Epigenetics in Obesity and Diabetes Mellitus: New Insights
Abstract
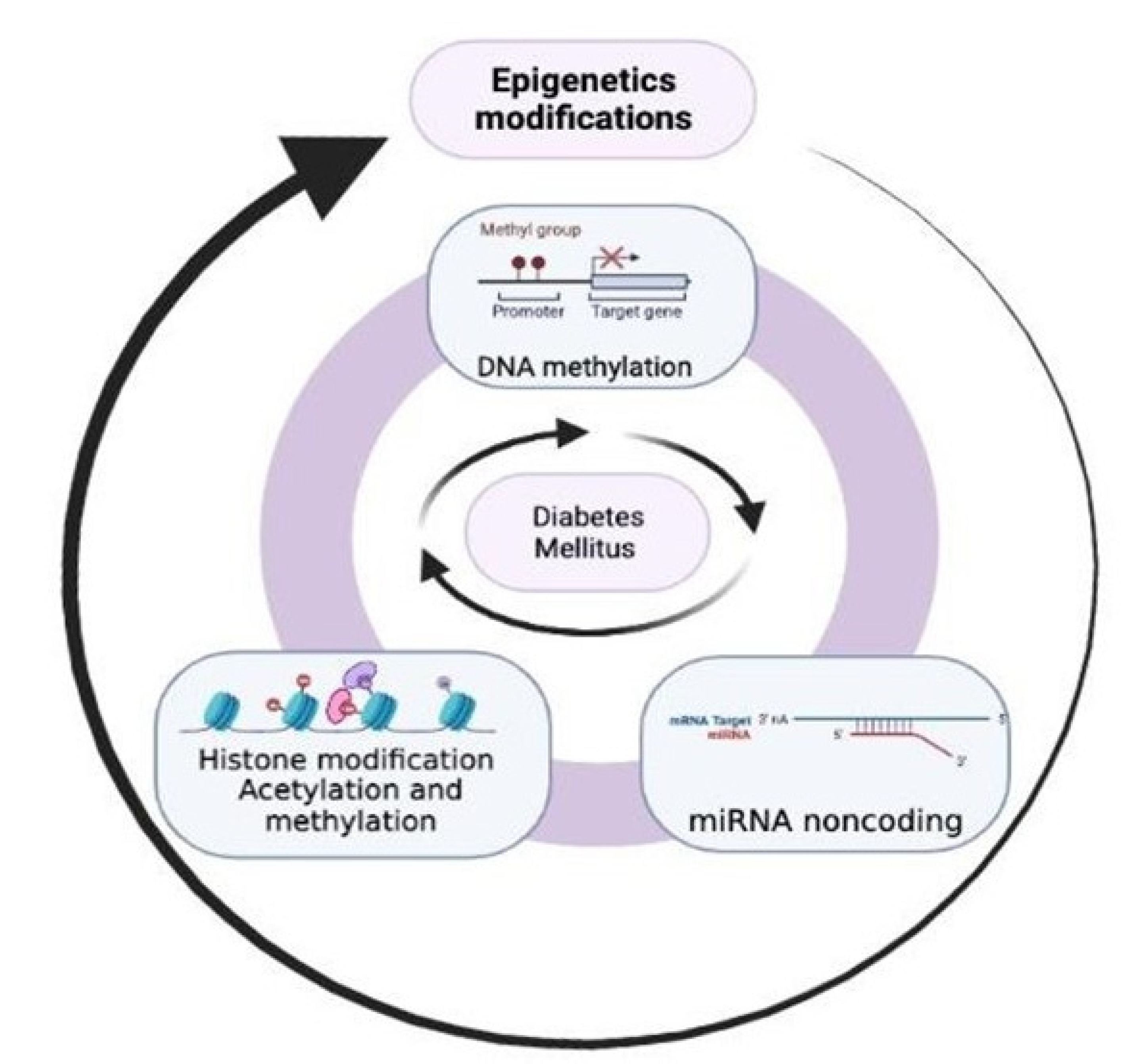
:1. Introduction
2. Obesity, Inflammation, and Epigenetics
3. Epigenetics in Diabetes Mellitus
4. From the Bench to the Bedside
4.1. Diabetic Nephropathy and Epigenetics
4.2. Diabetic Retinopathy and Epigenetics
4.3. Diabetic Neuropathy and Epigenetics
5. Is There a Possible Therapeutic Related to Epigenetics?
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Noble, D. Conrad Waddington and the Origin of Epigenetics. J. Exp. Biol. 2015, 218, 816–818. [Google Scholar] [CrossRef]
- Teschendorff, A.E.; West, J.; Beck, S. Age-Associated Epigenetic Drift: Implications, and a Case of Epigenetic Thrift? Hum. Mol. Genet. 2013, 22, R7–R15. [Google Scholar] [CrossRef]
- Vineis, P.; Chatziioannou, A.; Cunliffe, V.T.; Flanagan, J.M.; Hanson, M.; Kirsch-Volders, M.; Kyrtopoulos, S. Epigenetic Memory in Response to Environmental Stressors. FASEB J. 2017, 31, 2241–2251. [Google Scholar] [CrossRef]
- Rönn, T.; Ling, C. DNA Methylation as a Diagnostic and Therapeutic Target in the Battle against Type 2 Diabetes. Epigenomics 2015, 7, 451–460. [Google Scholar] [CrossRef]
- Sadakierska-Chudy, A.; Filip, M. A Comprehensive View of the Epigenetic Landscape. Part II: Histone Post-Translational Modification, Nucleosome Level, and Chromatin Regulation by NcRNAs. Neurotox. Res. 2015, 27, 172–197. [Google Scholar] [CrossRef]
- Chen, Z.; Miao, F.; Braffett, B.H.; Lachin, J.M.; Zhang, L.; Wu, X.; Roshandel, D.; Carless, M.; Li, X.A.; Tompkins, J.D.; et al. DNA Methylation Mediates Development of HbA1c-Associated Complications in Type 1 Diabetes. Nat. Metab. 2020, 2, 744–762. [Google Scholar] [CrossRef]
- Li, Y.; Zhang, Z.; Chen, J.; Liu, W.; Lai, W.; Liu, B.; Li, X.; Liu, L.; Xu, S.; Dong, Q.; et al. Stella Safeguards the Oocyte Methylome by Preventing de Novo Methylation Mediated by DNMT1. Nature 2018, 564, 136–140. [Google Scholar] [CrossRef]
- Kowluru, R.A.; Mohammad, G. Epigenetic Modifications in Diabetes. Metabolism 2022, 126, 154920. [Google Scholar] [CrossRef]
- Sarg, B.; Lopez, R.; Lindner, H.; Ponte, I.; Suau, P.; Roque, A. Identification of Novel Post-Translational Modifications in Linker Histones from Chicken Erythrocytes. J. Proteom. 2015, 113, 162–177. [Google Scholar] [CrossRef]
- Singh, R.; Chandel, S.; Dey, D.; Ghosh, A.; Roy, S.; Ravichandiran, V.; Ghosh, D. Epigenetic Modification and Therapeutic Targets of Diabetes Mellitus. Biosci. Rep. 2020, 40, BSR20202160. [Google Scholar] [CrossRef]
- Tanwar, V.S.; Reddy, M.A.; Natarajan, R. Emerging Role of Long Non-Coding RNAs in Diabetic Vascular Complications. Front. Endocrinol. 2021, 12, 665811. [Google Scholar] [CrossRef]
- Eliasson, L.; Esguerra, J.L.S. MicroRNA Networks in Pancreatic Islet Cells: Normal Function and Type 2 Diabetes. Diabetes 2020, 69, 804–812. [Google Scholar] [CrossRef]
- Ho, S.-M.; Cheong, A.; Adgent, M.A.; Veevers, J.; Suen, A.A.; Tam, N.N.C.; Leung, Y.-K.; Jefferson, W.N.; Williams, C.J. Environmental Factors, Epigenetics, and Developmental Origin of Reproductive Disorders. Reprod. Toxicol. 2017, 68, 85–104. [Google Scholar] [CrossRef]
- Sundar, I.K.; Rahman, I. Gene Expression Profiling of Epigenetic Chromatin Modification Enzymes and Histone Marks by Cigarette Smoke: Implications for COPD and Lung Cancer. Am. J. Physiol.-Lung Cell. Mol. Physiol. 2016, 311, L1245–L1258. [Google Scholar] [CrossRef]
- Sundar, I.K.; Yin, Q.; Baier, B.S.; Yan, L.; Mazur, W.; Li, D.; Susiarjo, M.; Rahman, I. DNA Methylation Profiling in Peripheral Lung Tissues of Smokers and Patients with COPD. Clin. Epigenet. 2017, 9, 38. [Google Scholar] [CrossRef]
- Biswas, S.; Rao, C.M. Epigenetics in Cancer: Fundamentals and Beyond. Pharmacol. Ther. 2017, 173, 118–134. [Google Scholar] [CrossRef]
- Jadhav, R.A.; Maiya, G.A.; Shivashankara, K.N.; Umakanth, S. Measurement of Visceral Fat for Early Prediction of Prediabetes-Cross-Sectional Study from Southern India. J. Taibah Univ. Med. Sci. 2022, 17, 983–990. [Google Scholar] [CrossRef]
- Hong, J.; Kim, Y.-H. Fatty Liver/Adipose Tissue Dual-Targeting Nanoparticles with Heme Oxygenase-1 Inducer for Amelioration of Obesity, Obesity-Induced Type 2 Diabetes, and Steatohepatitis. Adv. Sci. 2022, 9, e2203286. [Google Scholar] [CrossRef]
- Ling, C.; Rönn, T. Epigenetics in Human Obesity and Type 2 Diabetes. Cell Metab. 2019, 29, 1028–1044. [Google Scholar] [CrossRef]
- Parveen, N.; Dhawan, S. DNA Methylation Patterning and the Regulation of Beta Cell Homeostasis. Front. Endocrinol. 2021, 12, 651258. [Google Scholar] [CrossRef]
- Maude, H.; Sanchez-Cabanillas, C.; Cebola, I. Epigenetics of Hepatic Insulin Resistance. Front. Endocrinol. 2021, 12, 681356. [Google Scholar] [CrossRef]
- Edillor, C.R.; Parks, B.W.; Mehrabian, M.; Lusis, A.J.; Pellegrini, M. DNA Methylation Changes More Slowly Than Physiological States in Response to Weight Loss in Genetically Diverse Mouse Strains. Front. Endocrinol. 2019, 10, 882. [Google Scholar] [CrossRef]
- Arpón, A.; Milagro, F.I.; Santos, J.L.; García-Granero, M.; Riezu-Boj, J.I.; Martínez, J.A. Interaction among Sex, Aging, and Epigenetic Processes Concerning Visceral Fat, Insulin Resistance, and Dyslipidaemia. Front. Endocrinol. 2019, 10, 496. [Google Scholar] [CrossRef]
- Chambers, J.C.; Loh, M.; Lehne, B.; Drong, A.; Kriebel, J.; Motta, V.; Wahl, S.; Elliott, H.R.; Rota, F.; Scott, W.R.; et al. Epigenome-Wide Association of DNA Methylation Markers in Peripheral Blood from Indian Asians and Europeans with Incident Type 2 Diabetes: A Nested Case-Control Study. Lancet Diabetes Endocrinol. 2015, 3, 526–534. [Google Scholar] [CrossRef]
- Gutiérrez-Repiso, C.; Linares-Pineda, T.M.; Gonzalez-Jimenez, A.; Aguilar-Lineros, F.; Valdés, S.; Soriguer, F.; Rojo-Martínez, G.; Tinahones, F.J.; Morcillo, S. Epigenetic Biomarkers of Transition from Metabolically Healthy Obesity to Metabolically Unhealthy Obesity Phenotype: A Prospective Study. Int. J. Mol. Sci. 2021, 22, 10417. [Google Scholar] [CrossRef]
- Aslibekyan, S.; Demerath, E.W.; Mendelson, M.; Zhi, D.; Guan, W.; Liang, L.; Sha, J.; Pankow, J.S.; Liu, C.; Irvin, M.R.; et al. Epigenome-Wide Study Identifies Novel Methylation Loci Associated with Body Mass Index and Waist Circumference. Obesity 2015, 23, 1493–1501. [Google Scholar] [CrossRef]
- Kwon, E.J.; You, Y.-A.; Park, B.; Ha, E.H.; Kim, H.S.; Park, H.; Kim, Y.J. Association between the DNA Methylations of POMC, MC4R, and HNF4A and Metabolic Profiles in the Blood of Children Aged 7-9 Years. BMC Pediatr. 2018, 18, 121. [Google Scholar] [CrossRef]
- Obri, A.; Serra, D.; Herrero, L.; Mera, P. The Role of Epigenetics in the Development of Obesity. Biochem. Pharmacol. 2020, 177, 113973. [Google Scholar] [CrossRef]
- Jung, B.C.; Kang, S. Epigenetic Regulation of Inflammatory Factors in Adipose Tissue. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2021, 1866, 159019. [Google Scholar] [CrossRef]
- Naidoo, V.; Naidoo, M.; Ghai, M. Cell and Tissue-Specific Epigenetic Changes Associated with Chronic Inflammation in Insulin Resistance and Type 2 Diabetes Mellitus. Scand. J. Immunol. 2018, 88, e12723. [Google Scholar] [CrossRef]
- Ramos-Lopez, O.; Milagro, F.I.; Riezu-Boj, J.I.; Martinez, J.A. Epigenetic Signatures Underlying Inflammation: An Interplay of Nutrition, Physical Activity, Metabolic Diseases, and Environmental Factors for Personalized Nutrition. Inflamm. Res. 2021, 70, 29–49. [Google Scholar] [CrossRef]
- Ndisang, J.F.; Rastogi, S.; Vannacci, A. Immune and Inflammatory Processes in Obesity, Insulin Resistance, Diabetes, and Related Cardiometabolic Complications. J. Immunol. Res. 2014, 2014, 579560. [Google Scholar] [CrossRef]
- Dayeh, T.; Tuomi, T.; Almgren, P.; Perfilyev, A.; Jansson, P.A.; de Mello, V.D.; Pihlajamäki, J.; Vaag, A.; Groop, L.; Nilsson, E.; et al. DNA Methylation of Loci within ABCG1 and PHOSPHO1 in Blood DNA Is Associated with Future Type 2 Diabetes Risk. Epigenetics 2016, 11, 482–488. [Google Scholar] [CrossRef]
- Malodobra-Mazur, M.; Dziewulska, A.; Kozinski, K.; Dobrzyn, P.; Kolczynska, K.; Janikiewicz, J.; Dobrzyn, A. Stearoyl-CoA Desaturase Regulates Inflammatory Gene Expression by Changing DNA Methylation Level in 3T3 Adipocytes. Int. J. Biochem. Cell Biol. 2014, 55, 40–50. [Google Scholar] [CrossRef]
- You, D.; Nilsson, E.; Tenen, D.E.; Lyubetskaya, A.; Lo, J.C.; Jiang, R.; Deng, J.; Dawes, B.A.; Vaag, A.; Ling, C.; et al. Dnmt3a Is an Epigenetic Mediator of Adipose Insulin Resistance. eLife 2017, 6, e30766. [Google Scholar] [CrossRef]
- Castellano-Castillo, D.; Denechaud, P.-D.; Fajas, L.; Moreno-Indias, I.; Oliva-Olivera, W.; Tinahones, F.; Queipo-Ortuño, M.I.; Cardona, F. Human Adipose Tissue H3K4me3 Histone Mark in Adipogenic, Lipid Metabolism and Inflammatory Genes Is Positively Associated with BMI and HOMA-IR. PLoS ONE 2019, 14, e0215083. [Google Scholar] [CrossRef]
- Hex, N.; Bartlett, C.; Wright, D.; Taylor, M.; Varley, D. Estimating the Current and Future Costs of Type 1 and Type 2 Diabetes in the UK, Including Direct Health Costs and Indirect Societal and Productivity Costs. Diabet. Med. 2012, 29, 855–862. [Google Scholar] [CrossRef]
- Bommer, C.; Sagalova, V.; Heesemann, E.; Manne-Goehler, J.; Atun, R.; Bärnighausen, T.; Davies, J.; Vollmer, S. Global Economic Burden of Diabetes in Adults: Projections From 2015 to 2030. Diabetes Care 2018, 41, 963–970. [Google Scholar] [CrossRef]
- Meng, R.; Lv, J.; Yu, C.; Guo, Y.; Bian, Z.; Yang, L.; Chen, Y.; Zhang, H.; Chen, X.; Chen, J.; et al. Prenatal Famine Exposure, Adulthood Obesity Patterns and Risk of Type 2 Diabetes. Int. J. Epidemiol. 2018, 47, 399–408. [Google Scholar] [CrossRef]
- Vaiserman, A.; Lushchak, O. Developmental Origins of Type 2 Diabetes: Focus on Epigenetics. Ageing Res. Rev. 2019, 55, 100957. [Google Scholar] [CrossRef]
- Sommese, L.; Benincasa, G.; Lanza, M.; Sorriento, A.; Schiano, C.; Lucchese, R.; Alfano, R.; Nicoletti, G.F.; Napoli, C. Novel Epigenetic-Sensitive Clinical Challenges Both in Type 1 and Type 2 Diabetes. J. Diabetes Complicat. 2018, 32, 1076–1084. [Google Scholar] [CrossRef]
- Liang, Y.-Z.; Dong, J.; Zhang, J.; Wang, S.; He, Y.; Yan, Y.-X. Identification of Neuroendocrine Stress Response-Related Circulating MicroRNAs as Biomarkers for Type 2 Diabetes Mellitus and Insulin Resistance. Front. Endocrinol. 2018, 9, 132. [Google Scholar] [CrossRef]
- Martínez-Ramírez, O.C.; Salazar-Piña, A.; Cerón-Ramírez, X.; Rubio-Lightbourn, J.; Torres-Romero, F.; Casas-Avila, L.; Castro-Hernández, C. Effect of Inulin Intervention on Metabolic Control and Methylation of INS and IRS1 Genes in Patients with Type 2 Diabetes Mellitus. Nutrients 2022, 14, 5195. [Google Scholar] [CrossRef]
- Volkov, P.; Bacos, K.; Ofori, J.K.; Esguerra, J.L.S.; Eliasson, L.; Rönn, T.; Ling, C. Whole-Genome Bisulfite Sequencing of Human Pancreatic Islets Reveals Novel Differentially Methylated Regions in Type 2 Diabetes Pathogenesis. Diabetes 2017, 66, 1074–1085. [Google Scholar] [CrossRef]
- Olsson, A.H.; Volkov, P.; Bacos, K.; Dayeh, T.; Hall, E.; Nilsson, E.A.; Ladenvall, C.; Rönn, T.; Ling, C. Genome-Wide Associations between Genetic and Epigenetic Variation Influence MRNA Expression and Insulin Secretion in Human Pancreatic Islets. PLoS Genet. 2014, 10, e1004735. [Google Scholar] [CrossRef]
- Barajas-Olmos, F.; Centeno-Cruz, F.; Zerrweck, C.; Imaz-Rosshandler, I.; Martínez-Hernández, A.; Cordova, E.J.; Rangel-Escareño, C.; Gálvez, F.; Castillo, A.; Maydón, H.; et al. Altered DNA Methylation in Liver and Adipose Tissues Derived from Individuals with Obesity and Type 2 Diabetes. BMC Med. Genet. 2018, 19, 28. [Google Scholar] [CrossRef]
- Chen, Y.-T.; Liao, J.-W.; Tsai, Y.-C.; Tsai, F.-J. Inhibition of DNA Methyltransferase 1 Increases Nuclear Receptor Subfamily 4 Group A Member 1 Expression and Decreases Blood Glucose in Type 2 Diabetes. Oncotarget 2016, 7, 39162–39170. [Google Scholar] [CrossRef]
- Esau, C.; Kang, X.; Peralta, E.; Hanson, E.; Marcusson, E.G.; Ravichandran, L.V.; Sun, Y.; Koo, S.; Perera, R.J.; Jain, R.; et al. MicroRNA-143 Regulates Adipocyte Differentiation. J. Biol. Chem. 2004, 279, 52361–52365. [Google Scholar] [CrossRef]
- Li, X.; Li, C.; Sun, G. Histone Acetylation and Its Modifiers in the Pathogenesis of Diabetic Nephropathy. J. Diabetes Res. 2016, 2016, 4065382. [Google Scholar] [CrossRef]
- Morán, I.; Akerman, I.; van de Bunt, M.; Xie, R.; Benazra, M.; Nammo, T.; Arnes, L.; Nakić, N.; García-Hurtado, J.; Rodríguez-Seguí, S.; et al. Human β Cell Transcriptome Analysis Uncovers LncRNAs That Are Tissue-Specific, Dynamically Regulated, and Abnormally Expressed in Type 2 Diabetes. Cell Metab. 2012, 16, 435–448. [Google Scholar] [CrossRef]
- Akerman, I.; Tu, Z.; Beucher, A.; Rolando, D.M.Y.; Sauty-Colace, C.; Benazra, M.; Nakic, N.; Yang, J.; Wang, H.; Pasquali, L.; et al. Human Pancreatic β Cell LncRNAs Control Cell-Specific Regulatory Networks. Cell Metab. 2017, 25, 400–411. [Google Scholar] [CrossRef] [PubMed]
- Brennan, E.; Wang, B.; McClelland, A.; Mohan, M.; Marai, M.; Beuscart, O.; Derouiche, S.; Gray, S.; Pickering, R.; Tikellis, C.; et al. Protective Effect of Let-7 MiRNA Family in Regulating Inflammation in Diabetes-Associated Atherosclerosis. Diabetes 2017, 66, 2266–2277. [Google Scholar] [CrossRef]
- Voight, B.F.; Scott, L.J.; Steinthorsdottir, V.; Morris, A.P.; Dina, C.; Welch, R.P.; Zeggini, E.; Huth, C.; Aulchenko, Y.S.; Thorleifsson, G.; et al. Twelve Type 2 Diabetes Susceptibility Loci Identified through Large-Scale Association Analysis. Nat. Genet. 2010, 42, 579–589. [Google Scholar] [CrossRef] [PubMed]
- Kooner, J.S.; Saleheen, D.; Sim, X.; Sehmi, J.; Zhang, W.; Frossard, P.; Been, L.F.; Chia, K.-S.; Dimas, A.S.; Hassanali, N.; et al. Genome-Wide Association Study in Individuals of South Asian Ancestry Identifies Six New Type 2 Diabetes Susceptibility Loci. Nat. Genet. 2011, 43, 984–989. [Google Scholar] [CrossRef] [PubMed]
- Nathan, D.M.; Cleary, P.A.; Backlund, J.-Y.C.; Genuth, S.M.; Lachin, J.M.; Orchard, T.J.; Raskin, P.; Zinman, B. Intensive Diabetes Treatment and Cardiovascular Disease in Patients with Type 1 Diabetes. N. Engl. J. Med. 2005, 353, 2643–2653. [Google Scholar] [CrossRef] [PubMed]
- Bjornstad, P.; Drews, K.L.; Caprio, S.; Gubitosi-Klug, R.; Nathan, D.M.; Tesfaldet, B.; Tryggestad, J.; White, N.H.; Zeitler, P. Long-Term Complications in Youth-Onset Type 2 Diabetes. N. Engl. J. Med. 2021, 385, 416–426. [Google Scholar] [CrossRef]
- Lachin, J.M.; Nathan, D.M. Understanding Metabolic Memory: The Prolonged Influence of Glycemia During the Diabetes Control and Complications Trial (DCCT) on Future Risks of Complications During the Study of the Epidemiology of Diabetes Interventions and Complications (EDIC). Diabetes Care 2021, 44, 2216–2224. [Google Scholar] [CrossRef]
- Qie, R.; Chen, Q.; Wang, T.; Chen, X.; Wang, J.; Cheng, R.; Lin, J.; Zhao, Y.; Liu, D.; Qin, P.; et al. Association of ABCG1 Gene Methylation and Its Dynamic Change Status with Incident Type 2 Diabetes Mellitus: The Rural Chinese Cohort Study. J. Hum. Genet. 2021, 66, 347–357. [Google Scholar] [CrossRef] [PubMed]
- Tung, C.-W.; Hsu, Y.-C.; Shih, Y.-H.; Chang, P.-J.; Lin, C.-L. Glomerular Mesangial Cell and Podocyte Injuries in Diabetic Nephropathy. Nephrology 2018, 23 (Suppl. 4), 32–37. [Google Scholar] [CrossRef] [PubMed]
- Sugita, E.; Hayashi, K.; Hishikawa, A.; Itoh, H. Epigenetic Alterations in Podocytes in Diabetic Nephropathy. Front. Pharmacol. 2021, 12, 759299. [Google Scholar] [CrossRef]
- Hishikawa, A.; Hayashi, K.; Abe, T.; Kaneko, M.; Yokoi, H.; Azegami, T.; Nakamura, M.; Yoshimoto, N.; Kanda, T.; Sakamaki, Y.; et al. Decreased KAT5 Expression Impairs DNA Repair and Induces Altered DNA Methylation in Kidney Podocytes. Cell Rep. 2019, 26, 1318–1332.e4. [Google Scholar] [CrossRef] [PubMed]
- Smyth, L.J.; Kilner, J.; Nair, V.; Liu, H.; Brennan, E.; Kerr, K.; Sandholm, N.; Cole, J.; Dahlström, E.; Syreeni, A.; et al. Assessment of Differentially Methylated Loci in Individuals with End-Stage Kidney Disease Attributed to Diabetic Kidney Disease: An Exploratory Study. Clin. Epigenet. 2021, 13, 99. [Google Scholar] [CrossRef] [PubMed]
- Brasacchio, D.; Okabe, J.; Tikellis, C.; Balcerczyk, A.; George, P.; Baker, E.K.; Calkin, A.C.; Brownlee, M.; Cooper, M.E.; El-Osta, A. Hyperglycemia Induces a Dynamic Cooperativity of Histone Methylase and Demethylase Enzymes Associated with Gene-Activating Epigenetic Marks That Coexist on the Lysine Tail. Diabetes 2009, 58, 1229–1236. [Google Scholar] [CrossRef] [PubMed]
- Sun, G.; Reddy, M.A.; Yuan, H.; Lanting, L.; Kato, M.; Natarajan, R. Epigenetic Histone Methylation Modulates Fibrotic Gene Expression. J. Am. Soc. Nephrol. 2010, 21, 2069–2080. [Google Scholar] [CrossRef] [PubMed]
- Milas, O.; Gadalean, F.; Vlad, A.; Dumitrascu, V.; Gluhovschi, C.; Gluhovschi, G.; Velciov, S.; Popescu, R.; Bob, F.; Matusz, P.; et al. Deregulated Profiles of Urinary MicroRNAs May Explain Podocyte Injury and Proximal Tubule Dysfunction in Normoalbuminuric Patients with Type 2 Diabetes Mellitus. J. Investig. Med. 2018, 66, 747–754. [Google Scholar] [CrossRef]
- Petrica, L.; Hogea, E.; Gadalean, F.; Vlad, A.; Vlad, M.; Dumitrascu, V.; Velciov, S.; Gluhovschi, C.; Bob, F.; Ursoniu, S.; et al. Long Noncoding RNAs May Impact Podocytes and Proximal Tubule Function through Modulating MiRNAs Expression in Early Diabetic Kidney Disease of Type 2 Diabetes Mellitus Patients. Int. J. Med. Sci. 2021, 18, 2093–2101. [Google Scholar] [CrossRef]
- Dai, X.; Liao, R.; Liu, C.; Liu, S.; Huang, H.; Liu, J.; Jin, T.; Guo, H.; Zheng, Z.; Xia, M.; et al. Epigenetic Regulation of TXNIP-Mediated Oxidative Stress and NLRP3 Inflammasome Activation Contributes to SAHH Inhibition-Aggravated Diabetic Nephropathy. Redox Biol. 2021, 45, 102033. [Google Scholar] [CrossRef]
- Andersen, N.; Hjortdal, J.Ø.; Schielke, K.C.; Bek, T.; Grauslund, J.; Laugesen, C.S.; Lund-Andersen, H.; Cerqueira, C.; Andresen, J. The Danish Registry of Diabetic Retinopathy. Clin. Epidemiol. 2016, 8, 613–619. [Google Scholar] [CrossRef]
- Zhong, Q.; Kowluru, R.A. Role of Histone Acetylation in the Development of Diabetic Retinopathy and the Metabolic Memory Phenomenon. J. Cell. Biochem. 2010, 110, 1306–1313. [Google Scholar] [CrossRef]
- Liu, J.-Y.; Yao, J.; Li, X.-M.; Song, Y.-C.; Wang, X.-Q.; Li, Y.-J.; Yan, B.; Jiang, Q. Pathogenic Role of LncRNA-MALAT1 in Endothelial Cell Dysfunction in Diabetes Mellitus. Cell Death Dis. 2014, 5, e1506. [Google Scholar] [CrossRef] [Green Version]
- Kang, Q.; Yang, C. Oxidative Stress and Diabetic Retinopathy: Molecular Mechanisms, Pathogenetic Role and Therapeutic Implications. Redox Biol. 2020, 37, 101799. [Google Scholar] [CrossRef] [PubMed]
- Lund, J.; Ouwens, D.M.; Wettergreen, M.; Bakke, S.S.; Thoresen, G.H.; Aas, V. Increased Glycolysis and Higher Lactate Production in Hyperglycemic Myotubes. Cells 2019, 8, 1101. [Google Scholar] [CrossRef] [PubMed]
- Kowluru, R.A. Retinopathy in a Diet-Induced Type 2 Diabetic Rat Model and Role of Epigenetic Modifications. Diabetes 2020, 69, 689–698. [Google Scholar] [CrossRef]
- Katagiri, M.; Shoji, J.; Inada, N.; Kato, S.; Kitano, S.; Uchigata, Y. Evaluation of Vitreous Levels of Advanced Glycation End Products and Angiogenic Factors as Biomarkers for Severity of Diabetic Retinopathy. Int. Ophthalmol. 2018, 38, 607–615. [Google Scholar] [CrossRef] [PubMed]
- Lorenzi, M. The Polyol Pathway as a Mechanism for Diabetic Retinopathy: Attractive, Elusive, and Resilient. Exp. Diabetes Res. 2007, 2007, 61038. [Google Scholar] [CrossRef]
- Asnaghi, V.; Gerhardinger, C.; Hoehn, T.; Adeboje, A.; Lorenzi, M. A Role for the Polyol Pathway in the Early Neuroretinal Apoptosis and Glial Changes Induced by Diabetes in the Rat. Diabetes 2003, 52, 506–511. [Google Scholar] [CrossRef]
- Li, W.; Chen, S.; Mei, Z.; Zhao, F.; Xiang, Y. Polymorphisms in Sorbitol-Aldose Reductase (Polyol) Pathway Genes and Their Influence on Risk of Diabetic Retinopathy Among Han Chinese. Med. Sci. Monit. Int. Med. J. Exp. Clin. Res. 2019, 25, 7073–7078. [Google Scholar] [CrossRef]
- Jankovic, M.; Novakovic, I.; Nikolic, D.; Mitrovic Maksic, J.; Brankovic, S.; Petronic, I.; Cirovic, D.; Ducic, S.; Grajic, M.; Bogicevic, D. Genetic and Epigenomic Modifiers of Diabetic Neuropathy. Int. J. Mol. Sci. 2021, 22, 4887. [Google Scholar] [CrossRef]
- Fachrul, M.; Utomo, D.H.; Parikesit, A.A. LncRNA-Based Study of Epigenetic Regulations in Diabetic Peripheral Neuropathy. Silico Pharmacol. 2018, 6, 7. [Google Scholar] [CrossRef]
- Karki, D.B.; Yadava, S.K.; Pant, S.; Thusa, N.; Dangol, E.; Ghimire, S. Prevalence of Sensory Neuropathy in Type 2 Diabetes Mellitus and Its Correlation with Duration of Disease. Kathmandu Univ. Med. J. 2016, 14, 120–124. [Google Scholar]
- Huang, Y.; Liu, Y.; Li, L.; Su, B.; Yang, L.; Fan, W.; Yin, Q.; Chen, L.; Cui, T.; Zhang, J.; et al. Involvement of Inflammation-Related MiR-155 and MiR-146a in Diabetic Nephropathy: Implications for Glomerular Endothelial Injury. BMC Nephrol. 2014, 15, 142. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Y.-B.; Wu, Q.; Liu, J.; Fan, Y.-Z.; Yu, K.-F.; Cai, Y. MiR-199a-3p Is Involved in the Pathogenesis and Progression of Diabetic Neuropathy through Downregulation of SerpinE2. Mol. Med. Rep. 2017, 16, 2417–2424. [Google Scholar] [CrossRef] [PubMed]
- Ciccacci, C.; Latini, A.; Greco, C.; Politi, C.; D’Amato, C.; Lauro, D.; Novelli, G.; Borgiani, P.; Spallone, V. Association between a MIR499A Polymorphism and Diabetic Neuropathy in Type 2 Diabetes. J. Diabetes Complicat. 2018, 32, 11–17. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Song, C.; Liu, J.; Bi, Y.; Li, H. Inhibition of MiR-25 Aggravates Diabetic Peripheral Neuropathy. Neuroreport 2018, 29, 945–953. [Google Scholar] [CrossRef] [PubMed]
- Xourgia, E.; Papazafiropoulou, A.; Melidonis, A. Circulating MicroRNAs as Biomarkers for Diabetic Neuropathy: A Novel Approach. World J. Exp. Med. 2018, 8, 18–23. [Google Scholar] [CrossRef]
- Kato, M.; Natarajan, R. Epigenetics and Epigenomics in Diabetic Kidney Disease and Metabolic Memory. Nat. Rev. Nephrol. 2019, 15, 327–345. [Google Scholar] [CrossRef]
- Bansal, A.; Pinney, S.E. DNA Methylation and Its Role in the Pathogenesis of Diabetes. Pediatr. Diabetes 2017, 18, 167–177. [Google Scholar] [CrossRef]
- Arguelles, A.O.; Meruvu, S.; Bowman, J.D.; Choudhury, M. Are Epigenetic Drugs for Diabetes and Obesity at Our Door Step? Drug Discov. Today 2016, 21, 499–509. [Google Scholar] [CrossRef]
- Devaiah, B.N.; Case-Borden, C.; Gegonne, A.; Hsu, C.H.; Chen, Q.; Meerzaman, D.; Dey, A.; Ozato, K.; Singer, D.S. BRD4 Is a Histone Acetyltransferase That Evicts Nucleosomes from Chromatin. Nat. Struct. Mol. Biol. 2016, 23, 540–548. [Google Scholar] [CrossRef]
- Nicholls, S.J.; Schwartz, G.G.; Buhr, K.A.; Ginsberg, H.N.; Johansson, J.O.; Kalantar-Zadeh, K.; Kulikowski, E.; Toth, P.P.; Wong, N.; Sweeney, M.; et al. Apabetalone and Hospitalization for Heart Failure in Patients Following an Acute Coronary Syndrome: A Prespecified Analysis of the BETonMACE Study. Cardiovasc. Diabetol. 2021, 20, 13. [Google Scholar] [CrossRef]
- Ray, K.K.; Nicholls, S.J.; Buhr, K.A.; Ginsberg, H.N.; Johansson, J.O.; Kalantar-Zadeh, K.; Kulikowski, E.; Toth, P.P.; Wong, N.; Sweeney, M.; et al. Effect of Apabetalone Added to Standard Therapy on Major Adverse Cardiovascular Events in Patients With Recent Acute Coronary Syndrome and Type 2 Diabetes: A Randomized Clinical Trial. JAMA 2020, 323, 1565–1573. [Google Scholar] [CrossRef] [PubMed]
- Ray, K.K.; Nicholls, S.J.; Ginsberg, H.D.; Johansson, J.O.; Kalantar-Zadeh, K.; Kulikowski, E.; Toth, P.P.; Wong, N.; Cummings, J.L.; Sweeney, M.; et al. Effect of Selective BET Protein Inhibitor Apabetalone on Cardiovascular Outcomes in Patients with Acute Coronary Syndrome and Diabetes: Rationale, Design, and Baseline Characteristics of the BETonMACE Trial. Am. Heart J. 2019, 217, 72–83. [Google Scholar] [CrossRef] [PubMed]
- Kaimala, S.; Kumar, C.A.; Allouh, M.Z.; Ansari, S.A.; Emerald, B.S. Epigenetic Modifications in Pancreas Development, Diabetes, and Therapeutics. Med. Res. Rev. 2022, 42, 1343–1371. [Google Scholar] [CrossRef] [PubMed]
- The International Hypoglycaemia Study Group. Hypoglycaemia, Cardiovascular Disease, and Mortality in Diabetes: Epidemiology, Pathogenesis, and Management. Lancet Diabetes Endocrinol. 2019, 7, 385–396. [Google Scholar] [CrossRef] [PubMed]
- Hajmirza, A.; Emadali, A.; Gauthier, A.; Casasnovas, O.; Gressin, R.; Callanan, M.B. BET Family Protein BRD4: An Emerging Actor in NFκB Signaling in Inflammation and Cancer. Biomedicines 2018, 6, 16. [Google Scholar] [CrossRef] [PubMed]
- Gilham, D.; Tsujikawa, L.M.; Sarsons, C.D.; Halliday, C.; Wasiak, S.; Stotz, S.C.; Jahagirdar, R.; Sweeney, M.; Johansson, J.O.; Wong, N.C.W.; et al. Apabetalone Downregulates Factors and Pathways Associated with Vascular Calcification. Atherosclerosis 2019, 280, 75–84. [Google Scholar] [CrossRef]
- Wasiak, S.; Dzobo, K.E.; Rakai, B.D.; Kaiser, Y.; Versloot, M.; Bahjat, M.; Stotz, S.C.; Fu, L.; Sweeney, M.; Johansson, J.O.; et al. BET Protein Inhibitor Apabetalone (RVX-208) Suppresses pro-Inflammatory Hyper-Activation of Monocytes from Patients with Cardiovascular Disease and Type 2 Diabetes. Clin. Epigenet. 2020, 12, 166. [Google Scholar] [CrossRef]
- Haarhaus, M.; Ray, K.K.; Nicholls, S.J.; Schwartz, G.G.; Kulikowski, E.; Johansson, J.O.; Sweeney, M.; Halliday, C.; Lebioda, K.; Wong, N.; et al. Apabetalone Lowers Serum Alkaline Phosphatase and Improves Cardiovascular Risk in Patients with Cardiovascular Disease. Atherosclerosis 2019, 290, 59–65. [Google Scholar] [CrossRef]
- Charles, C.; Ferris, A.H. Chronic Kidney Disease. Prim. Care 2020, 47, 585–595. [Google Scholar] [CrossRef]
- Huang, D.; Refaat, M.; Mohammedi, K.; Jayyousi, A.; Al Suwaidi, J.; Abi Khalil, C. Macrovascular Complications in Patients with Diabetes and Prediabetes. BioMed Res. Int. 2017, 2017, 7839101. [Google Scholar] [CrossRef]
- Stratton, I.M.; Adler, A.I.; Neil, H.A.; Matthews, D.R.; Manley, S.E.; Cull, C.A.; Hadden, D.; Turner, R.C.; Holman, R.R. Association of Glycaemia with Macrovascular and Microvascular Complications of Type 2 Diabetes (UKPDS 35): Prospective Observational Study. BMJ 2000, 321, 405–412. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gæde, P.; Oellgaard, J.; Carstensen, B.; Rossing, P.; Lund-Andersen, H.; Parving, H.-H.; Pedersen, O. Years of Life Gained by Multifactorial Intervention in Patients with Type 2 Diabetes Mellitus and Microalbuminuria: 21 Years Follow-up on the Steno-2 Randomised Trial. Diabetologia 2016, 59, 2298–2307. [Google Scholar] [CrossRef]
- Kalantar-Zadeh, K.; Schwartz, G.G.; Nicholls, S.J.; Buhr, K.A.; Ginsberg, H.N.; Johansson, J.O.; Kulikowski, E.; Lebioda, K.; Toth, P.P.; Wong, N.; et al. Effect of Apabetalone on Cardiovascular Events in Diabetes, CKD, and Recent Acute Coronary Syndrome: Results from the BETonMACE Randomized Controlled Trial. Clin. J. Am. Soc. Nephrol. 2021, 16, 705–716. [Google Scholar] [CrossRef] [PubMed]
- Toth, P.P.; Schwartz, G.G.; Nicholls, S.J.; Khan, A.; Szarek, M.; Ginsberg, H.N.; Johansson, J.O.; Kalantar-Zadeh, K.; Kulikowski, E.; Lebioda, K.; et al. Reduction in the Risk of Major Adverse Cardiovascular Events with the BET Protein Inhibitor Apabetalone in Patients with Recent Acute Coronary Syndrome, Type 2 Diabetes, and Moderate to High Likelihood of Non-Alcoholic Fatty Liver Disease. Am. J. Prev. Cardiol. 2022, 11, 100372. [Google Scholar] [CrossRef] [PubMed]
| Diabetic Nephropathy | Diabetic Retinopathy | Diabetic Neuropathy | |
|---|---|---|---|
| DNA Methylation | AFF3, ARID5B, CUX1, ELMO1, FKBP5, HDAC4, ITGAL, LY9, PIM1, RUNX3, SEPTING9, UTF3A | ||
| Histone acetylation/methylation | H3K4, H3K27, H3K9 | ||
| Non-coding RNA | miRNA192, miRNA124, miRNA21, LncRNA MALAT1, LncRNA NEAT1 | LncRNA | mir-199a-3p, mir-499a, miRNA-29c, lncRNA MTHFSD, lncRNA, LMAN2L |
| eGRF < 60 mL/min per 1.73 m2 | eGRF > 60 mL/min per 1.73 m2 | |||
|---|---|---|---|---|
| Variable | Placebo | Apabetalone | Placebo | Apabetalone |
| Significant reduction of primary outcome MACE | None | Positive | None | None |
| Composite events MACE and CHF | None | Positive | None | None |
| Components (# of events) | ||||
| CV death | 10 | 5 | 4 | 4 |
| Non-fatal MI | 12 | 7 | 7 | 6 |
| Non-fatal stroke | 4 | 2 | 1 | 1 |
| CHF hospitalization | 9 | 2 | 3 | 2 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Suárez, R.; Chapela, S.P.; Álvarez-Córdova, L.; Bautista-Valarezo, E.; Sarmiento-Andrade, Y.; Verde, L.; Frias-Toral, E.; Sarno, G. Epigenetics in Obesity and Diabetes Mellitus: New Insights. Nutrients 2023, 15, 811. https://doi.org/10.3390/nu15040811
Suárez R, Chapela SP, Álvarez-Córdova L, Bautista-Valarezo E, Sarmiento-Andrade Y, Verde L, Frias-Toral E, Sarno G. Epigenetics in Obesity and Diabetes Mellitus: New Insights. Nutrients. 2023; 15(4):811. https://doi.org/10.3390/nu15040811
Chicago/Turabian StyleSuárez, Rosario, Sebastián P. Chapela, Ludwig Álvarez-Córdova, Estefanía Bautista-Valarezo, Yoredy Sarmiento-Andrade, Ludovica Verde, Evelyn Frias-Toral, and Gerardo Sarno. 2023. "Epigenetics in Obesity and Diabetes Mellitus: New Insights" Nutrients 15, no. 4: 811. https://doi.org/10.3390/nu15040811
APA StyleSuárez, R., Chapela, S. P., Álvarez-Córdova, L., Bautista-Valarezo, E., Sarmiento-Andrade, Y., Verde, L., Frias-Toral, E., & Sarno, G. (2023). Epigenetics in Obesity and Diabetes Mellitus: New Insights. Nutrients, 15(4), 811. https://doi.org/10.3390/nu15040811








