Sanguisorba officinalis L. Ameliorates Hepatic Steatosis and Fibrosis by Modulating Oxidative Stress, Fatty Acid Oxidation, and Gut Microbiota in CDAHFD-Induced Mice
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials and Extraction
2.2. Cell Culture
2.3. Animal Experiment
2.4. Biochemical Analysis
2.5. Histological Examination
2.6. Quantitative Reverse Transcription–Polymerase Chain Reaction (qRT-PCR)
2.7. Western Blotting
2.8. 16S rRNA Gene Sequencing of the Bacterial Community in Feces
2.9. Statistical Analysis
3. Results
3.1. Effects of SOEE on Plasma Lipid Profiling and Liver Injury Markers in CDAHFD-Fed Mice
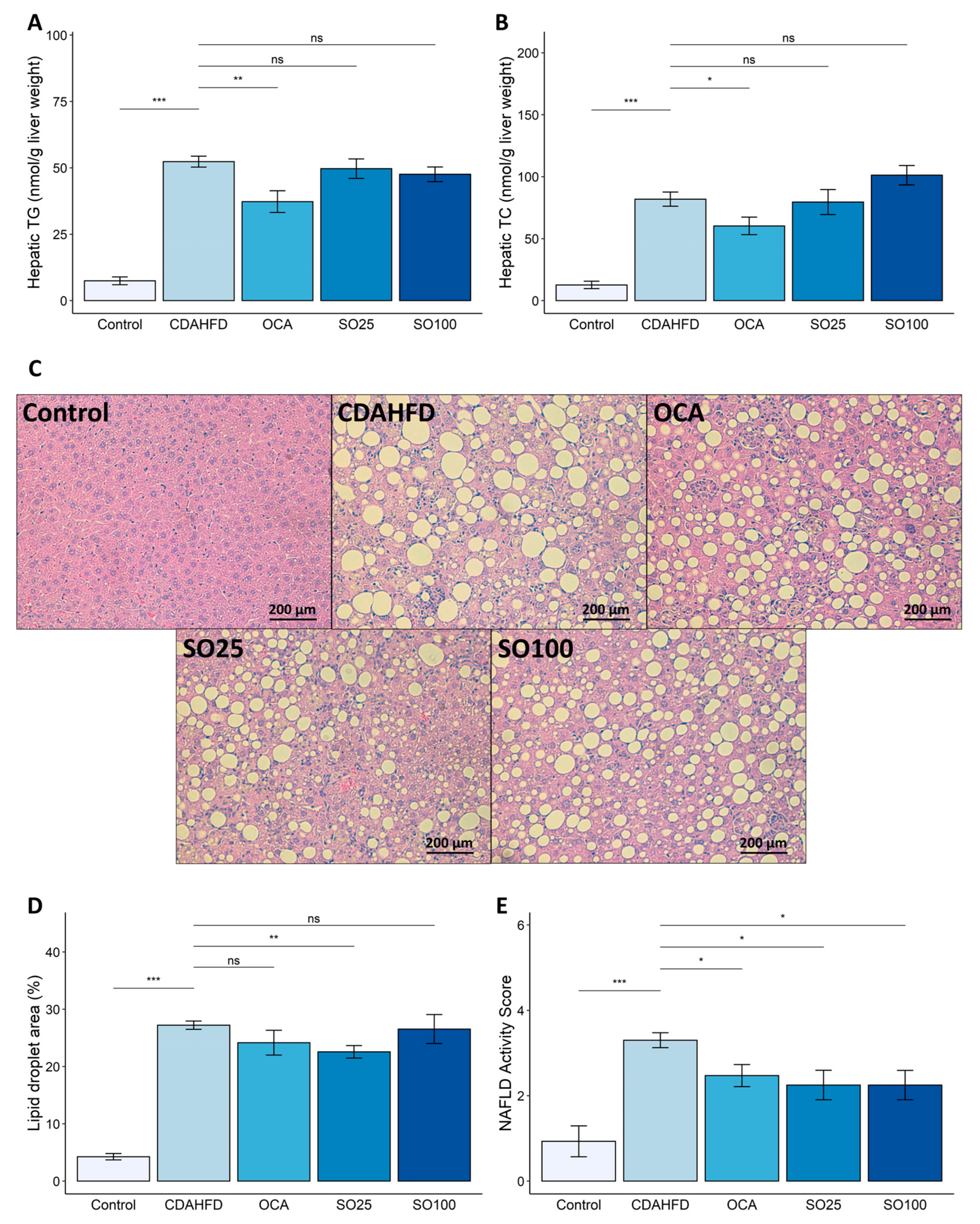
3.2. Effect of SOEE on NAFLD-Related Traits in CDAHFD-Fed Mice
3.3. Effects of SOEE on Oxidative-Stress- and Fatty-Acid-Oxidation-Related Markers in the Liver of CDAHFD-Fed Mice
3.4. Effects of SOEE on Hepatic Fibrosis in CDAHFD-Fed Mice
3.5. Effects of SOEE on Fibrosis-Related Gene Expression and Signaling Pathway in LX-2 Cells
3.6. Effects of SOEE on the Diversity and Composition of the Gut Microbiota
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Riazi, K.; Azhari, H.; Charette, J.H.; Underwood, F.E.; King, J.A.; Afshar, E.E.; Swain, M.G.; Congly, S.E.; Kaplan, G.G.; Shaheen, A.-A. The prevalence and incidence of NAFLD worldwide: A systematic review and meta-analysis. Lancet Gastroenterol. Hepatol. 2022, 7, 851–861. [Google Scholar] [CrossRef]
- Estes, C.; Razavi, H.; Loomba, R.; Younossi, Z.; Sanyal, A.J. Modeling the epidemic of nonalcoholic fatty liver disease demonstrates an exponential increase in burden of disease. Hepatol. Baltim. Md. 2018, 67, 123–133. [Google Scholar] [CrossRef]
- Anstee, Q.M.; Reeves, H.L.; Kotsiliti, E.; Govaere, O.; Heikenwalder, M. From NASH to HCC: Current concepts and future challenges. Nat. Rev. Gastroenterol. Hepatol. 2019, 16, 411–428. [Google Scholar] [CrossRef]
- Cobbina, E.; Akhlaghi, F. Non-Alcoholic Fatty Liver Disease (NAFLD)—Pathogenesis, Classification, and Effect on Drug Metabolizing Enzymes and Transporters. Drug Metab. Rev. 2017, 49, 197–211. [Google Scholar] [CrossRef]
- Ekstedt, M.; Franzén, L.E.; Mathiesen, U.L.; Thorelius, L.; Holmqvist, M.; Bodemar, G.; Kechagias, S. Long-term follow-up of patients with NAFLD and elevated liver enzymes. Hepatology 2006, 44, 865–873. [Google Scholar] [CrossRef]
- Sugasawa, T.; Ono, S.; Yonamine, M.; Fujita, S.-I.; Matsumoto, Y.; Aoki, K.; Nakano, T.; Tamai, S.; Yoshida, Y.; Kawakami, Y.; et al. One Week of CDAHFD Induces Steatohepatitis and Mitochondrial Dysfunction with Oxidative Stress in Liver. Int. J. Mol. Sci. 2021, 22, 5851. [Google Scholar] [CrossRef]
- Sutti, S.; Jindal, A.; Locatelli, I.; Vacchiano, M.; Gigliotti, L.; Bozzola, C.; Albano, E. Adaptive immune responses triggered by oxidative stress contribute to hepatic inflammation in NASH. Hepatol. Baltim. Md. 2014, 59, 886–897. [Google Scholar] [CrossRef]
- Caballero, F.; Fernández, A.; Matías, N.; Martínez, L.; Fucho, R.; Elena, M.; Caballeria, J.; Morales, A.; Fernández-Checa, J.C.; García-Ruiz, C. Specific contribution of methionine and choline in nutritional nonalcoholic steatohepatitis: Impact on mitochondrial S-adenosyl-L-methionine and glutathione. J. Biol. Chem. 2010, 285, 18528–18536. [Google Scholar] [CrossRef]
- Matsumoto, M.; Hada, N.; Sakamaki, Y.; Uno, A.; Shiga, T.; Tanaka, C.; Ito, T.; Katsume, A.; Sudoh, M. An improved mouse model that rapidly develops fibrosis in non-alcoholic steatohepatitis. Int. J. Exp. Pathol. 2013, 94, 93–103. [Google Scholar] [CrossRef]
- Weltman, M.; Farrell, G.; Liddle, C. Increased hepatocyte CYP2E1 expression in a rat nutritional model of hepatic steatosis with inflammation. Gastroenterology 1996, 111, 1645–1653. [Google Scholar] [CrossRef]
- Machado, M.V.; Michelotti, G.A.; Xie, G.; de Almeida, T.P.; Boursier, J.; Bohnic, B.; Guy, C.D.; Diehl, A.M. Mouse Models of Diet-Induced Nonalcoholic Steatohepatitis Reproduce the Heterogeneity of the Human Disease. PLoS ONE 2015, 10, e0127991. [Google Scholar] [CrossRef] [PubMed]
- Trevaskis, J.L.; Griffin, P.S.; Wittmer, C.; Neuschwander-Tetri, B.A.; Brunt, E.M.; Dolman, C.S.; Erickson, M.R.; Napora, J.; Parkes, D.G.; Roth, J.D. Glucagon-like peptide-1 receptor agonism improves metabolic, biochemical, and histopathological indices of nonalcoholic steatohepatitis in mice. Am. J. Physiol.-Gastrointest. Liver Physiol. 2012, 302, G762–G772. [Google Scholar] [CrossRef]
- Abe, N.; Tsuchida, T.; Yasuda, S.-I.; Oka, K. Dietary iron restriction leads to a reduction in hepatic fibrosis in a rat model of non-alcoholic steatohepatitis. Biol. Open 2019, 8, bio040519. [Google Scholar] [CrossRef] [PubMed]
- Lee, N.; Heo, Y.J.; Choi, S.-E.; Jeon, J.Y.; Han, S.J.; Kim, D.J.; Kang, Y.; Lee, K.W.; Kim, H.J. Hepatoprotective effects of gemigliptin and empagliflozin in a murine model of diet-induced non-alcoholic fatty liver disease. Biochem. Biophys. Res. Commun. 2022, 588, 154–160. [Google Scholar] [CrossRef] [PubMed]
- Suzuki-Kemuriyama, N.; Abe, A.; Nakane, S.; Uno, K.; Ogawa, S.; Watanabe, A.; Sano, R.; Yuki, M.; Miyajima, K.; Nakae, D. Extract of Siraitia grosvenorii (Luo Han Guo) protects against hepatic fibrosis in mice on a choline-deficient, methionine-lowered, L-amino acid-defined, high-fat diet without trans fatty acids. Fundam. Toxicol. Sci. 2021, 8, 135–145. [Google Scholar] [CrossRef]
- Zhang, H.; Chen, J.; Cen, Y. Burn wound healing potential of a polysaccharide from Sanguisorba officinalis L. in mice. Int. J. Biol. Macromol. 2018, 112, 862–867. [Google Scholar] [CrossRef]
- Yasueda, A.; Kayama, H.; Murohashi, M.; Nishimura, J.; Wakame, K.; Komatsu, K.-i.; Ogino, T.; Miyoshi, N.; Takahashi, H.; Uemura, M.; et al. Sanguisorba officinalis L. derived from herbal medicine prevents intestinal inflammation by inducing autophagy in macrophages. Sci. Rep. 2020, 10, 9972. [Google Scholar] [CrossRef]
- Zhang, L.; Koyyalamudi, S.R.; Jeong, S.C.; Reddy, N.; Smith, P.T.; Ananthan, R.; Longvah, T. Antioxidant and immunomodulatory activities of polysaccharides from the roots of Sanguisorba officinalis. Int. J. Biol. Macromol. 2012, 51, 1057–1062. [Google Scholar] [CrossRef]
- Yang, J.-H.; Hwang, Y.-H.; Gu, M.-J.; Cho, W.-K.; Ma, J.Y. Ethanol extracts of Sanguisorba officinalis L. suppress TNF-α/IFN-γ-induced pro-inflammatory chemokine production in HaCaT cells. Phytomedicine 2015, 22, 1262–1268. [Google Scholar] [CrossRef]
- Jung, D.-W.; Lee, O.-H.; Kang, I.-J. Sanguisorba officinalis L. Extracts Exert Antiobesity Effects in 3T3-L1 Adipocytes and C57BL/6J Mice Fed High-Fat Diets. J. Med. Food 2016, 19, 768–779. [Google Scholar] [CrossRef]
- Fernandes, F.H.A.; Batista, R.S.d.A.; de Medeiros, F.D.; Santos, F.S.; Medeiros, A.C.D. Development of a rapid and simple HPLC-UV method for determination of gallic acid in Schinopsis brasiliensis. Rev. Bras. Farmacogn. 2015, 25, 208–211. [Google Scholar] [CrossRef]
- Jang, E.; Kim, S.; Lee, N.-R.; Kim, H.; Chae, S.; Han, C.-W.; Kim, Y.; Lee, K.-T.; Kim, B.-J.; Inn, K.-S.; et al. Sanguisorba officinalis extract, ziyuglycoside I, and II exhibit antiviral effects against hepatitis B virus. Eur. J. Integr. Med. 2018, 20, 165–172. [Google Scholar] [CrossRef]
- Sun, H.; Wang, M.; Su, T.; Guo, P.; Tai, Y.; Cheng, H.; Zhu, Z.; Jiang, C.; Yan, S.; Wei, W.; et al. Ziyuglycoside I attenuates collagen-induced arthritis through inhibiting plasma cell expansion. J. Ethnopharmacol. 2022, 294, 115348. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.; Wang, K.; Zhang, K.; Huang, B.; Zhang, J.; Zhang, Y.; Zhu, L.; Zhou, B.; Zhou, F. Ziyuglycoside II inhibits the growth of human breast carcinoma MDA-MB-435 cells via cell cycle arrest and induction of apoptosis through the mitochondria dependent pathway. Int. J. Mol. Sci. 2013, 14, 18041–18055. [Google Scholar] [CrossRef]
- Reagan-Shaw, S.; Nihal, M.; Ahmad, N. Dose translation from animal to human studies revisited. FASEB J. Off. Publ. Fed. Am. Soc. Exp. Biol. 2008, 22, 659–661. [Google Scholar] [CrossRef] [PubMed]
- Heinemann, F.; Birk, G.; Stierstorfer, B. Deep learning enables pathologist-like scoring of NASH models. Sci. Rep. 2019, 9, 18454. [Google Scholar] [CrossRef]
- Kleiner, D.E.; Brunt, E.M.; Van Natta, M.; Behling, C.; Contos, M.J.; Cummings, O.W.; Ferrell, L.D.; Liu, Y.C.; Torbenson, M.S.; Unalp-Arida, A.; et al. Design and validation of a histological scoring system for nonalcoholic fatty liver disease. Hepatology 2005, 41, 1313–1321. [Google Scholar] [CrossRef]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef]
- Benjamini, Y.; Hochberg, Y. Controlling the False Discovery Rate—A Practical and Powerful Approach to Multiple Testing. J. R. Statst. Soc. B 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Kolde, R.; Franzosa, E.A.; Rahnavard, G.; Hall, A.B.; Vlamakis, H.; Stevens, C.; Daly, M.J.; Xavier, R.J.; Huttenhower, C. Host genetic variation and its microbiome interactions within the Human Microbiome Project. Genome Med. 2018, 10, 6. [Google Scholar] [CrossRef] [PubMed]
- Fujii, J.; Homma, T.; Kobayashi, S.; Seo, H.G. Mutual interaction between oxidative stress and endoplasmic reticulum stress in the pathogenesis of diseases specifically focusing on non-alcoholic fatty liver disease. World J. Biol. Chem. 2018, 9, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Tian, R.; She, Z.; Cai, J.; Li, H. Role of oxidative stress in the pathogenesis of nonalcoholic fatty liver disease. Free Radic. Biol. Med. 2020, 152, 116–141. [Google Scholar] [CrossRef]
- Masarone, M.; Rosato, V.; Dallio, M.; Gravina, A.G.; Aglitti, A.; Loguercio, C.; Federico, A.; Persico, M. Role of Oxidative Stress in Pathophysiology of Nonalcoholic Fatty Liver Disease. Oxidative Med. Cell. Longev. 2018, 2018, 9547613. [Google Scholar] [CrossRef]
- Tilg, H.; Moschen, A.R. Evolution of inflammation in nonalcoholic fatty liver disease: The multiple parallel hits hypothesis. Hepatology 2010, 52, 1836–1846. [Google Scholar] [CrossRef]
- Ighodaro, O.M.; Akinloye, O.A. First line defence antioxidants-superoxide dismutase (SOD), catalase (CAT) and glutathione peroxidase (GPX): Their fundamental role in the entire antioxidant defence grid. Alex. J. Med. 2018, 54, 287–293. [Google Scholar] [CrossRef]
- Rao, M.S.; Reddy, J.K. Peroxisomal beta-oxidation and steatohepatitis. Semin. Liver Dis. 2001, 21, 43–55. [Google Scholar] [CrossRef]
- Reddy, J.K.; Hashimoto, T. Peroxisomal beta-oxidation and peroxisome proliferator-activated receptor alpha: An adaptive metabolic system. Annu. Rev. Nutr. 2001, 21, 193–230. [Google Scholar] [CrossRef]
- Pawlak, M.; Lefebvre, P.; Staels, B. Molecular mechanism of PPARα action and its impact on lipid metabolism, inflammation and fibrosis in non-alcoholic fatty liver disease. J. Hepatol. 2015, 62, 720–733. [Google Scholar] [CrossRef]
- Fondevila, M.F.; Fernandez, U.; Heras, V.; Parracho, T.; Gonzalez-Rellan, M.J.; Novoa, E.; Porteiro, B.; Alonso, C.; Mayo, R.; da Silva Lima, N.; et al. Inhibition of carnitine palmitoyltransferase 1A in hepatic stellate cells protects against fibrosis. J. Hepatol. 2022, 77, 15–28. [Google Scholar] [CrossRef]
- Mead, J.; Irvine, S.; Ramji, D. Lipoprotein lipase: Structure, function, regulation, and role in disease. J. Mol. Med. 2002, 80, 753–769. [Google Scholar] [CrossRef]
- Ratziu, V. A position statement on NAFLD/NASH based on the EASL 2009 special conference. J. Hepatol. 2010, 53, 372–384. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Han, X.; Yin, L.; Xu, L.; Qi, Y.; Xu, Y.; Sun, H.; Lin, Y.; Liu, K.; Peng, J. Potent effects of dioscin against liver fibrosis. Sci. Rep. 2015, 5, 9713. [Google Scholar] [CrossRef]
- Inagaki, Y.; Okazaki, I. Emerging insights into Transforming growth factor β Smad signal in hepatic fibrogenesis. Gut 2007, 56, 284–292. [Google Scholar] [CrossRef] [PubMed]
- Breitkopf, K.; Godoy, P.; Ciuclan, L.; Singer, M.V.; Dooley, S. TGF-beta/Smad signaling in the injured liver. Z. Fur Gastroenterol. 2006, 44, 57–66. [Google Scholar] [CrossRef] [PubMed]
- de Vos, W.M.; Tilg, H.; Van Hul, M.; Cani, P.D. Gut microbiome and health: Mechanistic insights. Gut 2022, 71, 1020–1032. [Google Scholar] [CrossRef]
- Loomba, R.; Seguritan, V.; Li, W.; Long, T.; Klitgord, N.; Bhatt, A.; Dulai, P.S.; Caussy, C.; Bettencourt, R.; Highlander, S.K.; et al. Gut Microbiome-Based Metagenomic Signature for Non-invasive Detection of Advanced Fibrosis in Human Nonalcoholic Fatty Liver Disease. Cell Metab. 2019, 30, 607. [Google Scholar] [CrossRef]
- Neuschwander-Tetri, B.A.; Loomba, R.; Sanyal, A.J.; Lavine, J.E.; Van Natta, M.L.; Abdelmalek, M.F.; Chalasani, N.; Dasarathy, S.; Diehl, A.M.; Hameed, B.; et al. Farnesoid X nuclear receptor ligand obeticholic acid for non-cirrhotic, non-alcoholic steatohepatitis (FLINT): A multicentre, randomised, placebo-controlled trial. Lancet 2015, 385, 956–965. [Google Scholar] [CrossRef]
- Staels, B.; Rubenstrunk, A.; Noel, B.; Rigou, G.; Delataille, P.; Millatt, L.J.; Baron, M.; Lucas, A.; Tailleux, A.; Hum, D.W.; et al. Hepatoprotective effects of the dual peroxisome proliferator-activated receptor alpha/delta agonist, GFT505, in rodent models of nonalcoholic fatty liver disease/nonalcoholic steatohepatitis. Hepatology 2013, 58, 1941–1952. [Google Scholar] [CrossRef]
- Lefere, S.; Puengel, T.; Hundertmark, J.; Penners, C.; Frank, A.K.; Guillot, A.; de Muynck, K.; Heymann, F.; Adarbes, V.; Defrêne, E.; et al. Differential effects of selective- and pan-PPAR agonists on experimental steatohepatitis and hepatic macrophages☆. J. Hepatol. 2020, 73, 757–770. [Google Scholar] [CrossRef]
- Day, C.P.; James, O.F.W. Steatohepatitis: A tale of two “hits”? Gastroenterology 1998, 114, 842–845. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.-r.; Wang, H.-f.; Zuo, T.-c.; Guan, L.-l.; Dai, N. Salidroside alleviates oxidative stress in the liver with non- alcoholic steatohepatitis in rats. BMC Pharmacol. Toxicol. 2016, 17, 16. [Google Scholar] [CrossRef] [PubMed]
- Pryor, W.A.; Stanley, J.P. Suggested mechanism for the production of malonaldehyde during the autoxidation of polyunsaturated fatty acids. Nonenzymic production of prostaglandin endoperoxides during autoxidation. J. Org. Chem. 1975, 40, 3615–3617. [Google Scholar] [CrossRef] [PubMed]
- Hwang, I.; Uddin, M.J.; Pak, E.S.; Kang, H.; Jin, E.-J.; Jo, S.; Kang, D.; Lee, H.; Ha, H. The impaired redox balance in peroxisomes of catalase knockout mice accelerates nonalcoholic fatty liver disease through endoplasmic reticulum stress. Free. Radic. Biol. Med. 2020, 148, 22–32. [Google Scholar] [CrossRef] [PubMed]
- Fujita, K.; Nozaki, Y.; Wada, K.; Yoneda, M.; Fujimoto, Y.; Fujitake, M.; Endo, H.; Takahashi, H.; Inamori, M.; Kobayashi, N.; et al. Dysfunctional very-low-density lipoprotein synthesis and release is a key factor in nonalcoholic steatohepatitis pathogenesis. Hepatology 2009, 50, 772–780. [Google Scholar] [CrossRef]
- Xu, J.; Xiao, G.; Trujillo, C.; Chang, V.; Blanco, L.; Joseph, S.B.; Bassilian, S.; Saad, M.F.; Tontonoz, P.; Lee, W.N.P.; et al. Peroxisome Proliferator-activated Receptor α (PPARα) Influences Substrate Utilization for Hepatic Glucose Production. J. Biol. Chem. 2002, 277, 50237–50244. [Google Scholar] [CrossRef]
- Régnier, M.; Polizzi, A.; Smati, S.; Lukowicz, C.; Fougerat, A.; Lippi, Y.; Fouché, E.; Lasserre, F.; Naylies, C.; Bétoulières, C.; et al. Hepatocyte-specific deletion of Pparα promotes NAFLD in the context of obesity. Sci. Rep. 2020, 10, 6489. [Google Scholar] [CrossRef]
- Reeves, H.L.; Friedman, S.L. Activation of hepatic stellate cells--a key issue in liver fibrosis. Front. Biosci. A J. Virtual Libr. 2002, 7, d808–d826. [Google Scholar] [CrossRef]
- Puche, J.E.; Saiman, Y.; Friedman, S.L. Hepatic stellate cells and liver fibrosis. Compr. Physiol. 2013, 3, 1473–1492. [Google Scholar] [CrossRef]
- Yin, C.; Evason, K.J.; Asahina, K.; Stainier, D.Y.R. Hepatic stellate cells in liver development, regeneration, and cancer. J. Clin. Investig. 2013, 123, 1902–1910. [Google Scholar] [CrossRef]
- Yang, C.; Zeisberg, M.; Mosterman, B.; Sudhakar, A.; Yerramalla, U.; Holthaus, K.; Xu, L.; Eng, F.; Afdhal, N.; Kalluri, R. Liver fibrosis: Insights into migration of hepatic stellate cells in response to extracellular matrix and growth factors. Gastroenterology 2003, 124, 147–159. [Google Scholar] [CrossRef] [PubMed]
- Ender, F.; Godl, K.; Wenzl, S.; Sumper, M. Evidence for Autocatalytic Cross-Linking of Hydroxyproline-Rich Glycoproteins during Extracellular Matrix Assembly in Volvox. Plant Cell 2002, 14, 1147–1160. [Google Scholar] [CrossRef]
- Di Lullo, G.A.; Sweeney, S.M.; Körkkö, J.; Ala-Kokko, L.; San Antonio, J.D. Mapping the Ligand-binding Sites and Disease-associated Mutations on the Most Abundant Protein in the Human, Type I Collagen. J. Biol. Chem. 2002, 277, 4223–4231. [Google Scholar] [CrossRef] [PubMed]
- Dooley, S.; Delvoux, B.; Streckert, M.; Bonzel, L.; Stopa, M.; ten Dijke, P.; Gressner, A.M. Transforming growth factor β signal transduction in hepatic stellate cells via Smad2/3 phosphorylation, a pathway that is abrogated during in vitro progression to myofibroblasts. FEBS Lett. 2001, 502, 4–10. [Google Scholar] [CrossRef] [PubMed]
- Gressner, O.A.; Lahme, B.; Demirci, I.; Gressner, A.M.; Weiskirchen, R. Differential effects of TGF-β on connective tissue growth factor (CTGF/CCN2) expression in hepatic stellate cells and hepatocytes. J. Hepatol. 2007, 47, 699–710. [Google Scholar] [CrossRef]
- Jung, H.J.; Cho, K.; Kim, S.Y.; Seong, J.K.; Oh, S.H. Ethanol extract of Pharbitis nil ameliorates liver fibrosis through regulation of the TGFbeta1-SMAD2/3 pathway. J. Ethnopharmacol. 2022, 294, 115370. [Google Scholar] [CrossRef]
- Liu, Z.; Xu, B.; Ding, Y.; Ding, X.; Yang, Z. Guizhi Fuling pill attenuates liver fibrosis in vitro and in vivo via inhibiting TGF-beta1/Smad2/3 and activating IFN-gamma/Smad7 signaling pathways. Bioengineered 2022, 13, 9357–9368. [Google Scholar] [CrossRef]
- Ahn, S.B.; Jun, D.W.; Kang, B.K.; Lim, J.H.; Lim, S.; Chung, M.J. Randomized, Double-blind, Placebo-controlled Study of a Multispecies Probiotic Mixture in Nonalcoholic Fatty Liver Disease. Sci. Rep. 2019, 9, 5688. [Google Scholar] [CrossRef]
- Lee, N.Y.; Shin, M.J.; Youn, G.S.; Yoon, S.J.; Choi, Y.R.; Kim, H.S.; Gupta, H.; Han, S.H.; Kim, B.K.; Lee, D.Y.; et al. Lactobacillus attenuates progression of nonalcoholic fatty liver disease by lowering cholesterol and steatosis. Clin. Mol. Hepatol. 2021, 27, 110–124. [Google Scholar] [CrossRef]
- Jang, H.R.; Park, H.J.; Kang, D.; Chung, H.; Nam, M.H.; Lee, Y.; Park, J.H.; Lee, H.Y. A protective mechanism of probiotic Lactobacillus against hepatic steatosis via reducing host intestinal fatty acid absorption. Exp. Mol. Med. 2019, 51, 1–14. [Google Scholar] [CrossRef]
- Nguyen, H.T.; Gu, M.; Werlinger, P.; Cho, J.H.; Cheng, J.; Suh, J.W. Lactobacillus sakei MJM60958 as a Potential Probiotic Alleviated Non-Alcoholic Fatty Liver Disease in Mice Fed a High-Fat Diet by Modulating Lipid Metabolism, Inflammation, and Gut Microbiota. Int. J. Mol. Sci. 2022, 23, 3436. [Google Scholar] [CrossRef]
- Cao, F.; Ding, Q.; Zhuge, H.; Lai, S.; Chang, K.; Le, C.; Yang, G.; Valencak, T.G.; Li, S.; Ren, D. Lactobacillus plantarum ZJUIDS14 alleviates non-alcoholic fatty liver disease in mice in association with modulation in the gut microbiota. Front. Nutr. 2022, 9, 1071284. [Google Scholar] [CrossRef] [PubMed]
- Okubo, H.; Sakoda, H.; Kushiyama, A.; Fujishiro, M.; Nakatsu, Y.; Fukushima, T.; Matsunaga, Y.; Kamata, H.; Asahara, T.; Yoshida, Y.; et al. Lactobacillus casei strain Shirota protects against nonalcoholic steatohepatitis development in a rodent model. Am. J. Physiol. Gastrointest. Liver Physiol. 2013, 305, G911–G918. [Google Scholar] [CrossRef]
- Yuan, G.-h.; Zhang, Z.; Gao, X.-s.; Zhu, J.; Guo, W.-h.; Wang, L.; Ding, P.; Jiang, P.; Li, L. Gut microbiota-mediated tributyltin-induced metabolic disorder in rats. RSC Adv. 2020, 10, 43619–43628. [Google Scholar] [CrossRef] [PubMed]
- Raman, M.; Ahmed, I.; Gillevet, P.M.; Probert, C.S.; Ratcliffe, N.M.; Smith, S.; Greenwood, R.; Sikaroodi, M.; Lam, V.; Crotty, P.; et al. Fecal Microbiome and Volatile Organic Compound Metabolome in Obese Humans with Nonalcoholic Fatty Liver Disease. Clin. Gastroenterol. Hepatol. 2013, 11, 868–875. [Google Scholar] [CrossRef] [PubMed]





| Genes | Direction | Primer (5′→3′) | Species |
|---|---|---|---|
| Col1a1 | Forward | AGC ACG TCT GGT TTG GAG AG | Mouse |
| Col1a1 | Reverse | GAC ATT AGG CGC AGG AAG GT | Mouse |
| Col3a1 | Forward | GTG GAC ATT GGC CCT GTT TG | Mouse |
| Col3a1 | Reverse | AGT TGG TCA CTT GCA CTG GT | Mouse |
| Tgfb1 | Forward | GTG GCT GAA CCA AGG AGA CG | Mouse |
| Tgfb1 | Reverse | GTT TGG GGC TGA TCC CGT TG | Mouse |
| Timp1 | Forward | TTA TTC TCC ACT GTG CAG CCC | Mouse |
| Timp1 | Reverse | ACA AGA GGA TGC CAG ATG CC | Mouse |
| Acta2 | Forward | TCC AGC CAT CTT TCA TTG GGA | Mouse |
| Acta2 | Reverse | CCC CTG ACA GGA CGT TGT TA | Mouse |
| Ppara | Forward | GAA CTG ACG TTT GTG GCT GG | Mouse |
| Ppara | Reverse | GCT CTC TGT GTC CAC CAT GT | Mouse |
| Cpt1a | Forward | ACT CCG CTC GCT CAT TCC | Mouse |
| Cpt1a | Reverse | GAC TGT GAA CTG GAA GGC CA | Mouse |
| Lpl | Forward | GTG GAC ATC GGA GAA CTG CT | Mouse |
| Lpl | Reverse | CCT CTC GAT GAC GAA GCT GG | Mouse |
| Sod1 | Forward | GGG AAG CAT GGC GAT GAA AG | Mouse |
| Sod1 | Reverse | GCC TTC TGC TCG AAG TGG AT | Mouse |
| Cat | Forward | CAA GAT TGC CTT CTC CGG GT | Mouse |
| Cat | Reverse | ATG GTG TAG GAT TGC GGA GC | Mouse |
| Gpx1 | Forward | AGT CCA CCG TGT ATG CCT TC | Mouse |
| Gpx1 | Reverse | CCT CAG AGA GAC GCG ACA TT | Mouse |
| β-actin | Forward | CAT TGC TGA CAG GAT GCA GAA GG | Mouse |
| β-actin | Reverse | TGC TGG AAG GTG GAC AGT GAG G | Mouse |
| ACTA2 | Forward | GCC AAG CAC TGT CAG GAA | Human |
| ACTA2 | Reverse | ATT GTC ACA CAC CAA GGC A | Human |
| COL1A1 | Forward | ATG GAG CTC CTG GTC AGA T | Human |
| COL1A1 | Reverse | GTA GCA CCA TCA TTT CCA CG | Human |
| COL3A1 | Forward | GCT CTG CTT CAT CCC ACT AT | Human |
| COL3A1 | Reverse | CGC ATA GGA CTG ACC AAG AT | Human |
| TIMP1 | Forward | CTC TGA AAA GGG CTT CCA GTC | Human |
| TIMP1 | Reverse | AGG ATT CAG GCT ATC TGG GAC | Human |
| GAPDH | Forward | CAG CCG CAT CTT CTT TTG CG | Human |
| GAPDH | Reverse | TCC GTT GAC TCC GAC CTT CA | Human |
| Group | Body Weight (g) | Liver Weight (g) | ALT (IU/L) | AST (IU/L) | Plasma TG (mM/L) | Plasma TC (mM/L) |
|---|---|---|---|---|---|---|
| Control | *** 30.87 ± 3.38 | *** 1.15 ± 0.18 | *** 1.12 ± 0.56 | *** 5.49 ± 2.09 | 0.36 ± 0.11 | * 0.99 ± 0.14 |
| CDAHFD | 24.16 ± 2.34 | 1.67 ± 0.26 | 17.85 ± 2.58 | 13.13 ± 2.50 | 0.40 ± 0.10 | 0.83 ± 0.17 |
| OCA | 23.32 ± 1.99 | ** 2.04 ± 0.20 | # 24.45 ± 10.28 | * 25.69 ± 15.03 | *** 0.23 ± 0.07 | # 0.67 ± 0.17 |
| SO25 | 24.19 ± 1.81 | 1.68 ± 0.29 | 15.84 ± 4.41 | # 10.36 ± 3.48 | * 0.30 ± 0.06 | 0.77 ± 0.15 |
| SO100 | 24.50 ± 1.15 | 1.67 ± 0.19 | 18.24 ± 3.52 | 11.31 ± 3.11 | ** 0.27 ± 0.07 | 0.72 ± 0.11 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nam, Y.; Kim, M.; Erdenebileg, S.; Cha, K.H.; Ryu, D.H.; Kim, H.Y.; Lee, S.H.; Jung, J.H.; Nho, C.W. Sanguisorba officinalis L. Ameliorates Hepatic Steatosis and Fibrosis by Modulating Oxidative Stress, Fatty Acid Oxidation, and Gut Microbiota in CDAHFD-Induced Mice. Nutrients 2023, 15, 3779. https://doi.org/10.3390/nu15173779
Nam Y, Kim M, Erdenebileg S, Cha KH, Ryu DH, Kim HY, Lee SH, Jung JH, Nho CW. Sanguisorba officinalis L. Ameliorates Hepatic Steatosis and Fibrosis by Modulating Oxidative Stress, Fatty Acid Oxidation, and Gut Microbiota in CDAHFD-Induced Mice. Nutrients. 2023; 15(17):3779. https://doi.org/10.3390/nu15173779
Chicago/Turabian StyleNam, Yunseong, Myungsuk Kim, Saruul Erdenebileg, Kwang Hyun Cha, Da Hye Ryu, Ho Youn Kim, Su Hyeon Lee, Je Hyeong Jung, and Chu Won Nho. 2023. "Sanguisorba officinalis L. Ameliorates Hepatic Steatosis and Fibrosis by Modulating Oxidative Stress, Fatty Acid Oxidation, and Gut Microbiota in CDAHFD-Induced Mice" Nutrients 15, no. 17: 3779. https://doi.org/10.3390/nu15173779
APA StyleNam, Y., Kim, M., Erdenebileg, S., Cha, K. H., Ryu, D. H., Kim, H. Y., Lee, S. H., Jung, J. H., & Nho, C. W. (2023). Sanguisorba officinalis L. Ameliorates Hepatic Steatosis and Fibrosis by Modulating Oxidative Stress, Fatty Acid Oxidation, and Gut Microbiota in CDAHFD-Induced Mice. Nutrients, 15(17), 3779. https://doi.org/10.3390/nu15173779









