Quinoa Polyphenol Extract Alleviates Non-Alcoholic Fatty Liver Disease via Inhibiting Lipid Accumulation, Inflammation and Oxidative Stress
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Extraction of QPE
2.3. UPLC Q-TOF-MS Analysis of Phenolic Compounds in QPE
2.4. Cell Culture and Treatment
2.5. Cell Viability
2.6. Oil Red O Staining
2.7. Measurement of Intracellular TC and TG
2.8. Measurement of Oxidative Stress
2.9. Real-Time Quantitative Polymerase Chain Reaction (RT-qPCR) Analysis
2.10. Western Blot Analysis
2.11. Network Pharmacology Study
2.11.1. Identification of Potential Targets of Ferulic Acid (FA) and Protocatechuic Acid (PCA)
2.11.2. Collection of Core Targets of NAFLD
2.11.3. Construction of the Protein–Protein Interaction (PPI) Network
2.11.4. Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) Pathway Enrichment Analysis
2.12. Molecular Docking
2.13. Statistical Analysis
3. Results
3.1. Polyphenolic Composition of QPE
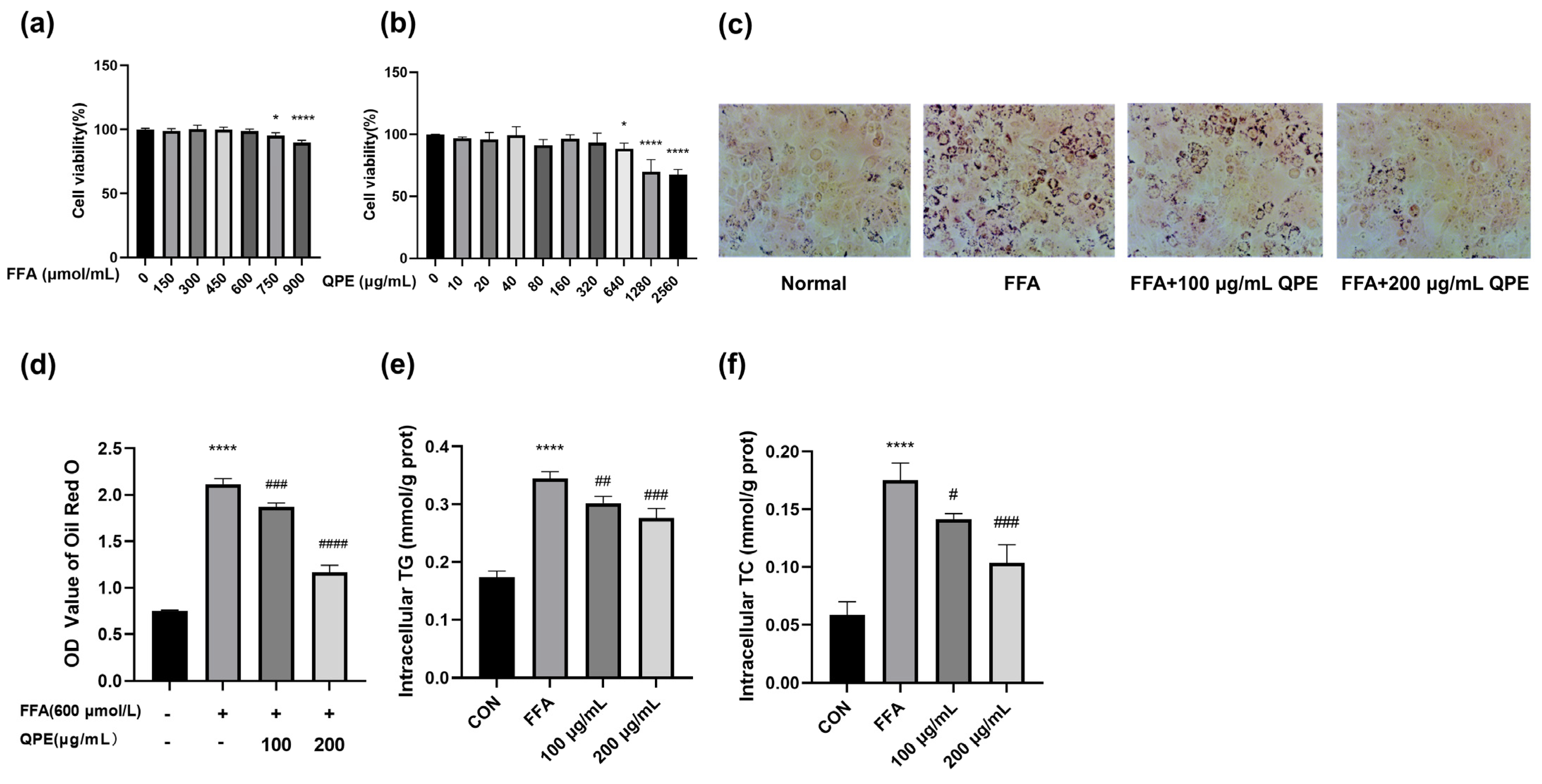
3.2. QPE Attenuated FFA-Induced Lipid Accumulation in HepG2 Cells
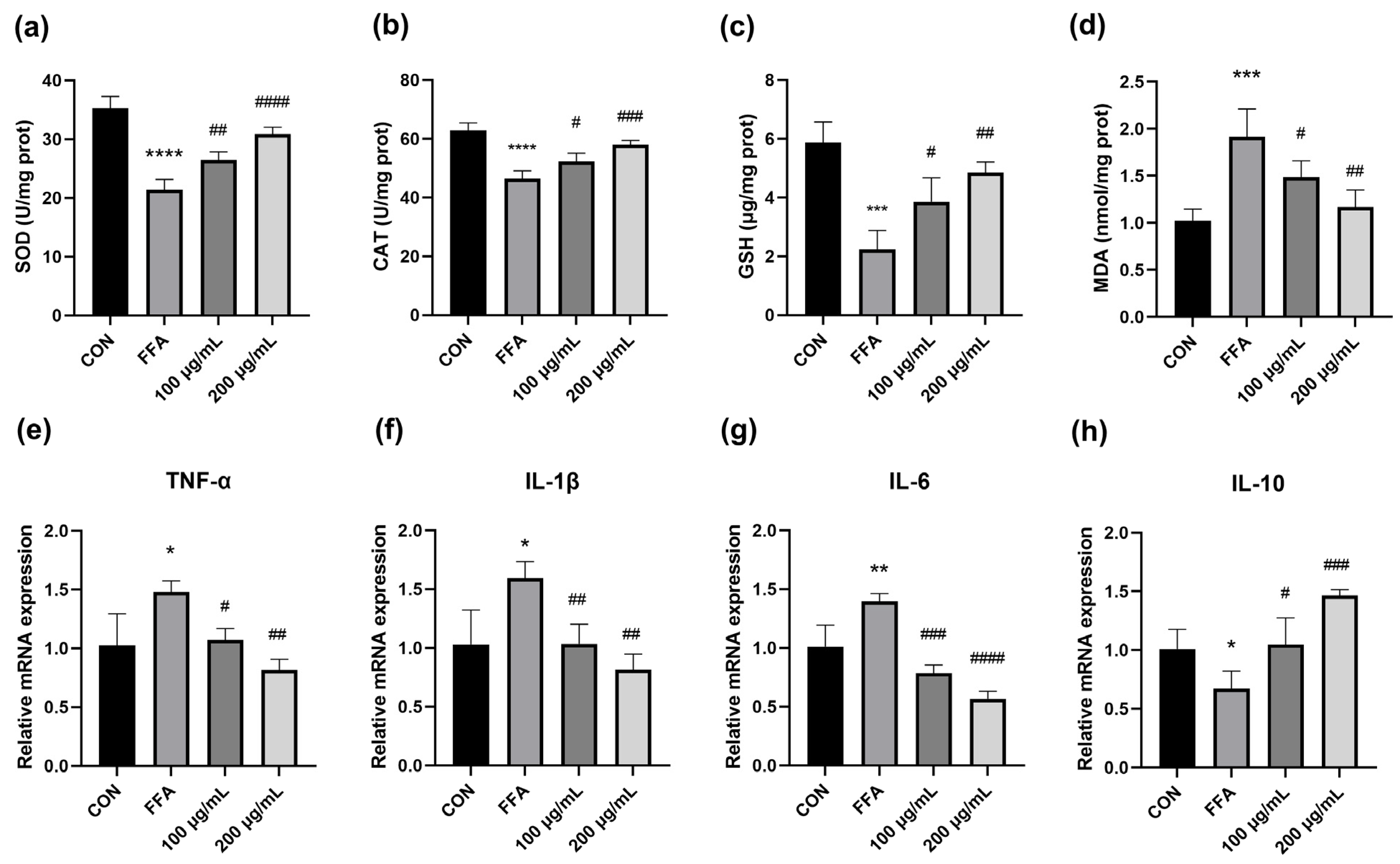
3.3. QPE Attenuated Oxidative Stress Caused by Lipid Accumulation in HepG2 Cells
3.4. QPE Alleviated FFA-Induced Inflammatory Response
3.5. Effect of QPE on mRNA Expression of Lipid Metabolism-Related Enzymes in NAFLD Cell Models
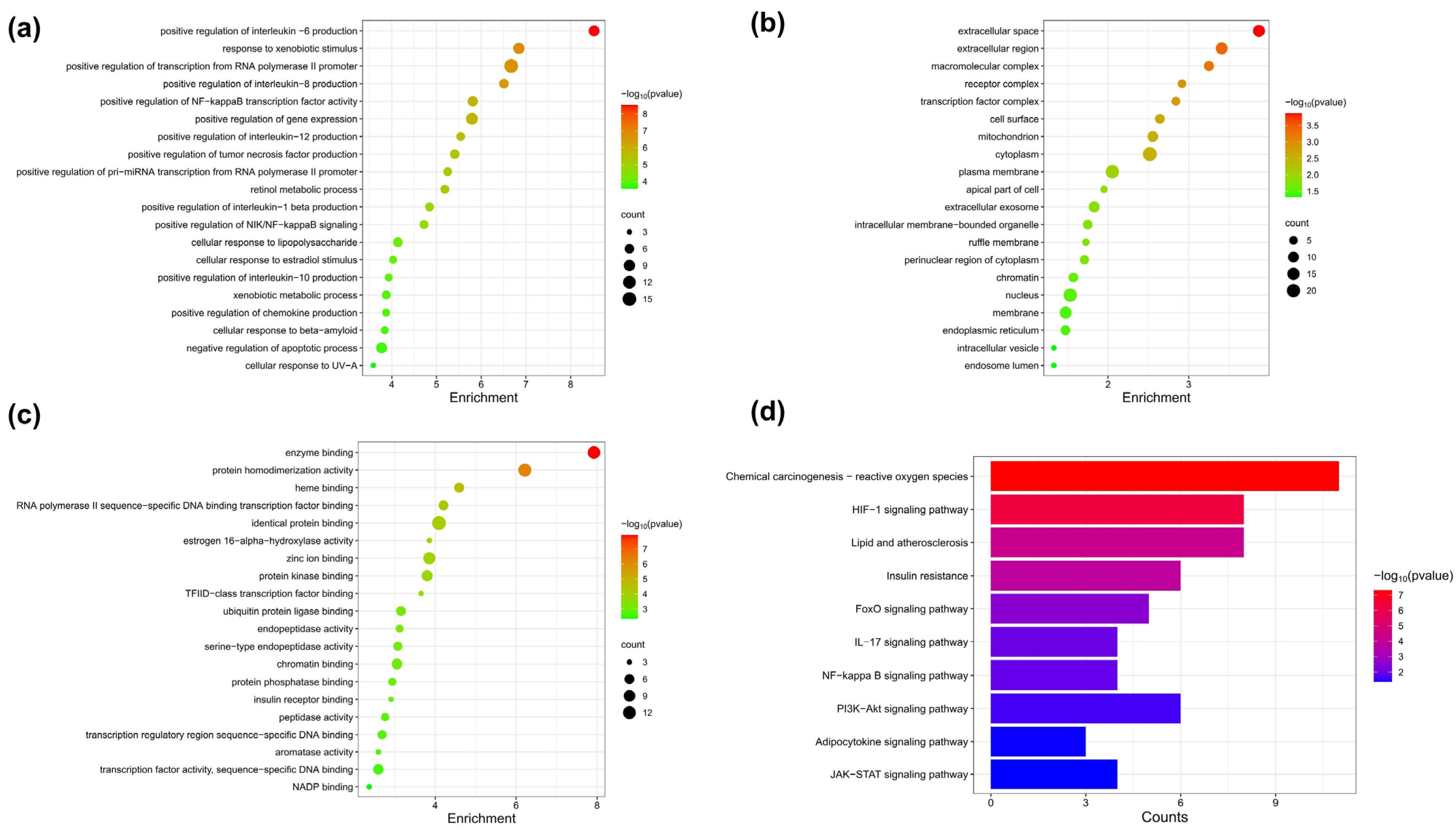
3.6. Target Network Analysis
3.7. Pathway Enrichment Analysis of FA Target Genes
3.8. Molecular Docking of FA with Core Targets
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Rinella, M.E. Nonalcoholic fatty liver disease: A systematic review. JAMA-J. Am. Med. Assoc. 2015, 314, 1521. [Google Scholar] [CrossRef] [PubMed]
- Younossi, Z.; Anstee, Q.M.; Marietti, M.; Hardy, T.; Henry, L.; Eslam, M.; George, J.; Bugianesi, E. Global burden of NAFLD and NASH: Trends, predictions, risk factors and prevention. Nat. Rev. Gastroenterol. Hepatol. 2018, 15, 11–20. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.S.; Tongson, J.; Kim, K.H.; Park, Y. Piceatannol attenuates fat accumulation and oxidative stress in steatosis-induced HepG2 cells. Curr. Res. Food Sci. 2020, 3, 92–99. [Google Scholar] [CrossRef]
- Batchuluun, B.; Pinkosky, S.L.; Steinberg, G.R. Lipogenesis inhibitors: Therapeutic opportunities and challenges. Nat. Rev. Drug Discov. 2022, 21, 283–305. [Google Scholar] [CrossRef]
- Ahmad, B.; Serpell, C.J.; Fong, I.L.; Wong, E.H. Molecular Mechanisms of Adipogenesis: The Anti-adipogenic Role of AMP-Activated Protein Kinase. Front. Mol. Biosci. 2020, 7, 76. [Google Scholar] [CrossRef]
- Rajan, P.; Natraj, P.; Ranaweera, S.S.; Dayarathne, L.A.; Lee, Y.J.; Han, C.H. Anti-adipogenic effect of the flavonoids through the activation of AMPK in palmitate (PA)-treated HepG2 cells. J. Vet. Sci. 2022, 23, e42022. [Google Scholar] [CrossRef] [PubMed]
- Lee, G.H.; Peng, C.; Park, S.A.; Hoang, T.H.; Lee, H.Y.; Kim, J.; Kang, S.I.; Lee, C.H.; Lee, J.S.; Chae, H.J. Citrus Peel Extract Ameliorates High-Fat Diet-Induced NAFLD via Activation of AMPK Signaling. Nutrients 2020, 12, 673. [Google Scholar] [CrossRef]
- Noratto, G.D.; Murphy, K.; Chew, B.P. Quinoa intake reduces plasma and liver cholesterol, lessens obesity-associated inflammation, and helps to prevent hepatic steatosis in obese db/db mouse. Food Chem. 2019, 287, 107–114. [Google Scholar] [CrossRef]
- Ren, G.X.; Teng, C.; Fan, X.; Guo, S.Y.; Zhao, G.; Zhang, L.Z.; Liang, Z.; Qin, P.Y. Nutrient composition, functional activity and industrial applications of quinoa (Chenopodium quinoa Willd.). Food Chem. 2023, 410, 135290. [Google Scholar] [CrossRef]
- Park, J.H.; Lee, Y.J.; Kim, Y.H.; Yoon, K.S. Antioxidant and Antimicrobial Activities of Quinoa (Chenopodium quinoa Willd.) Seeds Cultivated in Korea. Prev. Nutr. Food Sci. 2017, 22, 195–202. [Google Scholar]
- Marmouzi, I.; Madani, N.; Charrouf, Z.; Cherrah, Y.; Faouzi, M.Y.A. Analyse approximative et composition en acides gras et minéraux de Chenopodium quinoa Willd. Marocain, et propriétés antioxydantes selon la polarité. Phytothérapie 2015, 13, 110–117. [Google Scholar] [CrossRef]
- Noratto, G.; Carrion-Rabanal, R.; Medina, G.; Mencia, A.; Mohanty, I.; Gonzalez, D.; Murphy, K. Quinoa protective effects against obesity-induced intestinal inflammation. FASEB J. 2015, 29, 602–609. [Google Scholar] [CrossRef]
- Chen, S.N.; Che, S.Y.; Li, S.Q.; Ruan, Z. The combined impact of decabromodiphenyl ether and high fat exposure on non-alcoholic fatty liver disease in vivo and in vitro. Toxicology 2021, 464, 153015. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.R.; Park, K.I.; Ma, J.Y. Leonurus japonicus Houtt Attenuates Non-alcoholic Fatty Liver Disease in Free Fatty Acid-Induced HepG2 Cells and Mice Fed a High-Fat Diet. Nutrients 2017, 10, 20. [Google Scholar] [CrossRef]
- Liu, M.J.; Zhu, K.L.; Yao, Y.; Chen, Y.H.; Guo, H.M.; Ren, G.X.; Yang, X.S.; Li, J.C. Antioxidant, anti-inflammatory, and antitumor activities of phenolic compounds from white, red, and black Chenopodium quinoa seed. Cereal Chem. 2020, 97, 703–713. [Google Scholar] [CrossRef]
- Qian, G.T.; Li, X.Y.; Zhang, H.; Zhang, H.L.; Zhou, J.W.; Ma, X.H.; Sun, W.; Yang, W.; He, R.K.; Wahab, A.T.; et al. Metabolomics analysis reveals the accumulation patterns of flavonoids and phenolic acids in quinoa (Chenopodium quinoa Willd.) grains of different colors. Food Chem.-X 2023, 17, 100594. [Google Scholar] [CrossRef]
- Duan, Y.M.; Pan, X.F.; Luo, J.Y.; Xiao, X.; Li, J.Y.; Bestman, P.L.; Luo, M.Y. Association of Inflammatory Cytokines with Non-Alcoholic Fatty Liver Disease. Front. Immunol. 2022, 13, 880298. [Google Scholar] [CrossRef] [PubMed]
- Arai, N.; Miura, K.; Aizawa, K.; Sekiya, M.; Nagayama, M.; Sakamoto, H.; Maeda, H.; Morimoto, N.; Iwamoto, S.; Yamamoto, H. Probiotics suppress non-alcoholic steatohepatitis and carcinogenesis progression in hepatocyte-specific PTEN knockout mice. Sci. Rep. 2022, 12, 16206. [Google Scholar] [CrossRef]
- Li, H.; Hu, D.; Fan, H.M.; Zhang, Y.; LeSage, G.D.; Caudle, Y.; Stuart, C.; Liu, Z.M.; Yin, D.L. β-Arrestin 2 Negatively Regulates Toll-like Receptor 4 (TLR4)-triggered Inflammatory Signaling via Targeting p38 MAPK and Interleukin 10. J. Biol. Chem. 2014, 289, 23075–23085. [Google Scholar] [CrossRef]
- Bonte, M.A.; El Idrissi, F.; Gressier, B.; Devos, D.; Belarbi, K. Protein network exploration prioritizes targets for modulating neuroinflammation in Parkinson’s disease. Int. Immunopharmacol. 2021, 95, 107526. [Google Scholar] [CrossRef]
- Pasko, P.; Barton, H.; Zagrodzki, P.; Izewska, A.; Krosniak, M.; Gawlik, M.; Gawlik, M.; Gorinstein, S. Effect of Diet Supplemented with Quinoa Seeds on Oxidative Status in Plasma and Selected Tissues of High Fructose-Fed Rats. Plant Foods Hum. Nutr. 2010, 65, 146–151. [Google Scholar] [CrossRef] [PubMed]
- Al-Qabba, M.M.; El-Mowafy, M.A.; Althwab, S.A.; Alfheeaid, H.A.; Aljutaily, T.; Barakat, H. Phenolic Profile, Antioxidant Activity, and Ameliorating Efficacy of Chenopodium quinoa Sprouts against CCl4-Induced Oxidative Stress in Rats. Nutrients 2020, 12, 2904. [Google Scholar] [CrossRef]
- Holan, V.; Zajicova, A.; Javorkova, E.; Trosan, P.; Chudickova, M.; Pavlikova, M.; Krulova, M. Distinct cytokines balance the development of regulatory T cells and interleukin-10-producing regulatory B cells. Immunology 2014, 141, 577–586. [Google Scholar] [CrossRef] [PubMed]
- Ke, R.; Xu, Q.C.; Li, C.; Luo, L.Y.; Huang, D.Q. Mechanisms of AMPK in the maintenance of ATP balance during energy metabolism. Cell Biol. Int. 2018, 42, 384–392. [Google Scholar] [CrossRef]
- Tejada, S.; Capo, X.; Mascaro, C.M.; Monserrat-Mesquida, M.; Quetglas-Llabres, M.M.; Pons, A.; Tur, J.A.; Sureda, A. Hepatoprotective Effects of Resveratrol in Non-Alcoholic Fatty Live Disease. Curr. Pharm. Des. 2021, 27, 2558–2570. [Google Scholar] [CrossRef]
- Shan, M.Y.; Dai, Y.; Ren, X.D.; Zheng, J.; Zhang, K.B.; Chen, B.; Yan, J.; Xu, Z.H. Berberine mitigates nonalcoholic hepatic steatosis by downregulating SIRT1-FoxO1-SREBP2 pathway for cholesterol synthesis. J. Integr. Med.-JIM 2021, 19, 545–554. [Google Scholar] [CrossRef] [PubMed]
- An, J.P.; Choi, J.H.; Huh, J.; Lee, H.J.; Han, S.; Noh, J.R.; Kim, Y.H.; Lee, C.H.; Oh, W.K. Anti-hepatic steatosis activity of Sicyos angulatus extract in high-fat diet-fed mice and chemical profiling study using UHPLC-qTOF-MS/MS spectrometry. Phytomedicine 2019, 63, 152999. [Google Scholar] [CrossRef]
- Fang, K.; Wu, F.; Chen, G.; Dong, H.; Li, J.B.; Zhao, Y.; Xu, L.J.; Zou, X.; Lu, F.E. Diosgenin ameliorates palmitic acid-induced lipid accumulation via AMPK/ACC/CPT-1A and SREBP-1c/FAS signaling pathways in LO2 cells. BMC Complement. Altern. Med. 2019, 19, 255. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Sun, Q.; Wang, S.; Zhang, M.; Lei, Y.; Wu, J.; Wang, X.; Hu, W.; Meng, H.; Li, Z.; et al. Corydalis saxicola Bunting total alkaloids improve NAFLD by suppressing de novo lipogenesis through the AMPK-SREBP1 axis. J. Ethnopharmacol. 2023, 319 Pt 1, 117162. [Google Scholar] [CrossRef]
- Zduńska, K.; Dana, A.; Kolodziejczak, A.; Rotsztejn, H. Antioxidant Properties of Ferulic Acid and Its Possible Application. Ski. Pharmacol. Physiol. 2018, 31, 332–336. [Google Scholar] [CrossRef]
- Cui, K.L.; Zhang, L.C.; La, X.; Wu, H.L.; Yang, R.P.; Li, H.Q.; Li, Z.Y. Ferulic Acid and P-Coumaric Acid Synergistically Attenuate Non-Alcoholic Fatty Liver Disease through HDAC1/PPARG-Mediated Free Fatty Acid Uptake. Int. J. Mol. Sci. 2022, 23, 15297. [Google Scholar] [CrossRef] [PubMed]
- Yeerjiang, A.; Maimaitiyiming, D.; Yang, T.; Fang, L.; Aihemaitituoheti, A.; Aimaiti, M.; Zhang, X.Y.; Nijiati, Y. Pomegranate Peel in the Amelioration of High-Altitude Disease: A Network Pharmacology and Molecular Docking Study of Underlying Mechanisms. J. Food Biochem. 2023, 2023, 7186747. [Google Scholar] [CrossRef]
- Ljumovic, D.; Diamantis, I.; Alegakis, A.K.; Kouroumalis, E.A. Differential expression of matrix metalloproteinases in viral and non-viral chronic liver diseases. Clin. Chim. Acta 2004, 349, 203–211. [Google Scholar] [CrossRef] [PubMed]
- Coilly, A.; Desterke, C.; Guettier, C.; Samuel, D.; Chiappini, F. FABP4 and MMP9 levels identified as predictive factors for poor prognosis in patients with nonalcoholic fatty liver using data mining approaches and gene expression analysis. Sci. Rep. 2019, 9, 19785. [Google Scholar] [CrossRef]
- Liang, D.D.; Chen, H.J.; Zhao, L.P.; Zhang, W.X.; Hu, J.; Liu, Z.G.; Zhong, P.; Wang, W.; Wang, J.Y.; Liang, G. Inhibition of EGFR attenuates fibrosis and stellate cell activation in diet-induced model of nonalcoholic fatty liver disease. Biochim. Biophys. Acta (BBA) Mol. Basis Dis. 2018, 1864, 133–142. [Google Scholar] [CrossRef]



| No. | Compound | RT (min) | [M−H]− (m/z) | Fragment Ions (m/z) |
|---|---|---|---|---|
| 1 | Vanillic acid | 4.50 | 167.0391 | 152.0116, 108.0229 |
| 2 | Protocatechuic acid | 4.53 | 153.0188 | 109.0297, 108.0218 |
| 3 | Caffeic acid | 5.48 | 179.0422 | 135.0458 |
| 4 | p-Hydroxybenzoic acid | 6.07 | 137.0302 | 93.0341 |
| 5 | p-Coumaric acid | 6.78 | 163.0401 | 119.0509 |
| 6 | Ferulic acid | 7.60 | 193.0555 | 178.0267, 149.0601, 134.0368 |
| 7 | (-)-Catechin | 8.06 | 289.0862 | 175.0351, 137.0212, 134.0440, 123.0479 |
| 8 | (-)-Epicatechin | 8.07 | 289.0862 | 137.0243, 123.0448, 109.0295 |
| 9 | Sinapic acid | 8.15 | 223.0689 | 194.0116, 165.0168, 147.0482 |
| Targets | PDBID | Binding Energy (kcal·mol−1) |
|---|---|---|
| APP | 1AAP | −5.2 |
| EGFR | 3POZ | −6.2 |
| IFNG | 3BES | −5.5 |
| MMP2 | 1CK7 | −6.4 |
| MMP9 | 5UE3 | −7.3 |
| RELA | 1NFI | −5.8 |
| STAT3 | 6NJS | −5.8 |
| TP53 | 2BIM | −5.0 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yao, W.; Fan, M.; Qian, H.; Li, Y.; Wang, L. Quinoa Polyphenol Extract Alleviates Non-Alcoholic Fatty Liver Disease via Inhibiting Lipid Accumulation, Inflammation and Oxidative Stress. Nutrients 2024, 16, 2276. https://doi.org/10.3390/nu16142276
Yao W, Fan M, Qian H, Li Y, Wang L. Quinoa Polyphenol Extract Alleviates Non-Alcoholic Fatty Liver Disease via Inhibiting Lipid Accumulation, Inflammation and Oxidative Stress. Nutrients. 2024; 16(14):2276. https://doi.org/10.3390/nu16142276
Chicago/Turabian StyleYao, Wenjun, Mingcong Fan, Haifeng Qian, Yan Li, and Li Wang. 2024. "Quinoa Polyphenol Extract Alleviates Non-Alcoholic Fatty Liver Disease via Inhibiting Lipid Accumulation, Inflammation and Oxidative Stress" Nutrients 16, no. 14: 2276. https://doi.org/10.3390/nu16142276
APA StyleYao, W., Fan, M., Qian, H., Li, Y., & Wang, L. (2024). Quinoa Polyphenol Extract Alleviates Non-Alcoholic Fatty Liver Disease via Inhibiting Lipid Accumulation, Inflammation and Oxidative Stress. Nutrients, 16(14), 2276. https://doi.org/10.3390/nu16142276









