Nutritional Programming of the Lifespan of Male Drosophila by Activating FOXO on Larval Low-Nutrient Diet
Abstract
1. Introduction
2. Materials and Methods
2.1. Fly Stock and Husbandry
2.2. Lifespan Analysis
2.3. Body Weight Assay
2.4. Food Intake Assay
2.5. Fecundity Assay
2.6. Climbing Ability Assay
2.7. Starvation Assay
2.8. RNA Isolation and RT–qPCR
2.9. Western Blot
2.10. Carbohydrate and Triglyceride Assay
2.11. Oil Red O Staining of Gut Fat
3. Results
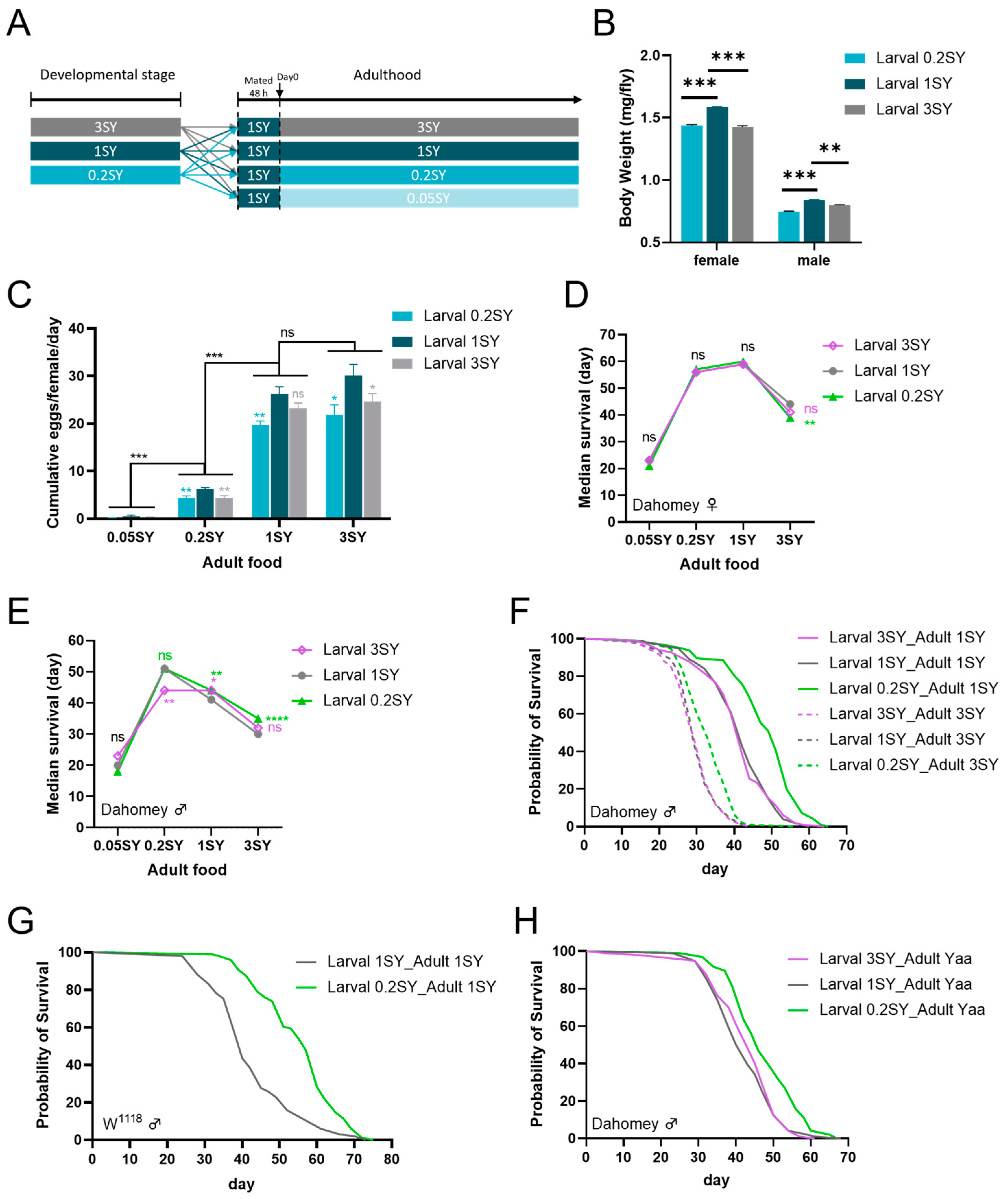
3.1. Developmental Diet Interacts with Adult Diet to Regulate Lifespan and Physiology in Adult Drosophila
3.2. Overlap between Nutritional Programming and Adult DR in Lifespan Extensions
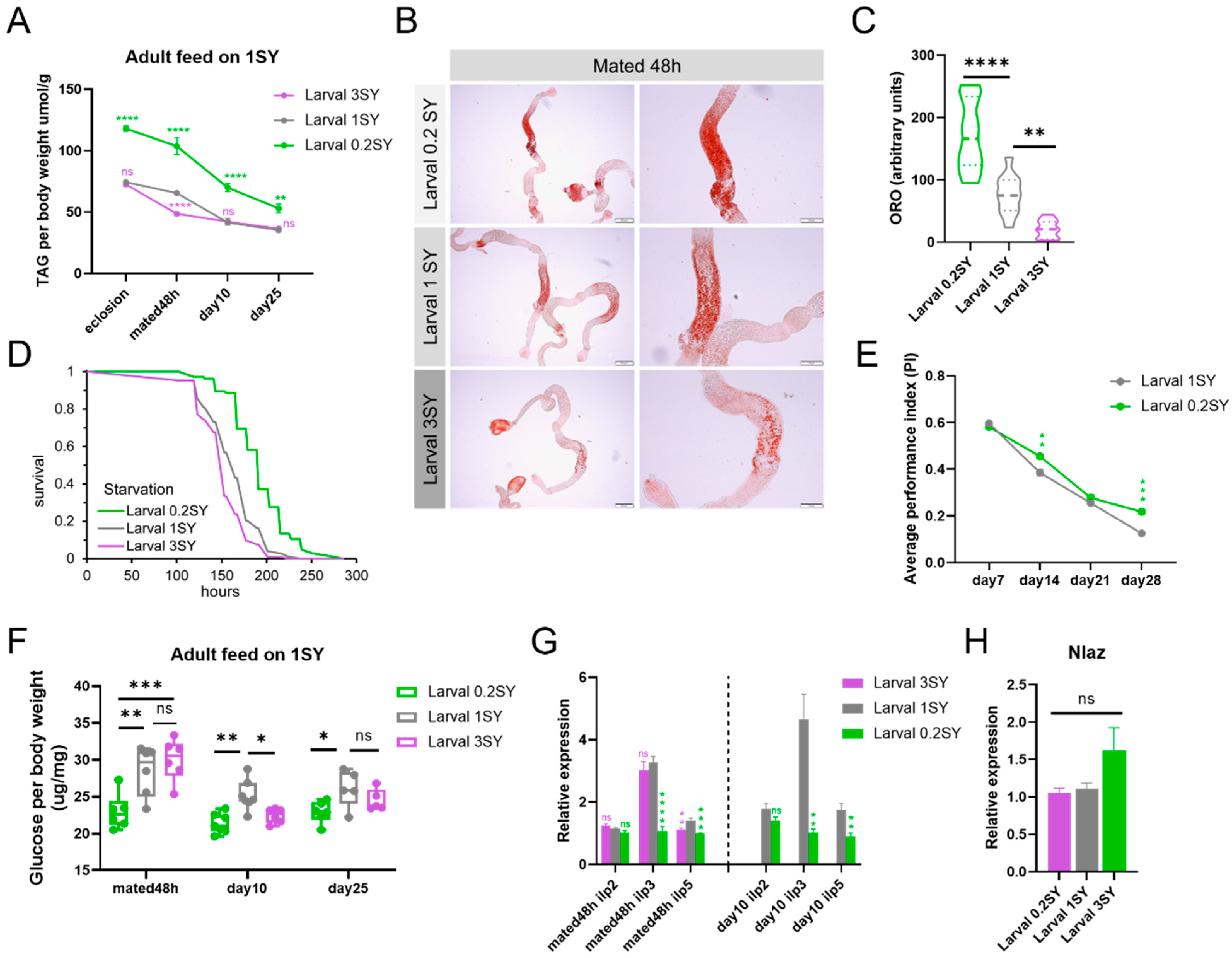
3.3. Low Nutrition during Development Promotes the Health Span of Male Drosophila
3.4. The Lifespan-Extending Effect of a Low-Yeast Developmental Diet Is Partially Dependent on the IIS and mTOR Signaling Pathways
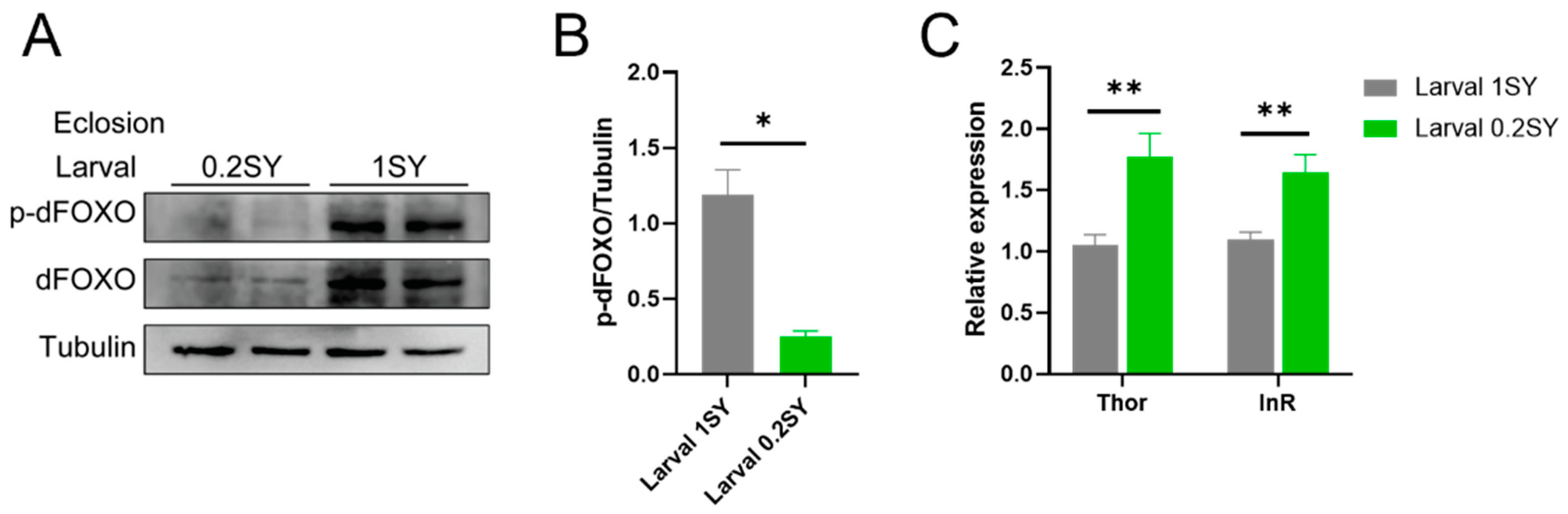
3.5. dFOXO Activity Is Enhanced with the Low-Yeast Diet during Development
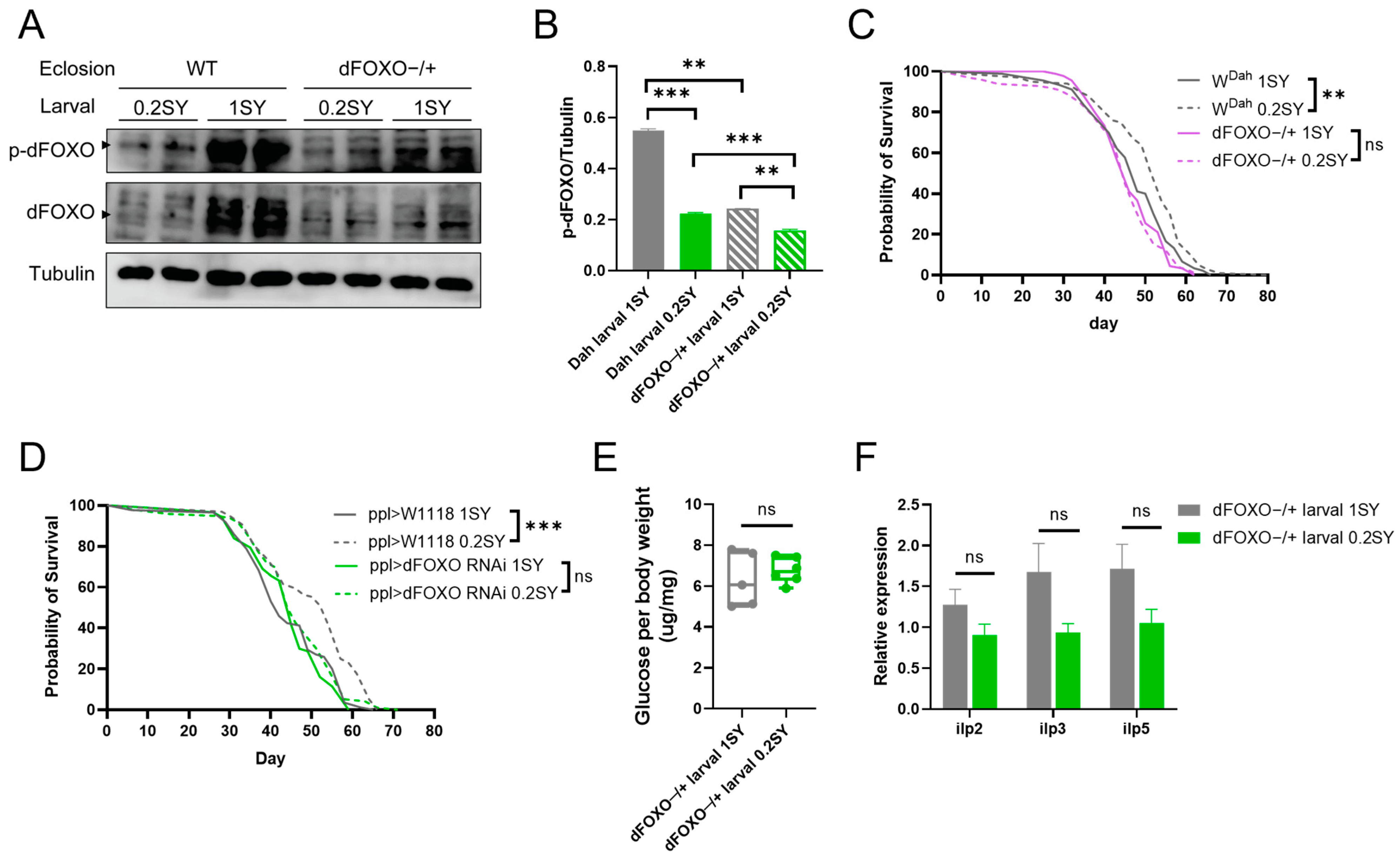
3.6. dFOXO Is Required for Nutritional Programming for Lifespan Extension with a Low-Yeast Diet during Development
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Le Couteur, D.G.; Solon-Biet, S.M.; Parker, B.L.; Pulpitel, T.; Brandon, A.E.; Hunt, N.J.; Wali, J.A.; Gokarn, R.; Senior, A.M.; Cooney, G.J.; et al. Nutritional reprogramming of mouse liver proteome is dampened by metformin, resveratrol, and rapamycin. Cell Metab. 2021, 33, 2367–2379.e4. [Google Scholar] [CrossRef] [PubMed]
- Fontana, L.; Partridge, L. Promoting health and longevity through diet: From model organisms to humans. Cell 2015, 161, 106–118. [Google Scholar] [CrossRef] [PubMed]
- Green, C.L.; Lamming, D.W.; Fontana, L. Molecular mechanisms of dietary restriction promoting health and longevity. Nat. Rev. Mol. Cell Biol. 2022, 23, 56–73. [Google Scholar] [CrossRef]
- Ulgherait, M.; Midoun, A.M.; Park, S.J.; Gatto, J.A.; Tener, S.J.; Siewert, J.; Klickstein, N.; Canman, J.C.; Ja, W.W.; Shirasu-Hiza, M. Circadian autophagy drives iTRF-mediated longevity. Nature 2021, 598, 353–358. [Google Scholar] [CrossRef]
- Hahn, O.; Drews, L.F.; Nguyen, A.; Tatsuta, T.; Gkioni, L.; Hendrich, O.; Zhang, Q.; Langer, T.; Pletcher, S.; Wakelam, M.J.O.; et al. A nutritional memory effect counteracts benefits of dietary restriction in old mice. Nat. Metab. 2019, 1, 1059–1073. [Google Scholar] [CrossRef]
- Van den Heuvel, J.; Zandveld, J.; Mulder, M.; Brakefield, P.M.; Kirkwood, T.B.; Shanley, D.P.; Zwaan, B.J. The plastic fly: The effect of sustained fluctuations in adult food supply on life-history traits. J. Evol. Biol. 2014, 27, 2322–2333. [Google Scholar] [CrossRef]
- Dobson, A.J.; Zandveld, J.; Mulder, M.; Brakefield, P.M.; Kirkwood, T.B.; Shanley, D.P.; Zwaan, B.J. Nutritional Programming of Lifespan by FOXO Inhibition on Sugar-Rich Diets. Cell Rep. 2017, 18, 299–306. [Google Scholar] [CrossRef]
- Obata, F.; Fons, C.O.; Gould, A.P. Early-life exposure to low-dose oxidants can increase longevity via microbiome remodelling in Drosophila. Nat. Commun. 2018, 9, 975. [Google Scholar] [CrossRef] [PubMed]
- Aiello, G.; Sabino, C.; Pernici, D.; Audano, M.; Antonica, F.; Gianesello, M.; Quattrone, A.; Mitro, N.; Romanel, A.; Soldano, A.; et al. Lifespan can be extended during a specific time window early in life. bioRxiv, 2022; preprint. [Google Scholar] [CrossRef]
- Lucas, A. Programming by early nutrition: An experimental approach. J. Nutr. 1998, 128 (Suppl. 2), 401s–406s. [Google Scholar] [CrossRef] [PubMed]
- Langley-Evans, S.C. Nutrition in early life and the programming of adult disease: A review. J. Hum. Nutr. Diet. 2015, 28 (Suppl. 1), 1–14. [Google Scholar] [CrossRef]
- Tarry-Adkins, J.L.; Ozanne, S.E. The impact of early nutrition on the ageing trajectory. Proc. Nutr. Soc. 2014, 73, 289–301. [Google Scholar] [CrossRef] [PubMed]
- Ozanne, S.E.; Hales, C.N. Lifespan: Catch-up growth and obesity in male mice. Nature 2004, 427, 411–412. [Google Scholar] [CrossRef] [PubMed]
- Tu, M.P.; Tatar, M. Juvenile diet restriction and the aging and reproduction of adult Drosophila melanogaster. Aging Cell 2003, 2, 327–333. [Google Scholar] [CrossRef] [PubMed]
- May, C.M.; Doroszuk, A.; Zwaan, B.J. The effect of developmental nutrition on life span and fecundity depends on the adult reproductive environment in Drosophila melanogaster. Ecol. Evol. 2015, 5, 1156–1168. [Google Scholar] [CrossRef]
- Stefana, M.I.; Driscoll, P.C.; Obata, F.; Pengelly, A.R.; Newell, C.L.; MacRae, J.I.; Gould, A.P. Developmental diet regulates Drosophila lifespan via lipid autotoxins. Nat. Commun. 2017, 8, 1384. [Google Scholar] [CrossRef]
- Snoeck, A.; Remacle, C.; Reusens, B.; Hoet, J.J. Effect of a low protein diet during pregnancy on the fetal rat endocrine pancreas. Biol. Neonate 1990, 57, 107–118. [Google Scholar] [CrossRef]
- Tarry-Adkins, J.L.; Chen, J.H.; Jones, R.H.; Smith, N.H.; Ozanne, S.E. Poor maternal nutrition leads to alterations in oxidative stress, antioxidant defense capacity, and markers of fibrosis in rat islets: Potential underlying mechanisms for development of the diabetic phenotype in later life. FASEB J. 2010, 24, 2762–2771. [Google Scholar] [CrossRef]
- Merlet-Bénichou, C.; Gilbert, T.; Muffat-Joly, M.; Lelièvre-Pégorier, M.; Leroy, B. Intrauterine growth retardation leads to a permanent nephron deficit in the rat. Pediatr. Nephrol. 1994, 8, 175–180. [Google Scholar] [CrossRef]
- Blackmore, H.L.; Niu, Y.; Fernandez-Twinn, D.S.; Tarry-Adkins, J.L.; Giussani, D.A.; Ozanne, S.E. Maternal diet-induced obesity programs cardiovascular dysfunction in adult male mouse offspring independent of current body weight. Endocrinology 2014, 155, 3970–3980. [Google Scholar] [CrossRef]
- Tarry-Adkins, J.L.; Chen, J.H.; Smith, N.S.; Jones, R.H.; Cherif, H.; Ozanne, S.E. Poor maternal nutrition followed by accelerated postnatal growth leads to telomere shortening and increased markers of cell senescence in rat islets. FASEB J. 2009, 23, 1521–1528. [Google Scholar] [CrossRef]
- Tarry-Adkins, J.L.; Fernandez-Twinn, D.S.; Chen, J.H.; Hargreaves, I.P.; Neergheen, V.; Aiken, C.E.; Ozanne, S.E. Poor maternal nutrition and accelerated postnatal growth induces an accelerated aging phenotype and oxidative stress in skeletal muscle of male rats. Dis. Model Mech. 2016, 9, 1221–1229. [Google Scholar] [CrossRef]
- He, Z.X.; Sun, Z.H.; Tan, Z.L.; Tang, S.X.; Zhou, C.S.; Han, X.F.; Wang, M.; Wu, D.Q.; Kang, J.H.; Beauchemin, K.A. Effects of maternal protein or energy restriction during late gestation on antioxidant status of plasma and immune tissues in postnatal goats. J. Anim. Sci. 2012, 90, 4319–4326. [Google Scholar] [CrossRef] [PubMed]
- Lillycrop, K.A.; Slater-Jefferies, J.L.; Hanson, M.A.; Godfrey, K.M.; Jackson, A.A.; Burdge, G.C. Induction of altered epigenetic regulation of the hepatic glucocorticoid receptor in the offspring of rats fed a protein-restricted diet during pregnancy suggests that reduced DNA methyltransferase-1 expression is involved in impaired DNA methylation and changes in histone modifications. Br. J. Nutr. 2007, 97, 1064–1073. [Google Scholar] [PubMed]
- Park, J.H.; Stoffers, D.A.; Nicholls, R.D.; Simmons, R.A. Development of type 2 diabetes following intrauterine growth retardation in rats is associated with progressive epigenetic silencing of Pdx1. J. Clin. Investig. 2008, 118, 2316–2324. [Google Scholar] [CrossRef] [PubMed]
- Plagemann, A.; Harder, T.; Brunn, M.; Harder, A.; Roepke, K.; Wittrock-Staar, M.; Ziska, T.; Schellong, K.; Rodekamp, E.; Melchior, K.; et al. Hypothalamic proopiomelanocortin promoter methylation becomes altered by early overfeeding: An epigenetic model of obesity and the metabolic syndrome. J. Physiol. 2009, 587 Pt 20, 4963–4976. [Google Scholar] [CrossRef]
- Johnson, C.; Kiefer, H.; Chaulot-Talmon, A.; Dance, A.; Sellem, E.; Jouneau, L.; Jammes, H.; Kastelic, J.; Thundathil, J. Prepubertal nutritional modulation in the bull and its impact on sperm DNA methylation. Cell Tissue Res. 2022, 389, 587–601. [Google Scholar] [CrossRef]
- Seals, D.R.; Melov, S. Translational geroscience: Emphasizing function to achieve optimal longevity. Aging 2014, 6, 718–730. [Google Scholar] [CrossRef]
- Cameron, K.M.; Miwa, S.; Walker, C.; von Zglinicki, T. Male mice retain a metabolic memory of improved glucose tolerance induced during adult onset, short-term dietary restriction. Longev. Healthspan 2012, 1, 3. [Google Scholar] [CrossRef]
- Selman, C.; Hempenstall, S. Evidence of a metabolic memory to early-life dietary restriction in male C57BL/6 mice. Longev. Healthspan 2012, 1, 2. [Google Scholar] [CrossRef]
- Anson, R.M.; Guo, Z.; de Cabo, R.; Iyun, T.; Rios, M.; Hagepanos, A.; Ingram, D.K.; Lane, M.A.; Mattson, M.P. Intermittent fasting dissociates beneficial effects of dietary restriction on glucose metabolism and neuronal resistance to injury from calorie intake. Proc. Natl. Acad. Sci. USA 2003, 100, 6216–6220. [Google Scholar] [CrossRef]
- Efeyan, A.; Comb, W.C.; Sabatini, D.M. Nutrient-sensing mechanisms and pathways. Nature 2015, 517, 302–310. [Google Scholar] [CrossRef] [PubMed]
- Kapahi, P.; Kaeberlein, M.; Hansen, M. Dietary restriction and lifespan: Lessons from invertebrate models. Ageing Res. Rev. 2017, 39, 3–14. [Google Scholar] [CrossRef] [PubMed]
- Tatar, M.; Kopelman, A.; Epstein, D.; Tu, M.P.; Yin, C.M.; Garofalo, R. A mutant Drosophila insulin receptor homolog that extends life-span and impairs neuroendocrine function. Science 2001, 292, 107–110. [Google Scholar] [CrossRef] [PubMed]
- Clancy, D.J.; Gems, D.; Harshman, L.G.; Oldham, S.; Stocker, H.; Hafen, E.; Leevers, S.J.; Partridge, L. Extension of life-span by loss of CHICO, a Drosophila insulin receptor substrate protein. Science 2001, 292, 104–106. [Google Scholar] [CrossRef]
- Kapahi, P.; Zid, B.M.; Harper, T.; Koslover, D.; Sapin, V.; Benzer, S. Regulation of lifespan in Drosophila by modulation of genes in the TOR signaling pathway. Curr. Biol. 2004, 14, 885–890. [Google Scholar] [CrossRef]
- Tatar, M.; Post, S.; Yu, K. Nutrient control of Drosophila longevity. Trends Endocrinol. Metab. 2014, 25, 509–517. [Google Scholar] [CrossRef]
- Taylor, J.R.; Wood, J.G.; Mizerak, E.; Hinthorn, S.; Liu, J.; Finn, M.; Gordon, S.; Zingas, L.; Chang, C.; Klein, M.A.; et al. Sirt6 regulates lifespan in Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 2022, 119, e2111176119. [Google Scholar] [CrossRef]
- Morris, B.J.; Willcox, D.C.; Donlon, T.A.; Willcox, B.J. FOXO3: A Major Gene for Human Longevity—A Mini-Review. Gerontology 2015, 61, 515–525. [Google Scholar] [CrossRef]
- Sun, X.; Chen, W.D.; Wang, Y.D. DAF-16/FOXO Transcription Factor in Aging and Longevity. Front. Pharmacol. 2017, 8, 548. [Google Scholar] [CrossRef]
- Shimokawa, I.; Komatsu, T.; Hayashi, N.; Kim, S.E.; Kawata, T.; Park, S.; Hayashi, H.; Yamaza, H.; Chiba, T.; Mori, R. The life-extending effect of dietary restriction requires Foxo3 in mice. Aging Cell 2015, 14, 707–709. [Google Scholar] [CrossRef]
- Puig, O.; Marr, M.T.; Ruhf, M.L.; Tjian, R. Control of cell number by Drosophila FOXO: Downstream and feedback regulation of the insulin receptor pathway. Genes Dev. 2003, 17, 2006–2020. [Google Scholar] [CrossRef] [PubMed]
- Slack, C.; Giannakou, M.E.; Foley, A.; Goss, M.; Partridge, L. dFOXO-independent effects of reduced insulin-like signaling in Drosophila. Aging Cell 2011, 10, 735–748. [Google Scholar] [CrossRef] [PubMed]
- Bjedov, I.; Toivonen, J.M.; Kerr, F.; Slack, C.; Jacobson, J.; Foley, A.; Partridge, L. Mechanisms of life span extension by rapamycin in the fruit fly Drosophila melanogaster. Cell Metab. 2010, 11, 35–46. [Google Scholar] [CrossRef] [PubMed]
- Weigelt, C.M.; Sehgal, R.; Tain, L.S.; Cheng, J.; Eßer, J.; Pahl, A.; Dieterich, C.; Grönke, S.; Partridge, L. An Insulin-Sensitive Circular RNA that Regulates Lifespan in Drosophila. Mol. Cell 2020, 79, 268–279.e5. [Google Scholar] [CrossRef]
- Bass, T.M.; Grandison, R.C.; Wong, R.; Martinez, P.; Partridge, L.; Piper, M.D. Optimization of dietary restriction protocols in Drosophila. J. Gerontol. Ser. A Biol. Sci. Med. Sci. 2007, 62, 1071–1081. [Google Scholar] [CrossRef]
- Piper, M.D.; Blanc, E.; Leitão-Gonçalves, R.; Yang, M.; He, X.; Linford, N.J.; Hoddinott, M.P.; Hopfen, C.; Soultoukis, G.A.; Niemeyer, C.; et al. A holidic medium for Drosophila melanogaster. Nat. Methods 2014, 11, 100–105. [Google Scholar] [CrossRef]
- Wu, Q.; Yu, G.; Park, S.J.; Gao, Y.; Ja, W.W.; Yang, M. Excreta Quantification (EX-Q) for Longitudinal Measurements of Food Intake in Drosophila. iScience 2020, 23, 100776. [Google Scholar] [CrossRef]
- Sofola, O.; Kerr, F.; Rogers, I.; Killick, R.; Augustin, H.; Gandy, C.; Allen, M.J.; Hardy, J.; Lovestone, S.; Partridge, L. Inhibition of GSK-3 ameliorates Abeta pathology in an adult-onset Drosophila model of Alzheimer’s disease. PLoS Genet. 2010, 6, e1001087. [Google Scholar] [CrossRef]
- Bos, M.; Burnet, B.; Farrow, R.; Woods, R.A. Development of Drosophila on sterol mutants of the yeast Saccharomyces cerevisiae. Genet. Res. 1976, 28, 163–176. [Google Scholar] [CrossRef]
- Fanson, B.G.; Taylor, P.W. Additive and interactive effects of nutrient classes on longevity, reproduction, and diet consumption in the Queensland fruit fly (Bactrocera tryoni). J. Insect Physiol. 2012, 58, 327–334. [Google Scholar] [CrossRef]
- Duxbury, E.M.L.; Chapman, T. Sex-Specific Responses of Life Span and Fitness to Variation in Developmental Versus Adult Diets in Drosophila melanogaster. J. Gerontol. Ser. A Biol. Sci. Med. Sci. 2020, 75, 1431–1438. [Google Scholar] [CrossRef]
- Klepsatel, P.; Knoblochová, D.; Girish, T.N.; Dircksen, H. The influence of developmental diet on reproduction and metabolism in Drosophila. BMC Evol. Biol. 2020, 20, 93. [Google Scholar] [CrossRef] [PubMed]
- Luis, N.M.; Wang, L.; Ortega, M.; Deng, H.; Katewa, S.D.; Li, P.W.; Karpac, J.; Jasper, H.; Kapahi, P. Intestinal IRE1 Is Required for Increased Triglyceride Metabolism and Longer Lifespan under Dietary Restriction. Cell Rep. 2016, 17, 1207–1216. [Google Scholar] [CrossRef] [PubMed]
- Pasco, M.Y.; Léopold, P. High sugar-induced insulin resistance in Drosophila relies on the lipocalin Neural Lazarillo. PLoS ONE 2012, 7, e36583. [Google Scholar] [CrossRef] [PubMed]
- Villanueva, J.E.; Livelo, C.; Trujillo, A.S.; Chandran, S.; Woodworth, B.; Andrade, L.; Le, H.D.; Manor, U.; Panda, S.; Melkani, G.C. Time-restricted feeding restores muscle function in Drosophila models of obesity and circadian-rhythm disruption. Nat. Commun. 2019, 10, 2700. [Google Scholar] [CrossRef]
- Ruiz, M.; Ganfornina, M.D.; Correnti, C.; Strong, R.K.; Sanchez, D. Ligand binding-dependent functions of the lipocalin NLaz: An in vivo study in Drosophila. FASEB J. 2014, 28, 1555–1567. [Google Scholar] [CrossRef]
- Layalle, S.; Arquier, N.; Léopold, P. The TOR pathway couples nutrition and developmental timing in Drosophila. Dev. Cell 2008, 15, 568–577. [Google Scholar] [CrossRef]
- Lin, X.; Smagghe, G. Roles of the insulin signaling pathway in insect development and organ growth. Peptides 2019, 122, 169923. [Google Scholar] [CrossRef]
- Kauwe, G.; Tsurudome, K.; Penney, J.; Mori, M.; Gray, L.; Calderon, M.R.; Elazouzzi, F.; Chicoine, N.; Sonenberg, N.; Haghighi, A.P. Acute Fasting Regulates Retrograde Synaptic Enhancement through a 4E-BP-Dependent Mechanism. Neuron 2016, 92, 1204–1212. [Google Scholar] [CrossRef]
- Martins, R.; Lithgow, G.J.; Link, W. Long live FOXO: Unraveling the role of FOXO proteins in aging and longevity. Aging Cell 2016, 15, 196–207. [Google Scholar] [CrossRef]
- Jünger, M.A.; Rintelen, F.; Stocker, H.; Wasserman, J.D.; Végh, M.; Radimerski, T.; Greenberg, M.E.; Hafen, E. The Drosophila forkhead transcription factor FOXO mediates the reduction in cell number associated with reduced insulin signaling. J. Biol. 2003, 2, 20. [Google Scholar] [CrossRef] [PubMed]
- Puig, O.; Tjian, R. Transcriptional feedback control of insulin receptor by dFOXO/FOXO1. Genes Dev. 2005, 19, 2435–2446. [Google Scholar] [CrossRef] [PubMed]
- Klepsatel, P.; Procházka, E.; Gáliková, M. Crowding of Drosophila larvae affects lifespan and other life-history traits via reduced availability of dietary yeast. Exp. Gerontol. 2018, 110, 298–308. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Sadighi Akha, A.A.; Miller, R.A.; Harper, J.M. Life-span extension in mice by preweaning food restriction and by methionine restriction in middle age. J. Gerontol. Ser. A Biol. Sci. Med. Sci. 2009, 64, 711–722. [Google Scholar] [CrossRef]
- May, C.M.; Zwaan, B.J. Relating past and present diet to phenotypic and transcriptomic variation in the fruit fly. BMC Genom. 2017, 18, 640. [Google Scholar] [CrossRef]
- Krittika, S.; Lenka, A.; Yadav, P. Evidence of dietary protein restriction regulating pupation height, development time and lifespan in Drosophila melanogaster. Biol. Open 2019, 8, bio042952. [Google Scholar] [CrossRef]
- López-Otín, C.; Blasco, M.A.; Partridge, L.; Serrano, M.; Kroemer, G. Hallmarks of aging: An expanding universe. Cell 2023, 186, 243–278. [Google Scholar] [CrossRef]
- Papadopoli, D.; Boulay, K.; Kazak, L.; Pollak, M.; Mallette, F.; Topisirovic, I.; Hulea, L. mTOR as a central regulator of lifespan and aging. F1000Research 2019, 8, F1000. [Google Scholar] [CrossRef]
- He, Y.; Jasper, H. Studying aging in Drosophila. Methods 2014, 68, 129–133. [Google Scholar] [CrossRef]
- Piper, M.D.; Partridge, L. Protocols to Study Aging in Drosophila. Methods Mol. Biol. 2016, 1478, 291–302. [Google Scholar]
- Evangelou, A.; Ignatiou, A.; Antoniou, C.; Kalanidou, S.; Chatzimatthaiou, S.; Shianiou, G.; Ellina, S.; Athanasiou, R.; Panagi, M.; Apidianakis, Y.; et al. Unpredictable Effects of the Genetic Background of Transgenic Lines in Physiological Quantitative Traits. G3 Genes Genomes Genet. 2019, 9, 3877–3890. [Google Scholar] [CrossRef] [PubMed]
- Ogienko, A.A.; Omelina, E.S.; Bylino, O.V.; Batin, M.A.; Georgiev, P.G.; Pindyurin, A.V. Drosophila as a Model Organism to Study Basic Mechanisms of Longevity. Int. J. Mol. Sci. 2022, 23, 11244. [Google Scholar] [CrossRef] [PubMed]
- Giannakou, M.E.; Goss, M.; Partridge, L. Role of dFOXO in lifespan extension by dietary restriction in Drosophila melanogaster: Not required, but its activity modulates the response. Aging Cell 2008, 7, 187–198. [Google Scholar] [CrossRef] [PubMed]


| Primer Name | Primer Sequence |
|---|---|
| Nlaz F: | GGACAACCCTCGAATGTAACT |
| Nlaz R: | GACGGCGTATGACTCGTAATC |
| dilp2 F: | CCTGCAGTTTGTCCAGGAGT |
| dilp2 R: | AGCCAGGGAATTGAGTACACC |
| dilp3 F: | GTATGGCTTCAACGCAATGAC |
| dilp3 R: | GAGCATCTGAACCCAACTATCAC |
| dilp5 F: | CGTGATCCCAGTTCTCCTGT |
| dilp5 R: | ACCCTCAGCATGTCCATCAA |
| Thor F: | CCAGGAAGGTTGTCATCTCG |
| Thor R: | TGAAAGCCCGCTCGTAGATA |
| InR F: | GGTGCTGGCATCATAGGTCT |
| InR R: | CCTGCCTCTGAGTGATAGAAGG |
| RP49 F: | GCCCAAGGGTATCGACAACA |
| RP49 R: | CTTGCGCTTCTTGGAGGAGA |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gao, Y.; Cheng, X.; Tian, Y.; Yuan, Z.; Fan, X.; Yang, D.; Yang, M. Nutritional Programming of the Lifespan of Male Drosophila by Activating FOXO on Larval Low-Nutrient Diet. Nutrients 2023, 15, 1840. https://doi.org/10.3390/nu15081840
Gao Y, Cheng X, Tian Y, Yuan Z, Fan X, Yang D, Yang M. Nutritional Programming of the Lifespan of Male Drosophila by Activating FOXO on Larval Low-Nutrient Diet. Nutrients. 2023; 15(8):1840. https://doi.org/10.3390/nu15081840
Chicago/Turabian StyleGao, Yue, Xingyi Cheng, Yao Tian, Zhixiao Yuan, Xiaolan Fan, Deying Yang, and Mingyao Yang. 2023. "Nutritional Programming of the Lifespan of Male Drosophila by Activating FOXO on Larval Low-Nutrient Diet" Nutrients 15, no. 8: 1840. https://doi.org/10.3390/nu15081840
APA StyleGao, Y., Cheng, X., Tian, Y., Yuan, Z., Fan, X., Yang, D., & Yang, M. (2023). Nutritional Programming of the Lifespan of Male Drosophila by Activating FOXO on Larval Low-Nutrient Diet. Nutrients, 15(8), 1840. https://doi.org/10.3390/nu15081840







