Weight Loss Promotion in Individuals with Obesity through Gut Microbiota Alterations with a Multiphase Modified Ketogenic Diet
Abstract
:1. Introduction
2. Materials and Methods
2.1. Subjects
2.2. Anthropometric Assessment and Blood Chemistry
2.3. Sample Collection
2.4. Metagenomic Data Processing and Quality Control
2.5. Analysis of Gut Microbiota Species and Functions
2.6. Machine Learning Analysis
2.7. Association Analysis of Gut Microbiota and Physiological Indicators
2.8. Differential Analysis of Metabolic Pathways of Gut Microbiota
2.9. Correlation Analysis between Metabolic Pathways of Gut Microbiota and Physiological Indicators
2.10. Statistical Analysis
3. Results
3.1. MDP-i-KD Changes Biochemical Measurements and Anthropometric Characteristics of Obese Subjects
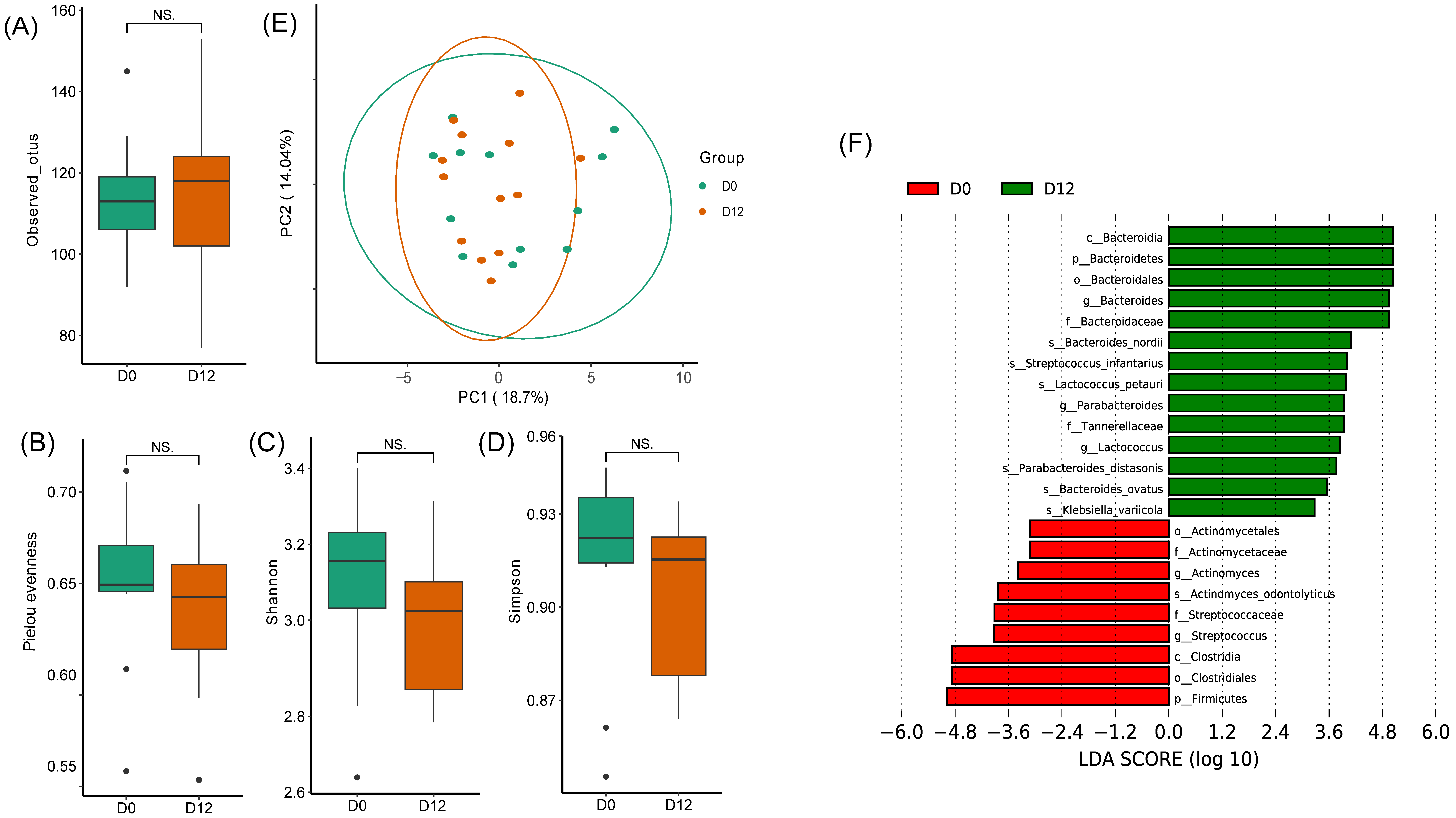
3.2. Effect of the MDP-i-KD on Gut Microbiota
3.3. Machine Learning Identifies Key Microbial Changes before and after MDP-i-KD Intervention
3.4. Species-Level Co-Occurrence Network Changes after the Intervention
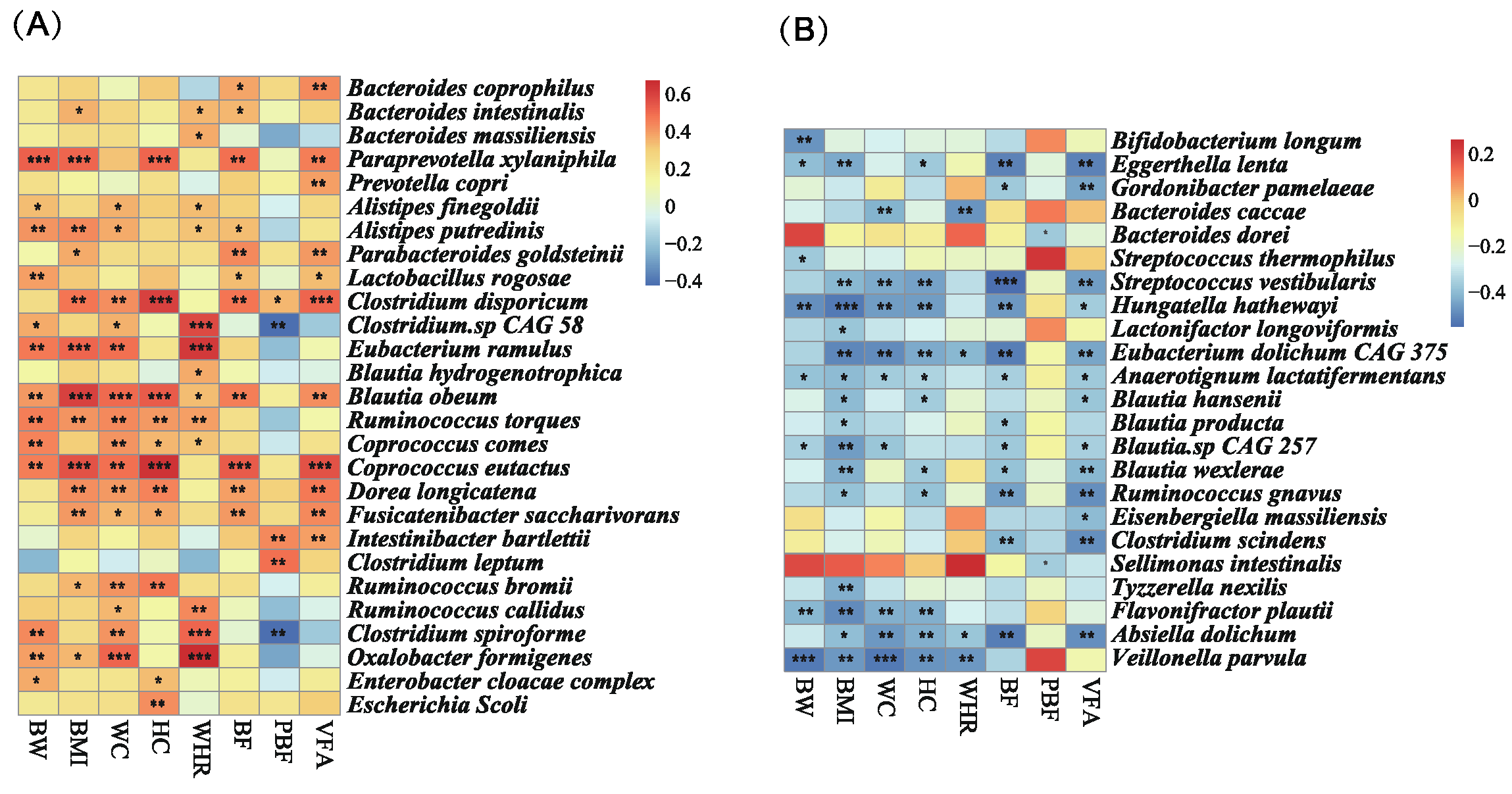
3.5. MDP-i-KD Alters the Correlation between Intestinal Microbiota Abundance and Physiological Indicators
3.6. The MDP-i-KD Changes the Gut Microbiota Profile of the Obese Subjects
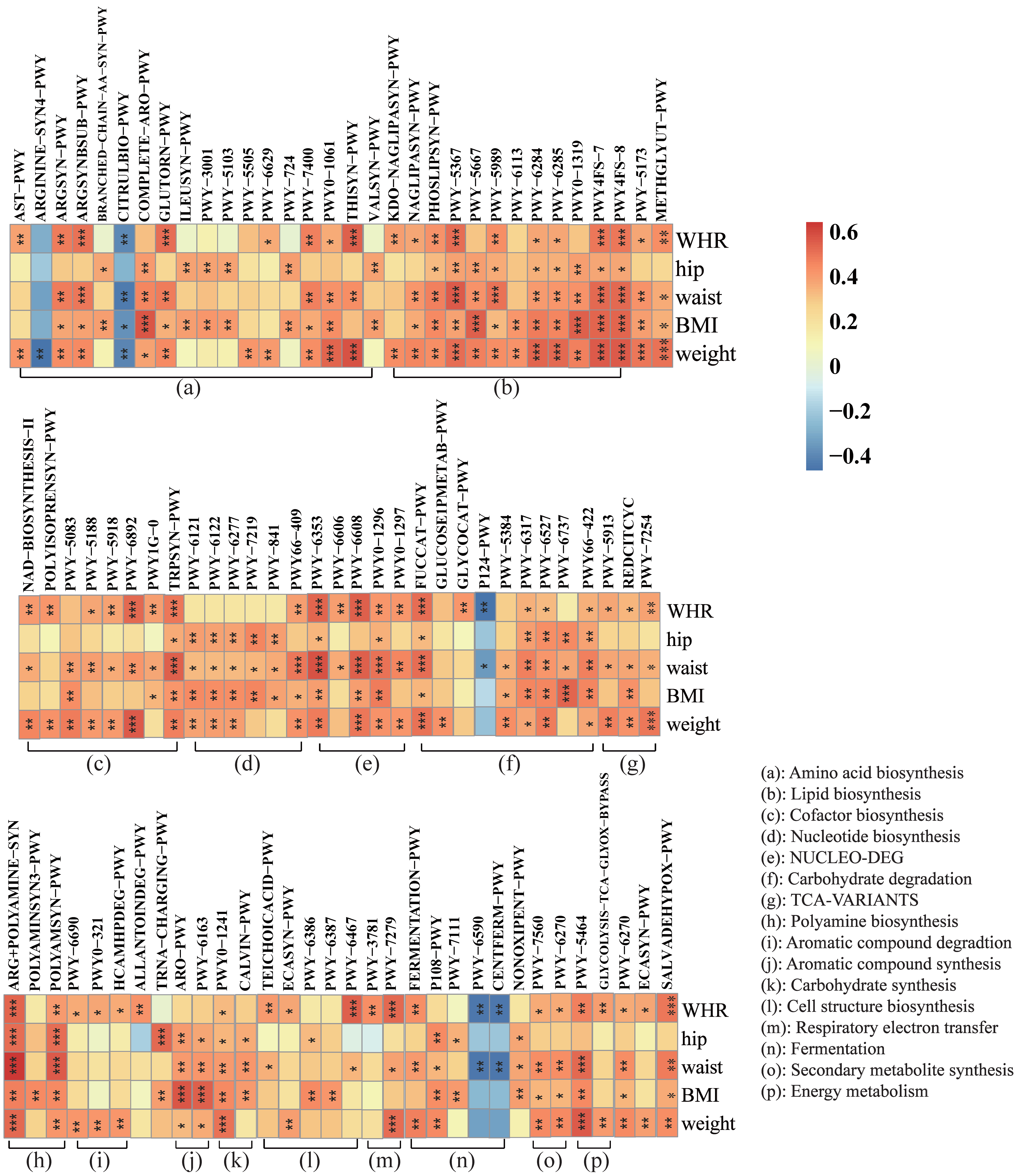
3.7. The MDP-i-KD Alters Metabolic Pathways and Correlates with Physiological Indicators Studied
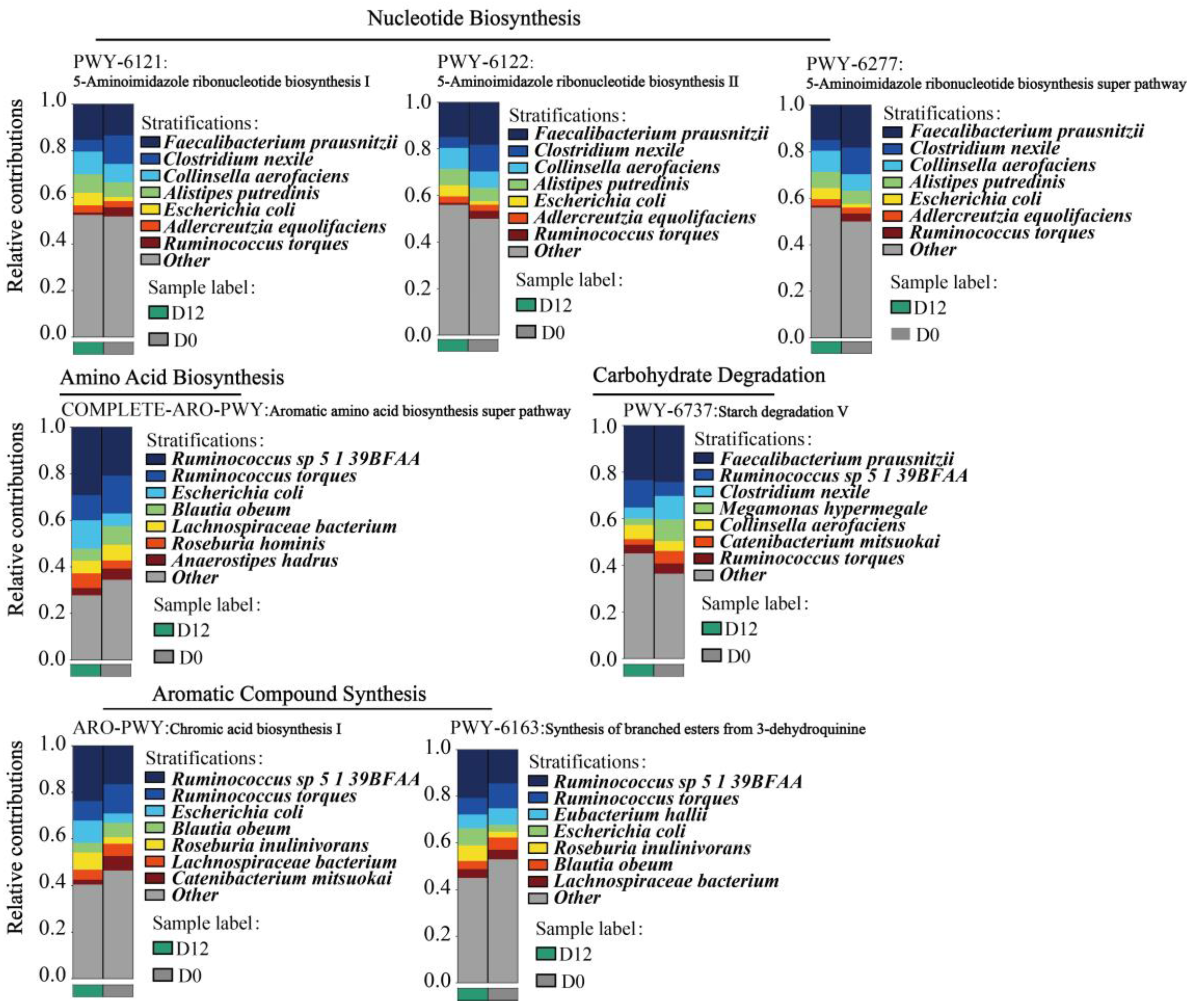
3.8. Species Contribution of the Metabolic Pathways of the Gut Microbiota
3.8.1. Analysis of Species Contribution to Key Metabolic Pathways
3.8.2. B. obeum and R. torques Are Involved in the Regulation of Various Metabolic Activities in the Gut of Subjects with Obesity
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Di Angelantonio, E.; Bhupathiraju, S.N.; Wormser, D.; Gao, P.; Kaptoge, S.; Berrington de Gonzalez, A.; Cairns, B.J.; Huxley, R.; Jackson, C.L.; Joshy, G.; et al. Body-mass index and all-cause mortality: Individual-participant-data meta-analysis of 239 prospective studies in four continents. Lancet 2016, 388, 776–786. [Google Scholar] [CrossRef] [PubMed]
- GBD 2015 Obesity Collaborators. Health Effects of Overweight and Obesity in 195 Countries over 25 Years. N. Engl. J. Med. 2017, 377, 13–27. [Google Scholar] [CrossRef] [PubMed]
- Hoffman, D.J.; Powell, T.L.; Barrett, E.S.; Hardy, D.B. Developmental origins of metabolic diseases. Physiol. Rev. 2021, 101, 739–795. [Google Scholar] [CrossRef]
- Ridaura, V.K.; Faith, J.J.; Rey, F.E.; Cheng, J.Y.; Duncan, A.E.; Kau, A.L.; Griffin, N.W.; Lombard, V.; Henrissat, B.; Bain, J.R.; et al. Gut Microbiota from Twins Discordant for Obesity Modulate Metabolism in Mice. Science 2013, 341, 1241214. [Google Scholar] [CrossRef] [PubMed]
- Perdomo, C.M.; Cohen, R.V.; Sumithran, P.; Clément, K.; Frühbeck, G. Contemporary medical, device, and surgical therapies for obesity in adults. Lancet 2023, 401, 1116–1130. [Google Scholar] [CrossRef] [PubMed]
- Bray, G.A.; Ryan, D.H. Evidence-based weight loss interventions: Individualized treatment options to maximize patient outcomes. Diabetes Obes. Metab. 2021, 23, 50–62. [Google Scholar] [CrossRef]
- Carmody, R.N.; Bisanz, J.E. Roles of the gut microbiome in weight management. Nat. Rev. Microbiol. 2023, 21, 535–550. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, M.; Chen, J.; Li, Y.; Kuang, Z.; Dende, C.; Raj, P.; Quinn, G.; Hu, Z.; Srinivasan, T.; et al. The gut microbiota reprograms intestinal lipid metabolism through long noncoding RNA Snhg9. Science 2023, 381, 851–857. [Google Scholar] [CrossRef]
- Anhê, F.F.; Zlitni, S.; Zhang, S.Y.; Choi, B.S.; Chen, C.Y.; Foley, K.P.; Barra, N.G.; Surette, M.G.; Biertho, L.; Richard, D.; et al. Human gut microbiota after bariatric surgery alters intestinal morphology and glucose absorption in mice independently of obesity. Gut 2023, 72, 460–471. [Google Scholar] [CrossRef]
- Qi, Q.; Li, J.; Yu, B.; Moon, J.Y.; Chai, J.C.; Merino, J.; Hu, J.; Ruiz-Canela, M.; Rebholz, C.; Wang, Z.; et al. Host and gut microbial tryptophan metabolism and type 2 diabetes: An integrative analysis of host genetics, diet, gut microbiome and circulating metabolites in cohort studies. Gut 2022, 71, 1095–1105. [Google Scholar] [CrossRef]
- Agus, A.; Clément, K.; Sokol, H. Gut microbiota-derived metabolites as central regulators in metabolic disorders. Gut 2021, 70, 1174. [Google Scholar] [CrossRef] [PubMed]
- Martin, A.M.; Sun, E.W.; Rogers, G.B.; Keating, D.J. The Influence of the Gut Microbiome on Host Metabolism Through the Regulation of Gut Hormone Release. Front. Physiol. 2019, 10, 428. [Google Scholar] [CrossRef] [PubMed]
- Fan, Y.; Pedersen, O. Gut microbiota in human metabolic health and disease. Nat. Rev. Microbiol. 2021, 19, 55–71. [Google Scholar] [CrossRef] [PubMed]
- Farhat, E.K.; Sher, E.K.; Džidić-Krivić, A.; Banjari, I.; Sher, F. Functional biotransformation of phytoestrogens by gut microbiota with impact on cancer treatment. J. Nutr. Biochem. 2023, 118, 109368. [Google Scholar] [CrossRef] [PubMed]
- von Schwartzenberg, R.J.; Bisanz, J.E.; Lyalina, S.; Spanogiannopoulos, P.; Ang, Q.Y.; Cai, J.; Dickmann, S.; Friedrich, M.; Liu, S.-Y.; Collins, S.L.; et al. Caloric restriction disrupts the microbiota and colonization resistance. Nature 2021, 595, 272–277. [Google Scholar] [CrossRef] [PubMed]
- Meslier, V.; Laiola, M.; Roager, H.M.; De Filippis, F.; Roume, H.; Quinquis, B.; Giacco, R.; Mennella, I.; Ferracane, R.; Pons, N.; et al. Mediterranean diet intervention in overweight and obese subjects lowers plasma cholesterol and causes changes in the gut microbiome and metabolome independently of energy intake. Gut 2020, 69, 1258–1268. [Google Scholar] [CrossRef]
- Zhang, S.; Wu, P.; Tian, Y.; Liu, B.; Huang, L.; Liu, Z.; Lin, N.; Xu, N.; Ruan, Y.; Zhang, Z.; et al. Gut Microbiota Serves a Predictable Outcome of Short-Term Low-Carbohydrate Diet (LCD) Intervention for Patients with Obesity. Microbiol. Spectr. 2021, 9, e0022321. [Google Scholar] [CrossRef]
- Zhu, F.; Ju, Y.; Wang, W.; Wang, Q.; Guo, R.; Ma, Q.; Sun, Q.; Fan, Y.; Xie, Y.; Yang, Z.; et al. Metagenome-wide association of gut microbiome features for schizophrenia. Nat. Commun. 2020, 11, 1612. [Google Scholar] [CrossRef]
- Gibbons, S.M. Microbial community ecology: Function over phylogeny. Nat. Ecol. Evol. 2017, 1, 0032. [Google Scholar] [CrossRef]
- Yannakoulia, M.; Poulimeneas, D.; Mamalaki, E.; Anastasiou, C.A. Dietary modifications for weight loss and weight loss maintenance. Metabolism 2019, 92, 153–162. [Google Scholar] [CrossRef]
- Zhu, H.; Bi, D.; Zhang, Y.; Kong, C.; Du, J.; Wu, X.; Wei, Q.; Qin, H. Ketogenic diet for human diseases: The underlying mechanisms and potential for clinical implementations. Signal Transduct. Target. Ther. 2022, 7, 11. [Google Scholar] [CrossRef] [PubMed]
- Amini, M.R.; Aminianfar, A.; Naghshi, S.; Larijani, B.; Esmaillzadeh, A. The effect of ketogenic diet on body composition and anthropometric measures: A systematic review and meta-analysis of randomized controlled trials. Crit. Rev. Food Sci. Nutr. 2022, 62, 3644–3657. [Google Scholar] [CrossRef]
- Castellana, M.; Conte, E.; Cignarelli, A.; Perrini, S.; Giustina, A.; Giovanella, L.; Giorgino, F.; Trimboli, P. Efficacy and safety of very low calorie ketogenic diet (VLCKD) in patients with overweight and obesity: A systematic review and meta-analysis. Rev. Endocr. Metab. Disord. 2020, 21, 5–16. [Google Scholar] [CrossRef]
- Cunha, G.M.; Guzman, G.; Correa De Mello, L.L.; Trein, B.; Spina, L.; Bussade, I.; Marques Prata, J.; Sajoux, I.; Countinho, W. Efficacy of a 2-Month Very Low-Calorie Ketogenic Diet (VLCKD) Compared to a Standard Low-Calorie Diet in Reducing Visceral and Liver Fat Accumulation in Patients with Obesity. Front. Endocrinol. 2020, 11, 607. [Google Scholar] [CrossRef] [PubMed]
- Van Hul, M.; Cani, P.D. The gut microbiota in obesity and weight management: Microbes as friends or foe? Nat. Rev. Endocrinol. 2023, 19, 258–271. [Google Scholar] [CrossRef] [PubMed]
- Ang, Q.Y.; Alexander, M.; Newman, J.C.; Tian, Y.; Cai, J.; Upadhyay, V.; Turnbaugh, J.A.; Verdin, E.; Hall, K.D.; Leibel, R.L.; et al. Ketogenic Diets Alter the Gut Microbiome Resulting in Decreased Intestinal Th17 Cells. Cell 2020, 181, 1263–1275.e1216. [Google Scholar] [CrossRef] [PubMed]
- Salim, F.; Mizutani, S.; Zolfo, M.; Yamada, T. Recent advances of machine learning applications in human gut microbiota study: From observational analysis toward causal inference and clinical intervention. Curr. Opin. Biotechnol. 2023, 79, 102884. [Google Scholar] [CrossRef]
- Yuan, W.W.; Lu, W.W.; Wang, H.C.; Wu, W.J.; Zhou, Q.Y.; Chen, Y.T.; Lee, Y.K.; Zhao, J.X.; Zhang, H.; Chen, W. A multiphase dietetic protocol incorporating an improved ketogenic diet enhances weight loss and alters the gut microbiome of obese people. Int. J. Food Sci. Nutr. 2022, 73, 238–250. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Li, H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv 2013, arXiv:1303.3997. [Google Scholar]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G. Genome Project Data Processing S: The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef] [PubMed]
- Quinlan, A.R.; Hall, I.M. BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics 2010, 26, 841–842. [Google Scholar] [CrossRef] [PubMed]
- Beghini, F.; McIver, L.J.; Blanco-Míguez, A.; Dubois, L.; Asnicar, F.; Maharjan, S.; Mailyan, A.; Manghi, P.; Scholz, M.; Thomas, A.M.; et al. Integrating taxonomic, functional, and strain-level profiling of diverse microbial communities with bioBakery 3. eLife 2021, 10, e65088. [Google Scholar] [CrossRef] [PubMed]
- Segata, N.; Waldron, L.; Ballarini, A.; Narasimhan, V.; Jousson, O.; Huttenhower, C. Metagenomic microbial community profiling using unique clade-specific marker genes. Nat. Methods 2012, 9, 811–814. [Google Scholar] [CrossRef] [PubMed]
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B.; Grisel, O.; Blondel, M.; Prettenhofer, P.; Weiss, R.; Dubourg, V.; et al. Scikit-learn: Machine Learning in Python. J. Mach. Learn. Res. 2011, 12, 2825–2830. [Google Scholar]
- Lundberg, S.M.; Erion, G.; Chen, H.; DeGrave, A.; Prutkin, J.M.; Nair, B.; Katz, R.; Himmelfarb, J.; Bansal, N.; Lee, S.-I. From local explanations to global understanding with explainable AI for trees. Nat. Mach. Intell. 2020, 2, 56–67. [Google Scholar] [CrossRef]
- Gallardo-Becerra, L.; Cornejo-Granados, F.; García-López, R.; Valdez-Lara, A.; Bikel, S.; Canizales-Quinteros, S.; López-Contreras, B.E.; Mendoza-Vargas, A.; Nielsen, H.; Ochoa-Leyva, A. Metatranscriptomic analysis to define the Secrebiome, and 16S rRNA profiling of the gut microbiome in obesity and metabolic syndrome of Mexican children. Microb. Cell Factories 2020, 19, 61. [Google Scholar] [CrossRef]
- Yan, P.; Sun, Y.; Luo, J.; Liu, X.; Wu, J.; Miao, Y. Integrating the serum proteomic and fecal metaproteomic to analyze the impacts of overweight/obesity on IBD: A pilot investigation. Clin. Proteom. 2023, 20, 6. [Google Scholar] [CrossRef]
- Haro, C.; Montes-Borrego, M.; Rangel-Zúñiga, O.A.; Alcalá-Díaz, J.F.; Gómez-Delgado, F.; Pérez-Martínez, P.; Delgado-Lista, J.; Quintana-Navarro, G.M.; Tinahones, F.J.; Landa, B.B.; et al. Two Healthy Diets Modulate Gut Microbial Community Improving Insulin Sensitivity in a Human Obese Population. J. Clin. Endocrinol. Metab. 2016, 101, 233–242. [Google Scholar] [CrossRef]
- You, H.J.; Si, J.; Kim, J.; Yoon, S.; Cha, K.H.; Yoon, H.S.; Lee, G.; Yu, J.; Choi, J.S.; Jung, M.; et al. Bacteroides vulgatus SNUG 40005 Restores Akkermansia Depletion by Metabolite Modulation. Gastroenterology 2023, 164, 103–116. [Google Scholar] [CrossRef]
- Liu, R.; Hong, J.; Xu, X.; Feng, Q.; Zhang, D.; Gu, Y.; Shi, J.; Zhao, S.; Liu, W.; Wang, X.; et al. Gut microbiome and serum metabolome alterations in obesity and after weight-loss intervention. Nat. Med. 2017, 23, 859–868. [Google Scholar] [CrossRef] [PubMed]
- Choi, Y.J.; Jeon, S.M.; Shin, S. Impact of a Ketogenic Diet on Metabolic Parameters in Patients with Obesity or Overweight and with or without Type 2 Diabetes: A Meta-Analysis of Randomized Controlled Trials. Nutrients 2020, 12, 2005. [Google Scholar] [CrossRef] [PubMed]
- Muscogiuri, G.; Barrea, L.; Campolo, F.; Sbardella, E.; Sciammarella, C.; Tarsitano, M.G.; Bottiglieri, F.; Colao, A.; Faggiano, A. Ketogenic diet: A tool for the management of neuroendocrine neoplasms? Crit. Rev. Food Sci. Nutr. 2022, 62, 1035–1045. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.J.; Zhou, S.Z.; Zhou, Y.F.; Yu, L.F.; Zhang, L.M.; Wang, Y. Altered gut microbiome composition in children with refractory epilepsy after ketogenic diet. Epilepsy Res. 2018, 145, 163–168. [Google Scholar] [CrossRef] [PubMed]
- Fan, Y.Y.; Wang, H.; Liu, X.Y.; Zhang, J.M.; Liu, G. Crosstalk between the Ketogenic Diet and Epilepsy: From the Perspective of Gut Microbiota. Mediat. Inflamm. 2019, 2019, 8373060. [Google Scholar] [CrossRef]
- Lindefeldt, M.; Eng, A.; Darban, H.; Bjerkner, A.; Zetterstrom, C.K.; Allander, T.; Andersson, B.; Borenstein, E.; Dahlin, M.; Prast-Nielsen, S. The ketogenic diet influences taxonomic and functional composition of the gut microbiota in children with severe epilepsy. NPJ Biofilms Microbiomes 2019, 5, 5. [Google Scholar] [CrossRef]
- Ma, D.; Wang, A.C.; Parikh, I.; Green, S.J.; Hoffman, J.D.; Chlipala, G.; Murphy, M.P.; Sokola, B.S.; Bauer, B.; Hartz, A.M.S.; et al. Ketogenic diet enhances neurovascular function with altered gut microbiome in young healthy mice. Sci. Rep. 2018, 8, 6670. [Google Scholar] [CrossRef]
- Nagpal, R.; Neth, B.J.; Wang, S.; Mishra, S.P.; Craft, S.; Yadav, H. Gut mycobiome and its interaction with diet, gut bacteria and alzheimer’s disease markers in subjects with mild cognitive impairment: A pilot study. EBioMedicine 2020, 59, 102950. [Google Scholar] [CrossRef]
- Dilmore, A.H.; Martino, C.; Neth, B.J.; West, K.A.; Zemlin, J.; Rahman, G.; Panitchpakdi, M.; Meehan, M.J.; Weldon, K.C.; Blach, C.; et al. Effects of a ketogenic and low-fat diet on the human metabolome, microbiome, and foodome in adults at risk for Alzheimer’s disease. Alzheimer’s Dementa 2023. [Google Scholar] [CrossRef]
- Nakamura, K.; Hagihara, K.; Nagai, N.; Egashira, R.; Takeuchi, M.; Nakano, M.; Saito, H.; Moriguchi, M.; Tonari, S.; Watanabe, S.; et al. Ketogenic Effects of Multiple Doses of a Medium Chain Triglycerides Enriched Ketogenic Formula in Healthy Men under the Ketogenic Diet: A Randomized, Double-Blinded, Placebo-Controlled Study. Nutrients 2022, 14, 1199. [Google Scholar] [CrossRef]
- Muscogiuri, G.; Barrea, L.; Laudisio, D.; Pugliese, G.; Salzano, C.; Savastano, S.; Colao, A. The management of very low-calorie ketogenic diet in obesity outpatient clinic: A practical guide. J. Transl. Med. 2019, 17, 356. [Google Scholar] [CrossRef] [PubMed]
- Basciani, S.; Costantini, D.; Contini, S.; Persichetti, A.; Watanabe, M.; Mariani, S.; Lubrano, C.; Spera, G.; Lenzi, A.; Gnessi, L. Safety and efficacy of a multiphase dietetic protocol with meal replacements including a step with very low calorie diet. Endocrine 2015, 48, 863–870. [Google Scholar] [CrossRef] [PubMed]
- Swidsinski, A.; Dörffel, Y.; Loening-Baucke, V.; Gille, C.; Göktas, Ö.; Reißhauer, A.; Neuhaus, J.; Weylandt, K.H.; Guschin, A.; Bock, M. Reduced Mass and Diversity of the Colonic Microbiome in Patients with Multiple Sclerosis and Their Improvement with Ketogenic Diet. Front. Microbiol. 2017, 8, 1141. [Google Scholar] [CrossRef]
- Hu, X.; Xia, K.; Dai, M.; Han, X.; Yuan, P.; Liu, J.; Liu, S.; Jia, F.; Chen, J.; Jiang, F.; et al. Intermittent fasting modulates the intestinal microbiota and improves obesity and host energy metabolism. NPJ Biofilms Microbiomes 2023, 9, 19. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Liao, M.; Zhou, N.; Bao, L.; Ma, K.; Zheng, Z.; Wang, Y.; Liu, C.; Wang, W.; Wang, J.; et al. Parabacteroides distasonis Alleviates Obesity and Metabolic Dysfunctions via Production of Succinate and Secondary Bile Acids. Cell Rep. 2019, 26, 222–235.e225. [Google Scholar] [CrossRef] [PubMed]
- Depommier, C.; Everard, A.; Druart, C.; Plovier, H.; Van Hul, M.; Vieira-Silva, S.; Falony, G.; Raes, J.; Maiter, D.; Delzenne, N.M.; et al. Supplementation with Akkermansia muciniphila in overweight and obese human volunteers: A proof-of-concept exploratory study. Nat. Med. 2019, 25, 1096–1103. [Google Scholar] [CrossRef]
- Wang, C.; Xiao, Y.; Yu, L.; Tian, F.; Zhao, J.; Zhang, H.; Chen, W.; Zhai, Q. Protective effects of different Bacteroides vulgatus strains against lipopolysaccharide-induced acute intestinal injury, and their underlying functional genes. J. Adv. Res. 2022, 36, 27–37. [Google Scholar] [CrossRef]
- Rakoff-Nahoum, S.; Foster, K.R.; Comstock, L.E. The evolution of cooperation within the gut microbiota. Nature 2016, 533, 255–259. [Google Scholar] [CrossRef]
- Joseph, N.; Clayton, J.B.; Hoops, S.L.; Linhardt, C.A.; Hashim, A.M.; Yusof, B.N.M.; Kumar, S.; Nordin, S.A. Alteration of the Gut Microbiome in Normal and Overweight School Children from Selangor with Lactobacillus Fermented Milk Administration. Evol. Bioinform. 2020, 16, 1176934320965943. [Google Scholar] [CrossRef]
- Jain, H.; Singh, G.; Eranki, A. Actinomyces odontolyticus causing meningitis and cervical abscess. In Baylor University Medical Center Proceedings; Taylor & Francis: Abingdon, UK, 2021. [Google Scholar] [CrossRef]
- Chen, L.M.; Collij, V.; Jaeger, M.; van den Munckhof, I.C.L.; Vila, A.V.; Kurilshikov, A.; Gacesa, R.; Sinha, T.; Oosting, M.; Joosten, L.A.B.; et al. Gut microbial co-abundance networks show specificity in inflammatory bowel disease and obesity. Nat. Commun. 2020, 11, 4018. [Google Scholar] [CrossRef]
- Arnoriaga-Rodríguez, M.; Mayneris-Perxachs, J.; Burokas, A.; Contreras-Rodríguez, O.; Blasco, G.; Coll, C.; Biarnés, C.; Miranda-Olivos, R.; Latorre, J.; Moreno-Navarrete, J.M.; et al. Obesity Impairs Short-Term and Working Memory through Gut Microbial Metabolism of Aromatic Amino Acids. Cell Metab. 2020, 32, 548–560.e547. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Fang, S.; Wei, H.; He, M.; Fu, H.; Xiong, X.; Zhou, Y.; Wu, J.; Gao, J.; Yang, H.; et al. Prevotella copri increases fat accumulation in pigs fed with formula diets. Microbiome 2021, 9, 175. [Google Scholar] [CrossRef] [PubMed]
- Kasai, C.; Sugimoto, K.; Moritani, I.; Tanaka, J.; Oya, Y.; Inoue, H.; Tameda, M.; Shiraki, K.; Ito, M.; Takei, Y.; et al. Comparison of the gut microbiota composition between obese and non-obese individuals in a Japanese population, as analyzed by terminal restriction fragment length polymorphism and next-generation sequencing. BMC Gastroenterol. 2015, 15, 100. [Google Scholar] [CrossRef] [PubMed]
- Roses, C.; Cuevas-Sierra, A.; Quintana, S.; Riezu-Boj, J.I.; Martinez, J.A.; Milagro, F.I.; Barcelo, A. Gut Microbiota Bacterial Species Associated with Mediterranean Diet-Related Food Groups in a Northern Spanish Population. Nutrients 2021, 13, 636. [Google Scholar] [CrossRef]
- You, A.; Li, Y.; Shen, C.; Fan, H.; He, J.; Liu, Z.; Xue, Q.; Zhang, Y.; Zheng, L. Associations of non-traditional cardiovascular risk factors and body mass index with metabolic syndrome in the Chinese elderly population. Diabetol. Metab. Syndr. 2023, 15, 129. [Google Scholar] [CrossRef]
- Li, S.; Feng, L.; Sun, X.; Ding, J.; Zhou, W. Association between serum uric acid and measures of adiposity in Chinese adults: A cross-sectional study. BMJ Open 2023, 13, e072317. [Google Scholar] [CrossRef]
- Su, S.Y.; Lin, T.H.; Liu, Y.H.; Wu, P.Y.; Huang, J.C.; Su, H.M.; Chen, S.C. Sex Difference in the Associations among Obesity-Related Indices with Hyperuricemia in a Large Taiwanese Population Study. Nutrients 2023, 15, 3419. [Google Scholar] [CrossRef]
- Andres-Hernando, A.; Cicerchi, C.; Kuwabara, M.; Orlicky, D.J.; Sanchez-Lozada, L.G.; Nakagawa, T.; Johnson, R.J.; Lanaspa, M.A. Umami-induced obesity and metabolic syndrome is mediated by nucleotide degradation and uric acid generation. Nat. Metab. 2021, 3, 1189–1201. [Google Scholar] [CrossRef]
- Kiana Ashley, W.; Chidimma, K.; Tanya, M.; Julie Anne Kathryn, M.; Jeremy, K.N.; Jia, V.L.; Mark, R.J.; Elaine, H.; Makrina, D.S. Longitudinal metabolic and gut bacterial profiling of pregnant women with previous bariatric surgery. Gut 2020, 69, 1452. [Google Scholar] [CrossRef]
- Depommier, C.; Everard, A.; Druart, C.; Maiter, D.; Thissen, J.P.; Loumaye, A.; Hermans, M.P.; Delzenne, N.M.; de Vos, W.M.; Cani, P.D. Serum metabolite profiling yields insights into health promoting effect of A. muciniphila in human volunteers with a metabolic syndrome. Gut Microbes 2021, 13, 1994270. [Google Scholar] [CrossRef]



| Characteristic | Before | After | p-Value | Change |
|---|---|---|---|---|
| Body composition | ||||
| Bodyweight, kg | 86.6 ± 14.7 | 78.4 ± 13.2 | 0.0017 | −8.2 ± 2.5 |
| BMI, kg/m2 | 31.0 ± 2.6 | 28.1 ± 2.3 * | 0.0002 | −2.9 ± 0.8 |
| Waist, cm | 99.3 ± 10.0 | 91.9 ± 8.9 | 0.0017 | −7.3 ± 3 |
| Hip, cm | 105.4 ± 5.0 | 100.2 ± 5.2 * | 0.0016 | −5.2 ± 1.7 |
| Blood pressure, mm Hg | ||||
| Systolic | 132.3 ± 14.1 | 123.5 ± 14.8 | 0.0118 | −8.8 ± 10.6 |
| diastolic | 78.4 ± 11.5 | 72.4 ± 11.2 | 0.0291 | −6.0 ± 10 |
| Liver function, U/L | ||||
| AST | 26.5 ± 10.3 | 21.2 ± 5.8 | 0.0357 | −5.2 ± 7.3 |
| ALP | 78.4 ± 26.4 | 84.0 ± 37.0 | 0.4214 | 5.6 ± 18.8 |
| ALT | 41.9 ± 30.7 | 27.5 ± 22.7 | 0.0134 | −14.4 ± 19.5 |
| Renal function | ||||
| Albumin, g/L | 48.3 ± 1.3 | 46.6 ± 2.1 | 0.0645 | −1.7 ± 2.5 |
| BUN, mmol/L | 4.7 ± 1.5 | 5.3 ± 0.8 | 0.0803 | 0.6 ± 1.5 |
| Creatinine, μmol/L | 71.5 ± 13.6 | 72.0 ± 13.2 | 0.7869 | 0.5 ± 5.0 |
| UA, μmol/L | 452.35 ± 29.73 | 438.05 ± 22.19 | 0.4973 | −14.3 ± 37.10 |
| Lipids, mmol/L | ||||
| Total cholesterol | 5.2 ± 0.9 | 5.2 ± 0.9 | 0.6848 | 0.0 ± 0.8 |
| LDL cholesterol | 3.4 ± 0.8 | 3.4 ± 0.9 | 0.5292 | −0.1 ± 0.7 |
| HDL cholesterol | 1.0 ± 0.2 | 1.1 ± 0.2 | 0.1259 | 0.1 ± 0.2 |
| Triglyceride | 2.3 ± 2.1 | 1.5 ± 0.7 | 0.0081 | −0.9 ± 1.6 |
| HbA1c [%] | 5.4 ± 0.5 | 5.0 ± 0.4 * | 0.0058 | −0.4 ± 0.5 |
| Variable | D0 | D12 |
|---|---|---|
| Number of edges | 760.000 | 901.000 |
| Number of positive edges | 722.000 | 885.000 |
| Number of negative edges | 38.000 | 16.000 |
| Number of vertices | 299.000 | 318.000 |
| Average degree | 5.084 | 5.667 |
| Average path length | 2.867 | 2.395 |
| Diameter | 8.000 | 10.000 |
| Average clustering coefficient | 0.905 | 0.920 |
| Centralization degree | 0.036 | 0.028 |
| Modularity | 0.941 | 0.888 |
| Number of modularity | 141.000 | 138.000 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, H.; Lv, X.; Zhao, S.; Yuan, W.; Zhou, Q.; Sadiq, F.A.; Zhao, J.; Lu, W.; Wu, W. Weight Loss Promotion in Individuals with Obesity through Gut Microbiota Alterations with a Multiphase Modified Ketogenic Diet. Nutrients 2023, 15, 4163. https://doi.org/10.3390/nu15194163
Wang H, Lv X, Zhao S, Yuan W, Zhou Q, Sadiq FA, Zhao J, Lu W, Wu W. Weight Loss Promotion in Individuals with Obesity through Gut Microbiota Alterations with a Multiphase Modified Ketogenic Diet. Nutrients. 2023; 15(19):4163. https://doi.org/10.3390/nu15194163
Chicago/Turabian StyleWang, Hongchao, Xinchen Lv, Sijia Zhao, Weiwei Yuan, Qunyan Zhou, Faizan Ahmed Sadiq, Jianxin Zhao, Wenwei Lu, and Wenjun Wu. 2023. "Weight Loss Promotion in Individuals with Obesity through Gut Microbiota Alterations with a Multiphase Modified Ketogenic Diet" Nutrients 15, no. 19: 4163. https://doi.org/10.3390/nu15194163
APA StyleWang, H., Lv, X., Zhao, S., Yuan, W., Zhou, Q., Sadiq, F. A., Zhao, J., Lu, W., & Wu, W. (2023). Weight Loss Promotion in Individuals with Obesity through Gut Microbiota Alterations with a Multiphase Modified Ketogenic Diet. Nutrients, 15(19), 4163. https://doi.org/10.3390/nu15194163






