Gut Dysbiosis Shaped by Cocoa Butter-Based Sucrose-Free HFD Leads to Steatohepatitis, and Insulin Resistance in Mice
Abstract
:1. Introduction
2. Materials and Methods
2.1. Animals
2.2. Oral Glucose Tolerance Test (OGTT)
2.3. Insulin Tolerance Test (ITT)
2.4. Plasma Measurements
2.5. Histological Analysis
2.6. Gene Expression Analysis by qRT-PCR
2.7. Microbiome Sequencing
2.8. 16S rRNA Microbiome Analysis and Bioinformatics Statistics
2.9. Biodiversity Analysis
2.10. Identification of Biomarker Microbiome
2.11. Microbial Metagenomic Functional Predictions
2.12. Prediction of Biomarker Microbiome-Epigenome Interactions
3. Results
3.1. C-HFD and F-HFD Have Differential Effects on the Gut Microbiome in Mice
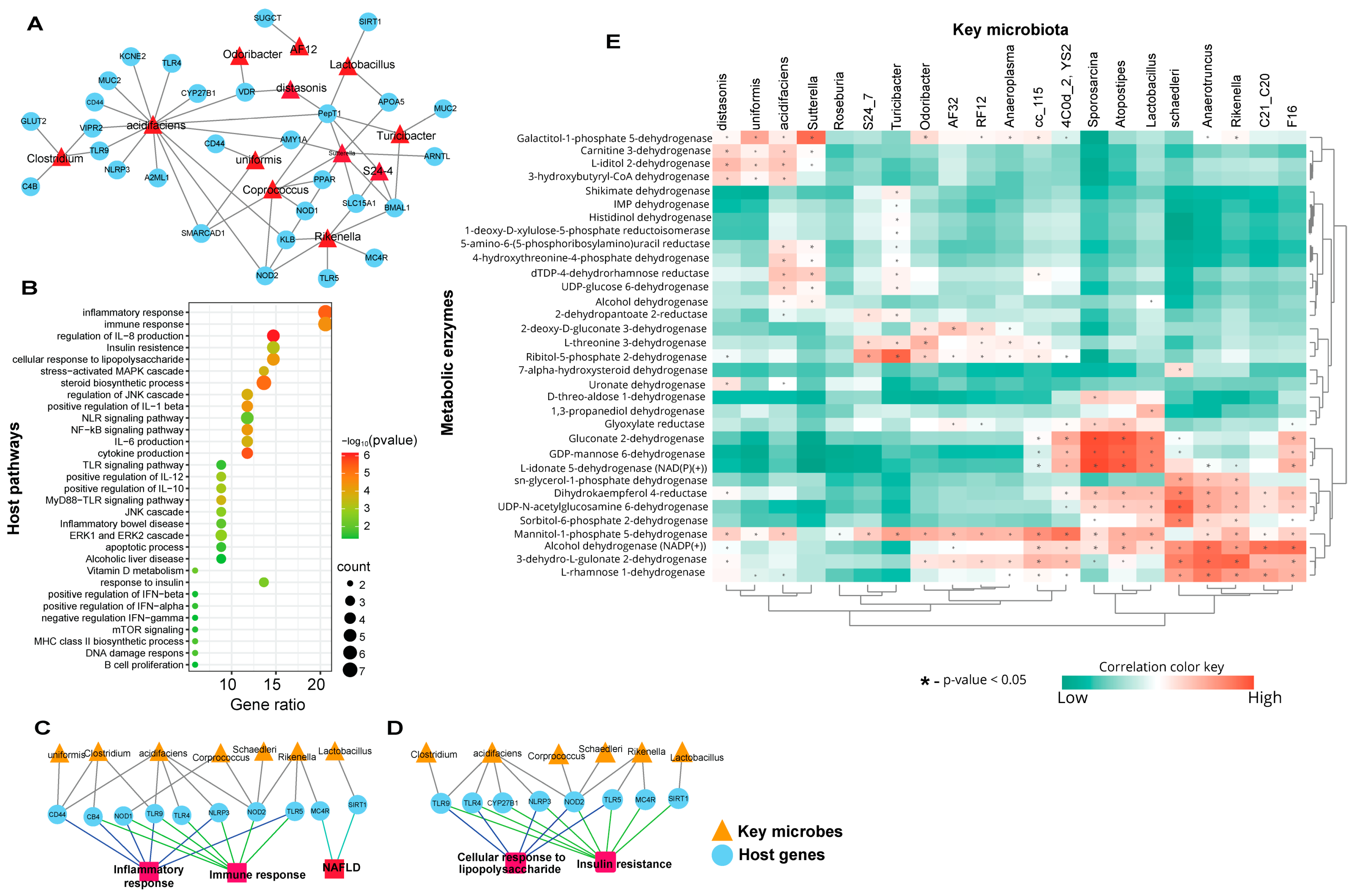
3.2. Biomarker Gut Microbiota Associate with Host Genes Involved in Immune-Metabolic Regulation
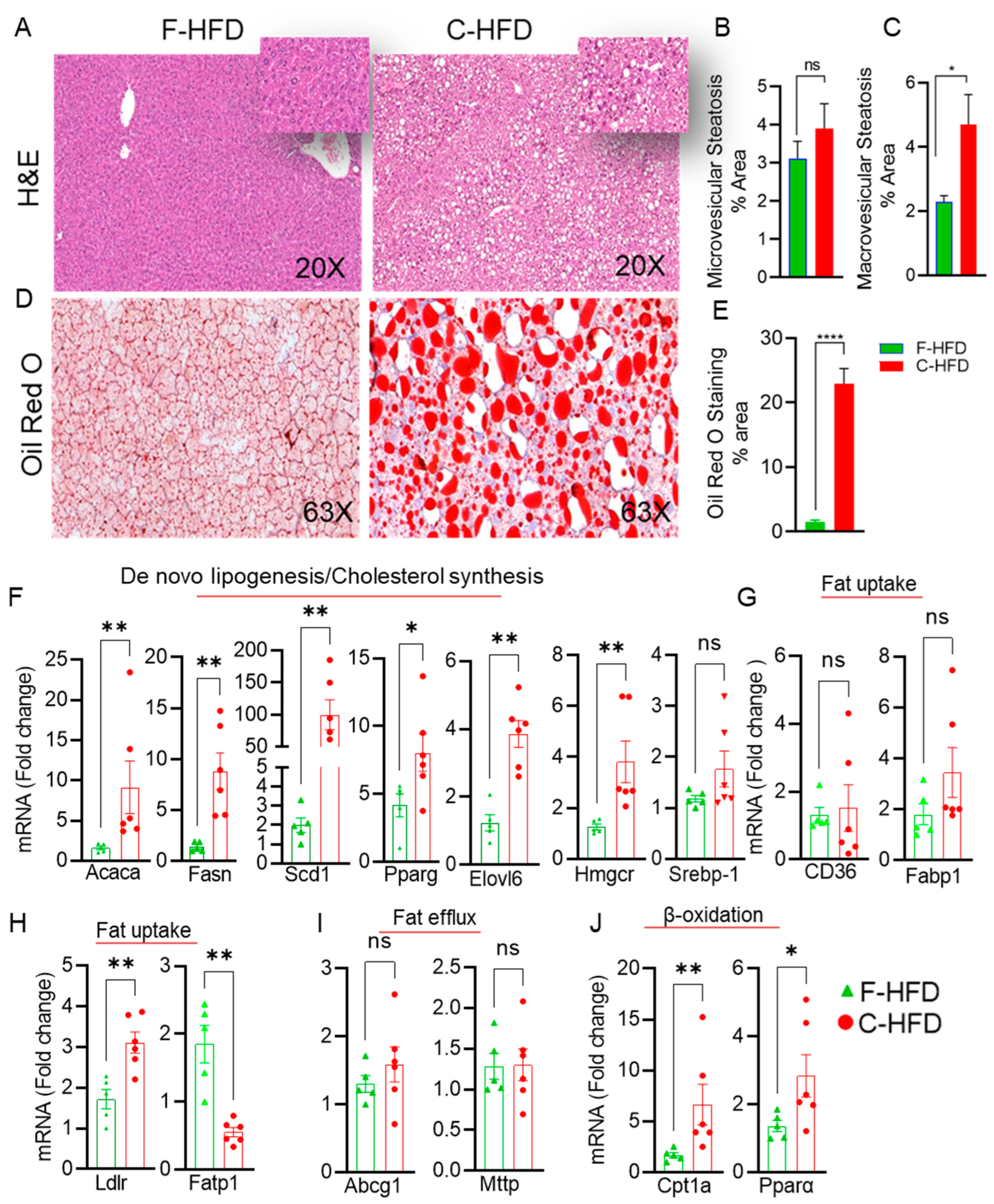
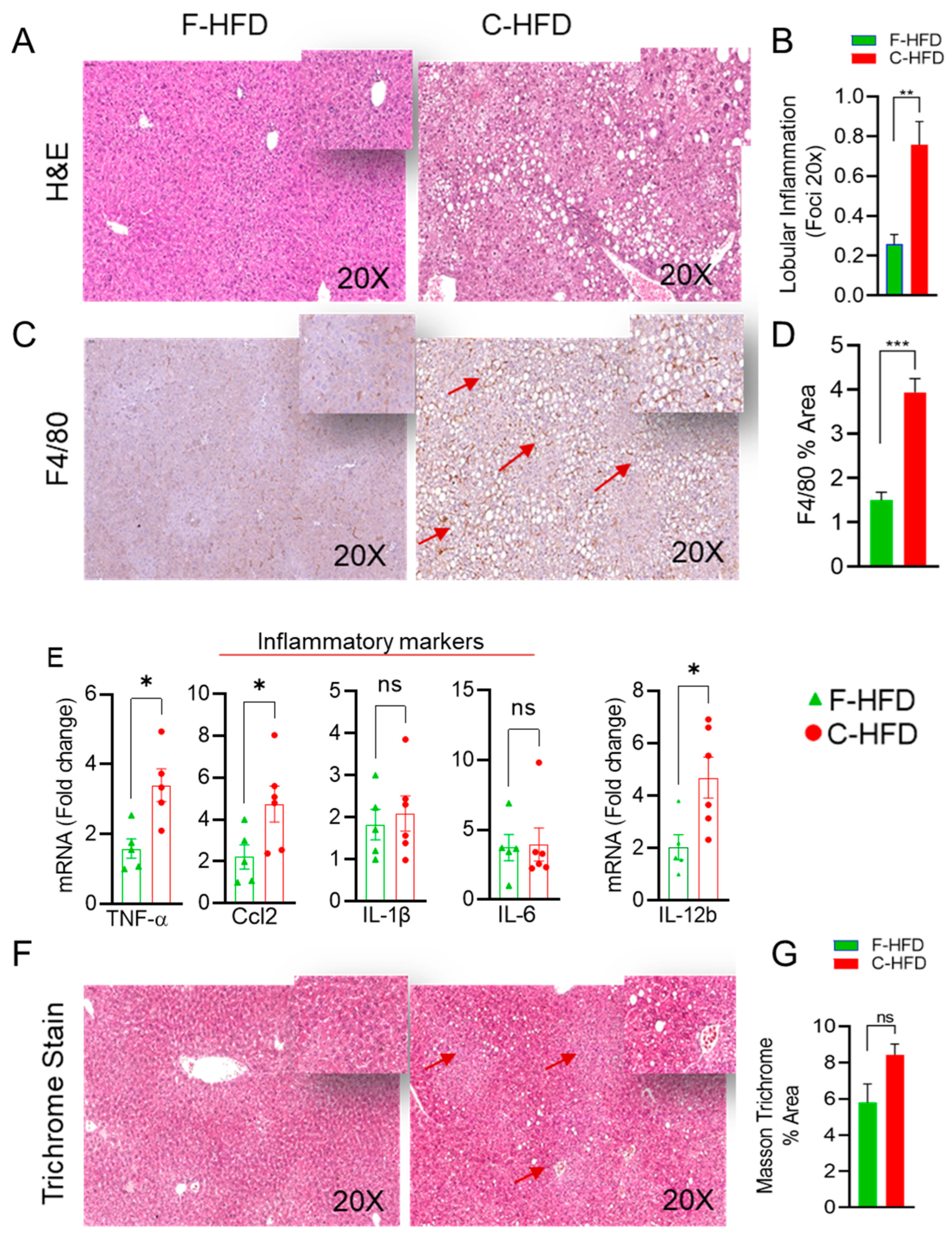
3.3. Mice Fed with C-HFD Display Liver Steatosis and Inflammation
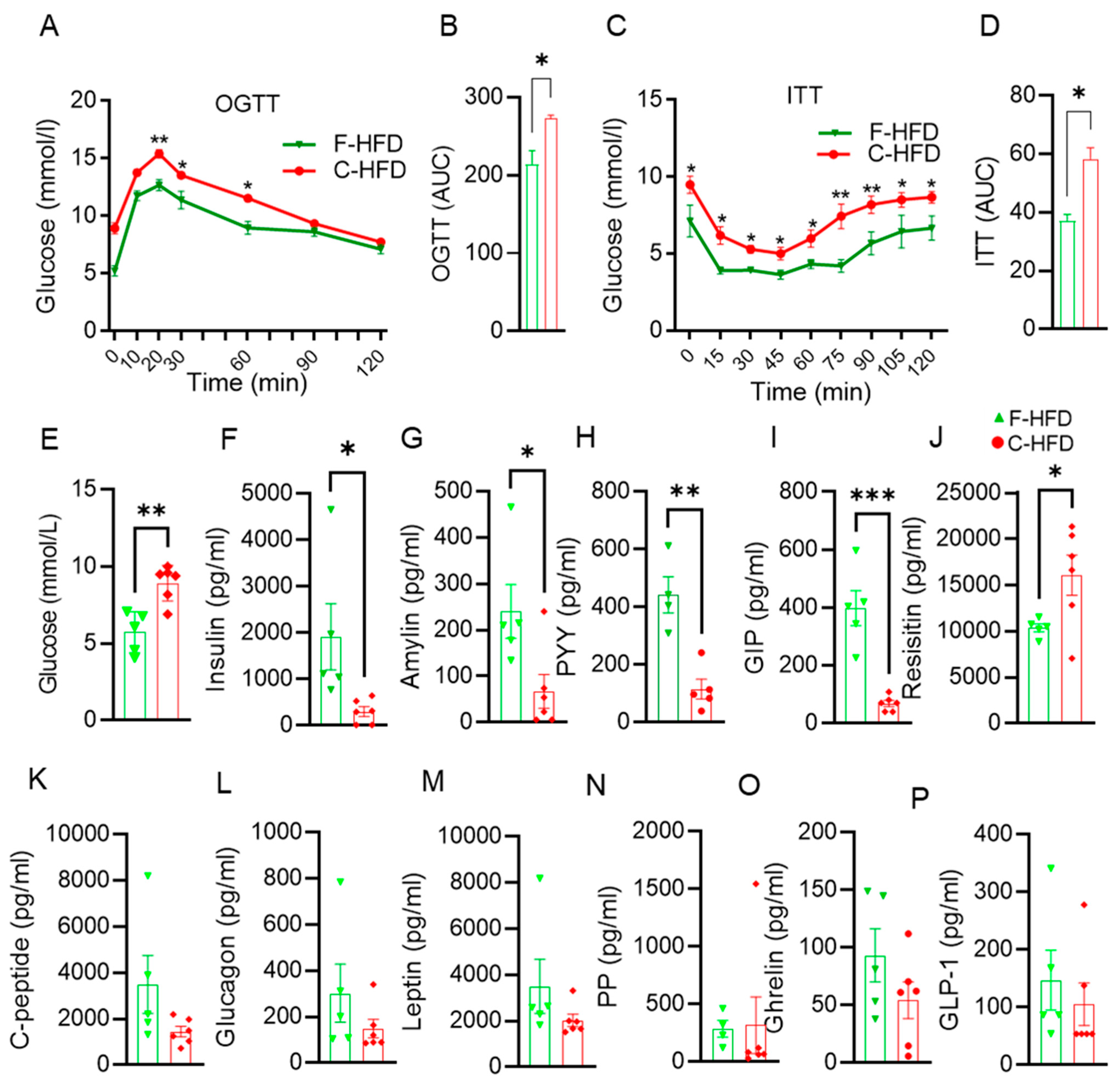
3.4. Impaired Glucose Tolerance in C-HFD Mice, Compared with F-HFD Mice
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Zhang, P. Influence of foods and nutrition on the gut microbiome and implications for intestinal health. Int. J. Mol. Sci. 2022, 23, 9588. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Tan, Y.; Cheng, H.; Zhang, D.; Feng, W.; Peng, C. Functions of gut microbiota metabolites, current status and future perspectives. Aging Dis. 2022, 13, 1106. [Google Scholar] [CrossRef] [PubMed]
- Cano, P.G.; Santacruz, A.; Trejo, F.M.; Sanz, Y. Bifidobacterium cect 7765 improves metabolic and immunological alterations associated with obesity in high-fat diet-fed mice. Obesity 2013, 21, 2310–2321. [Google Scholar] [CrossRef] [PubMed]
- Sugawara, Y.; Kanazawa, A.; Aida, M.; Yoshida, Y.; Yamashiro, Y.; Watada, H. Association of gut microbiota and inflammatory markers in obese patients with type 2 diabetes mellitus: Post hoc analysis of a synbiotic interventional study. Biosci. Microbiota Food Health 2022, 41, 103–111. [Google Scholar] [CrossRef] [PubMed]
- Martínez-López, Y.E.; Esquivel-Hernández, D.A.; Sánchez-Castañeda, J.P.; Neri-Rosario, D.; Guardado-Mendoza, R.; Resendis-Antonio, O. Type 2 diabetes, gut microbiome, and systems biology: A novel perspective for a new era. Gut Microbes 2022, 14, 2111952. [Google Scholar] [CrossRef] [PubMed]
- An, J.; Wang, Q.; Yi, S.; Liu, X.; Jin, H.; Xu, J.; Wen, G.; Zhu, J.; Tuo, B. The source of the fat significantly affects the results of high-fat diet intervention. Sci. Rep. 2022, 12, 4315. [Google Scholar] [CrossRef]
- Zhang, M.; Yang, X.-J. Effects of a high fat diet on intestinal microbiota and gastrointestinal diseases. World J. Gastroenterol. 2016, 22, 8905. [Google Scholar] [CrossRef]
- Trakman, G.L.; Fehily, S.; Basnayake, C.; Hamilton, A.L.; Russell, E.; Wilson-O’Brien, A.; Kamm, M.A. Diet and gut microbiome in gastrointestinal disease. J. Gastroenterol. Hepatol. 2022, 37, 237–245. [Google Scholar] [CrossRef]
- Thomas, F.; Hehemann, J.-H.; Rebuffet, E.; Czjzek, M.; Michel, G. Environmental and gut bacteroidetes: The food connection. Front. Microbiol. 2011, 2, 93. [Google Scholar] [CrossRef]
- Rinninella, E.; Raoul, P.; Cintoni, M.; Franceschi, F.; Miggiano, G.A.D.; Gasbarrini, A.; Mele, M.C. What is the healthy gut microbiota composition? A changing ecosystem across age, environment, diet, and diseases. Microorganisms 2019, 7, 14. [Google Scholar] [CrossRef]
- Cho, K.Y. Association of gut microbiota with obesity in children and adolescents. Clin. Exp. Pediatr. 2023, 66, 148–154. [Google Scholar] [CrossRef] [PubMed]
- Koliada, A.; Syzenko, G.; Moseiko, V.; Budovska, L.; Puchkov, K.; Perederiy, V.; Gavalko, Y.; Dorofeyev, A.; Romanenko, M.; Tkach, S.; et al. Association between body mass index and firmicutes/bacteroidetes ratio in an adult ukrainian population. BMC Microbiol. 2017, 17, 120. [Google Scholar] [CrossRef] [PubMed]
- Chanda, D.; De, D. Meta-analysis reveals obesity associated gut microbial alteration patterns and reproducible contributors of functional shift. bioRxiv 2022. [Google Scholar] [CrossRef]
- Kübeck, R.; Bonet-Ripoll, C.; Hoffmann, C.; Walker, A.; Müller, V.M.; Schüppel, V.L.; Lagkouvardos, I.; Scholz, B.; Engel, K.-H.; Daniel, H. Dietary fat and gut microbiota interactions determine diet-induced obesity in mice. Mol. Metab. 2016, 5, 1162–1174. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Zhu, Y.; Zhao, F.; Song, S.; Li, Y.; Xu, X.; Zhou, G.; Li, C. Fish oil, lard and soybean oil differentially shape gut microbiota of middle-aged rats. Sci. Rep. 2017, 7, 826. [Google Scholar] [CrossRef] [PubMed]
- Quek, R.Y.C.; Peh, E.W.Y.; Henry, C.J. Effects of cocoa butter and cocoa butter equivalent in a chocolate confectionery on human blood triglycerides, glucose and insulin. Foods 2020, 9, 455. [Google Scholar] [CrossRef] [PubMed]
- Patterson, W.L.; Georgel, P.T. Breaking the cycle: The role of omega-3 polyunsaturated fatty acids in inflammation-driven cancers. Biochem. Cell Biol. 2014, 92, 321–328. [Google Scholar] [CrossRef] [PubMed]
- Gustavsson, C.; Parini, P.; Ostojic, J.; Cheung, L.; Hu, J.; Zadjali, F.; Tahir, F.; Brismar, K.; Norstedt, G.; Tollet-Egnell, P. Cocoa butter and safflower oil elicit different effects on hepatic gene expression and lipid metabolism in rats. Lipids 2009, 44, 1011–1027. [Google Scholar] [CrossRef] [PubMed]
- Bäckhed, F.; Ding, H.; Wang, T.; Hooper, L.V.; Koh, G.Y.; Nagy, A.; Semenkovich, C.F.; Gordon, J.I. The gut microbiota as an environmental factor that regulates fat storage. Proc. Natl. Acad. Sci. USA 2004, 101, 15718–15723. [Google Scholar] [CrossRef]
- Bäckhed, F.; Manchester, J.K.; Semenkovich, C.F.; Gordon, J.I. Mechanisms underlying the resistance to diet-induced obesity in germ-free mice. Proc. Natl. Acad. Sci. USA 2007, 104, 979–984. [Google Scholar] [CrossRef]
- Jennison, E.; Byrne, C.D. The role of the gut microbiome and diet in the pathogenesis of non-alcoholic fatty liver disease. Clin. Mol. Hepatol. 2021, 27, 22. [Google Scholar] [CrossRef] [PubMed]
- Garcia, K.; Ferreira, G.; Reis, F.; Viana, S. Impact of dietary sugars on gut microbiota and metabolic health. Diabetology 2022, 3, 549–560. [Google Scholar] [CrossRef]
- Konstantopoulos, P.; Doulamis, I.P.; Tzani, A.; Korou, M.L.; Agapitos, E.; Vlachos, I.S.; Pergialiotis, V.; Verikokos, C.; Mastorakos, G.; Katsilambros, N.L.; et al. Metabolic effects of crocus sativus and protective action against non-alcoholic fatty liver disease in diabetic rats. Biomed. Rep. 2017, 6, 513–518. [Google Scholar] [CrossRef] [PubMed]
- Fengler, V.H.I.; Macheiner, T.; Kessler, S.M.; Czepukojc, B.; Gemperlein, K.; Müller, R.; Kiemer, A.K.; Magnes, C.; Haybaeck, J.; Lackner, C.; et al. Susceptibility of different mouse wild type strains to develop diet-induced nafld/afld-associated liver disease. PLoS ONE 2016, 11, e0155163. [Google Scholar] [CrossRef]
- Crowe, A.R.; Yue, W. Semi-quantitative determination of protein expression using immunohistochemistry staining and analysis: An integrated protocol. Bio-protocol 2019, 9, e3465. [Google Scholar] [CrossRef] [PubMed]
- Mehlem, A.; Hagberg, C.E.; Muhl, L.; Eriksson, U.; Falkevall, A. Imaging of neutral lipids by oil red o for analyzing the metabolic status in health and disease. Nat. Protoc. 2013, 8, 1149–1154. [Google Scholar] [CrossRef]
- Yen, K.; Le, T.T.; Bansal, A.; Narasimhan, S.D.; Cheng, J.X.; Tissenbaum, H.A. A comparative study of fat storage quantitation in nematode caenorhabditis elegans using label and label-free methods. PLoS ONE 2010, 5, e12810. [Google Scholar] [CrossRef]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, interactive, scalable and extensible microbiome data science using qiime 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Peña, A.G.; Goodrich, J.K.; Gordon, J.I.; et al. Qiime allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef]
- Dhariwal, A.; Chong, J.; Habib, S.; King, I.L.; Agellon, L.B.; Xia, J. Microbiomeanalyst: A web-based tool for comprehensive statistical, visual and meta-analysis of microbiome data. Nucleic Acids Res. 2017, 45, W180–W188. [Google Scholar] [CrossRef]
- Ombrello, A.K. Dada2. In Encyclopedia of Medical Immunology: Immunodeficiency Diseases; MacKay, I., Rose, N.R., Eds.; Springer: New York, NY, USA, 2020; pp. 1–7. [Google Scholar]
- DeSantis, T.Z.; Hugenholtz, P.; Larsen, N.; Rojas, M.; Brodie, E.L.; Keller, K.; Huber, T.; Dalevi, D.; Hu, P.; Andersen, G.L. Greengenes, a chimera-checked 16s rrna gene database and workbench compatible with arb. Appl. Environ. Microbiol. 2006, 72, 5069–5072. [Google Scholar] [CrossRef] [PubMed]
- Lozupone, C.; Knight, R. Unifrac: A new phylogenetic method for comparing microbial communities. Appl. Environ. Microbiol. 2005, 71, 8228–8235. [Google Scholar] [CrossRef] [PubMed]
- Qin, J.; Li, Y.; Cai, Z.; Li, S.; Zhu, J.; Zhang, F.; Liang, S.; Zhang, W.; Guan, Y.; Shen, D.; et al. A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature 2012, 490, 55–60. [Google Scholar] [CrossRef] [PubMed]
- Yatsunenko, T.; Rey, F.E.; Manary, M.J.; Trehan, I.; Dominguez-Bello, M.G.; Contreras, M.; Magris, M.; Hidalgo, G.; Baldassano, R.N.; Anokhin, A.P.; et al. Human gut microbiome viewed across age and geography. Nature 2012, 486, 222–227. [Google Scholar] [CrossRef] [PubMed]
- Faith, D.P. Conservation evaluation and phylogenetic diversity. Biol. Conserv. 1992, 61, 1–10. [Google Scholar] [CrossRef]
- Langille, M.G.I.; Zaneveld, J.; Caporaso, J.G.; McDonald, D.; Knights, D.; Reyes, J.A.; Clemente, J.C.; Burkepile, D.E.; Vega Thurber, R.L.; Knight, R.; et al. Predictive functional profiling of microbial communities using 16s rrna marker gene sequences. Nat. Biotechnol. 2013, 31, 814–821. [Google Scholar] [CrossRef] [PubMed]
- Caspi, R.; Altman, T.; Billington, R.; Dreher, K.; Foerster, H.; Fulcher, C.A.; Holland, T.A.; Keseler, I.M.; Kothari, A.; Kubo, A.; et al. The metacyc database of metabolic pathways and enzymes and the biocyc collection of pathway/genome databases. Nucleic Acids Res. 2014, 42, D459–D471. [Google Scholar] [CrossRef] [PubMed]
- Ma, W.; Zhang, L.; Zeng, P.; Huang, C.; Li, J.; Geng, B.; Yang, J.; Kong, W.; Zhou, X.; Cui, Q. An analysis of human microbe-disease associations. Brief. Bioinform. 2017, 18, 85–97. [Google Scholar] [CrossRef]
- Wang, L.; Zhang, W.; Wu, X.; Liang, X.; Cao, L.; Zhai, J.; Yang, Y.; Chen, Q.; Liu, H.; Zhang, J.; et al. Miaome: Human microbiome affect the host epigenome. Comput. Struct. Biotechnol. J. 2022, 20, 2455–2463. [Google Scholar] [CrossRef]
- Delpino, F.M.; Figueiredo, L.M.; da Silva, B.G.C. Effects of omega-3 supplementation on body weight and body fat mass: A systematic review. Clin. Nutr. ESPEN 2021, 44, 122–129. [Google Scholar] [CrossRef]
- Togo, J.; Hu, S.; Li, M.; Niu, C.; Speakman, J.R. Impact of dietary sucrose on adiposity and glucose homeostasis in c57bl/6j mice depends on mode of ingestion: Liquid or solid. Mol. Metab. 2019, 27, 22–32. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Zhang, B.; Peng, B.; Wang, J.; Hu, Y.; Wang, R.; Wang, S. Different dose of sucrose consumption divergently influences gut microbiota and ppar-γ/mapk/nf-κb pathway in dss-induced colitis mice. Nutrients 2022, 14, 2765. [Google Scholar] [CrossRef] [PubMed]
- Alasmar, R.M.; Varadharajan, K.; Shanmugakonar, M.; Al-Naemi, H.A. Early-life sugar consumption affects the microbiome in juvenile mice. Mol. Nutr. Food Res. 2023, 67, 2200322. [Google Scholar] [CrossRef] [PubMed]
- Sun, S.; Araki, Y.; Hanzawa, F.; Umeki, M.; Kojima, T.; Nishimura, N.; Ikeda, S.; Mochizuki, S.; Oda, H. High sucrose diet-induced dysbiosis of gut microbiota promotes fatty liver and hyperlipidemia in rats. J. Nutr. Biochem. 2021, 93, 108621. [Google Scholar] [CrossRef] [PubMed]
- Dalby, M.J.; Ross, A.W.; Walker, A.W.; Morgan, P.J. Dietary uncoupling of gut microbiota and energy harvesting from obesity and glucose tolerance in mice. Cell Rep. 2017, 21, 1521–1533. [Google Scholar] [CrossRef] [PubMed]
- Muegge, B.D.; Kuczynski, J.; Knights, D.; Clemente, J.C.; González, A.; Fontana, L.; Henrissat, B.; Knight, R.; Gordon, J.I. Diet drives convergence in gut microbiome functions across mammalian phylogeny and within humans. Science 2011, 332, 970–974. [Google Scholar] [CrossRef] [PubMed]
- De Filippo, C.; Cavalieri, D.; Di Paola, M.; Ramazzotti, M.; Poullet, J.B.; Massart, S.; Collini, S.; Pieraccini, G.; Lionetti, P. Impact of diet in shaping gut microbiota revealed by a comparative study in children from europe and rural africa. Proc. Natl. Acad. Sci. USA 2010, 107, 14691–14696. [Google Scholar] [CrossRef] [PubMed]
- Cronin, P.; Joyce, S.A.; O’Toole, P.W.; O’Connor, E.M. Dietary fibre modulates the gut microbiota. Nutrients 2021, 13, 1655. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Kong, Q.; Li, X.; Zhao, J.; Zhang, H.; Chen, W.; Wang, G. A high-fat diet increases gut microbiota biodiversity and energy expenditure due to nutrient difference. Nutrients 2020, 12, 3197. [Google Scholar] [CrossRef]
- Xiao, L.; Sonne, S.B.; Feng, Q.; Chen, N.; Xia, Z.; Li, X.; Fang, Z.; Zhang, D.; Fjære, E.; Midtbø, L.K.; et al. High-fat feeding rather than obesity drives taxonomical and functional changes in the gut microbiota in mice. Microbiome 2017, 5, 43. [Google Scholar] [CrossRef]
- Rowland, I.; Gibson, G.; Heinken, A.; Scott, K.; Swann, J.; Thiele, I.; Tuohy, K. Gut microbiota functions: Metabolism of nutrients and other food components. Eur. J. Nutr. 2018, 57, 1–24. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Xiao, Y.; Li, D.; Zhang, S.; Wu, Y.; Zhang, Q.; Bai, W. New insights into the mechanisms of high-fat diet mediated gut microbiota in chronic diseases. iMeta 2023, 2, e69. [Google Scholar] [CrossRef]
- De Bandt, J.P.; Waligora-Dupriet, A.J.; Butel, M.J. Intestinal microbiota in inflammation and insulin resistance: Relevance to humans. Curr. Opin. Clin. Nutr. Metab. Care 2011, 14, 334–340. [Google Scholar] [CrossRef] [PubMed]
- Zou, Y.; Ju, X.; Chen, W.; Yuan, J.; Wang, Z.; Aluko, R.E.; He, R. Rice bran attenuated obesity via alleviating dyslipidemia, browning of white adipocytes and modulating gut microbiota in high-fat diet-induced obese mice. Food Funct. 2020, 11, 2406–2417. [Google Scholar] [CrossRef]
- Shin, N.R.; Whon, T.W.; Bae, J.W. Proteobacteria: Microbial signature of dysbiosis in gut microbiota. Trends Biotechnol. 2015, 33, 496–503. [Google Scholar] [CrossRef]
- Rizzatti, G.; Lopetuso, L.R.; Gibiino, G.; Binda, C.; Gasbarrini, A. Proteobacteria: A common factor in human diseases. BioMed Res. Int. 2017, 2017, 9351507. [Google Scholar] [CrossRef]
- Anderson, J.R.; Carroll, I.; Azcarate-Peril, M.A.; Rochette, A.D.; Heinberg, L.J.; Peat, C.; Steffen, K.; Manderino, L.M.; Mitchell, J.; Gunstad, J. A preliminary examination of gut microbiota, sleep, and cognitive flexibility in healthy older adults. Sleep Med. 2017, 38, 104–107. [Google Scholar] [CrossRef] [PubMed]
- Bik, E.M.; Eckburg, P.B.; Gill, S.R.; Nelson, K.E.; Purdom, E.A.; Francois, F.; Perez-Perez, G.; Blaser, M.J.; Relman, D.A. Molecular analysis of the bacterial microbiota in the human stomach. Proc. Natl. Acad. Sci. USA 2006, 103, 732–737. [Google Scholar] [CrossRef]
- Chipashvili, O.; Utter, D.R.; Bedree, J.K.; Ma, Y.; Schulte, F.; Mascarin, G.; Alayyoubi, Y.; Chouhan, D.; Hardt, M.; Bidlack, F.; et al. Episymbiotic saccharibacteria suppresses gingival inflammation and bone loss in mice through host bacterial modulation. Cell Host Microbe 2021, 29, 1649–1662.e7. [Google Scholar] [CrossRef]
- Xiao, L.; Feng, Q.; Liang, S.; Sonne, S.B.; Xia, Z.; Qiu, X.; Li, X.; Long, H.; Zhang, J.; Zhang, D.; et al. A catalog of the mouse gut metagenome. Nat. Biotechnol. 2015, 33, 1103–1108. [Google Scholar] [CrossRef]
- Vieira, J.R.P.; Rezende, A.T.d.O.; Fernandes, M.R.; da Silva, N.A. Intestinal microbiota and active systemic lupus erythematosus: A systematic review. Adv. Rheumatol. 2021, 61, 42. [Google Scholar] [CrossRef]
- Le Roy, T.; Moens de Hase, E.; Van Hul, M.; Paquot, A.; Pelicaen, R.; Regnier, M.; Depommier, C.; Druart, C.; Everard, A.; Maiter, D.; et al. Dysosmobacter welbionis is a newly isolated human commensal bacterium preventing diet-induced obesity and metabolic disorders in mice. Gut 2022, 71, 534–543. [Google Scholar] [CrossRef] [PubMed]
- Daniel, H.; Gholami, A.M.; Berry, D.; Desmarchelier, C.; Hahne, H.; Loh, G.; Mondot, S.; Lepage, P.; Rothballer, M.; Walker, A.; et al. High-fat diet alters gut microbiota physiology in mice. ISME J. 2014, 8, 295–308. [Google Scholar] [CrossRef] [PubMed]
- Kameyama, K.; Itoh, K. Intestinal colonization by a lachnospiraceae bacterium contributes to the development of diabetes in obese mice. Microbes Environ. 2014, 29, 427–430. [Google Scholar] [CrossRef] [PubMed]
- Zheng, J.; Zhang, L.; Gao, Y.; Wu, H.; Zhang, J. The dynamic effects of maternal high-calorie diet on glycolipid metabolism and gut microbiota from weaning to adulthood in offspring mice. Front. Nutr. 2022, 9, 941969. [Google Scholar] [CrossRef] [PubMed]
- Sun, T.; Liu, S.; Zhou, Y.; Yao, Z.; Zhang, D.; Cao, S.; Wei, Z.; Tan, B.; Li, Y.; Lian, Z.; et al. Evolutionary biologic changes of gut microbiota in an ‘adenoma-carcinoma sequence’ mouse colorectal cancer model induced by 1,2-dimethylhydrazine. Oncotarget 2016, 8, 444–457. [Google Scholar] [CrossRef] [PubMed]
- Chu, M.; Zhang, X. Bacterial atlas of mouse gut microbiota. Cell. Microbiol. 2022, 2022, 5968814. [Google Scholar] [CrossRef]
- Yan, G.; Li, S.; Wen, Y.; Luo, Y.; Huang, J.; Chen, B.; Lv, S.; Chen, L.; He, L.; He, M.; et al. Characteristics of intestinal microbiota in c57bl/6 mice with non-alcoholic fatty liver induced by high-fat diet. Front. Microbiol. 2022, 13, 1051200. [Google Scholar] [CrossRef] [PubMed]
- Velázquez, K.T.; Enos, R.T.; Bader, J.E.; Sougiannis, A.T.; Carson, M.S.; Chatzistamou, I.; Carson, J.A.; Nagarkatti, P.S.; Nagarkatti, M.; Murphy, E.A. Prolonged high-fat-diet feeding promotes non-alcoholic fatty liver disease and alters gut microbiota in mice. World J. Hepatol. 2019, 11, 619–637. [Google Scholar] [CrossRef]
- Wang, M.; Li, L.; Qian, J.; Wang, N.; Bao, J.; Lu, J.; Chen, F.; Li, Y.; Zhang, Y.; Yan, F. Periodontitis salivary microbiota exacerbates nonalcoholic fatty liver disease in high-fat diet-induced obese mice. iScience 2023, 26, 106346. [Google Scholar] [CrossRef]
- Characteristics of intestinal bacteria with fatty liver diseases and cirrhosis. Ann. Hepatol. 2019, 18, 796–803. [CrossRef]
- Norheim, F.; Chella Krishnan, K.; Bjellaas, T.; Vergnes, L.; Pan, C.; Parks, B.W.; Meng, Y.; Lang, J.; Ward, J.A.; Reue, K.; et al. Genetic regulation of liver lipids in a mouse model of insulin resistance and hepatic steatosis. Mol. Syst. Biol. 2021, 17, e9684. [Google Scholar] [CrossRef]
- Weng, X.; Maxwell-Warburton, S.; Hasib, A.; Ma, L.; Kang, L. The membrane receptor cd44: Novel insights into metabolism. Trends Endocrinol. Metab. 2022, 33, 318–332. [Google Scholar] [CrossRef]
- Bauer, S.; Hezinger, L.; Rexhepi, F.; Ramanathan, S.; Kufer, T.A. Nod-like receptors—Emerging links to obesity and associated morbidities. Int. J. Mol. Sci. 2023, 24, 8595. [Google Scholar] [CrossRef] [PubMed]
- Itoh, M.; Suganami, T.; Nakagawa, N.; Tanaka, M.; Yamamoto, Y.; Kamei, Y.; Terai, S.; Sakaida, I.; Ogawa, Y. Melanocortin 4 receptor-deficient mice as a novel mouse model of nonalcoholic steatohepatitis. Am. J. Pathol. 2011, 179, 2454–2463. [Google Scholar] [CrossRef]
- Vachharajani, V.T.; Liu, T.; Wang, X.; Hoth, J.J.; Yoza, B.K.; McCall, C.E. Sirtuins link inflammation and metabolism. J. Immunol. Res. 2016, 2016, 8167273. [Google Scholar] [CrossRef]
- Fontes-Cal, T.C.M.; Mattos, R.T.; Medeiros, N.I.; Pinto, B.F.; Belchior-Bezerra, M.; Roque-Souza, B.; Dutra, W.O.; Ferrari, T.C.A.; Vidigal, P.V.T.; Faria, L.C.; et al. Crosstalk between plasma cytokines, inflammation, and liver damage as a new strategy to monitoring nafld progression. Front. Immunol. 2021, 12, 708959. [Google Scholar] [CrossRef] [PubMed]
- Meng, Y.; Ma, F.; Guan, M. Role of steroid hormones in the pathogenesis of nonalcoholic fatty liver disease. Metabolites 2021, 11, 320. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Poulsen, K.L.; Wu, L.; Liu, S.; Miyata, T.; Song, Q.; Wei, Q.; Zhao, C.; Lin, C.; Yang, J. Targeted therapeutics and novel signaling pathways in non-alcohol-associated fatty liver/steatohepatitis (nafl/nash). Signal Transduct. Target. Ther. 2022, 7, 287. [Google Scholar] [CrossRef]
- Cani, P.D.; Geurts, L.; Matamoros, S.; Plovier, H.; Duparc, T. Glucose metabolism: Focus on gut microbiota, the endocannabinoid system and beyond. Diabetes Metab. 2014, 40, 246–257. [Google Scholar] [CrossRef]
- Lee, J.; Song, X.; Hyun, B.; Jeon, C.O.; Hyun, S. Drosophila gut immune pathway suppresses host development-promoting effects of acetic acid bacteria. Mol. Cells 2023, 46, 637–653. [Google Scholar] [CrossRef] [PubMed]
- Thompson, K.N.; Bonham, K.S.; Ilott, N.E.; Britton, G.J.; Colmenero, P.; Bullers, S.J.; McIver, L.J.; Ma, S.; Nguyen, L.H.; Filer, A.; et al. Alterations in the gut microbiome implicate key taxa and metabolic pathways across inflammatory arthritis phenotypes. Sci. Transl. Med. 2023, 15, eabn4722. [Google Scholar] [CrossRef] [PubMed]
- Fan, S.; Guo, W.; Xiao, D.; Guan, M.; Liao, T.; Peng, S.; Feng, A.; Wang, Z.; Yin, H.; Li, M.; et al. Microbiota-gut-brain axis drives overeating disorders. Cell Metab. 2023, 35, 2011–2027.e2017. [Google Scholar] [CrossRef] [PubMed]
- Houghton, M.; Kerimi, A.; Mouly, V.; Tumova, S.; Williamson, G. Gut microbiome catabolites as novel modulators of muscle cell glucose metabolism. FASEB J. 2019, 33, 1887–1898. [Google Scholar] [CrossRef] [PubMed]
- Nishida, A.; Ando, Y.; Kimura, I.; Miyamoto, J. Involvement of gut microbial metabolites derived from diet on host energy homeostasis. Int. J. Mol. Sci. 2022, 23, 5562. [Google Scholar] [CrossRef] [PubMed]
- Leung, C.; Rivera, L.; Furness, J.B.; Angus, P.W. The role of the gut microbiota in nafld. Nat. Rev. Gastroenterol. Hepatol. 2016, 13, 412–425. [Google Scholar] [CrossRef] [PubMed]
- Piao, Y.; Yun, S.Y.; Fu, Z.; Jang, J.M.; Back, M.J.; Kim, H.H.; Kim, D.K. Recombinant human hapln1 mitigates pulmonary emphysema by increasing tgf-β receptor i and sirtuins levels in human alveolar epithelial cells. Mol. Cells 2023, 46, 558–572. [Google Scholar] [CrossRef] [PubMed]
- Gabbia, D.; De Martin, S. Targeting the adipose tissue-liver-gut microbiota crosstalk to cure masld. Biology 2023, 12, 1471. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.A.; Kwon, Y.W.; Kim, H.R.; Shin, N.; Son, H.J.; Cheong, C.S.; Kim, D.J.; Hwang, O. A novel pyrazolo[3,4-d]pyrimidine induces heme oxygenase-1 and exerts anti-inflammatory and neuroprotective effects. Mol. Cells 2022, 45, 134–147. [Google Scholar] [CrossRef]
- Han, H.; Wang, M.; Zhong, R.; Yi, B.; Schroyen, M.; Zhang, H. Depletion of gut microbiota inhibits hepatic lipid accumulation in high-fat diet-fed mice. Int. J. Mol. Sci. 2022, 23, 9350. [Google Scholar] [CrossRef]
- Cortez-Pinto, H.; Borralho, P.; Machado, J.; Lopes, M.T.; Gato, I.V.; Santos, A.M.; Guerreiro, A.S. Microbiota modulation with synbiotic decreases liver fibrosis in a high fat choline deficient diet mice model of non-alcoholic steatohepatitis (nash). GE-Port. J. Gastroenterol. 2016, 23, 132–141. [Google Scholar] [CrossRef] [PubMed]
- Hrncir, T.; Hrncirova, L.; Kverka, M.; Hromadka, R.; Machova, V.; Trckova, E.; Kostovcikova, K.; Kralickova, P.; Krejsek, J.; Tlaskalova-Hogenova, H. Gut microbiota and nafld: Pathogenetic mechanisms, microbiota signatures, and therapeutic interventions. Microorganisms 2021, 9, 957. [Google Scholar] [CrossRef] [PubMed]
- Yajima, M.; Karaki, S.I.; Tsuruta, T.; Kimura, S.; Nio-Kobayashi, J.; Kuwahara, A.; Yajima, T. Diversity of the intestinal microbiota differently affects non-neuronal and atropine-sensitive ileal contractile responses to short-chain fatty acids in mice. Biomed. Res. 2016, 37, 319–328. [Google Scholar] [CrossRef] [PubMed]
- Khan, A.; Ding, Z.; Ishaq, M.; Bacha, A.S.; Khan, I.; Hanif, A.; Li, W.; Guo, X. Understanding the effects of gut microbiota dysbiosis on nonalcoholic fatty liver disease and the possible probiotics role: Recent updates. Int. J. Biol. Sci. 2021, 17, 818–833. [Google Scholar] [CrossRef] [PubMed]
- Gurung, M.; Li, Z.; You, H.; Rodrigues, R.; Jump, D.B.; Morgun, A.; Shulzhenko, N. Role of gut microbiota in type 2 diabetes pathophysiology. EBioMedicine 2020, 51, 102590. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Liao, M.; Zhou, N.; Bao, L.; Ma, K.; Zheng, Z.; Wang, Y.; Liu, C.; Wang, W.; Wang, J.; et al. Parabacteroides distasonis alleviates obesity and metabolic dysfunctions via production of succinate and secondary bile acids. Cell Rep. 2019, 26, 222–235.e225. [Google Scholar] [CrossRef] [PubMed]
- Foley, K.P.; Zlitni, S.; Duggan, B.M.; Barra, N.G.; Anhê, F.F.; Cavallari, J.F.; Henriksbo, B.D.; Chen, C.Y.; Huang, M.; Lau, T.C.; et al. Gut microbiota impairs insulin clearance in obese mice. Mol. Metab. 2020, 42, 101067. [Google Scholar] [CrossRef] [PubMed]
- Roth, J.D.; Hughes, H.; Kendall, E.; Baron, A.D.; Anderson, C.M. Antiobesity effects of the β-cell hormone amylin in diet-induced obese rats: Effects on food intake, body weight, composition, energy expenditure, and gene expression. Endocrinology 2006, 147, 5855–5864. [Google Scholar] [CrossRef] [PubMed]
- Boey, D.; Lin, S.; Karl, T.; Baldock, P.; Lee, N.; Enriquez, R.; Couzens, M.; Slack, K.; Dallmann, R.; Sainsbury, A.; et al. Peptide yy ablation in mice leads to the development of hyperinsulinaemia and obesity. Diabetologia 2006, 49, 1360–1370. [Google Scholar] [CrossRef]
- El, K.; Gray, S.M.; Capozzi, M.E.; Knuth, E.R.; Jin, E.; Svendsen, B.; Clifford, A.; Brown, J.L.; Encisco, S.E.; Chazotte, B.M.; et al. Gip mediates the incretin effect and glucose tolerance by dual actions on α cells and β cells. Sci. Adv. 2021, 7, eabf1948. [Google Scholar] [CrossRef]
- Li, F.-P.; He, J.; Li, Z.-Z.; Luo, Z.-F.; Yan, L.; Li, Y. Effects of resistin expression on glucose metabolism and hepatic insulin resistance. Endocrine 2009, 35, 243–251. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kochumon, S.; Malik, M.Z.; Sindhu, S.; Arefanian, H.; Jacob, T.; Bahman, F.; Nizam, R.; Hasan, A.; Thomas, R.; Al-Rashed, F.; et al. Gut Dysbiosis Shaped by Cocoa Butter-Based Sucrose-Free HFD Leads to Steatohepatitis, and Insulin Resistance in Mice. Nutrients 2024, 16, 1929. https://doi.org/10.3390/nu16121929
Kochumon S, Malik MZ, Sindhu S, Arefanian H, Jacob T, Bahman F, Nizam R, Hasan A, Thomas R, Al-Rashed F, et al. Gut Dysbiosis Shaped by Cocoa Butter-Based Sucrose-Free HFD Leads to Steatohepatitis, and Insulin Resistance in Mice. Nutrients. 2024; 16(12):1929. https://doi.org/10.3390/nu16121929
Chicago/Turabian StyleKochumon, Shihab, Md. Zubbair Malik, Sardar Sindhu, Hossein Arefanian, Texy Jacob, Fatemah Bahman, Rasheeba Nizam, Amal Hasan, Reeby Thomas, Fatema Al-Rashed, and et al. 2024. "Gut Dysbiosis Shaped by Cocoa Butter-Based Sucrose-Free HFD Leads to Steatohepatitis, and Insulin Resistance in Mice" Nutrients 16, no. 12: 1929. https://doi.org/10.3390/nu16121929
APA StyleKochumon, S., Malik, M. Z., Sindhu, S., Arefanian, H., Jacob, T., Bahman, F., Nizam, R., Hasan, A., Thomas, R., Al-Rashed, F., Shenouda, S., Wilson, A., Albeloushi, S., Almansour, N., Alhamar, G., Al Madhoun, A., Alzaid, F., Thanaraj, T. A., Koistinen, H. A., ... Ahmad, R. (2024). Gut Dysbiosis Shaped by Cocoa Butter-Based Sucrose-Free HFD Leads to Steatohepatitis, and Insulin Resistance in Mice. Nutrients, 16(12), 1929. https://doi.org/10.3390/nu16121929









