Fingerprinting of Proteases, Protease Inhibitors and Indigenous Peptides in Human Milk
Abstract
:1. Introduction
2. Materials and Methods
2.1. Human Milk Samples
2.2. Sample Preparation of the Human Milk Proteome and Peptidome
2.3. Trypsinization of Intact Proteins in the Proteome
2.4. Desalting and Sample Cleaning of Trypsinated Proteome and Peptidome
2.5. Analyzing the Proteome and the Peptidome with the nLC-MS/MS timsTOF Pro II
2.6. Spectral Data Searching Using FragPipe
2.7. Post-Processing of the FragPipe Data
2.8. Extracting Proteases, Peptidases and Protease Inhibitors from the Proteome, and Cleavage Sites from the Peptidome
2.9. Bioactive Peptides
2.10. Statistics
3. Results
3.1. Most Abundant Human Milk Proteins in the Proteome and Peptidome
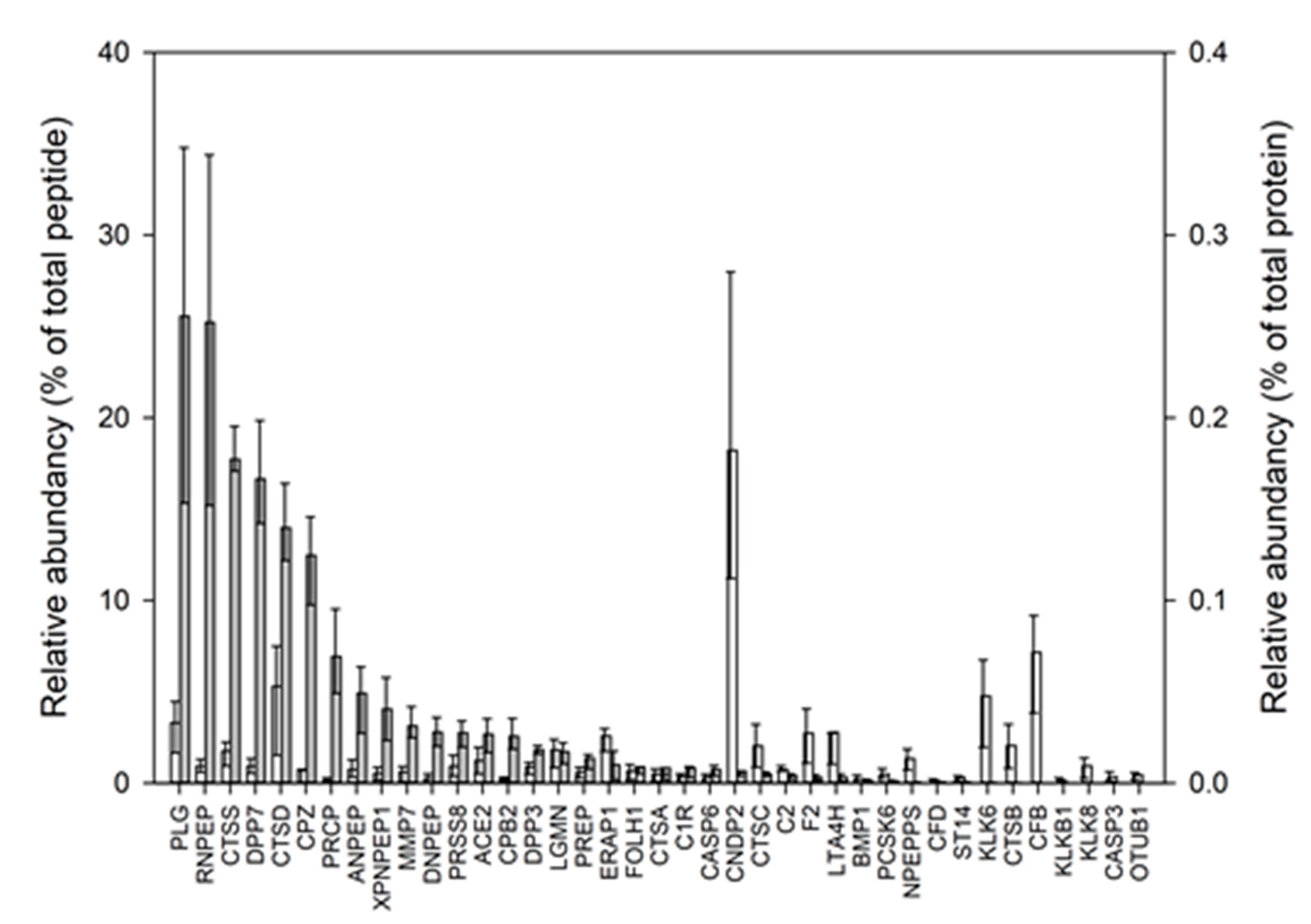
3.2. Proteases Identified and Quantified in the Human Milk Proteome
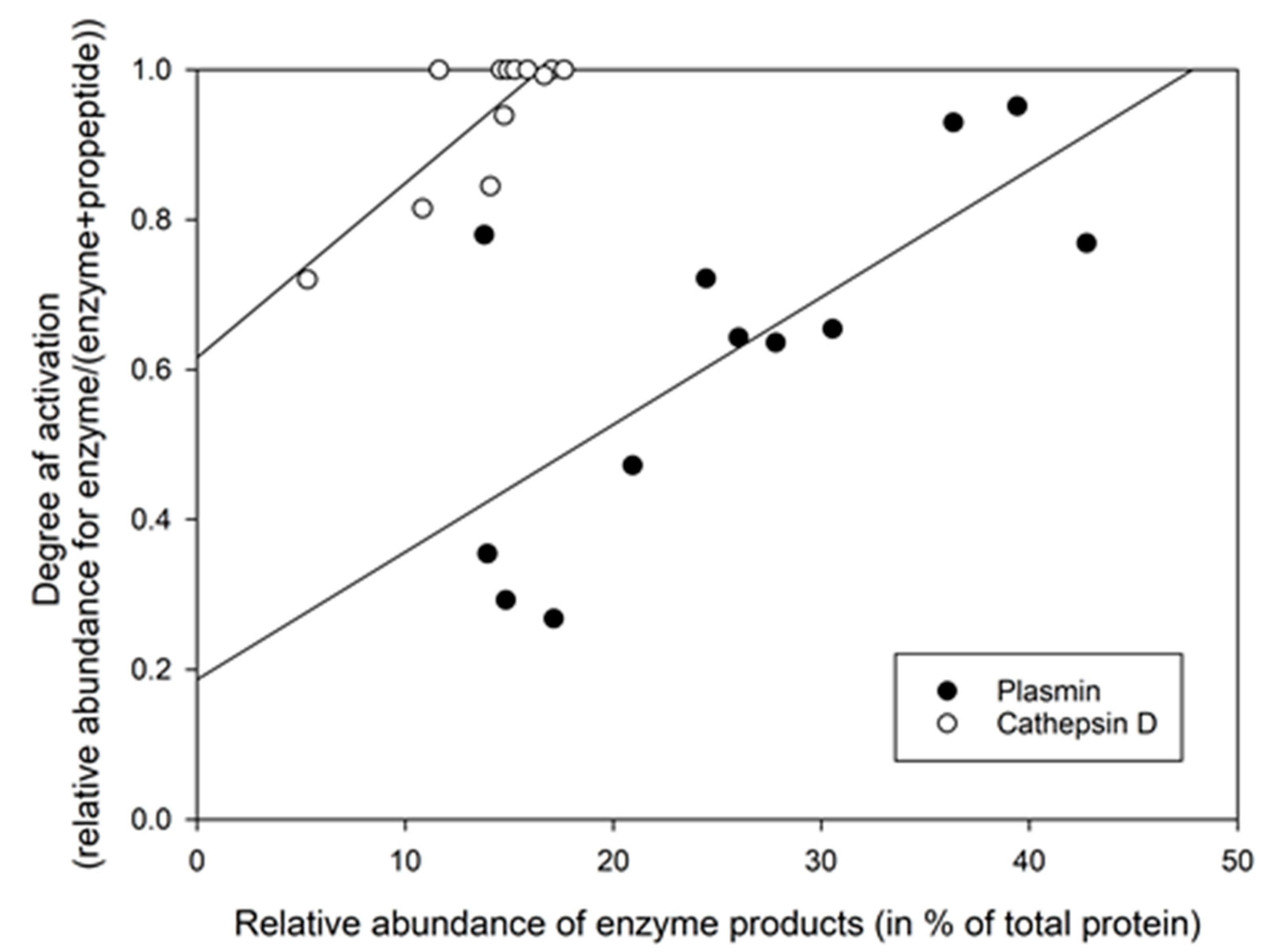
3.3. Using the Relative Abundance of Peptides to Calculate the Activation Degree of Proteases
3.4. Determining Protease Activity Based on the Relative Abundance of Their Ascribed Cleavage Products
3.5. Protease Inhibitors Identified and Quantified in the Human Milk Proteome
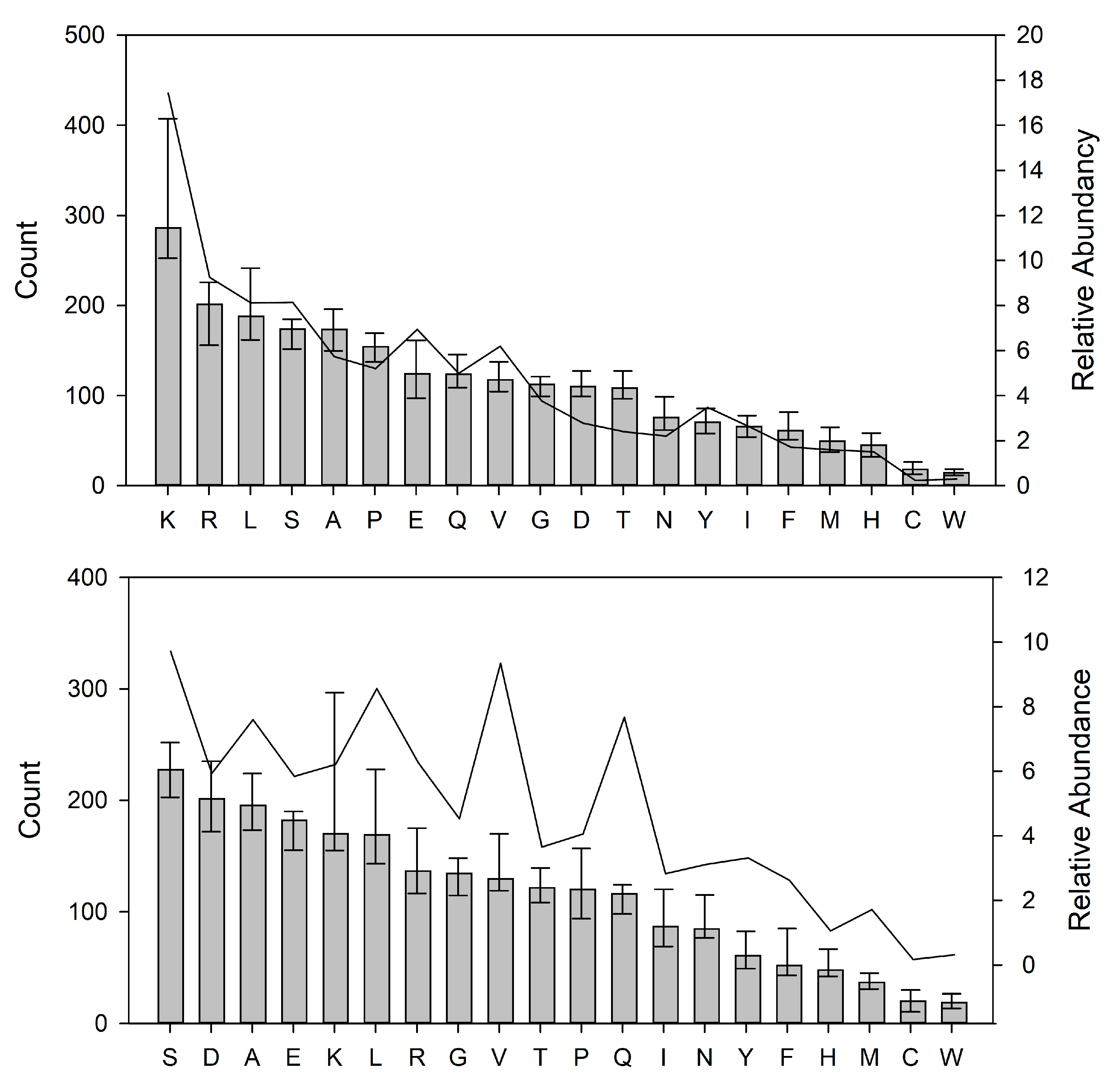
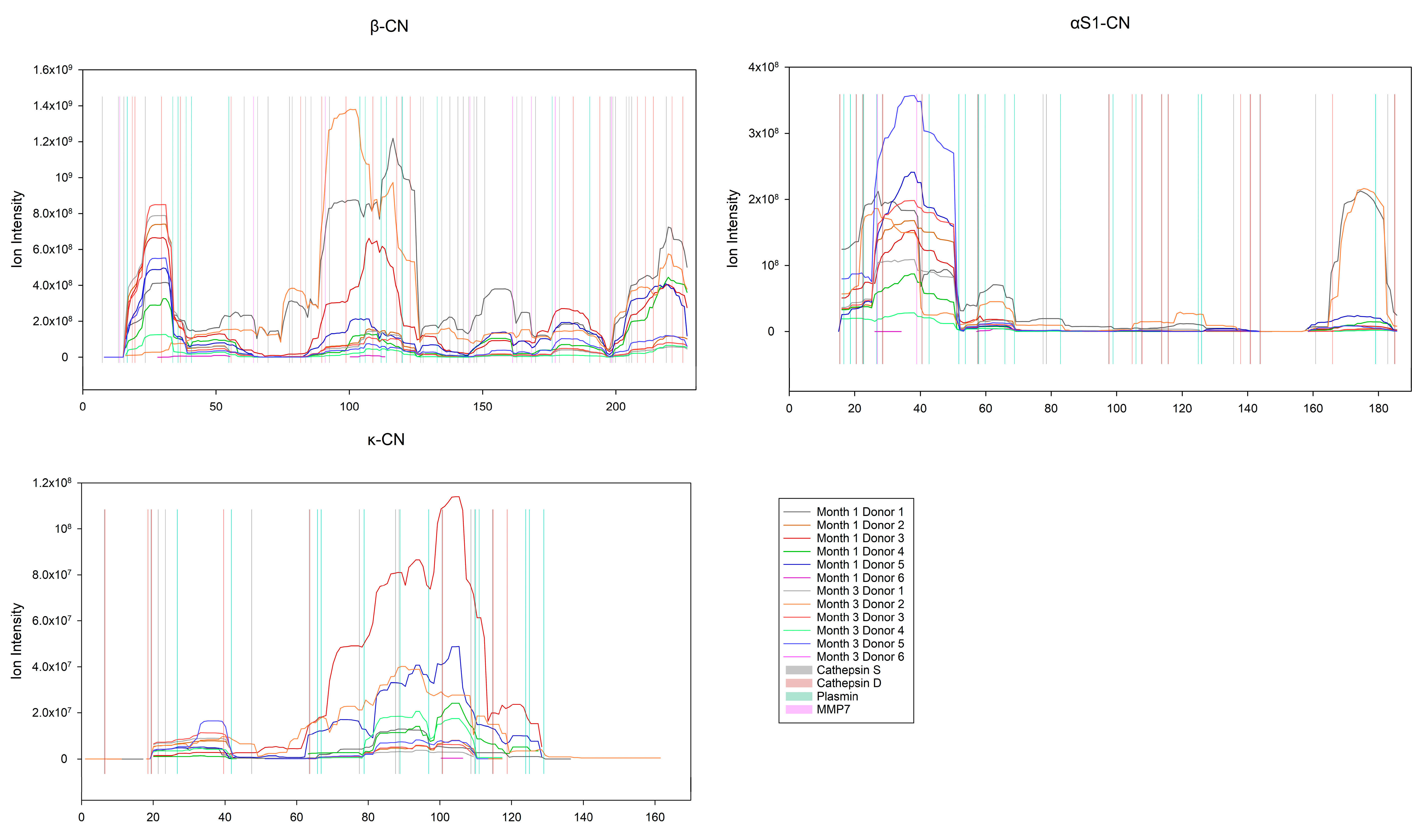
3.6. Preferred Cleavage Sites in Human Milk Proteins by the Indigenous Proteases
Enzyme Activity on the Caseins and Whey Proteins
3.7. Bioactive Peptides
4. Discussion
4.1. Identification and Abundance of Proteins and Peptides in the HM Proteome and Peptidome
4.2. Abundancies of Different Protease Systems in Human Milk
4.3. Abundance of Different Protease Inhibitors in Human Milk
4.4. Selective Hydrolysis and Preferred Cleavage Site Motifs in Human Milk
4.5. Bioactive Peptides
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lönnerdal, B. Bioactive Proteins in Human Milk: Health, Nutrition, and Implications for Infant Formulas. J. Pediatr. 2016, 173 (Suppl.), S4–S9. [Google Scholar] [CrossRef]
- Zhu, J.; Dingess, K.A.; Mank, M.; Stahl, B.; Heck, A.J.R. Personalized Profiling Reveals Donor- and Lactation-Specific Trends in the Human Milk Proteome and Peptidome. J. Nutr. 2021, 151, 826–839. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.; Dingess, K.A. The Functional Power of the Human Milk Proteome. Nutrients 2019, 11, 1834. [Google Scholar] [CrossRef] [PubMed]
- Dingess, K.A.; Gazi, I.; van den Toorn, H.W.P.; Mank, M.; Stahl, B.; Reiding, K.R.; Heck, A.J.R. Monitoring Human Milk β-Casein Phosphorylation and O-Glycosylation Over Lactation Reveals Distinct Differences between the Proteome and Endogenous Peptidome. Int. J. Mol. Sci. 2021, 22, 8140. [Google Scholar] [CrossRef] [PubMed]
- Jensen, R.G. CHAPTER 1—Introduction. In Handbook of Milk Composition; Academic Press: San Diego, CA, USA, 1995; pp. 1–3. ISBN 978-0-12-384430-9. [Google Scholar]
- Donovan, S.M. Human Milk Proteins: Composition and Physiological Significance. In Human Milk: Composition, Clinical Benefits and Future Opportunities: 90th Nestlé Nutrition Institute Workshop, Lausanne, October-November 2017; Donovan, S.M., German, J.B., Lönnerdal, B., Lucas, A., Eds.; S.Karger AG: Basel, Switzerland, 2019; Volume 90, pp. 93–101. ISBN 978-3-318-06340-0. [Google Scholar]
- Liao, Y.; Weber, D.; Xu, W.; Durbin-Johnson, B.P.; Phinney, B.S.; Lönnerdal, B. Absolute Quantification of Human Milk Caseins and the Whey/Casein Ratio during the First Year of Lactation. J. Proteome Res. 2017, 16, 4113–4121. [Google Scholar] [CrossRef]
- Michaelsen, K.F.; Skafte, L.; Badsberg, J.H.; Jørgensen, M. Variation in Macronutrients in Human Bank Milk: Influencing Factors and Implications for Human Milk Banking. J. Pediatr. Gastroenterol. Nutr. 1990, 11, 229–239. [Google Scholar] [CrossRef]
- Ballard, O.; Morrow, A.L. Human Milk Composition: Nutrients and Bioactive Factors. Pediatr. Clin. N. Am. 2013, 60, 49–74. [Google Scholar] [CrossRef]
- Dingess, K.A.; Li, C.; Zhu, J. Human Milk Proteome: What’s New? Curr. Opin. Clin. Nutr. Metab. Care 2021, 24, 252–258. [Google Scholar] [CrossRef]
- Dallas, D.C.; Guerrero, A.; Khaldi, N.; Castillo, P.A.; Martin, W.F.; Smilowitz, J.T.; Bevins, C.L.; Barile, D.; German, J.B.; Lebrilla, C.B. Extensive in Vivo Human Milk Peptidomics Reveals Specific Proteolysis Yielding Protective Antimicrobial Peptides. J. Proteome Res. 2013, 12, 2295–2304. [Google Scholar] [CrossRef]
- Nielsen, S.D.; Beverly, R.L.; Dallas, D.C. Milk Proteins Are Predigested Within the Human Mammary Gland. J. Mammary Gland Biol. Neoplasia 2017, 22, 251–261. [Google Scholar] [CrossRef]
- Dallas, D.C.; Guerrero, A.; Khaldi, N.; Borghese, R.; Bhandari, A.; Underwood, M.A.; Lebrilla, C.B.; German, J.B.; Barile, D. A Peptidomic Analysis of Human Milk Digestion in the Infant Stomach Reveals Protein-Specific Degradation Patterns. J. Nutr. 2014, 144, 815–820. [Google Scholar] [CrossRef] [PubMed]
- Koh, J.; Kim, B.J.; Qu, Y.; Huang, H.; Dallas, D.C. Top-Down Glycopeptidomics Reveals Intact Glycomacropeptide Is Digested to a Wide Array of Peptides in Human Jejunum. J. Nutr. 2021, 152, 429–438. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, S.D.-H.; Liang, N.; Rathish, H.; Kim, B.J.; Lueangsakulthai, J.; Koh, J.; Qu, Y.; Schulz, H.-J.; Dallas, D.C. Bioactive Milk Peptides: An Updated Comprehensive Overview and Database. Crit. Rev. Food Sci. Nutr. 2023. [Google Scholar] [CrossRef] [PubMed]
- Dallas, D.C.; Smink, C.J.; Robinson, R.C.; Tian, T.; Guerrero, A.; Parker, E.A.; Smilowitz, J.T.; Hettinga, K.A.; Underwood, M.A.; Lebrilla, C.B.; et al. Endogenous Human Milk Peptide Release Is Greater after Preterm Birth than Term Birth. J. Nutr. 2015, 145, 425–433. [Google Scholar] [CrossRef]
- Demers-Mathieu, V.; Nielsen, S.D.; Underwood, M.A.; Borghese, R.; Dallas, D.C. Analysis of Milk from Mothers Who Delivered Prematurely Reveals Few Changes in Proteases and Protease Inhibitors across Gestational Age at Birth and Infant Postnatal Age. J. Nutr. 2017, 147, 1152–1159. [Google Scholar] [CrossRef]
- Chan, L.E.; Beverly, R.L.; Dallas, D.C. The Enzymology of Human Milk BT. In Agents of Change: Enzymes in Milk and Dairy Products; Kelly, A.L., Larsen, L.B., Eds.; Springer International Publishing: Cham, Switzerland, 2021; pp. 209–243. ISBN 978-3-030-55482-8. [Google Scholar]
- D’Alessandro, A.; Scaloni, A.; Zolla, L. Human Milk Proteins: An Interactomics and Updated Functional Overview. J. Proteome Res. 2010, 9, 3339–3373. [Google Scholar] [CrossRef]
- Klein, J.; Eales, J.; Zürbig, P.; Vlahou, A.; Mischak, H.; Stevens, R. Proteasix: A Tool for Automated and Large-Scale Prediction of Proteases Involved in Naturally Occurring Peptide Generation. Proteomics 2013, 13, 1077–1082. [Google Scholar] [CrossRef]
- Song, J.; Tan, H.; Perry, A.J.; Akutsu, T.; Webb, G.I.; Whisstock, J.C.; Pike, R.N. PROSPER: An Integrated Feature-Based Tool for Predicting Protease Substrate Cleavage Sites. PLoS ONE 2012, 7, e50300. [Google Scholar] [CrossRef]
- Vijayakumar, V.; Guerrero, A.N.; Davey, N.; Lebrilla, C.B.; Shields, D.C.; Khaldi, N. EnzymePredictor: A Tool for Predicting and Visualizing Enzymatic Cleavages of Digested Proteins. J. Proteome Res. 2012, 11, 6056–6065. [Google Scholar] [CrossRef]
- Rawlings, N.D.; Barrett, A.J.; Thomas, P.D.; Huang, X.; Bateman, A.; Finn, R.D. The MEROPS Database of Proteolytic Enzymes, Their Substrates and Inhibitors in 2017 and a Comparison with Peptidases in the PANTHER Database. Nucleic Acids Res. 2018, 46, D624–D632. [Google Scholar] [CrossRef]
- Korycha-Dahl, M.; Dumas, B.R.; Chene, N.; Martal, J. Plasmin Activity in Milk. J. Dairy Sci. 1983, 66, 704–711. [Google Scholar] [CrossRef]
- Heegaard, C.W.; Larsen, L.B.; Rasmussen, L.K.; Højberg, K.E.; Petersen, T.E.; Andreasen, P.A. Plasminogen Activation System in Human Milk. J. Pediatr. Gastroenterol. Nutr. 1997, 25, 159–166. [Google Scholar] [CrossRef] [PubMed]
- Armaforte, E.; Curran, E.; Huppertz, T.; Ryan, C.A.; Caboni, M.F.; O’Connor, P.M.; Ross, R.P.; Hirtz, C.; Sommerer, N.; Chevalier, F.; et al. Proteins and Proteolysis in Pre-Term and Term Human Milk and Possible Implications for Infant Formulae. Int. Dairy J. 2010, 20, 715–723. [Google Scholar] [CrossRef]
- Zwirzitz, A.; Reiter, M.; Skrabana, R.; Ohradanova-Repic, A.; Majdic, O.; Gutekova, M.; Cehlar, O.; Petrovčíková, E.; Kutejova, E.; Stanek, G.; et al. Lactoferrin Is a Natural Inhibitor of Plasminogen Activation. J. Biol. Chem. 2018, 293, 8600–8613. [Google Scholar] [CrossRef] [PubMed]
- Lindberg, T. Protease Inhibitors in Human Milk. Pediatr. Res. 1979, 13, 969–972. [Google Scholar] [CrossRef]
- Monti, J.C.; Mermoud, A.-F.; Jollès, P. Trypsin in Human Milk. Experientia 1986, 42, 39–41. [Google Scholar] [CrossRef]
- Lubetzky, R.; Mandel, D.; Mimouni, F.B.; Herman, L.; Reich, R.; Reif, S. MMP-2 and MMP-9 and Their Tissue Inhibitor in Preterm Human Milk. J. Pediatr. Gastroenterol. Nutr. 2010, 51, 210–212. [Google Scholar] [CrossRef]
- Cheung, P.Y.; Sawicki, G.; Gross, S.; Van Aerde, J.; Radomski, M. Differential Expression of Matrix Metalloproteinases and the Tissue Inhibitor in Human Milk. Proc. West. Pharmacol. Soc. 2001, 44, 97–98. [Google Scholar]
- Lindberg, T.; Ohlsson, K.; Weström, B. Protease Inhibitors and Their Relation to Protease Activity in Human Milk. Pediatr. Res. 1982, 16, 479–483. [Google Scholar] [CrossRef]
- Hendrixson, D.R.; Qiu, J.; Shewry, S.C.; Fink, D.L.; Petty, S.; Baker, E.N.; Plaut, A.G.; St Geme, J.W. Human Milk Lactoferrin Is a Serine Protease That Cleaves Haemophilus Surface Proteins at Arginine-Rich Sites. Mol. Microbiol. 2003, 47, 607–617. [Google Scholar] [CrossRef]
- France, T.C.; O’Mahony, J.A.; Kelly, A.L. The Plasmin System in Milk and Dairy Products BT. In Agents of Change: Enzymes in Milk and Dairy Products; Kelly, A.L., Larsen, L.B., Eds.; Springer International Publishing: Cham, Switzerland, 2021; pp. 11–55. ISBN 978-3-030-55482-8. [Google Scholar]
- Larsen, L.B.; Nielsen, S.D.-H.; Paludetti, L.; Kelly, A.L. Lysosomal and Other Indigenous Non-Plasmin Proteases in Bovine Milk BT. In Agents of Change: Enzymes in Milk and Dairy Products; Kelly, A.L., Larsen, L.B., Eds.; Springer International Publishing: Cham, Switzerland, 2021; pp. 57–84. ISBN 978-3-030-55482-8. [Google Scholar]
- Wittlin, S.; Rösel, J.; Hofmann, F.; Stover, D.R. Mechanisms and Kinetics of Procathepsin D Activation. Eur. J. Biochem. 1999, 265, 384–393. [Google Scholar] [CrossRef] [PubMed]
- Poulsen, K.O.; Astono, J.; Jakobsen, R.R.; Uldbjerg, N.; Fuglsang, J.; Nielsen, D.S.; Sundekilde, U.K. Influence of Maternal Body Mass Index on Human Milk Composition and Associations to Infant Metabolism and Gut Colonisation: MAINHEALTH—A Study Protocol for an Observational Birth Cohort. BMJ Open 2022, 12, e059552. [Google Scholar] [CrossRef]
- Dallas, D.C.; Nielsen, S.D. Milk Peptidomics to Identify Functional Peptides and for Quality Control of Dairy Products. In Methods in Molecular Biology; Schrader, M., Fricker, L., Eds.; Springer: New York, NY, USA, 2018; pp. 223–240. ISBN 978-1-4939-7537-2. [Google Scholar]
- Chambers, M.C.; Maclean, B.; Burke, R.; Amodei, D.; Ruderman, D.L.; Neumann, S.; Gatto, L.; Fischer, B.; Pratt, B.; Egertson, J.; et al. A Cross-Platform Toolkit for Mass Spectrometry and Proteomics. Nat. Biotechnol. 2012, 30, 918–920. [Google Scholar] [CrossRef]
- Kong, A.T.; Leprevost, F.V.; Avtonomov, D.M.; Mellacheruvu, D.; Nesvizhskii, A.I. MSFragger: Ultrafast and Comprehensive Peptide Identification in Mass Spectrometry-Based Proteomics. Nat. Methods 2017, 14, 513–520. [Google Scholar] [CrossRef]
- Guerrero, A.; Dallas, D.C.; Contreras, S.; Chee, S.; Parker, E.A.; Sun, X.; Dimapasoc, L.; Barile, D.; German, J.B.; Lebrilla, C.B. Mechanistic Peptidomics: Factors That Dictate Specificity in the Formation of Endogenous Peptides in Human Milk. Mol. Cell. Proteom. 2014, 13, 3343–3351. [Google Scholar] [CrossRef]
- Nielsen, S.D.; Beverly, R.L.; Underwood, M.A.; Dallas, D.C. Release of Functional Peptides from Mother’s Milk and Fortifier Proteins in the Premature Infant Stomach. PLoS ONE 2018, 13, e0208204. [Google Scholar] [CrossRef] [PubMed]
- Turula, H.; Wobus, C.E. The Role of the Polymeric Immunoglobulin Receptor and Secretory Immunoglobulins during Mucosal Infection and Immunity. Viruses 2018, 10, 237. [Google Scholar] [CrossRef] [PubMed]
- Bondt, A.; Dingess, K.A.; Hoek, M.; van Rijswijck, D.M.H.; Heck, A.J.R. A Direct MS-Based Approach to Profile Human Milk Secretory Immunoglobulin A (IgA1) Reveals Donor-Specific Clonal Repertoires with High Longitudinal Stability. Front. Immunol. 2021, 12, 789748. [Google Scholar] [CrossRef]
- Virca, G.D.; Salvesen, G.S.; Travis, J. Human Neutrophil Elastase and Cathepsin G Cleavage Sites in the Bait Region of Alpha 2-Macroglobulin. Proposed Structural Limits of the Bait Region. Hoppe. Seylers. Z. Physiol. Chem. 1983, 364, 1297–1302. [Google Scholar] [CrossRef]
- Hall, P.K.; Nelles, L.P.; Travis, J.; Roberts, R.C. Proteolytic Cleavage Sites on Alpha 2-Macroglobulin Resulting in Proteinase Binding Are Different for Trypsin and Staphylococcus Aureus V-8 Proteinase. Biochem. Biophys. Res. Commun. 1981, 100, 8–16. [Google Scholar] [CrossRef]
- Christensen, B.; Schack, L.; Kläning, E.; Sørensen, E.S. Osteopontin Is Cleaved at Multiple Sites Close to Its Integrin-Binding Motifs in Milk and Is a Novel Substrate for Plasmin and Cathepsin D. J. Biol. Chem. 2010, 285, 7929–7937. [Google Scholar] [CrossRef] [PubMed]
- Hernández-Ledesma, B.; Quirós, A.; Amigo, L.; Recio, I. Identification of Bioactive Peptides after Digestion of Human Milk and Infant Formula with Pepsin and Pancreatin. Int. Dairy J. 2007, 17, 42–49. [Google Scholar] [CrossRef]



| Time | Month 1 | Month 3 | p-Value | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Donor | 1 | 2 | 3 | 4 | 5 | 6 | Average | 1 | 2 | 3 | 4 | 5 | 6 | Average | |
| Proteins identified | |||||||||||||||
| Proteome | 1996 | 1697 | 1350 | 2001 | 1601 | 2772 | 1903 ± 445 | 2137 | 1755 | 1550 | 1324 | 1623 | 2262 | 1775 ± 328 | 0.62 |
| Peptidome | 302 | 445 | 307 | 292 | 315 | 294 | 326 ± 54 | 60 | 306 | 351 | 319 | 274 | 306 | 269 ± 96 | 0.29 |
| Unique proteins 1 | 2132 | 1973 | 1516 | 2160 | 1771 | 2889 | 2074 ± 426 | 2164 | 1904 | 1761 | 1486 | 1750 | 2390 | 1909 ± 295 | 0.50 |
| Overlap 2 | 166 | 169 | 141 | 133 | 145 | 177 | 155 ± 16 | 33 | 157 | 140 | 157 | 147 | 178 | 135 ± 47 | 0.41 |
| Overlap in% of unique 3 | 7.79 | 8.57 | 9.30 | 6.16 | 8.19 | 6.13 | 7.69 ± 1.18 | 1.52 | 8.25 | 7.95 | 10.57 | 8.40 | 7.45 | 7.36 ± 2.79 | 0.81 |
| Peptidome in% of unique 4 | 14.17 | 22.55 | 20.25 | 13.52 | 17.79 | 10.18 | 16.41 ± 4.22 | 2.77 | 16.07 | 19.93 | 21.47 | 15.66 | 12.80 | 13.35 ± 6.20 | 0.64 |
| Peptides identified | |||||||||||||||
| Proteome | 13,065 | 10,424 | 6848 | 12,490 | 8978 | 18,396 | 11,700 ± 3649 | 14,127 | 9693 | 8808 | 7775 | 11,036 | 16,255 | 11,282 ± 2995 | 0.85 |
| Peptidome | 2464 | 5039 | 2603 | 4295 | 2747 | 4316 | 3577 ± 1006 | 208 | 2560 | 4627 | 2868 | 1830 | 3355 | 2575 ± 1358 | 0.22 |
| Unique peptides 1 | 15,479 | 15,402 | 9380 | 16,719 | 11,687 | 22,640 | 15,218 ± 4163 | 14,334 | 12,164 | 13,403 | 10,553 | 12,828 | 19,525 | 13,801 ± 2810 | 0.54 |
| Overlap 2 | 60 | 72 | 82 | 75 | 45 | 80 | 69 ± 13 | 7 | 100 | 39 | 99 | 44 | 95 | 64 ± 36 | 0.78 |
| Overlap in% of unique 3 | 0.39 | 0.47 | 0.87 | 0.45 | 0.39 | 0.35 | 0.49 ± 0.18 | 0.05 | 0.82 | 0.29 | 0.94 | 0.34 | 0.49 | 0.49 ± 0.31 | 0.99 |
| Peptidome in% of unique 4 | 15.92 | 32.72 | 27.75 | 25.69 | 23.50 | 19.06 | 24.11 ± 5.52 | 1.45 | 21.05 | 34.52 | 27.18 | 14.27 | 17.18 | 19.92 ± 10.67 | 0.39 |
| Protein ID 1 | Gene Name | Protease | Average ± St.dev 2 (%) | Type 1 | Class 1 |
|---|---|---|---|---|---|
| Q96KP4 | CNDP2 | Cytosolic non-specific dipeptidase | 0.18 ± 0.09 | D | M |
| P00751 | CFB | Complement factor B | 0.07 ± 0.05 | P | S |
| P07339 | CTSD | Cathepsin D | 0.05 ± 0.07 | P | A |
| Q92876 | KLK6 | Kallikrein-6 | 0.05 ± 0.03 | P | S |
| P00747 | PLG | Plasminogen | 0.03 ± 0.02 | P | S |
| P09960 | LTA4H | Leukotriene | 0.03 ± 0.02 | A | M |
| P00734 | F2 | Prothrombin | 0.03 ± 0.02 | P | S |
| Q9NZ08 | ERAP1 | Endoplasmic reticulum aminopeptidase 1 | 0.03 ± 0.02 | A | M |
| P07858 | CTSB | Cathepsin B | 0.02 ± 0.02 | C, D, P 3 | T |
| P53634 | CTSC | Dipeptidyl peptidase 1 | 0.02 ± 0.01 | A | T |
| Q99538 | LGMN | Legumain | 0.02 ± 0.01 | P | T |
| P25774 | CTSS | Cathepsin S | 0.02 ± 0.01 | P | T |
| P55786 | NPEPPS | Puromycin-sensitive aminopeptidase | 0.01 ± 0.01 | A | M |
| Q9BYF1 | ACE2 | Angiotensin-converting enzyme 2 | 0.01 ± 0.01 | C, P | M |
| O60259 | KLK8 | Kallikrein-8 | 0.01 ± 0.01 | P | S |
| Q16651 | PRSS8 | Prostasin | 0.01 ± 0.01 | P | S |
| Q9H4A4 | RNPEP | Aminopeptidase B | 0.01 ± 4.47 × 10−3 | A | M |
| Q9UHL4 | DPP7 | Dipeptidyl peptidase 2 | 0.01 ± 0.01 | A | S |
| Q9NY33 | DPP3 | Dipeptidyl peptidase 3 | 0.01 ± 0.01 | A | M |
| P15144 | ANPEP | Aminopeptidase N | 0.01 ± 0.01 | A | M |
| P06681 | C2 | Complement C2 | 0.01 ± 0.01 | P | S |
| Q66K79 | CPZ | Carboxypeptidase Z | 0.01 ± 0.01 | C | M |
| Q04609 | FOLH1 | Glutamate carboxypeptidase 2 | 0.01 ± 4.23 × 10−3 | C | M |
| P09237 | MMP7 | Matrilysin | 0.01 ± 0.01 | P | M |
| P48147 | PREP | Prolyl endopeptidase | 0.01 ± 0.01 | P | S |
| Q9NQW7 | XPNPEP1 | Xaa-Pro aminopeptidase 1 | 0.01 ± 3.57 × 10−3 | A | M |
| P10619 | CTSA | Lysosomal protective protein | 0.01 ± 4.56 × 10−3 | C, P | S |
| P29122 | PCSK6 | Proprotein convertase subtilisin/kexin type 6 | <0.01 | P | S |
| Q96FW1 | OTUB1 | Ubiquitin thioesterase OTUB1 | <0.01 | P | T |
| P00736 | C1R | Complement C1r subcomponent | <0.01 | P | S |
| P42574 | CASP3 | Caspase-3 | <0.01 | P | T |
| Q9Y5Y6 | ST14 | Suppressor of tumorigenicity 14 protein | <0.01 | P | S |
| P55212 | CASP6 | Caspase-6 | <0.01 | P | T |
| Q9ULA0 | DNPEP | Aspartyl aminopeptidase | <0.01 | A | M |
| P13497 | BMP1 | Bone morphogenetic protein 1 | <0.01 | P | M |
| Q96IY4 | CPB2 | Carboxypeptidase B2 | <0.01 | C | M |
| P03952 | KLKB1 | Plasma kallikrein | <0.01 | P | S |
| P42785 | PRCP | Lysosomal Pro-X carboxypeptidase | <0.01 | C | S |
| P00746 | CFD | Complement factor D | <0.01 | P | S |
| Protein ID 1 | Gene | Inhibitor Name | Average ± St.dev. (in%) | Class of Protease Inhibitor |
|---|---|---|---|---|
| P29622 | SERPINA4 | Kallistatin | 1.39 ± 2.23 | S |
| P01011 | SERPINA3 | α-1-antichymotrypsin | 0.54 ± 0.47 | S |
| P01009 | SERPINA1 | α-1-antitrypsin | 0.40 ± 0,12 | S |
| P01034 | CST3 | Cystatin-C | 0.19 ± 0.07 | T |
| P01033 | TIMP1 | Metalloproteinase inhibitor 1 | 0.11 ± 0.05 | M |
| P01042 | KNG1 | Kininogen-1 | 0.10 ± 0.03 | T |
| P01008 | SERPINC1 | Antithrombin-III | 0.06 ± 0.04 | S |
| P05155 | SERPING1 | Plasma protease C1 inhibitor | 0.05 ± 0.02 | S |
| P01023 | A2M | α-2-macroglobulin | 0.04 ± 0.02 | S, T, M 2 |
| P08697 | SERPINF2 | α-2-antiplasmin | 0.04 ± 0.01 | S |
| P05067 | APP | Amyloid-beta precursor protein | 0.02 ± 0.02 | S |
| P30086 | PEBP1 | Phosphatidylethanolamine-binding protein 1 | 0.02 ± 0.02 | S |
| P02760 | AMBP | Protein AMBP | 0.02 ± 0.01 | S |
| P04080 | CSTB | Cystatin-B | 0.02 ± 0.01 | T |
| P19827 | ITIH1 | Inter- α -trypsin inhibitor heavy chain H1 | 0.01 ± 0.02 | S |
| P35237 | SERPINB6 | Serpin B6 | 0.01 ± 0.01 | S |
| P19823 | ITIH2 | Inter- α -trypsin inhibitor heavy chain H2 | 0.01 ± 0.01 | S |
| Q99727 | TIMP4 | Metalloproteinase inhibitor 4 | 0.01 ± 7.89 × 10−3 | M |
| P05546 | SERPIND1 | Heparin cofactor 2 | 0.01 ± 6.68 × 10−3 | S |
| Q14508 | WFDC2 | WAP four-disulfide core domain protein 2 | 0.01 ± 8.06 × 10−3 | S, T |
| P30740 | SERPINB1 | Leukocyte elastase inhibitor | <0.01 | S |
| Q99574 | SERPINI1 | Neuroserpin | <0.01 | S |
| Q06481 | APLP2 | Amyloid-beta precursor-like protein 2 | <0.01 | S |
| P20810 | CAST | Calpastatin | <0.01 | T |
| P05154 | SERPINA5 | Plasma serine protease inhibitor | <0.01 | S |
| O95980 | RECK | Reversion-inducing cysteine-rich protein with Kazal motifs | <0.01 | S |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Thesbjerg, M.N.; Nielsen, S.D.-H.; Sundekilde, U.K.; Poulsen, N.A.; Larsen, L.B. Fingerprinting of Proteases, Protease Inhibitors and Indigenous Peptides in Human Milk. Nutrients 2023, 15, 4169. https://doi.org/10.3390/nu15194169
Thesbjerg MN, Nielsen SD-H, Sundekilde UK, Poulsen NA, Larsen LB. Fingerprinting of Proteases, Protease Inhibitors and Indigenous Peptides in Human Milk. Nutrients. 2023; 15(19):4169. https://doi.org/10.3390/nu15194169
Chicago/Turabian StyleThesbjerg, Martin Nørmark, Søren Drud-Heydary Nielsen, Ulrik Kræmer Sundekilde, Nina Aagaard Poulsen, and Lotte Bach Larsen. 2023. "Fingerprinting of Proteases, Protease Inhibitors and Indigenous Peptides in Human Milk" Nutrients 15, no. 19: 4169. https://doi.org/10.3390/nu15194169
APA StyleThesbjerg, M. N., Nielsen, S. D.-H., Sundekilde, U. K., Poulsen, N. A., & Larsen, L. B. (2023). Fingerprinting of Proteases, Protease Inhibitors and Indigenous Peptides in Human Milk. Nutrients, 15(19), 4169. https://doi.org/10.3390/nu15194169








