Proteomic Analysis Reveals Changes in Tight Junctions in the Small Intestinal Epithelium of Mice Fed a High-Fat Diet
(This article belongs to the Section Nutrition and Metabolism)
Abstract
:1. Introduction
2. Materials and Methods
2.1. Murine Model
2.2. Intestinal Permeability
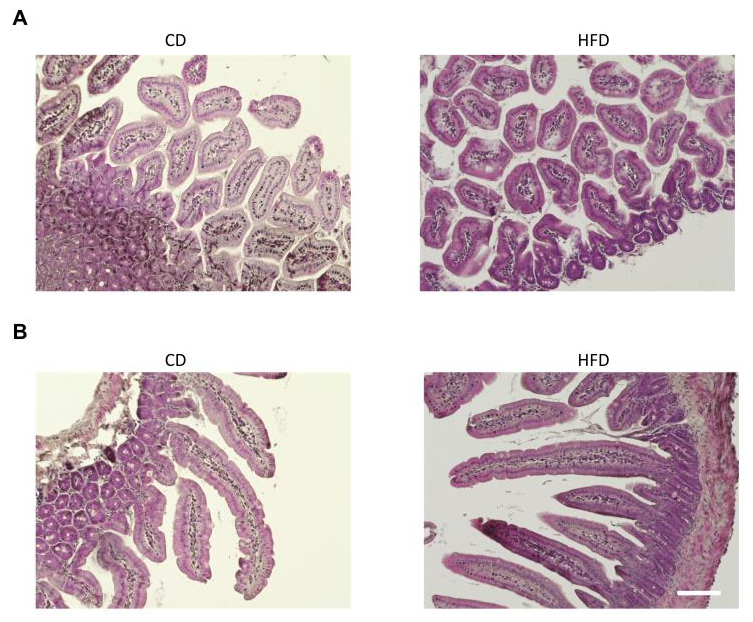
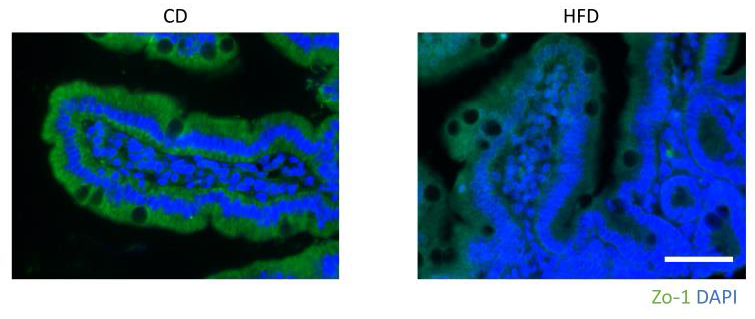
2.3. Histological and Immunohistochemical Analyses
2.4. Isolation of Intestinal Epithelial Cells
2.5. Proteomic Assay
2.6. Statistical Analysis
3. Results
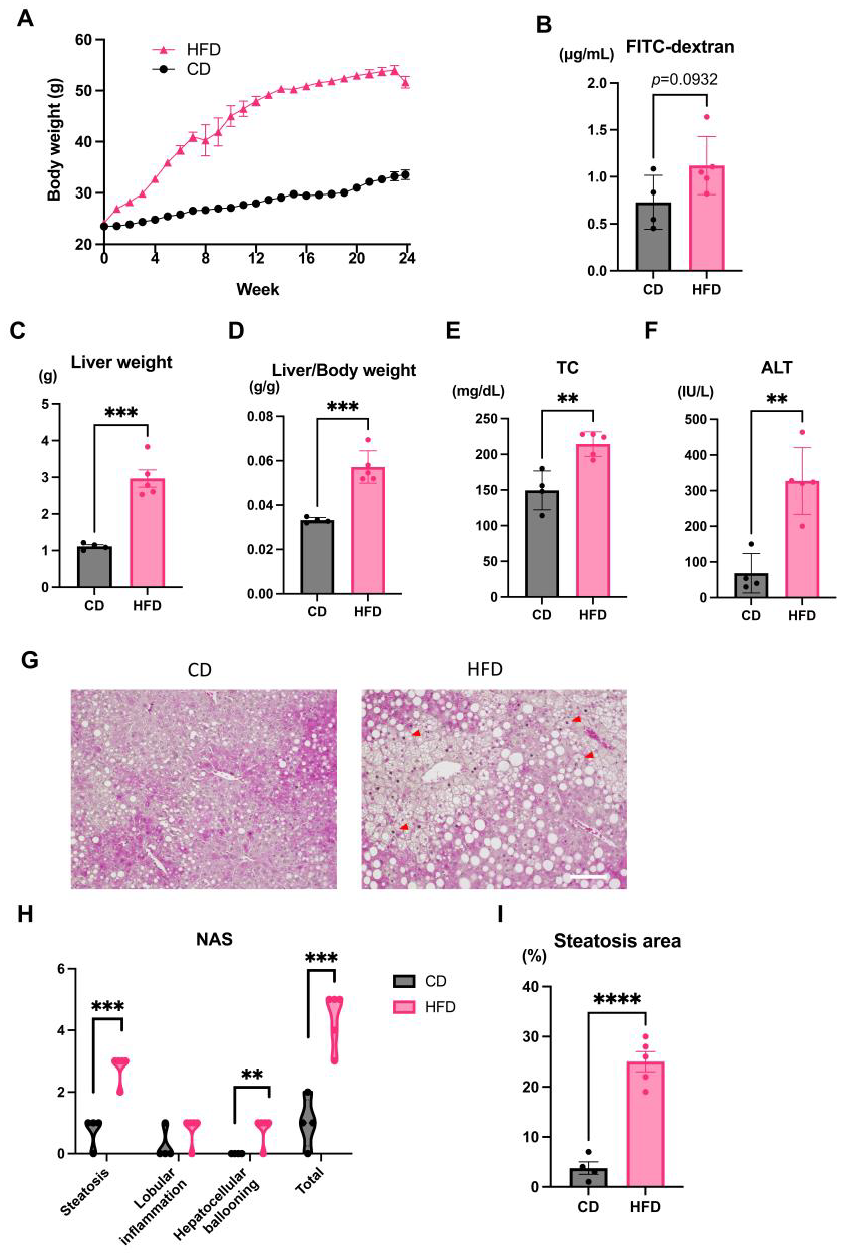
3.1. Evaluation of the Liver and Changes in Intestinal Permeability Induced by an HFD in Mice
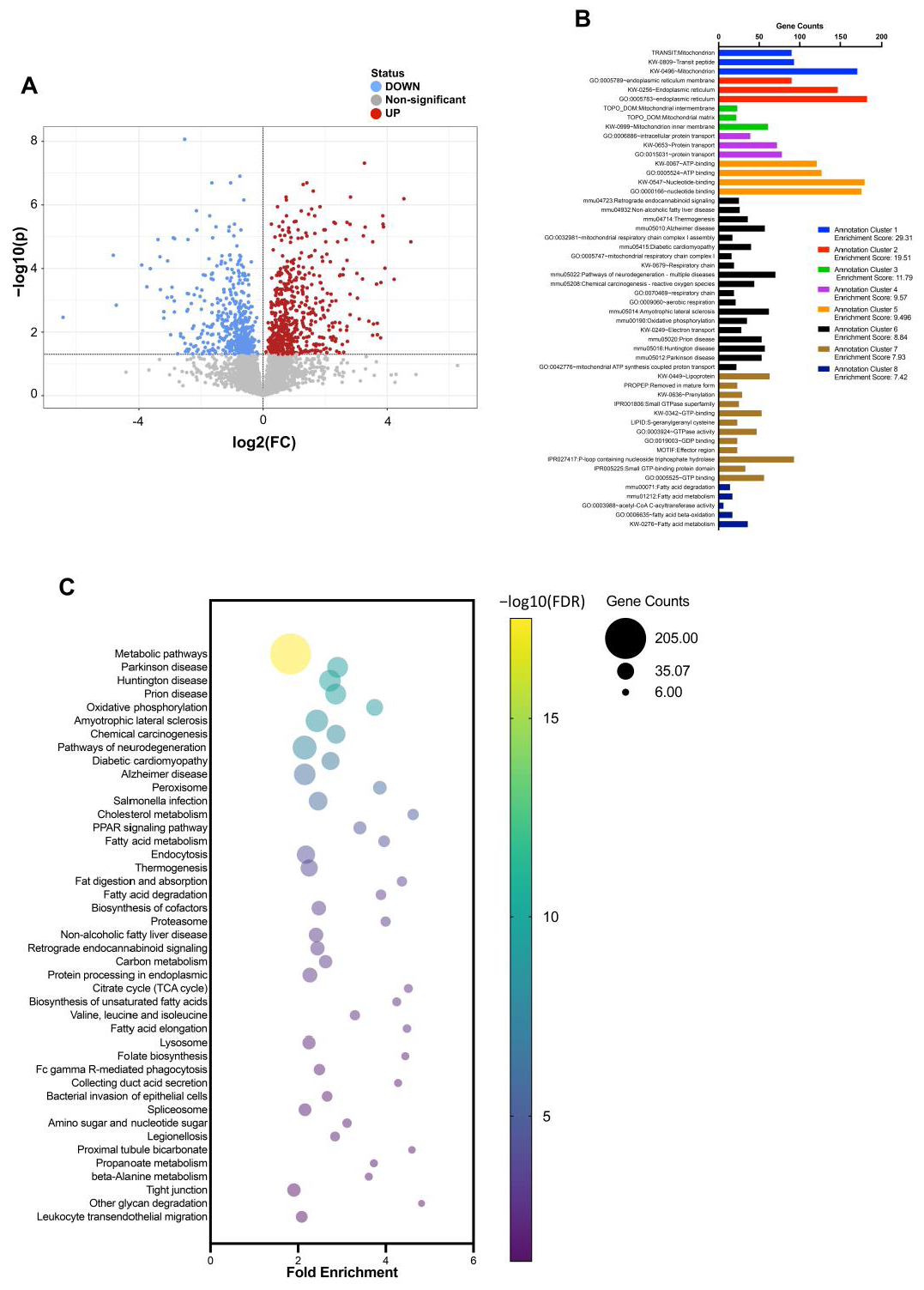
3.2. Proteomic Changes in the Upper and Lower Small Intestines Induced by an HFD
3.3. Gene Ontology and KEGG Pathway Enrichment Analysis of Differentially Expressed Proteins in the Upper Small Intestine
3.4. Correlations of TJ Proteins and Intestinal Permeability
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Younossi, Z.M.; Koenig, A.B.; Abdelatif, D.; Fazel, Y.; Henry, L.; Wymer, M. Global Epidemiology of Nonalcoholic Fatty Liver Disease—Meta-Analytic Assessment of Prevalence, Incidence, and Outcomes. Hepatology 2016, 64, 73–84. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ito, T.; Ishigami, M.; Zou, B.; Tanaka, T.; Takahashi, H.; Kurosaki, M.; Maeda, M.; Thin, K.N.; Tanaka, K.; Takahashi, Y.; et al. The Epidemiology of NAFLD and Lean NAFLD in Japan: A Meta-Analysis with Individual and Forecasting Analysis, 1995–2040. Hepatol. Int. 2021, 15, 366–379. [Google Scholar] [CrossRef]
- Tateishi, R.; Uchino, K.; Fujiwara, N.; Takehara, T.; Okanoue, T.; Seike, M.; Yoshiji, H.; Yatsuhashi, H.; Shimizu, M.; Torimura, T.; et al. A Nationwide Survey on Non-B, Non-C Hepatocellular Carcinoma in Japan: 2011–2015 Update. J. Gastroenterol. 2019, 54, 367–376. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, J.; Shen, J.; Sun, T.T.; Zhang, X.; Wong, N. Obesity, Insulin Resistance, NASH and Hepatocellular Carcinoma. Semin. Cancer Biol. 2013, 23, 483–491. [Google Scholar] [CrossRef]
- Smith, B.W.; Adams, L.A. Non-Alcoholic Fatty Liver Disease. Crit. Rev. Clin. Lab. Sci. 2011, 48, 97–113. [Google Scholar] [CrossRef] [PubMed]
- Marra, F.; Svegliati-Baroni, G. Lipotoxicity and the Gut-Liver Axis in NASH Pathogenesis. J. Hepatol. 2018, 68, 280–295. [Google Scholar] [CrossRef] [PubMed]
- Albillos, A.; de Gottardi, A.; Rescigno, M. The Gut-Liver Axis in Liver Disease: Pathophysiological Basis for Therapy. J. Hepatol. 2020, 72, 558–577. [Google Scholar] [CrossRef] [Green Version]
- Volynets, V.; Küper, M.A.; Strahl, S.; Maier, I.B.; Spruss, A.; Wagnerberger, S.; Königsrainer, A.; Bischoff, S.C.; Bergheim, I. Nutrition, Intestinal Permeability, and Blood Ethanol Levels Are Altered in Patients with Nonalcoholic Fatty Liver Disease (NAFLD). Dig. Dis. Sci. 2012, 57, 1932–1941. [Google Scholar] [CrossRef]
- Miele, L.; Valenza, V.; la Torre, G.G.; Montalto, M.; Cammarota, G.; Ricci, R.; Mascianà, R.; Forgione, A.; Gabrieli, M.L.; Perotti, G.; et al. Increased Intestinal Permeability and Tight Junction Alterations in Nonalcoholic Fatty Liver Disease. Hepatology 2009, 49, 1877–1887. [Google Scholar] [CrossRef] [PubMed]
- Mouries, J.; Brescia, P.; Silvestri, A.; Spadoni, I.; Sorribas, M.; Wiest, R.; Mileti, E.; Galbiati, M.; Invernizzi, P.; Adorini, L.; et al. Microbiota-Driven Gut Vascular Barrier Disruption Is a Prerequisite for Non-Alcoholic Steatohepatitis Development. J. Hepatol. 2019, 71, 1216–1228. [Google Scholar] [CrossRef] [Green Version]
- Ferolla, S.M.; Armiliato, G.N.A.; Couto, C.A.; Ferrari, T.C.A. The Role of Intestinal Bacteria Overgrowth in Obesity-Related Nonalcoholic Fatty Liver Disease. Nutrients 2014, 6, 5583–5599. [Google Scholar] [CrossRef]
- Serino, M.; Luche, E.; Gres, S.; Baylac, A.; Bergé, M.; Cenac, C.; Waget, A.; Klopp, P.; Iacovoni, J.; Klopp, C.; et al. Original Article: Metabolic Adaptation to a High-Fat Diet Is Associated with a Change in the Gut Microbiota. Gut 2012, 61, 543. [Google Scholar] [CrossRef] [PubMed]
- Gudan, A.; Jamioł-Milc, D.; Hawryłkowicz, V.; Skonieczna-Żydecka, K.; Stachowska, E. The Prevalence of Small Intestinal Bacterial Overgrowth in Patients with Non-Alcoholic Liver Diseases: NAFLD, NASH, Fibrosis, Cirrhosis-A Systematic Review, Meta-Analysis and Meta-Regression. Nutrients 2022, 14, 5261. [Google Scholar] [CrossRef] [PubMed]
- de La Serre, C.B.; Ellis, C.L.; Lee, J.; Hartman, A.L.; Rutledge, J.C.; Raybould, H.E. Propensity to High-Fat Diet-Induced Obesity in Rats Is Associated with Changes in the Gut Microbiota and Gut Inflammation. Am. J. Physiol. Gastrointest. Liver Physiol. 2010, 299, G440. [Google Scholar] [CrossRef]
- Brun, P.; Castagliuolo, I.; di Leo, V.; Buda, A.; Pinzani, M.; Palù, G.; Martines, D. Increased Intestinal Permeability in Obese Mice: New Evidence in the Pathogenesis of Nonalcoholic Steatohepatitis. Am. J. Physiol. Gastrointest Liver Physiol. 2007, 292, G518–G525. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meroni, M.; Longo, M.; Dongiovanni, P. The Role of Probiotics in Nonalcoholic Fatty Liver Disease: A New Insight into Therapeutic Strategies. Nutrients 2019, 11, 2642. [Google Scholar] [CrossRef] [Green Version]
- Parnell, J.A.; Raman, M.; Rioux, K.P.; Reimer, R.A. The Potential Role of Prebiotic Fibre for Treatment and Management of Non-Alcoholic Fatty Liver Disease and Associated Obesity and Insulin Resistance. Liver Int. 2012, 32, 701–711. [Google Scholar] [CrossRef] [PubMed]
- Carpi, R.Z.; Barbalho, S.M.; Sloan, K.P.; Laurindo, L.F.; Gonzaga, H.F.; Grippa, P.C.; Zutin, T.L.M.; Girio, R.J.S.; Repetti, C.S.F.; Detregiachi, C.R.P.; et al. The Effects of Probiotics, Prebiotics and Synbiotics in Non-Alcoholic Fat Liver Disease (NAFLD) and Non-Alcoholic Steatohepatitis (NASH): A Systematic Review. Int. J. Mol. Sci. 2022, 23, 8850. [Google Scholar] [CrossRef] [PubMed]
- Turner, J.R.; Buschmann, M.M.; Romero-Calvo, I.; Sailer, A.; Shen, L. The Role of Molecular Remodeling in Differential Regulation of Tight Junction Permeability. Semin. Cell Dev. Biol. 2014, 36, 204–212. [Google Scholar] [CrossRef] [Green Version]
- Sacco, F.; Silvestri, A.; Posca, D.; Pirrò, S.; Gherardini, P.F.; Castagnoli, L.; Mann, M.; Cesareni, G. Deep Proteomics of Breast Cancer Cells Reveals That Metformin Rewires Signaling Networks Away from a Pro-Growth State. Cell Syst. 2016, 2, 159–171. [Google Scholar] [CrossRef] [Green Version]
- Zhao, H.; Zhang, H.; Wu, H.; Li, H.; Liu, L.; Guo, J.; Li, C.; Shih, D.Q.; Zhang, X. Protective Role of 1,25(OH)2vitamin D3 in the Mucosal Injury and Epithelial Barrier Disruption in DSS-Induced Acute Colitis in Mice. BMC Gastroenterol. 2012, 12, 57. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kleiner, D.E.; Brunt, E.M.; Van Natta, M.; Behling, C.; Contos, M.J.; Cummings, O.W.; Ferrell, L.D.; Liu, Y.C.; Torbenson, M.S.; Unalp-Arida, A.; et al. Design and Validation of a Histological Scoring System for Nonalcoholic Fatty Liver Disease. Hepatology 2005, 41, 1313–1321. [Google Scholar] [CrossRef]
- Sato, T.; Vries, R.G.; Snippert, H.J.; van de Wetering, M.; Barker, N.; Stange, D.E.; van Es, J.H.; Abo, A.; Kujala, P.; Peters, P.J.; et al. Single Lgr5 Stem Cells Build Crypt-Villus Structures in Vitro without a Mesenchymal Niche. Nature 2009, 459, 262–265. [Google Scholar] [CrossRef]
- Kojima, H.; Yagi, H.; Kushige, H.; Toda, Y.; Takayama, K.; Masuda, S.; Morisaku, T.; Tsuchida, T.; Kuroda, K.; Hirukawa, K.; et al. Decellularized Organ-Derived Scaffold Is a Promising Carrier for Human Induced Pluripotent Stem Cells-Derived Hepatocytes. Cells 2022, 11, 1258. [Google Scholar] [CrossRef] [PubMed]
- Pang, Z.; Zhou, G.; Ewald, J.; Chang, L.; Hacariz, O.; Basu, N.; Xia, J. Using MetaboAnalyst 5.0 for LC–HRMS Spectra Processing, Multi-Omics Integration and Covariate Adjustment of Global Metabolomics Data. Nat. Protoc. 2022, 17, 1735–1761. [Google Scholar] [CrossRef]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Systematic and Integrative Analysis of Large Gene Lists Using DAVID Bioinformatics Resources. Nat. Protoc. 2009, 4, 44–57. [Google Scholar] [CrossRef] [PubMed]
- Sherman, B.T.; Hao, M.; Qiu, J.; Jiao, X.; Baseler, M.W.; Lane, H.C.; Imamichi, T.; Chang, W. DAVID: A Web Server for Functional Enrichment Analysis and Functional Annotation of Gene Lists (2021 Update). Nucleic Acids Res. 2022, 50, W216. [Google Scholar] [CrossRef]
- Homma, Y.; Hiragi, S.; Fukuda, M. Rab Family of Small GTPases: An Updated View on Their Regulation and Functions. FEBS J 2021, 288, 36–55. [Google Scholar] [CrossRef]
- Ding, L.; Lu, Z.; Foreman, O.; Tatum, R.; Lu, Q.; Renegar, R.; Cao, J.; Chen, Y. Inflammation and Disruption of the Mucosal Architecture in Claudin-7-Deficient Mice. Gastroenterology 2012, 142, 305–315. [Google Scholar] [CrossRef] [Green Version]
- Barth, A.I.M.; Kim, H.; Riedel-Kruse, I.H. Regulation of Epithelial Migration by Epithelial Cell Adhesion Molecule Requires Its Claudin-7 Interaction Domain. PLoS ONE 2018, 13, e0204957. [Google Scholar] [CrossRef]
- Zeisel, M.B.; Dhawan, P.; Baumert, T.F. Tight Junction Proteins in Gastrointestinal and Liver Disease. Gut 2019, 68, 547. [Google Scholar] [CrossRef]
- Cho, Y.E.; Kim, D.K.; Seo, W.; Gao, B.; Yoo, S.H.; Song, B.J. Fructose Promotes Leaky Gut, Endotoxemia and Liver Fibrosis through CYP2E1-Mediated Oxidative and Nitrative Stress. Hepatology 2021, 73, 2180. [Google Scholar] [CrossRef] [PubMed]
- Ohtani, N.; Hara, E. Gut-liver Axis-mediated Mechanism of Liver Cancer: A Special Focus on the Role of Gut Microbiota. Cancer Sci. 2021, 112, 4433. [Google Scholar] [CrossRef] [PubMed]
- Genser, L.; Aguanno, D.; Soula, H.A.; Dong, L.; Trystram, L.; Assmann, K.; Salem, J.E.; Vaillant, J.C.; Oppert, J.M.; Laugerette, F.; et al. Increased Jejunal Permeability in Human Obesity Is Revealed by a Lipid Challenge and Is Linked to Inflammation and Type 2 Diabetes. J. Pathol. 2018, 246, 217–230. [Google Scholar] [CrossRef]
- Dekker, M.J.; Su, Q.; Baker, C.; Rutledge, A.C.; Adeli, K. Fructose: A Highly Lipogenic Nutrient Implicated in Insulin Resistance, Hepatic Steatosis, and the Metabolic Syndrome. Am. J. Physiol. Endocrinol. Metab. 2010, 299, 685–694. [Google Scholar] [CrossRef] [Green Version]
- Chassaing, B.; Etienne-Mesmin, L.; Gewirtz, A.T. Microbiota-Liver Axis in Hepatic Disease. Hepatology 2014, 59, 328–339. [Google Scholar] [CrossRef] [Green Version]
- Everard, A.; Belzer, C.; Geurts, L.; Ouwerkerk, J.P.; Druart, C.; Bindels, L.B.; Guiot, Y.; Derrien, M.; Muccioli, G.G.; Delzenne, N.M.; et al. Cross-Talk between Akkermansia Muciniphila and Intestinal Epithelium Controls Diet-Induced Obesity. Proc. Natl. Acad. Sci. USA 2013, 110, 9066–9071. [Google Scholar] [CrossRef] [Green Version]
- de Wit, N.J.; Bosch-Vermeulen, H.; de Groot, P.J.; Hooiveld, G.J.; Bromhaar, M.M.G.; Jansen, J.; Müller, M.; van der Meer, R. The Role of the Small Intestine in the Development of Dietary Fat-Induced Obesity and Insulin Resistance in C57BL/6J Mice. BMC Med. Genom. 2008, 1, 14. [Google Scholar] [CrossRef]
- Stamatovic, S.M.; Johnson, A.M.; Sladojevic, N.; Keep, R.F.; Andjelkovic, A.V. Endocytosis of Tight Junction Proteins and the Regulation of Degradation and Recycling. Ann. N. Y. Acad. Sci. 2017, 1397, 54–65. [Google Scholar] [CrossRef] [PubMed]
- Shen, L. Tight Junctions on the Move: Molecular Mechanisms for Epithelial Barrier Regulation. Ann. N. Y. Acad. Sci. 2012, 1258, 9–18. [Google Scholar] [CrossRef] [Green Version]
- Agola, J.; Jim, P.; Ward, H.; BasuRay, S.; Wandinger-Ness, A. Rab GTPases as Regulators of Endocytosis, Targets of Disease and Therapeutic Opportunities. Clin. Genet. 2011, 80, 305–318. [Google Scholar] [CrossRef] [Green Version]
- Schuppan, D.; Surabattula, R.; Wang, X.Y. Determinants of Fibrosis Progression and Regression in NASH. J. Hepatol. 2018, 68, 238–250. [Google Scholar] [CrossRef]
- Ladwein, M.; Pape, U.F.; Schmidt, D.S.; Schnölzer, M.; Fiedler, S.; Langbein, L.; Franke, W.W.; Moldenhauer, G.; Zöller, M. The Cell-Cell Adhesion Molecule EpCAM Interacts Directly with the Tight Junction Protein Claudin-7. Exp. Cell Res. 2005, 309, 345–357. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.; Yang, Y.; Yang, F.; Liu, S.; Zhu, Z.; Lei, Z.; Guo, J. Functions of EpCAM in Physiological Processes and Diseases (Review). Int. J. Mol. Med. 2018, 42, 1771–1785. [Google Scholar] [CrossRef] [Green Version]
- Lei, Z.; Maeda, T.; Tamura, A.; Nakamura, T.; Yamazaki, Y.; Shiratori, H.; Yashiro, K.; Tsukita, S.; Hamada, H. EpCAM Contributes to Formation of Functional Tight Junction in the Intestinal Epithelium by Recruiting Claudin Proteins. Dev. Biol. 2012, 371, 136–145. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mueller, J.L.; McGeough, M.D.; Peña, C.A.; Sivagnanam, M. Functional Consequences of EpCam Mutation in Mice and Men. Am. J. Physiol. Gastrointest Liver Physiol. 2014, 306, 278–288. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, C.-J.; Mannan, P.; Lu, M.; Udey, M.C. Epithelial Cell Adhesion Molecule (EpCAM) Regulates Claudin Dynamics and Tight Junctions. J. Biol. Chem. 2013, 288, 12253–12268. [Google Scholar] [CrossRef] [Green Version]




| Protein | Pearson r | 95% Confidence Interval | R Squared | p (Two-Tailed) | p Value Summary |
|---|---|---|---|---|---|
| F11r | −0.1358 | −0.7338 to 0.5807 | 0.01845 | 0.7275 | ns |
| Llgl2 | −0.4785 | −0.8671 to 0.2720 | 0.229 | 0.1925 | ns |
| Rab8a | −0.7657 | −0.9478 to −0.2066 | 0.5862 | 0.0162 | * |
| Rab8b | −0.447 | −0.8568 to 0.3088 | 0.1998 | 0.2277 | ns |
| Rap2c | −0.7321 | −0.9395 to −0.1324 | 0.536 | 0.0249 | * |
| Arpc1a | 0.8983 | 0.5805 to 0.9786 | 0.8069 | 0.001 | *** |
| Arpc1b | −0.1386 | −0.7351 to 0.5788 | 0.01921 | 0.7221 | ns |
| Arpc2 | −0.07812 | −0.7056 to 0.6181 | 0.006102 | 0.8417 | ns |
| Arpc5 | 0.7178 | 0.1025 to 0.9358 | 0.5152 | 0.0295 | * |
| Actb | −0.4112 | −0.8446 to 0.3480 | 0.1691 | 0.2716 | ns |
| Afdn | −0.1252 | −0.8026 to 0.6932 | 0.01568 | 0.7891 | ns |
| Cdc42 | −0.009934 | −0.6696 to 0.6585 | 9.87 × 10−5 | 0.9798 | ns |
| Cgn | −0.692 | −0.9291 to −0.05163 | 0.4789 | 0.0389 | * |
| Cldn15 | −0.0411 | −0.7703 to 0.7347 | 0.001689 | 0.9303 | ns |
| Cldn7 | −0.7494 | −0.9438 to −0.1698 | 0.5616 | 0.0201 | * |
| Dlg1 | 0.1179 | −0.5926 to 0.7252 | 0.0139 | 0.7626 | ns |
| Hspa4 | 0.8191 | 0.3399 to 0.9606 | 0.6709 | 0.0069 | ** |
| Myh14 | −0.8424 | −0.9660 to −0.4046 | 0.7096 | 0.0044 | ** |
| Myl12b | −0.8336 | −0.9640 to −0.3797 | 0.6949 | 0.0052 | ** |
| Nedd4l | −0.4019 | −0.8414 to 0.3577 | 0.1615 | 0.2836 | ns |
| Mpp5 | −0.7453 | −0.9507 to −0.08549 | 0.5554 | 0.0338 | * |
| Prkaa1 | −0.4497 | −0.8577 to 0.3057 | 0.2022 | 0.2246 | ns |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Muto, H.; Honda, T.; Tanaka, T.; Yokoyama, S.; Yamamoto, K.; Ito, T.; Imai, N.; Ishizu, Y.; Maeda, K.; Ishikawa, T.; et al. Proteomic Analysis Reveals Changes in Tight Junctions in the Small Intestinal Epithelium of Mice Fed a High-Fat Diet. Nutrients 2023, 15, 1473. https://doi.org/10.3390/nu15061473
Muto H, Honda T, Tanaka T, Yokoyama S, Yamamoto K, Ito T, Imai N, Ishizu Y, Maeda K, Ishikawa T, et al. Proteomic Analysis Reveals Changes in Tight Junctions in the Small Intestinal Epithelium of Mice Fed a High-Fat Diet. Nutrients. 2023; 15(6):1473. https://doi.org/10.3390/nu15061473
Chicago/Turabian StyleMuto, Hisanori, Takashi Honda, Taku Tanaka, Shinya Yokoyama, Kenta Yamamoto, Takanori Ito, Norihiro Imai, Yoji Ishizu, Keiko Maeda, Tetsuya Ishikawa, and et al. 2023. "Proteomic Analysis Reveals Changes in Tight Junctions in the Small Intestinal Epithelium of Mice Fed a High-Fat Diet" Nutrients 15, no. 6: 1473. https://doi.org/10.3390/nu15061473







