482Ser variant in PPARGC1A in type 2 diabetes and related phenotypes. Diabetologia 2006, 49, 501–505. [Google Scholar] [CrossRef] [Green Version]
- Maciejewska, A.; Sawczuk, M.; Cieszczyk, P.; Mozhayskaya, I.A.; Ahmetov, I.I. The PPARGC1A gene Gly482Ser in Polish and Russian athletes. J. Sports Sci. 2012, 30, 101–113. [Google Scholar] [CrossRef]
- Eynon, N.; Meckel, Y.; Sagiv, M.; Yamin, C.; Amir, R.; Sagiv, M.; Goldhammer, E.; Duarte, J.A.; Oliveira, J. Do PPARGC1A and PPARalpha polymorphisms influence sprint or endurance phenotypes? Scand. J. Med. Sci. Sports 2010, 20, e145–e150. [Google Scholar] [CrossRef]
- Eynon, N.; Meckel, Y.; Alves, A.J.; Yamin, C.; Sagiv, M.; Goldhammer, E.; Sagiv, M. Is there an interaction between PPARD T294C and PPARGC1A Gly482Ser polymorphisms and human endurance performance? Exp. Physiol. 2009, 94, 1147–1152. [Google Scholar] [CrossRef] [PubMed]
- Lucia, A.; Gómez-Gallego, F.; Barroso, I.; Rabadán, M.; Bandrés, F.; San Juan, A.F.; Chicharro, J.L.; Ekelund, U.; Brage, S.; Earnest, C.P.; et al. PPARGC1A genotype (Gly482Ser) predicts exceptional endurance capacity in European men. J. Appl. Physiol. 2005, 99, 344–348. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Petr, M.; Maciejewska-Skrendo, A.; Zajac, A.; Chycki, J.; Stastny, P. Association of elite sports status with gene variants of peroxisome proliferator activated receptors and their transcriptional coactivator. Int. J. Mol. Sci. 2020, 21, 162. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hong, M.S.; Kim, H.K.; Shin, D.H.; Song, D.K.; Ban, J.Y.; Kim, B.S.; Chung, J.-H. Integrative Study on PPARGC1A: Hypothalamic Expression of Ppargc1a in ob/ob Mice and Association between PPARGC1A and Obesity in Korean Population. Mol. Cell. Toxicol. 2008, 4, 318–322. [Google Scholar]
- Franks, P.W.; Ekelund, U.; Brage, S.; Luan, J.; Schafer, A.J.; O’Rahilly, S.; Barroso, I.; Wareham, N.J. PPARGC1A coding variation may initiate impaired NEFA clearance during glucose challenge. Diabetologia 2007, 50, 569–573. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pérusse, L.; Rice, T.; Chagnon, Y.C.; Després, J.P.; Lemieux, S.; Roy, S.; Lacaille, M.; Ho-Kim, M.A.; Chagnon, M.; Province, M.A.; et al. A genome-wide scan for abdominal fat assessed by computed tomography in the Québec Family Study. Diabetes 2001, 50, 614–621. [Google Scholar] [CrossRef] [Green Version]
- Arya, R.; Duggirala, R.; Jenkinson, C.P.; Almasy, L.; Blangero, J.; O’Connell, P.; Stern, M.P. Evidence of a novel quantitative-trait locus for obesity on chromosome 4p in Mexican Americans. Am. J. Hum. Genet. 2004, 74, 272–282. [Google Scholar] [CrossRef] [Green Version]
- Pihlajamäki, J.; Kinnunen, M.; Ruotsalainen, E.; Salmenniemi, U.; Vauhkonen, I.; Kuulasmaa, T.; Kainulainen, S.; Laakso, M. Haplotypes of PPARGC1A are associated with glucose tolerance, body mass index and insulin sensitivity in offspring of patients with type 2 diabetes. Diabetologia 2005, 48, 1331–1334. [Google Scholar] [CrossRef] [Green Version]
- Povel, C.M.; Feskens, E.J.M.; Imholz, S.; Blaak, E.E.; Boer, J.M.A.; Dollé, M.E.T. Glucose levels and genetic variants across transcriptional pathways: Interaction effects with BMI. Int. J. Obes. 2010, 34, 840–845. [Google Scholar] [CrossRef] [Green Version]
- Da Fonseca, A.C.P.; Da Fonseca, G.P.; Marchesini, B.; Voigt, D.D.; Campos Junior, M.; Zembrzuski, V.M.; Carneiro, J.R.I.; Nogueira Neto, J.F.; Cabello, P.H.; Cabello, G.M.K. Genetic Variants in the Activation of the Brown-Like Adipocyte Pathway and the Risk for Severe Obesity. Obes. Facts 2020, 13, 130–143. [Google Scholar] [CrossRef]
- Semple, R.K.; Crowley, V.C.; Sewter, C.P.; Laudes, M.; Christodoulides, C.; Considine, R.V.; Vidal-Puig, A.; O’Rahilly, S. Expression of the thermogenic nuclear hormone receptor coactivator PGC-1alpha is reduced in the adipose tissue of morbidly obese subjects. Int. J. Obes. Relat. Metab. Disord. 2004, 28, 176–179. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hoeks, J.; Hesselink, M.K.C.; Russell, A.P.; Mensink, M.; Saris, W.H.M.; Mensink, R.P.; Schrauwen, P. Peroxisome proliferator-activated receptor-gamma coactivator-1 and insulin resistance: Acute effect of fatty acids. Diabetologia 2006, 49, 2419–2426. [Google Scholar] [CrossRef] [PubMed]
- Albuquerque, D.; Nóbrega, C.; Rodríguez-López, R.; Manco, L. Association study of common polymorphisms in MSRA, TFAP2B, MC4R, NRXN3, PPARGC1A, TMEM18, SEC16B, HOXB5 and OLFM4 genes with obesity-related traits among Portuguese children. J. Hum. Genet. 2014, 59, 307–313. [Google Scholar] [CrossRef]
- Lin, J.; Handschin, C.; Spiegelman, B.M. Metabolic control through the PGC-1 family of transcription coactivators. Cell Metab. 2005, 1, 361–370. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Spiegelman, B.M.; Puigserver, P.; Wu, Z. Regulation of adipogenesis and energy balance by PPARgamma and PGC-1. Int. J. Obes. Relat. Metab. Disord. 2000, 24 (Suppl. S4), S8–S10. [Google Scholar] [CrossRef] [PubMed]

| Protein | Gene | RsID | Alleles (Variants) | Molecular Significance | Selected Phenotypic Effects |
|---|---|---|---|---|---|
| PPARγ | PPARG | rs1801282 | C (Pro12) | Normal binding affinity of the PPARγ2 protein to the target genes | Lower BMI values in obese individuals or when diet is rich in saturated fatty acids |
| G (12Ala) | Reduced binding affinity of the PPARγ2 protein to the target genes | Lower BMI values in lean individuals or when diet is rich in polyunsaturated fatty acids | |||
| PPARδ | PPARD | rs2016520 | T | Normal transcriptional activity of the PPARD gene | |
| C | Higher transcriptional activity of the PPARD gene | Lower total cholesterol and blood triglycerides after training, lower risk of metabolic syndrome with moderate fat consumption | |||
| rs2267668 | A | Not known | Lower triglyceride levels after training | ||
| G | Not known | Lower total cholesterol after training, weaker response to physical training as measured by individual anaerobic threshold and peak aerobic capacity | |||
| rs1053049 | T | Not known | Higher triglyceride levels after training | ||
| C | Not known | Lower triglyceride levels after training | |||
| PPARα | PPARA | rs1800206 | C (Leu162) | Higher transcriptional activity of the PPARA gene in the absence or at low concentrations of ligands | Lower levels of triglycerides and Apo-CIII when diet is low in polyunsaturated fatty acids |
| G (162Val) | Higher transcriptional activity of the PPARA gene at high ligand concentrations | Lower total cholesterol and Apo-AI levels when diet is rich in polyunsaturated fatty acids | |||
| rs1800234 | T (Val227) | Normal transactivation activity of the PPARα protein in the presence of PPARα-specific ligands | higher cholesterol levels in individuals who do not drink alcohol | ||
| C (227Ala) | Lower transactivation activity of the PPARα protein in the presence of PPARα-specific ligands | Reduced levels of cholesterol, LDL-C, and triglycerides, increased gamma-glutamyl transpeptidase activity in alcoholic drinkers | |||
| rs4253778 | G | Normal expression of the PPARA gene | |||
| C | Lower expression of the PPARA gene, weaker stimulation of expression of genes controlled by PPARα | Higher total and LDL cholesterol levels; positive association with dyslipidemia, lower TG and VLDL levels, and higher HDL-C levels in populations of different ethnic origins | |||
| rs4253747 | T | Not known | Weaker appetite suppression response in high-altitude appetite loss | ||
| A | Not known | Stronger appetite suppression response in high-altitude appetite loss | |||
| rs6008259 | G | Not known | Higher levels of total cholesterol and LDL-C at a high daily intake of linolenic acid | ||
| A | Not known | Lower levels of total cholesterol and LDL-C at a high daily intake of linolenic acid | |||
| rs3892755 | C | Not known | Higher levels of total cholesterol and LDL-C at a high daily intake of eicosapentaenoic acid and docosahexaenoic acid | ||
| T | Not known | Lower levels of total cholesterol and LDL-C at a high daily intake of eicosapentaenoic acid and docosahexaenoic acid | |||
| PGC-1α | PPARGC1A | rs8192678 | G (Gly482) | Normal transcriptional activity of the PPARGC1A gene, normal binding affinity of the PGC-1α protein to the target transcriptional factors | Normal insulin secretion; normal insulin-dependent response; lower risk of dyslipidemia, obesity and type 2 diabetes; normal glucose uptake by cells; improvement of aerobic capacity |
| A (482Ser) | Lower transcriptional activity of the PPARGC1A gene, reduced binding affinity of the PGC-1α protein to the target transcriptional factors | Decreased insulin secretion; weaker insulin-dependent response; higher risk of dyslipidemia, obesity and type 2 diabetes; decrease in glucose uptake by cells; impairment of aerobic capacity |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Maciejewska-Skrendo, A.; Massidda, M.; Tocco, F.; Leźnicka, K. The Influence of the Differentiation of Genes Encoding Peroxisome Proliferator-Activated Receptors and Their Coactivators on Nutrient and Energy Metabolism. Nutrients 2022, 14, 5378. https://doi.org/10.3390/nu14245378
Maciejewska-Skrendo A, Massidda M, Tocco F, Leźnicka K. The Influence of the Differentiation of Genes Encoding Peroxisome Proliferator-Activated Receptors and Their Coactivators on Nutrient and Energy Metabolism. Nutrients. 2022; 14(24):5378. https://doi.org/10.3390/nu14245378
Chicago/Turabian StyleMaciejewska-Skrendo, Agnieszka, Myosotis Massidda, Filippo Tocco, and Katarzyna Leźnicka. 2022. "The Influence of the Differentiation of Genes Encoding Peroxisome Proliferator-Activated Receptors and Their Coactivators on Nutrient and Energy Metabolism" Nutrients 14, no. 24: 5378. https://doi.org/10.3390/nu14245378
APA StyleMaciejewska-Skrendo, A., Massidda, M., Tocco, F., & Leźnicka, K. (2022). The Influence of the Differentiation of Genes Encoding Peroxisome Proliferator-Activated Receptors and Their Coactivators on Nutrient and Energy Metabolism. Nutrients, 14(24), 5378. https://doi.org/10.3390/nu14245378





