Influence of Aging, Macronutrient Composition and Time-Restricted Feeding on the Fischer344 x Brown Norway Rat Gut Microbiota
Abstract
:1. Introduction
2. Materials and Methods
2.1. Subjects
2.2. Tissue Collection
2.3. Nutritional Ketosis
2.4. Cytokine Analysis
2.5. Fecal Microbiome Taxonomy
2.6. Metagenomics
2.7. Statistical Analysis
3. Results
3.1. Confirmation of Nutritional Ketosis
3.2. Cytokine Analysis
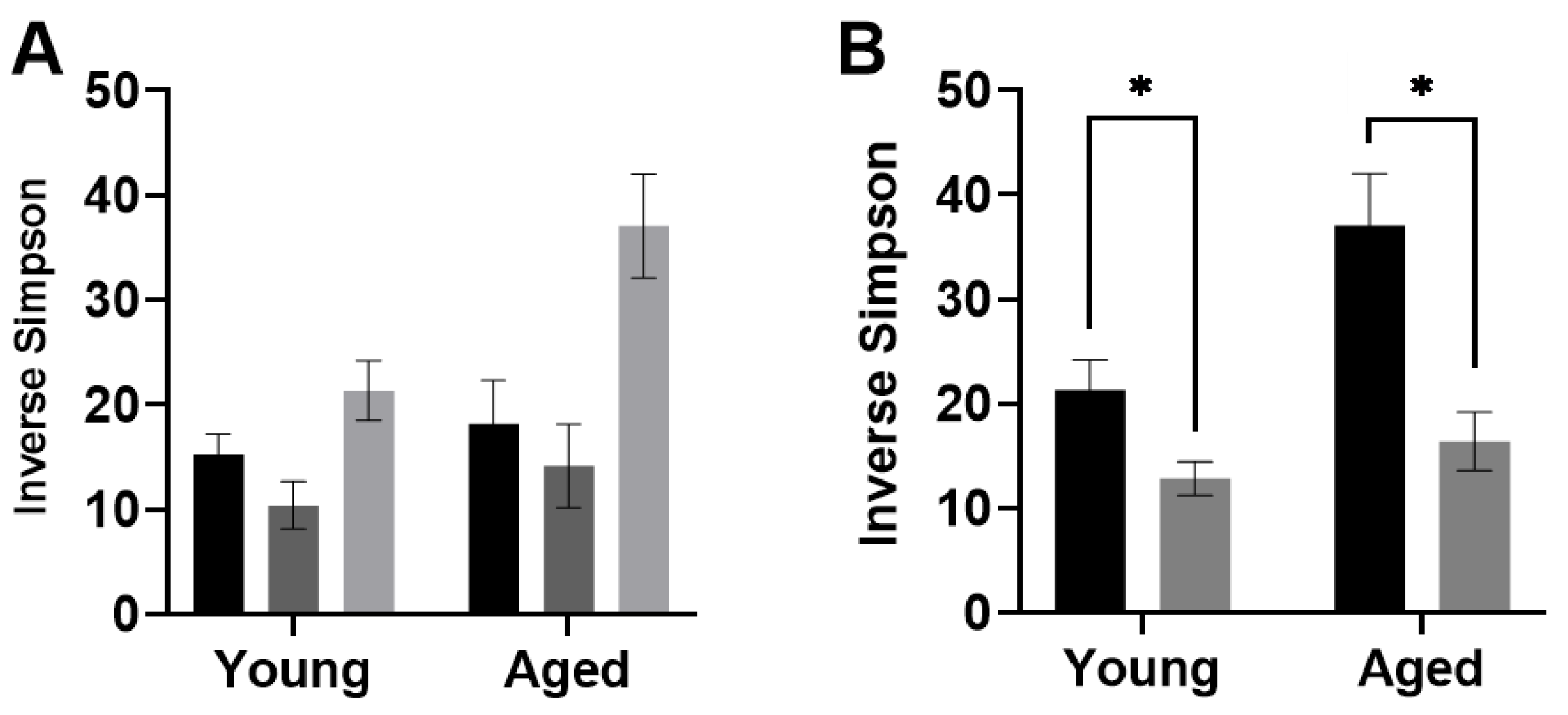
3.3. 16S Microbiome Analysis
3.3.1. Diversity
3.3.2. Differential Abundance
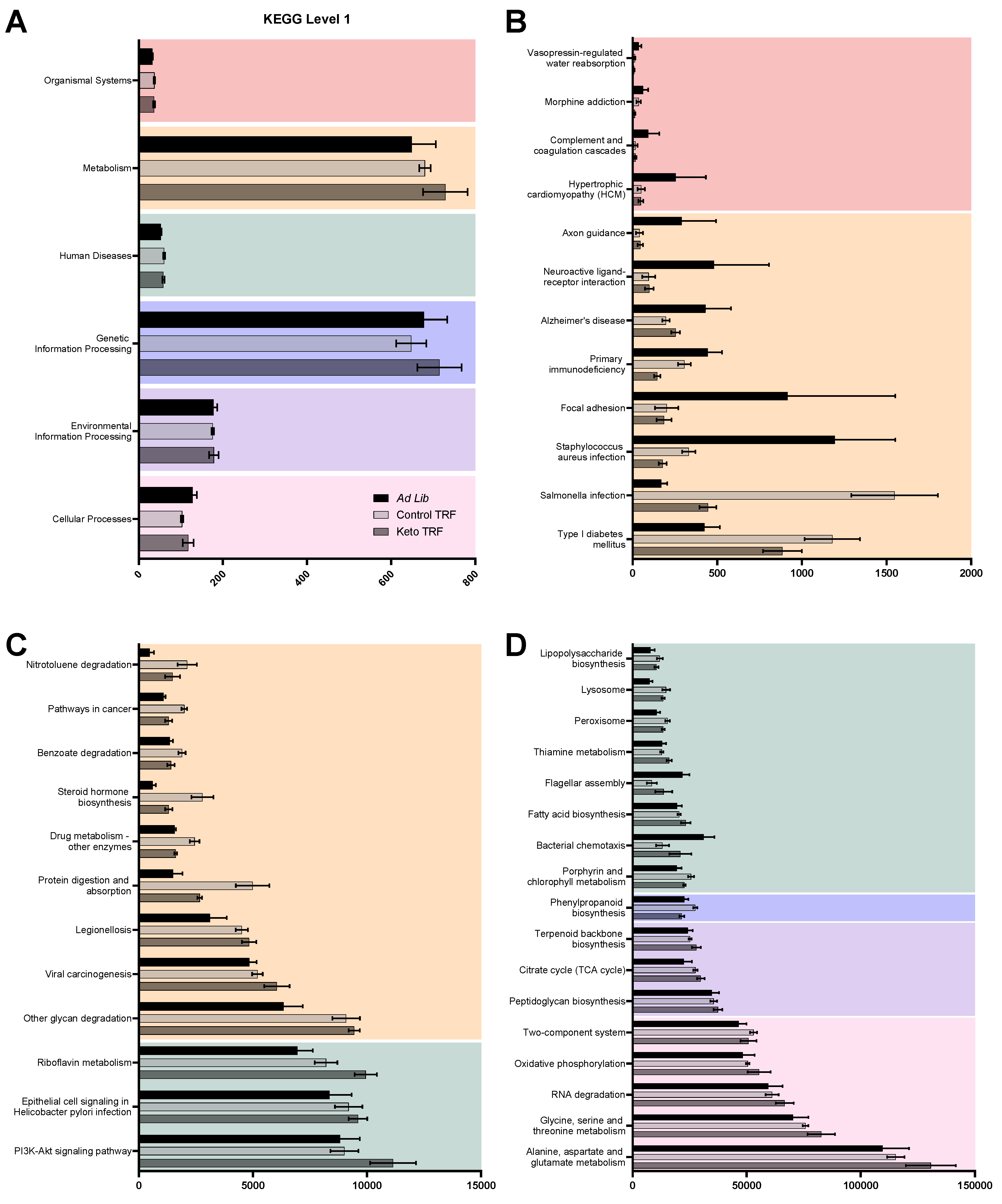
3.3.3. Metagenomics
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Klein, P.; Tyrlikova, I.; Mathews, G.C. Dietary Treatment in Adults with Refractory Epilepsy: A Review. Neurology 2014, 83, 1978–1985. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ye, F.; Li, X.-J.; Jiang, W.-L.; Sun, H.-B.; Liu, J. Efficacy of and Patient Compliance with a Ketogenic Diet in Adults with Intractable Epilepsy: A Meta-Analysis. J. Clin. Neurol. 2015, 11, 26. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hernandez, A.R.; Hernandez, C.M.; Campos, K.; Truckenbrod, L.; Federico, Q.; Moon, B.; McQuail, J.A.; Maurer, A.P.; Bizon, J.L.; Burke, S.N. A Ketogenic Diet Improves Cognition and Has Biochemical Effects in Prefrontal Cortex That Are Dissociable From Hippocampus. Front. Aging Neurosci. 2018, 10, 391. [Google Scholar] [CrossRef] [PubMed]
- Gopinath, B.; Russell, J.; Kifley, A.; Flood, V.M.; Mitchell, P. Adherence to Dietary Guidelines and Successful Aging Over 10 Years. J. Gerontol. Ser. A 2016, 71, 349–355. [Google Scholar] [CrossRef] [Green Version]
- Tinsley, G.M.; La Bounty, P.M. Effects of Intermittent Fasting on Body Composition and Clinical Health Markers in Humans. Nutr. Rev. 2015, 73, 661–674. [Google Scholar] [CrossRef]
- Lee, S.A.; Sypniewski, C.; Bensadon, B.A.; McLaren, C.; Donahoo, W.T.; Sibille, K.T.; Anton, S. Determinants of Adherence in Time-Restricted Feeding in Older Adults: Lessons from a Pilot Study. Nutrients 2020, 12, 874. [Google Scholar] [CrossRef] [Green Version]
- Villareal, D.T.; Fontana, L.; Das, S.K.; Redman, L.; Smith, S.R.; Saltzman, E.; Bales, C.; Rochon, J.; Pieper, C.; Huang, M.; et al. Effect of Two-Year Caloric Restriction on Bone Metabolism and Bone Mineral Density in Non-Obese Younger Adults: A Randomized Clinical Trial. J. Bone Miner. Res. Off. J. Am. Soc. Bone Miner. Res. 2016, 31, 40–51. [Google Scholar] [CrossRef]
- Cienfuegos, S.; Gabel, K.; Kalam, F.; Ezpeleta, M.; Wiseman, E.; Pavlou, V.; Lin, S.; Oliveira, M.L.; Varady, K.A. Effects of 4- and 6-h Time-Restricted Feeding on Weight and Cardiometabolic Health: A Randomized Controlled Trial in Adults with Obesity. Cell Metab. 2020, 32, 366–378.e3. [Google Scholar] [CrossRef]
- Newell, C.; Bomhof, M.R.; Reimer, R.A.; Hittel, D.S.; Rho, J.M.; Shearer, J. Ketogenic Diet Modifies the Gut Microbiota in a Murine Model of Autism Spectrum Disorder. Mol. Autism 2016, 7, 37. [Google Scholar] [CrossRef] [Green Version]
- Swidsinski, A.; Dörffel, Y.; Loening-Baucke, V.; Gille, C.; Göktas, Ö.; Reißhauer, A.; Neuhaus, J.; Weylandt, K.-H.; Guschin, A.; Bock, M. Reduced Mass and Diversity of the Colonic Microbiome in Patients with Multiple Sclerosis and Their Improvement with Ketogenic Diet. Front. Microbiol. 2017, 8, 1141. [Google Scholar] [CrossRef]
- Olson, C.A.; Vuong, H.E.; Yano, J.M.; Liang, Q.Y.; Nusbaum, D.J.; Hsiao, E.Y. The Gut Microbiota Mediates the Anti-Seizure Effects of the Ketogenic Diet. Cell 2018, 173, 1728–1741.e13. [Google Scholar] [CrossRef] [Green Version]
- Gutiérrez-Repiso, C.; Hernández-García, C.; García-Almeida, J.M.; Bellido, D.; Martín-Núñez, G.M.; Sánchez-Alcoholado, L.; Alcaide-Torres, J.; Sajoux, I.; Tinahones, F.J.; Moreno-Indias, I. Effect of Synbiotic Supplementation in a Very-Low-Calorie Ketogenic Diet on Weight Loss Achievement and Gut Microbiota: A Randomized Controlled Pilot Study. Mol. Nutr. Food Res. 2019, 63, 1900167. [Google Scholar] [CrossRef]
- Le, H.H.; Johnson, E.L. Going Keto? Say ΒHB-Ye Bye to Your Gut Bifidobacteria. Cell Host Microbe 2020, 28, 3–5. [Google Scholar] [CrossRef]
- Yuan, W.; Lu, W.; Wang, H.; Wu, W.; Zhou, Q.; Chen, Y.; Lee, Y.K.; Zhao, J.; Zhang, H.; Chen, W. A Multiphase Dietetic Protocol Incorporating an Improved Ketogenic Diet Enhances Weight Loss and Alters the Gut Microbiome of Obese People. Int. J. Food Sci. Nutr. 2021, 73, 238–250. [Google Scholar] [CrossRef]
- Xu, C.; Zhu, H.; Qiu, P. Aging Progression of Human Gut Microbiota. BMC Microbiol. 2019, 19, 236. [Google Scholar] [CrossRef] [Green Version]
- Buford, T.W. (Dis)Trust Your Gut: The Gut Microbiome in Age-Related Inflammation, Health, and Disease. Microbiome 2017, 5, 80. [Google Scholar] [CrossRef] [Green Version]
- Peeke, P.M.; Greenway, F.L.; Billes, S.K.; Zhang, D.; Fujioka, K. Effect of Time Restricted Eating on Body Weight and Fasting Glucose in Participants with Obesity: Results of a Randomized, Controlled, Virtual Clinical Trial. Nutr. Diabetes 2021, 11, 6. [Google Scholar] [CrossRef]
- Lettieri Barbato, D.; Tatulli, G.; Aquilano, K.; Ciriolo, M.R. Mitochondrial Hormesis Links Nutrient Restriction to Improved Metabolism in Fat Cell. Aging 2015, 7, 869–881. [Google Scholar] [CrossRef] [Green Version]
- Brandhorst, S.; Choi, I.Y.; Wei, M.; Cheng, C.W.; Sedrakyan, S.; Navarrete, G.; Dubeau, L.; Yap, L.P.; Park, R.; Vinciguerra, M.; et al. A Periodic Diet That Mimics Fasting Promotes Multi-System Regeneration, Enhanced Cognitive Performance, and Healthspan. Cell Metab. 2015, 22, 86–99. [Google Scholar] [CrossRef] [Green Version]
- Moro, T.; Tinsley, G.; Bianco, A.; Marcolin, G.; Pacelli, Q.F.; Battaglia, G.; Palma, A.; Gentil, P.; Neri, M.; Paoli, A. Effects of Eight Weeks of Time-Restricted Feeding (16/8) on Basal Metabolism, Maximal Strength, Body Composition, Inflammation, and Cardiovascular Risk Factors in Resistance-Trained Males. J. Transl. Med. 2016, 14, 290. [Google Scholar] [CrossRef]
- Gabel, K.; Marcell, J.; Cares, K.; Kalam, F.; Cienfuegos, S.; Ezpeleta, M.; Varady, K.A. Effect of Time Restricted Feeding on the Gut Microbiome in Adults with Obesity: A Pilot Study. Nutr. Health 2020, 26, 79–85. [Google Scholar] [CrossRef]
- Zeb, F.; Wu, X.; Chen, L.; Fatima, S.; Ijaz-ul-Haq; Chen, A.; Xu, C.; Jianglei, R.; Feng, Q.; Li, M. Time-Restricted Feeding Is Associated with Changes in Human Gut Microbiota Related to Nutrient Intake. Nutrition 2020, 78, 110797. [Google Scholar] [CrossRef]
- Zeb, F.; Wu, X.; Chen, L.; Fatima, S.; Haq, I.-U.; Chen, A.; Majeed, F.; Feng, Q.; Li, M. Effect of Time-Restricted Feeding on Metabolic Risk and Circadian Rhythm Associated with Gut Microbiome in Healthy Males. Br. J. Nutr. 2020, 123, 1216–1226. [Google Scholar] [CrossRef]
- Zarrinpar, A.; Chaix, A.; Yooseph, S.; Panda, S. Diet and Feeding Pattern Affect the Diurnal Dynamics of the Gut Microbiome. Cell Metab. 2014, 20, 1006–1017. [Google Scholar] [CrossRef] [Green Version]
- Beli, E.; Yan, Y.; Moldovan, L.; Vieira, C.P.; Gao, R.; Duan, Y.; Prasad, R.; Bhatwadekar, A.; White, F.A.; Townsend, S.D.; et al. Restructuring of the Gut Microbiome by Intermittent Fasting Prevents Retinopathy and Prolongs Survival in Db/Db Mice. Diabetes 2018, 67, 1867–1879. [Google Scholar] [CrossRef] [Green Version]
- Liu, Z.; Dai, X.; Zhang, H.; Shi, R.; Hui, Y.; Jin, X.; Zhang, W.; Wang, L.; Wang, Q.; Wang, D.; et al. Gut Microbiota Mediates Intermittent-Fasting Alleviation of Diabetes-Induced Cognitive Impairment. Nat. Commun. 2020, 11, 855. [Google Scholar] [CrossRef] [Green Version]
- Cignarella, F.; Cantoni, C.; Ghezzi, L.; Salter, A.; Dorsett, Y.; Chen, L.; Phillips, D.; Weinstock, G.M.; Fontana, L.; Cross, A.H.; et al. Intermittent Fasting Confers Protection in CNS Autoimmunity by Altering the Gut Microbiota. Cell Metab. 2018, 27, 1222–1235.e6. [Google Scholar] [CrossRef] [Green Version]
- Shi, H.; Zhang, B.; Abo-Hamzy, T.; Nelson, J.W.; Ambati, C.S.R.; Petrosino, J.F.; Bryan, R.M.; Durgan, D.J. Restructuring the Gut Microbiota by Intermittent Fasting Lowers Blood Pressure. Circ. Res. 2021, 128, 1240–1254. [Google Scholar] [CrossRef]
- Hoyer, S. Causes and Consequences of Disturbances of Cerebral Glucose Metabolism in Sporadic Alzheimer Disease: Therapeutic Implications. In Proceedings of the Frontiers in Clinical Neuroscience; Vécsei, L., Ed.; Springer US: Boston, MA, USA, 2004; pp. 135–152. [Google Scholar]
- Ryan, C.M.; Freed, M.I.; Rood, J.A.; Cobitz, A.R.; Waterhouse, B.R.; Strachan, M.W.J. Improving Metabolic Control Leads to Better Working Memory in Adults With Type 2 Diabetes. Diabetes Care 2006, 29, 345–351. [Google Scholar] [CrossRef] [Green Version]
- Taylor, V.H.; MacQueen, G.M. Cognitive Dysfunction Associated with Metabolic Syndrome. Obes. Rev. 2007, 8, 409–418. [Google Scholar] [CrossRef]
- Proctor, C.; Thiennimitr, P.; Chattipakorn, N.; Chattipakorn, S.C. Diet, Gut Microbiota and Cognition. Metab. Brain Dis. 2017, 32, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Naveed, M.; Zhou, Q.-G.; Xu, C.; Taleb, A.; Meng, F.; Ahmed, B.; Zhang, Y.; Fukunaga, K.; Han, F. Gut-Brain Axis: A Matter of Concern in Neuropsychiatric Disorders…! Prog. Neuropsychopharmacol. Biol. Psychiatry 2021, 104, 110051. [Google Scholar] [CrossRef] [PubMed]
- Lichtash, C.; Fung, J.; Ostoich, K.C.; Ramos, M. Therapeutic Use of Intermittent Fasting and Ketogenic Diet as an Alternative Treatment for Type 2 Diabetes in a Normal Weight Woman: A 14-Month Case Study. BMJ Case Rep. 2020, 13, e234223. [Google Scholar] [CrossRef] [PubMed]
- Arbour, M.W.; Stec, M.; Walker, K.C.; Wika, J.C. Clinical Implications for Women of a Low-Carbohydrate or Ketogenic Diet With Intermittent Fasting. Nurs. Womens Health 2021, 25, 139–151. [Google Scholar] [CrossRef]
- Hartman, A.L.; Rubenstein, J.E.; Kossoff, E.H. Intermittent Fasting: A “New” Historical Strategy for Controlling Seizures? Epilepsy Res. 2013, 104, 275–279. [Google Scholar] [CrossRef] [Green Version]
- Özkul, C.; Yalınay, M.; Karakan, T. Islamic Fasting Leads to an Increased Abundance of Akkermansia Muciniphila and Bacteroides Fragilis Group: A Preliminary Study on Intermittent Fasting. Turk. J. Gastroenterol. 2019, 30, 1030–1035. [Google Scholar] [CrossRef]
- Ozkul, C.; Yalinay, M.; Karakan, T. Structural Changes in Gut Microbiome after Ramadan Fasting: A Pilot Study. Benef. Microbes 2020, 11, 227–233. [Google Scholar] [CrossRef]
- Su, J.; Wang, Y.; Zhang, X.; Ma, M.; Xie, Z.; Pan, Q.; Ma, Z.; Peppelenbosch, M.P. Remodeling of the Gut Microbiome during Ramadan-Associated Intermittent Fasting. Am. J. Clin. Nutr. 2021, 113, 1332–1342. [Google Scholar] [CrossRef]
- Larrick, J.W.; Mendelsohn, A.R.; Larrick, J.W. Beneficial Gut Microbiome Remodeled During Intermittent Fasting in Humans. Rejuvenation Res. 2021, 24, 234–237. [Google Scholar] [CrossRef]
- Hernandez, A.R.; Hernandez, C.M.; Campos, K.T.; Truckenbrod, L.M.; Sakarya, Y.; McQuail, J.A.; Carter, C.S.; Bizon, J.L.; Maurer, A.P.; Burke, S.N. The Antiepileptic Ketogenic Diet Alters Hippocampal Transporter Levels and Reduces Adiposity in Aged Rats. J. Gerontol. Ser. A 2018, 73, 450–458. [Google Scholar] [CrossRef]
- Lynch, S.; Johnston, J.D.; Robertson, M.D. Early versus Late Time-Restricted Feeding in Adults at Increased Risk of Developing Type 2 Diabetes: Is There an Optimal Time to Eat for Metabolic Health? Nutr. Bull. 2021, 46, 69–76. [Google Scholar] [CrossRef]
- Kumar, R.; Eipers, P.; Little, R.B.; Crowley, M.; Crossman, D.K.; Lefkowitz, E.J.; Morrow, C.D. Getting Started with Microbiome Analysis: Sample Acquisition to Bioinformatics. Curr. Protoc. Hum. Genet. 2014, 82, 18.8.1–18.8.29. [Google Scholar] [CrossRef] [Green Version]
- Buford, T.W.; Carter, C.S.; VanDerPol, W.J.; Chen, D.; Lefkowitz, E.J.; Eipers, P.; Morrow, C.D.; Bamman, M.M. Composition and Richness of the Serum Microbiome Differ by Age and Link to Systemic Inflammation. GeroScience 2018, 40, 257–268. [Google Scholar] [CrossRef] [Green Version]
- Woese, C.R.; Gutell, R.R. Evidence for Several Higher Order Structural Elements in Ribosomal RNA. Proc. Natl. Acad. Sci. USA 1989, 86, 3119–3122. [Google Scholar] [CrossRef] [Green Version]
- Kumar, H.; Lund, R.; Laiho, A.; Lundelin, K.; Ley, R.E.; Isolauri, E.; Salminen, S. Gut Microbiota as an Epigenetic Regulator: Pilot Study Based on Whole-Genome Methylation Analysis. mBio 2014, 5, e02113-14. [Google Scholar] [CrossRef] [Green Version]
- Caporaso, J.G.; Lauber, C.L.; Walters, W.A.; Berg-Lyons, D.; Lozupone, C.A.; Turnbaugh, P.J.; Fierer, N.; Knight, R. Global Patterns of 16S RRNA Diversity at a Depth of Millions of Sequences per Sample. Proc. Natl. Acad. Sci. USA 2011, 108, 4516–4522. [Google Scholar] [CrossRef] [Green Version]
- Kozich, J.J.; Westcott, S.L.; Baxter, N.T.; Highlander, S.K.; Schloss, P.D. Development of a Dual-Index Sequencing Strategy and Curation Pipeline for Analyzing Amplicon Sequence Data on the MiSeq Illumina Sequencing Platform. Appl. Environ. Microbiol. 2013, 79, 5112–5120. [Google Scholar] [CrossRef] [Green Version]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA Ribosomal RNA Gene Database Project: Improved Data Processing and Web-Based Tools. Nucleic Acids Res. 2012, 41, D590–D596. [Google Scholar] [CrossRef]
- Shetty, S.A.; Lahti, L. Microbiome Data Science. J. Biosci. 2019, 44, 115. [Google Scholar] [CrossRef]
- Chao, A.; Chiu, C.-H. Species Richness: Estimation and Comparison. In Wiley StatsRef: Statistics Reference Online; John Wiley & Sons, Ltd.: Hoboken, NJ, USA, 2016. [Google Scholar]
- Simpson, E.H. Measurement of Diversity. Nature 1949, 163, 688. [Google Scholar] [CrossRef]
- Locey, K.J.; Lennon, J.T. Scaling Laws Predict Global Microbial Diversity. Proc. Natl. Acad. Sci. USA 2016, 113, 5970–5975. [Google Scholar] [CrossRef] [Green Version]
- Magurran, A.; McGill, B.J. (Eds.) Biological Diversity: Frontiers in Measurement and Assessment; Oxford University Press: Oxford, UK, 2010. [Google Scholar]
- Lozupone, C.; Hamady, M.; Knight, R. UniFrac—An Online Tool for Comparing Microbial Community Diversity in a Phylogenetic Context. BMC Bioinform. 2006, 7, 371. [Google Scholar] [CrossRef] [Green Version]
- McMurdie, P.J.; Holmes, S. Phyloseq: An R Package for Reproducible Interactive Analysis and Graphics of Microbiome Census Data. PLoS ONE 2013, 8, e61217. [Google Scholar] [CrossRef] [Green Version]
- Oksanen, J.; Blanchet, F.; Friendly, M.; Kindt, R.; Legendre, P.; McGlinn, D.; Minchin, P.; Ohara, R.; Simpson, G.; Solymos, P.; et al. Vegan: Commmunity Ecology Package. 2017. Available online: https://cran.r-project.org/web/packages/vegan/index.html (accessed on 7 March 2022).
- Mandal, S.; Van Treuren, W.; White, R.A.; Eggesbø, M.; Knight, R.; Peddada, S.D. Analysis of Composition of Microbiomes: A Novel Method for Studying Microbial Composition. Microb. Ecol. Health Dis. 2015, 26. [Google Scholar] [CrossRef] [Green Version]
- Kemp, K.M.; Colson, J.; Lorenz, R.G.; Maynard, C.L.; Pollock, J.S. Early Life Stress in Mice Alters Gut Microbiota Independent of Maternal Microbiota Inheritance. Am. J. Physiol.-Regul. Integr. Comp. Physiol. 2021, 320, R663–R674. [Google Scholar] [CrossRef]
- Meyer, F.; Paarmann, D.; D’Souza, M.; Olson, R.; Glass, E.; Kubal, M.; Paczian, T.; Rodriguez, A.; Stevens, R.; Wilke, A.; et al. The Metagenomics RAST Server—A Public Resource for the Automatic Phylogenetic and Functional Analysis of Metagenomes. BMC Bioinform. 2008, 9, 386. [Google Scholar] [CrossRef] [Green Version]
- Benjamini, Y.; Hochberg, Y. Controlling The False Discovery Rate—A Practical And Powerful Approach To Multiple Testing. Ournal R. Stat. Soc. Ser. B Methodol. 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Osaki, M.; Oshimura, M.; Ito, H. PI3K-Akt Pathway: Its Functions and Alterations in Human Cancer. Apoptosis 2004, 9, 667–676. [Google Scholar] [CrossRef]
- Paoli, A.; Mancin, L.; Bianco, A.; Thomas, E.; Mota, J.F.; Piccini, F. Ketogenic Diet and Microbiota: Friends or Enemies? Genes 2019, 10, 534. [Google Scholar] [CrossRef] [Green Version]
- Perry, R.J.; Peng, L.; Barry, N.A.; Cline, G.W.; Zhang, D.; Cardone, R.L.; Petersen, K.F.; Kibbey, R.G.; Goodman, A.L.; Shulman, G.I. Acetate Mediates a Microbiome-Brain-β-Cell Axis to Promote Metabolic Syndrome. Nature 2016, 534, 213–217. [Google Scholar] [CrossRef] [Green Version]
- Si, X.; Shang, W.; Zhou, Z.; Strappe, P.; Wang, B.; Bird, A.; Blanchard, C. Gut Microbiome-Induced Shift of Acetate to Butyrate Positively Manages Dysbiosis in High Fat Diet. Mol. Nutr. Food Res. 2018, 62, 1700670. [Google Scholar] [CrossRef] [PubMed]
- Naito, Y.; Uchiyama, K.; Takagi, T. A Next-Generation Beneficial Microbe: Akkermansia Muciniphila. J. Clin. Biochem. Nutr. 2018, 63, 33–35. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, M. Tu1852—Metformin Ameliorated Inflammatory Bowel Disease by Enhancing Akkermansia Muciniphila. Gastroenterology 2019, 156, S-1148. [Google Scholar] [CrossRef]
- Samaras, K.; Makkar, S.; Crawford, J.D.; Kochan, N.A.; Wen, W.; Draper, B.; Trollor, J.N.; Brodaty, H.; Sachdev, P.S. Metformin Use Is Associated With Slowed Cognitive Decline and Reduced Incident Dementia in Older Adults With Type 2 Diabetes: The Sydney Memory and Ageing Study. Diabetes Care 2020, 43, 2691–2701. [Google Scholar] [CrossRef]
- O’Brien, R.J.; Wong, P.C. Amyloid Precursor Protein Processing and Alzheimer’s Disease. Annu. Rev. Neurosci. 2011, 34, 185–204. [Google Scholar] [CrossRef] [Green Version]
- Taylor, M.K.; Sullivan, D.K.; Mahnken, J.D.; Burns, J.M.; Swerdlow, R.H. Feasibility and Efficacy Data from a Ketogenic Diet Intervention in Alzheimer’s Disease. Alzheimers Dement. Transl. Res. Clin. Interv. 2018, 4, 28–36. [Google Scholar] [CrossRef]
- Broom, G.M.; Shaw, I.C.; Rucklidge, J.J. The Ketogenic Diet as a Potential Treatment and Prevention Strategy for Alzheimer’s Disease. Nutrition 2019, 60, 118–121. [Google Scholar] [CrossRef]
- Rusek, M.; Pluta, R.; Ułamek-Kozioł, M.; Czuczwar, S.J. Ketogenic Diet in Alzheimer’s Disease. Int. J. Mol. Sci. 2019, 20, 3892. [Google Scholar] [CrossRef] [Green Version]
- Lilamand, M.; Mouton-Liger, F.; Paquet, C. Ketogenic Diet Therapy in Alzheimer’s Disease: An Updated Review. Curr. Opin. Clin. Nutr. Metab. Care 2021, 24, 372–378. [Google Scholar] [CrossRef]
- Liang, W.S.; Reiman, E.M.; Valla, J.; Dunckley, T.; Beach, T.G.; Grover, A.; Niedzielko, T.L.; Schneider, L.E.; Mastroeni, D.; Caselli, R.; et al. Alzheimer’s Disease Is Associated with Reduced Expression of Energy Metabolism Genes in Posterior Cingulate Neurons. Proc. Natl. Acad. Sci. USA 2008, 105, 4441–4446. [Google Scholar] [CrossRef] [Green Version]
- Mastroeni, D.; Khdour, O.M.; Delvaux, E.; Nolz, J.; Olsen, G.; Berchtold, N.; Cotman, C.; Hecht, S.M.; Coleman, P.D. Nuclear but Not Mitochondrial-Encoded Oxidative Phosphorylation Genes Are Altered in Aging, Mild Cognitive Impairment, and Alzheimer’s Disease. Alzheimers Dement. J. Alzheimers Assoc. 2017, 13, 510–519. [Google Scholar] [CrossRef] [Green Version]
- Duckworth, W.C.; Bennett, R.G.; Hamel, F.G. Insulin Degradation: Progress and Potential. Endocr. Rev. 1998, 19, 608–624. [Google Scholar] [CrossRef] [Green Version]
- Currenti, W.; Godos, J.; Castellano, S.; Mogavero, M.P.; Ferri, R.; Caraci, F.; Grosso, G.; Galvano, F. Time Restricted Feeding and Mental Health: A Review of Possible Mechanisms on Affective and Cognitive Disorders. Int. J. Food Sci. Nutr. 2021, 72, 723–733. [Google Scholar] [CrossRef]
- Newman, J.C.; Covarrubias, A.J.; Zhao, M.; Yu, X.; Gut, P.; Ng, C.-P.; Huang, Y.; Haldar, S.; Verdin, E. Ketogenic Diet Reduces Midlife Mortality and Improves Memory in Aging Mice. Cell Metab. 2017, 26, 547–557.e8. [Google Scholar] [CrossRef] [Green Version]
- Hernandez, A.; Truckenbrod, L.; Federico, Q.; Campos, K.; Moon, B.; Ferekides, N.; Hoppe, M.; D’Agostino, D.; Burke, S. Metabolic Switching Is Impaired by Aging and Facilitated by Ketosis Independent of Glycogen. Aging 2020, 12, 7963–7984. [Google Scholar] [CrossRef]
- Browning, J.D.; Baxter, J.; Satapati, S.; Burgess, S.C. The Effect of Short-Term Fasting on Liver and Skeletal Muscle Lipid, Glucose, and Energy Metabolism in Healthy Women and Men. J. Lipid Res. 2012, 53, 577–586. [Google Scholar] [CrossRef] [Green Version]
- Cani, P.D.; Van Hul, M.; Lefort, C.; Depommier, C.; Rastelli, M.; Everard, A. Microbial Regulation of Organismal Energy Homeostasis. Nat. Metab. 2019, 1, 34–46. [Google Scholar] [CrossRef]
- Rastelli, M.; Cani, P.D.; Knauf, C. The Gut Microbiome Influences Host Endocrine Functions. Endocr. Rev. 2019, 40, 1271–1284. [Google Scholar] [CrossRef]
- Park, S.; Zhang, T.; Wu, X.; Yi Qiu, J. Ketone Production by Ketogenic Diet and by Intermittent Fasting Has Different Effects on the Gut Microbiota and Disease Progression in an Alzheimer’s Disease Rat Model. J. Clin. Biochem. Nutr. 2020, 67, 188–198. [Google Scholar] [CrossRef] [Green Version]
- Wehrens, S.M.T.; Christou, S.; Isherwood, C.; Middleton, B.; Gibbs, M.A.; Archer, S.N.; Skene, D.J.; Johnston, J.D. Meal Timing Regulates the Human Circadian System. Curr. Biol. 2017, 27, 1768–1775.e3. [Google Scholar] [CrossRef] [Green Version]
- Kaczmarek, J.L.; Musaad, S.M.; Holscher, H.D. Time of Day and Eating Behaviors Are Associated with the Composition and Function of the Human Gastrointestinal Microbiota. Am. J. Clin. Nutr. 2017, 106, 1220–1231. [Google Scholar] [CrossRef] [Green Version]
- Daas, M.C.; de Roos, N.M. Intermittent Fasting Contributes to Aligned Circadian Rhythms through Interactions with the Gut Microbiome. Benef. Microbes 2021, 12, 147–161. [Google Scholar] [CrossRef]
- Davis, C.D. The Gut Microbiome and Its Role in Obesity. Nutr. Today 2016, 51, 167–174. [Google Scholar] [CrossRef] [Green Version]
- Li, G.; Xie, C.; Lu, S.; Nichols, R.G.; Tian, Y.; Li, L.; Patel, D.; Ma, Y.; Brocker, C.N.; Yan, T.; et al. Intermittent Fasting Promotes White Adipose Browning and Decreases Obesity by Shaping the Gut Microbiota. Cell Metab. 2017, 26, 672–685.e4. [Google Scholar] [CrossRef] [Green Version]




| 3 Diet Comparison x Age | Feeding Paradigm x Age | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| F (df, Error) | p | t | DF | p | F (df, Error) | p | t | DF | p | |||||
| Chao1 | Interaction | F (2, 27) = 1.339 | 0.2791 | TRF Control | 0.461 | 27 | 0.9566 | Interaction | F (1, 29) = 2.432 | 0.1297 | Young | 1.448 | 29 | 0.2917 |
| Age | F (1, 27) = 1.211 | 0.2808 | TRF Keto | 0.4362 | 27 | 0.9628 | Age | F (1, 29) = 2.512 | 0.1238 | Aged | 3.185 | 29 | 0.0069 | |
| Diet | F (2, 27) = 5.507 | 0.0099 | Ad libitum Chow | 1.836 | 27 | 0.2147 | Feeding Paradigm | F (1, 29) = 11.46 | 0.0021 | |||||
| Diversity (Inverse Simpson) | Interaction | F (2, 27) = 2.385 | 0.1112 | TRF Control | 1.413 | 27 | 0.426 | Interaction | F (1, 29) = 4.531 | 0.0419 | Young | 2.378 | 29 | 0.0479 |
| Age | F (1, 27) = 8.091 | 0.0084 | TRF Keto | 3.909 | 27 | 0.0017 | Age | F (1, 29) = 11.44 | 0.0021 | Aged | 4.673 | 29 | 0.0001 | |
| Diet | F (2, 27) = 14.15 | <0.0001 | Ad libitum Chow | 5.134 | 27 | <0.0001 | Feeding Paradigm | F (1, 29) = 26.28 | <0.0001 | |||||
| Evenness (Simpson) | Interaction | F (2, 27) = 0.2374 | 0.7903 | TRF Control | 1.469 | 27 | 0.393 | Interaction | F (1, 29) = 0.3164 | 0.5781 | Young | 1.374 | 29 | 0.3276 |
| Age | F (1, 27) = 1.989 | 0.1699 | TRF Keto | 1.367 | 27 | 0.4544 | Age | F (1, 29) = 2.327 | 0.138 | Aged | 1.84 | 29 | 0.1462 | |
| Diet | F (2, 27) = 3.718 | 0.0375 | Ad libitum Chow | 2.725 | 27 | 0.0331 | Feeding Paradigm | F (1, 29) = 5.265 | 0.0292 | |||||
| Dominance (Simpson) | Interaction | F (2, 27) = 0.4912 | 0.6173 | TRF Control | 2.065 | 27 | 0.1388 | Interaction | F (1, 29) = 0.009902 | 0.9214 | Young | 2.089 | 29 | 0.089 |
| Age | F (1, 27) = 1.947 | 0.1743 | TRF Keto | 1.432 | 27 | 0.4148 | Age | F (1, 29) = 1.460 | 0.2367 | Aged | 1.568 | 29 | 0.239 | |
| Diet | F (2, 27) = 5.718 | 0.0085 | Ad libitum Chow | 3.356 | 27 | 0.0071 | Feeding Paradigm | F (1, 29) = 6.425 | 0.0169 | |||||
| Rarity | Interaction | F (2, 27) = 0.1713 | 0.8435 | TRF Control | 0.6629 | 27 | 0.8845 | Interaction | F (1, 29) = 0.3796 | 0.5426 | Young | 1.512 | 29 | 0.2625 |
| Age | F (1, 27) = 2.187 | 0.1508 | TRF Keto | 2.493 | 27 | 0.0563 | Age | F (1, 29) = 1.495 | 0.2313 | Aged | 2.022 | 29 | 0.1023 | |
| Diet | F (2, 27) = 3.253 | 0.0543 | Ad libitum Chow | 1.782 | 27 | 0.2363 | Feeding Paradigm | F (1, 29) = 6.365 | 0.0174 | |||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hernandez, A.R.; Kemp, K.M.; Burke, S.N.; Buford, T.W.; Carter, C.S. Influence of Aging, Macronutrient Composition and Time-Restricted Feeding on the Fischer344 x Brown Norway Rat Gut Microbiota. Nutrients 2022, 14, 1758. https://doi.org/10.3390/nu14091758
Hernandez AR, Kemp KM, Burke SN, Buford TW, Carter CS. Influence of Aging, Macronutrient Composition and Time-Restricted Feeding on the Fischer344 x Brown Norway Rat Gut Microbiota. Nutrients. 2022; 14(9):1758. https://doi.org/10.3390/nu14091758
Chicago/Turabian StyleHernandez, Abbi R., Keri M. Kemp, Sara N. Burke, Thomas W. Buford, and Christy S. Carter. 2022. "Influence of Aging, Macronutrient Composition and Time-Restricted Feeding on the Fischer344 x Brown Norway Rat Gut Microbiota" Nutrients 14, no. 9: 1758. https://doi.org/10.3390/nu14091758
APA StyleHernandez, A. R., Kemp, K. M., Burke, S. N., Buford, T. W., & Carter, C. S. (2022). Influence of Aging, Macronutrient Composition and Time-Restricted Feeding on the Fischer344 x Brown Norway Rat Gut Microbiota. Nutrients, 14(9), 1758. https://doi.org/10.3390/nu14091758






