Leaf Extract of Perilla frutescens (L.) Britt Promotes Adipocyte Browning via the p38 MAPK Pathway and PI3K-AKT Pathway
Abstract
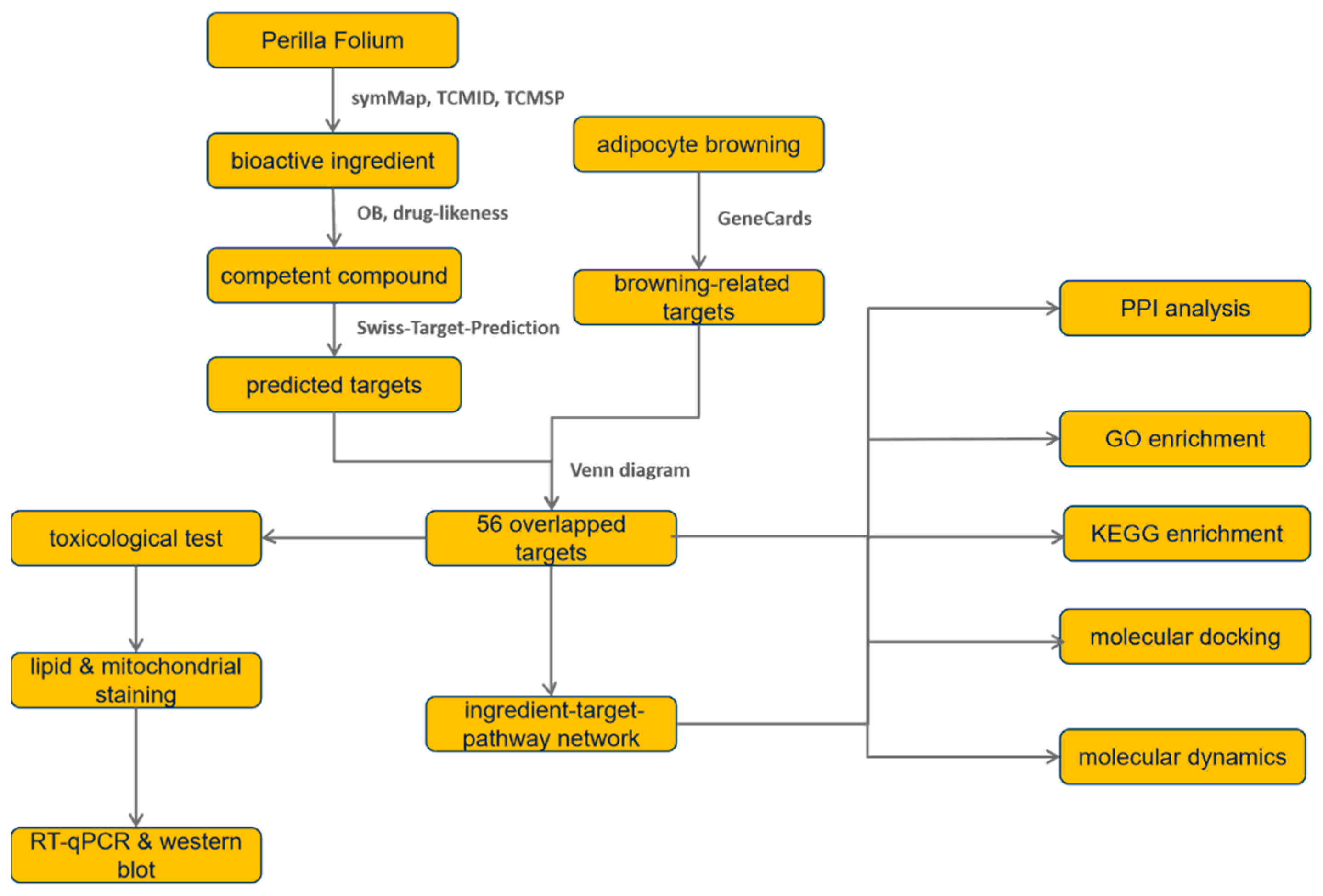
:1. Introduction
2. Materials and Methods
2.1. Leaf of Perilla frutescens (L.) Britt (PF) Bioactive Ingredient Acquisition
2.2. Prediction of Protein-Encoding Target Genes Based on Compound Structures
2.3. Search for Browning-Related Targets
2.4. Topological Analysis for the Interactions among Overlapped Target Genes
2.5. Enrichment Analysis Based on GO and KEGG Database
2.6. Exploration of the Interactions among Ingredients, Targets, and Pathways
2.7. Molecular Docking Simulation
2.8. Molecule Dynamics of Ligand and Target Combination
2.9. Preparation of PF Extract
2.10. Cell Culture and Differentiation
2.11. Cell Viability Assay
2.12. Real-Time Quantitative Polymerase Chain Reaction (RT-qPCR)
2.13. Western Blot Analysis
2.14. Determination of Lipid Accumulation
2.15. Mitochondrial Mass Measurement
2.16. Statistical Analysis
3. Results
3.1. Putative Targets of PF Activating Browning
3.2. Enrichment Analysis
3.3. Protein–Protein Interaction (PPI) Network of Targets for ZYS Promoting Adipocyte Browning
3.4. Ingredient–Target–Pathway (I-T-P) Network Construction
3.5. Molecular Docking Demonstrating Compound–Protein Interaction
3.6. Molecular Dynamics Simulation of Ligand Complex
3.7. Effect of PF on Bone Marrow Mesenchymal Stem Cell (BMSC) Viability
3.8. PF Inhibited Lipid Accumulation and Downregulated Adipogenesis-Related Gene Expression
3.9. PF Promoted Mitochondrial Biogenesis and Upregulated Brite Adipocyte-Related Gene Expression
3.10. Effect of PF on Browning Could Be Mediated by the p38 MAPK Pathway as Well as PI3K-AKT Pathway
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Nakanishi, H.; Matar, R.H.; Vahibe, A.; Abu Dayyeh, B.K.; Galvani, C.; Pullatt, R.; Davis, S.S., Jr.; Clapp, B.; Ghanem, O.M. Single Versus Double Anastomosis Duodenal Switch in the Management of Obesity: A Meta-analysis and Systematic Review. Surg. Laparosc. Endosc. Percutan. Tech. 2022, 32, 595–605. [Google Scholar] [CrossRef] [PubMed]
- Dickerson, R.N.; Andromalos, L.; Brown, J.C.; Correia, M.; Pritts, W.; Ridley, E.J.; Robinson, K.N.; Rosenthal, M.D.; van Zanten, A.R.H. Obesity and critical care nutrition: Current practice gaps and directions for future research. Crit. Care 2022, 26, 283. [Google Scholar] [CrossRef] [PubMed]
- Klein Hazebroek, M.; Keipert, S. Obesity-resistance of UCP1-deficient mice associates with sustained FGF21 sensitivity in inguinal adipose tissue. Front. Endocrinol. 2022, 13, 909621. [Google Scholar] [CrossRef] [PubMed]
- Sakaguchi, M.; Okagawa, S.; Okubo, Y.; Otsuka, Y.; Fukuda, K.; Igata, M.; Kondo, T.; Sato, Y.; Yoshizawa, T.; Fukuda, T.; et al. Phosphatase protector alpha4 (alpha4) is involved in adipocyte maintenance and mitochondrial homeostasis through regulation of insulin signaling. Nat. Commun. 2022, 13, 6092. [Google Scholar] [CrossRef] [PubMed]
- Lee, D.K.; Kim, T.; Byeon, J.; Park, M.; Kim, S.; Kim, J.; Choi, S.; Lee, G.; Park, C.; Lee, K.W.; et al. REDD1 promotes obesity-induced metabolic dysfunction via atypical NF-kappaB activation. Nat. Commun. 2022, 13, 6303. [Google Scholar] [CrossRef]
- Herzig, S.; Shaw, R.J. AMPK: Guardian of metabolism and mitochondrial homeostasis. Nat. Rev. Mol. Cell Biol. 2018, 19, 121–135. [Google Scholar] [CrossRef] [Green Version]
- Kwan, H.Y.; Wu, J.; Su, T.; Chao, X.J.; Liu, B.; Fu, X.; Chan, C.L.; Lau, R.H.Y.; Tse, A.K.W.; Han, Q.B.; et al. Cinnamon induces browning in subcutaneous adipocytes. Sci. Rep. 2017, 7, 2447. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Zhang, L.; Dong, L.; Hu, X.; Feng, F.; Chen, F. 6-Gingerol, a Functional Polyphenol of Ginger, Promotes Browning through an AMPK-Dependent Pathway in 3T3-L1 Adipocytes. J. Agric. Food Chem. 2019, 67, 14056–14065. [Google Scholar] [CrossRef] [PubMed]
- Thomas, S.S.; Kim, M.; Lee, S.J.; Cha, Y.S. Antiobesity Effects of Purple Perilla (Perilla frutescens var. acuta) on Adipocyte Differentiation and Mice Fed a High-fat Diet. J. Food Sci. 2018, 83, 2384–2393. [Google Scholar] [CrossRef] [PubMed]
- Zheng, F.; Quan, H.-Y.; Quan, J.; Yin, X. Effect of Perillae Folium Extract on 3T3-L1 Adipocytes. Lishizhen Med. Mater. Med. Res. 2017, 29, 281–284. [Google Scholar]
- Ying, P.; Fei, H.Y.; Quan, H.Y. Effects and mechanism research on Perillae folium extract in obese mice. Zhong Hua Zhong Yi Yao Za Zhi 2017, 32, 3993–3996. [Google Scholar]
- Cheng, Y.; Liu, H.; Li, J.; Ma, Y.; Song, C.; Wang, Y.; Li, P.; Chen, Y.; Zhang, Z. Monascin abrogates RANKL-mediated osteoclastogenesis in RAW264.7 cells via regulating MAPKs signaling pathways. Front. Pharmacol. 2022, 13, 950122. [Google Scholar] [CrossRef] [PubMed]
- He, Q.; Yang, J.; Chen, D.; Li, Y.; Gong, D.; Ge, H.; Wang, Z.; Wang, H.; Chen, P. 12-Deoxyphorbol-13-Hexadecanoate Abrogates OVX-Induced Bone Loss in Mice and Osteoclastogenesis via Inhibiting ROS Level and Regulating RANKL-Mediated NFATc1 Activation. Front. Pharm. 2022, 13, 899776. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Zhong, H.; Xu, P.; Zhou, A.; Ding, L.; Qiu, J.; Wu, H.; Dai, M. Deciphering the combination mechanisms of Gualou-Xiebai herb pair against atherosclerosis by network pharmacology and HPLC-Q-TOF-MS technology. Front. Pharmacol. 2022, 13, 941400. [Google Scholar] [CrossRef]
- Wang, Y.X.; Yang, Z.; Wang, W.X.; Huang, Y.X.; Zhang, Q.; Li, J.J.; Tang, Y.P.; Yue, S.J. Methodology of network pharmacology for research on Chinese herbal medicine against COVID-19: A review. J. Integr. Med. 2022, 20, 477–487. [Google Scholar] [CrossRef]
- Zhang, G.B.; Li, Q.Y.; Chen, Q.L.; Su, S.B. Network pharmacology: A new approach for chinese herbal medicine research. Evid. Based Complement. Alternat. Med. 2013, 2013, 621423. [Google Scholar] [CrossRef] [Green Version]
- Wu, Q.; Li, X.; Jiang, X.W.; Yao, D.; Zhou, L.J.; Xu, Z.H.; Wang, N.; Zhao, Q.C.; Zhang, Z. Yuan-Zhi decoction in the treatment of Alzheimer’s disease: An integrated approach based on chemical profiling, network pharmacology, molecular docking and experimental evaluation. Front. Pharmacol. 2022, 13, 893244. [Google Scholar] [CrossRef]
- Sharma, B.; Bhattacherjee, D.; Zyryanov, G.V.; Purohit, R. An insight from computational approach to explore novel, high-affinity phosphodiesterase 10A inhibitors for neurological disorders. J. Biomol. Struct. Dyn. 2022, 40, 1–13. [Google Scholar] [CrossRef]
- Bhardwaj, V.K.; Purohit, R. A lesson for the maestro of the replication fork: Targeting the protein-binding interface of proliferating cell nuclear antigen for anticancer therapy. J. Cell Biochem. 2022, 123, 1091–1102. [Google Scholar] [CrossRef]
- Singh, R.; Bhardwaj, V.K.; Purohit, R. Computational targeting of allosteric site of MEK1 by quinoline-based molecules. Cell Biochem. Funct. 2022, 40, 481–490. [Google Scholar] [CrossRef]
- Que, W.; Wu, Z.; Chen, M.; Zhang, B.; You, C.; Lin, H.; Zhao, Z.; Liu, M.; Qiu, H.; Cheng, Y. Molecular Mechanism of Gelsemium elegans (Gardner and Champ.) Benth. Against Neuropathic Pain Based on Network Pharmacology and Experimental Evidence. Front. Pharmacol. 2021, 12, 792932. [Google Scholar] [CrossRef] [PubMed]
- An, J.; Fan, H.; Han, M.; Peng, C.; Xie, J.; Peng, F. Exploring the mechanisms of neurotoxicity caused by fuzi using network pharmacology and molecular docking. Front. Pharmacol. 2022, 13, 961012. [Google Scholar] [CrossRef]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef] [Green Version]
- Ling, J.; Huang, Y.; Sun, Z.; Guo, X.; Chang, A.; Pan, J.; Zhuo, X. Exploration of the effect of Celastrol on protein targets in nasopharyngeal carcinoma: Network pharmacology, molecular docking and experimental evaluations. Front. Pharmacol. 2022, 13, 996728. [Google Scholar] [CrossRef] [PubMed]
- Liao, Y.; Ding, Y.; Yu, L.; Xiang, C.; Yang, M. Exploring the mechanism of Alisma orientale for the treatment of pregnancy induced hypertension and potential hepato-nephrotoxicity by using network pharmacology, network toxicology, molecular docking and molecular dynamics simulation. Front. Pharmacol. 2022, 13, 1027112. [Google Scholar] [CrossRef] [PubMed]
- Nakano, M.; Champagne, B. Nonlinear optical properties in open-shell molecular systems. Wiley Interdiscip. Rev. Comput. Mol. Sci. 2016, 6, 198–210. [Google Scholar] [CrossRef] [Green Version]
- Mottola, D.M.; Laiter, S.; Watts, V.J.; Tropsha, A.; Wyrick, S.D.; Nichols, D.E.; Mailman, R.B. Conformational analysis of D1 dopamine receptor agonists: Pharmacophore assessment and receptor mapping. J. Med. Chem. 1996, 39, 285–296. [Google Scholar] [CrossRef]
- Maier, J.A.; Martinez, C.; Kasavajhala, K.; Wickstrom, L.; Hauser, K.E.; Simmerling, C. ff14SB: Improving the Accuracy of Protein Side Chain and Backbone Parameters from ff99SB. J. Chem. Theory Comput. 2015, 11, 3696–3713. [Google Scholar] [CrossRef] [Green Version]
- Yang, H.; Sun, W.; Ma, P.; Yao, C.; Fan, Y.; Li, S.; Yuan, J.; Zhang, Z.; Li, X.; Lin, M.; et al. Multiple Components Rapidly Screened from Perilla Leaves Attenuate Asthma Airway Inflammation by Synergistic Targeting on Syk. J. Inflamm. Res. 2020, 13, 897–911. [Google Scholar] [CrossRef]
- Bae, J.S.; Han, M.; Shin, H.S.; Kim, M.K.; Shin, C.Y.; Lee, D.H.; Chung, J.H. Perilla frutescens leaves extract ameliorates ultraviolet radiation-induced extracellular matrix damage in human dermal fibroblasts and hairless mice skin. J. Ethnopharmacol. 2017, 195, 334–342. [Google Scholar] [CrossRef]
- Zhao, Y.; Kong, H.; Zhang, X.; Hu, X.; Wang, M. The effect of Perilla (Perilla frutescens) leaf extracts on the quality of surimi fish balls. Food Sci. Nutr. 2019, 7, 2083–2090. [Google Scholar] [CrossRef] [Green Version]
- Lin, Z.; He, H.; Wang, M.; Liang, J. MicroRNA-130a controls bone marrow mesenchymal stem cell differentiation towards the osteoblastic and adipogenic fate. Cell Prolif. 2019, 52, e12688. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Xie, H.; Liu, W.; Hu, R.; Huang, B.; Tan, Y.F.; Xu, K.; Sheng, Z.F.; Zhou, H.D.; Wu, X.P.; et al. A novel microRNA targeting HDAC5 regulates osteoblast differentiation in mice and contributes to primary osteoporosis in humans. J. Clin. Investig. 2009, 119, 3666–3677. [Google Scholar] [CrossRef] [Green Version]
- Chen, C.C.; Kuo, C.H.; Leu, Y.L.; Wang, S.H. Corylin reduces obesity and insulin resistance and promotes adipose tissue browning through SIRT-1 and beta3-AR activation. Pharmacol. Res. 2021, 164, 105291. [Google Scholar] [CrossRef]
- Jung, T.W.; Hwang, E.J.; Pyun, D.H.; Kim, T.J.; Lee, H.J.; Abd El-Aty, A.M.; Bang, J.S.; Kim, H.C.; Jeong, J.H. 3-hydroxymorphinan enhances mitochondrial biogenesis and adipocyte browning through AMPK-dependent pathway. Biochem. Biophys. Res. Commun. 2021, 577, 17–23. [Google Scholar] [CrossRef] [PubMed]
- Hsin, K.Y.; Ghosh, S.; Kitano, H. Combining machine learning systems and multiple docking simulation packages to improve docking prediction reliability for network pharmacology. PLoS ONE 2013, 8, e83922. [Google Scholar] [CrossRef] [Green Version]
- Kolotkin, R.L.; Meter, K.; Williams, G.R. Quality of life and obesity. Obes. Rev. 2001, 2, 219–229. [Google Scholar] [CrossRef] [PubMed]
- Vecchie, A.; Dallegri, F.; Carbone, F.; Bonaventura, A.; Liberale, L.; Portincasa, P.; Fruhbeck, G.; Montecucco, F. Obesity phenotypes and their paradoxical association with cardiovascular diseases. Eur. J. Intern. Med. 2018, 48, 6–17. [Google Scholar] [CrossRef] [PubMed]
- Filippatos, T.D.; Derdemezis, C.S.; Gazi, I.F.; Nakou, E.S.; Mikhailidis, D.P.; Elisaf, M.S. Orlistat-associated adverse effects and drug interactions: A critical review. Drug Saf. 2008, 31, 53–65. [Google Scholar] [CrossRef]
- Kelly, A.S.; Auerbach, P.; Barrientos-Perez, M.; Gies, I.; Hale, P.M.; Marcus, C.; Mastrandrea, L.D.; Prabhu, N.; Arslanian, S.; Investigators, N.N.T. A Randomized, Controlled Trial of Liraglutide for Adolescents with Obesity. N. Engl. J. Med. 2020, 382, 2117–2128. [Google Scholar] [CrossRef]
- Cameron, F.; Whiteside, G.; McKeage, K. Phentermine and topiramate extended release (Qsymia): First global approval. Drugs 2012, 72, 2033–2042. [Google Scholar] [CrossRef]
- Ghaben, A.L.; Scherer, P.E. Adipogenesis and metabolic health. Nat. Rev. Mol. Cell Biol. 2019, 20, 242–258. [Google Scholar] [CrossRef] [PubMed]
- Li, G.; Xie, C.; Lu, S.; Nichols, R.G.; Tian, Y.; Li, L.; Patel, D.; Ma, Y.; Brocker, C.N.; Yan, T.; et al. Intermittent Fasting Promotes White Adipose Browning and Decreases Obesity by Shaping the Gut Microbiota. Cell Metab. 2017, 26, 672–685.e674. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, J.; Zhang, Q.; Zhuo, Y.; Fang, Z.; Che, L.; Xu, S.; Feng, B.; Lin, Y.; Jiang, X.; Zhao, X.; et al. Effects of Multi-Strain Probiotics and Perilla frutescens Seed Extract Supplementation Alone or Combined on Growth Performance, Antioxidant Indices, and Intestinal Health of Weaned Piglets. Animals 2022, 12, 2246. [Google Scholar] [CrossRef]
- Maeda, A.; Fujimura, T.; Hirakawa, N.; Baba, K.; Kawamoto, S. A Methoxyflavanone from Perilla frutescens Induces Cellular Senescence in A549 Human Lung Adenocarcinoma Cells but Not in Normal Human Bronchial Epithelial Cells. Biol. Pharm. Bull. 2022, 45, 1581–1584. [Google Scholar] [CrossRef]
- Wang, Z.; Jin, X.; Zhang, X.; Xie, X.; Tu, Z.; He, X. From Function to Metabolome: Metabolomic Analysis Reveals the Effect of Probiotic Fermentation on the Chemical Compositions and Biological Activities of Perilla frutescens Leaves. Front. Nutr. 2022, 9, 933193. [Google Scholar] [CrossRef]
- Wang, Z.X.; Lin, Q.Q.; Tu, Z.C.; Zhang, L. The influence of in vitro gastrointestinal digestion on the Perilla frutescens leaf extract: Changes in the active compounds and bioactivities. J. Food Biochem. 2020, 44, e13530. [Google Scholar] [CrossRef] [PubMed]
- Xia, T.; Ma, J.; Sun, Y.; Sun, Y. Androgen receptor suppresses inflammatory response of airway epithelial cells in allergic asthma through MAPK1 and MAPK14. Hum. Exp. Toxicol. 2022, 41, 9603271221121320. [Google Scholar] [CrossRef]
- Ding, Q.Y.; Zhang, Y.; Ma, L.; Chen, Y.G.; Wu, J.H.; Zhang, H.F.; Wang, X. Inhibiting MAPK14 showed anti-prolactinoma effect. BMC Endocr. Disord. 2020, 20, 138. [Google Scholar] [CrossRef]
- Lee, N.H.; Choi, M.J.; Yu, H.; Kim, J.I.; Cheon, H.G. Adapalene induces adipose browning through the RARbeta-p38 MAPK-ATF2 pathway. Arch. Pharm. Res. 2022, 45, 340–351. [Google Scholar] [CrossRef]
- Mukherjee, S.; Yun, J.W. Prednisone stimulates white adipocyte browning via beta3-AR/p38 MAPK/ERK signaling pathway. Life Sci. 2022, 288, 120204. [Google Scholar] [CrossRef] [PubMed]
- Wu, T.; Liu, Q.; Li, Y.; Li, H.; Chen, L.; Yang, X.; Tang, Q.; Pu, S.; Kuang, J.; Li, R.; et al. Feeding-induced hepatokine, Manf, ameliorates diet-induced obesity by promoting adipose browning via p38 MAPK pathway. J. Exp. Med. 2021, 218, e20201203. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Qin, R. Vitamin D3 affects browning of white adipocytes by regulating autophagy via PI3K/Akt/mTOR/p53 signaling in vitro and in vivo. Apoptosis 2022, 27, 992–1003. [Google Scholar] [CrossRef]
- Lu, Y.; Xu, Y.; Yuan, W.; Wang, M.; Zhou, Y.; Chen, K.; Huang, Q. Downregulation of osteopontin inhibits browning of white adipose tissues through PI3K-AKT pathway in C57BL/6 mice. Eur. J. Pharmacol. 2020, 866, 172822. [Google Scholar] [CrossRef] [PubMed]
- Bargut, T.C.L.; Souza-Mello, V.; Aguila, M.B.; Mandarim-de-Lacerda, C.A. Browning of white adipose tissue: Lessons from experimental models. Horm. Mol. Biol. Clin. Investig. 2017, 31, 1. [Google Scholar] [CrossRef] [PubMed]



 indicates known interactions from curated databases.
indicates known interactions from curated databases.  indicates known interactions experimentally determined.
indicates known interactions experimentally determined.  represents predicted interactions with gene neighborhood.
represents predicted interactions with gene neighborhood.  represents predicted interactions with gene fusions.
represents predicted interactions with gene fusions.  represents predicted interactions with gene co-occurrence.
represents predicted interactions with gene co-occurrence.  indicates interaction from textmining.
indicates interaction from textmining.  indicates interaction from co-expression.
indicates interaction from co-expression.  indicates interaction from protein homology. (B). Ranking of the top 30 nodes from the PPI network according to the number of adjacent nodes. (C). Cluster 1 filtered by MCODE plugin. (D). Cluster 2 filtered by MCODE plugin. (E). Cluster 3 filtered by MCODE plugin.
indicates interaction from protein homology. (B). Ranking of the top 30 nodes from the PPI network according to the number of adjacent nodes. (C). Cluster 1 filtered by MCODE plugin. (D). Cluster 2 filtered by MCODE plugin. (E). Cluster 3 filtered by MCODE plugin.
 indicates known interactions from curated databases.
indicates known interactions from curated databases.  indicates known interactions experimentally determined.
indicates known interactions experimentally determined.  represents predicted interactions with gene neighborhood.
represents predicted interactions with gene neighborhood.  represents predicted interactions with gene fusions.
represents predicted interactions with gene fusions.  represents predicted interactions with gene co-occurrence.
represents predicted interactions with gene co-occurrence.  indicates interaction from textmining.
indicates interaction from textmining.  indicates interaction from co-expression.
indicates interaction from co-expression.  indicates interaction from protein homology. (B). Ranking of the top 30 nodes from the PPI network according to the number of adjacent nodes. (C). Cluster 1 filtered by MCODE plugin. (D). Cluster 2 filtered by MCODE plugin. (E). Cluster 3 filtered by MCODE plugin.
indicates interaction from protein homology. (B). Ranking of the top 30 nodes from the PPI network according to the number of adjacent nodes. (C). Cluster 1 filtered by MCODE plugin. (D). Cluster 2 filtered by MCODE plugin. (E). Cluster 3 filtered by MCODE plugin.













| Compound | PubChem CID | Label |
|---|---|---|
| 1-(2,4,5-Triethoxyphenyl)propan-2-amine | 44719557 | C1 |
| 19435-97-3 | 3084311 | C2 |
| Cumic Acid | 10820 | C3 |
| Dibutyl phthalate | 3026 | C4 |
| Diisobutyl phthalate | 6782 | C5 |
| Eugenol | 3314 | C6 |
| Isoeugenol | 853433 | C7 |
| Luteolin | 5280445 | C8 |
| Methyl Caffeate | 689075 | C9 |
| Methyl Protocatechuate | 287064 | C10 |
| Nerolidyl Acetate | 5363426 | C11 |
| Nonanoic Acid | 8158 | C12 |
| Patchouli Alcohol | 146159297 | C13 |
| Safrol | 5144 | C14 |
| Tau-Cadinol | 160799 | C15 |
| Vanillic Acid | 8468 | C16 |
| Zoomaric Acid | 445638 | C17 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, F.; Wu, S.; Li, D.; Dong, J.; Huang, X. Leaf Extract of Perilla frutescens (L.) Britt Promotes Adipocyte Browning via the p38 MAPK Pathway and PI3K-AKT Pathway. Nutrients 2023, 15, 1487. https://doi.org/10.3390/nu15061487
Chen F, Wu S, Li D, Dong J, Huang X. Leaf Extract of Perilla frutescens (L.) Britt Promotes Adipocyte Browning via the p38 MAPK Pathway and PI3K-AKT Pathway. Nutrients. 2023; 15(6):1487. https://doi.org/10.3390/nu15061487
Chicago/Turabian StyleChen, Fancheng, Silin Wu, Dejian Li, Jian Dong, and Xiaowei Huang. 2023. "Leaf Extract of Perilla frutescens (L.) Britt Promotes Adipocyte Browning via the p38 MAPK Pathway and PI3K-AKT Pathway" Nutrients 15, no. 6: 1487. https://doi.org/10.3390/nu15061487
APA StyleChen, F., Wu, S., Li, D., Dong, J., & Huang, X. (2023). Leaf Extract of Perilla frutescens (L.) Britt Promotes Adipocyte Browning via the p38 MAPK Pathway and PI3K-AKT Pathway. Nutrients, 15(6), 1487. https://doi.org/10.3390/nu15061487






