Long-Term Dietary Consumption of Grapes Affects Kidney Health in C57BL/6J Mice
Abstract
1. Introduction
2. Results
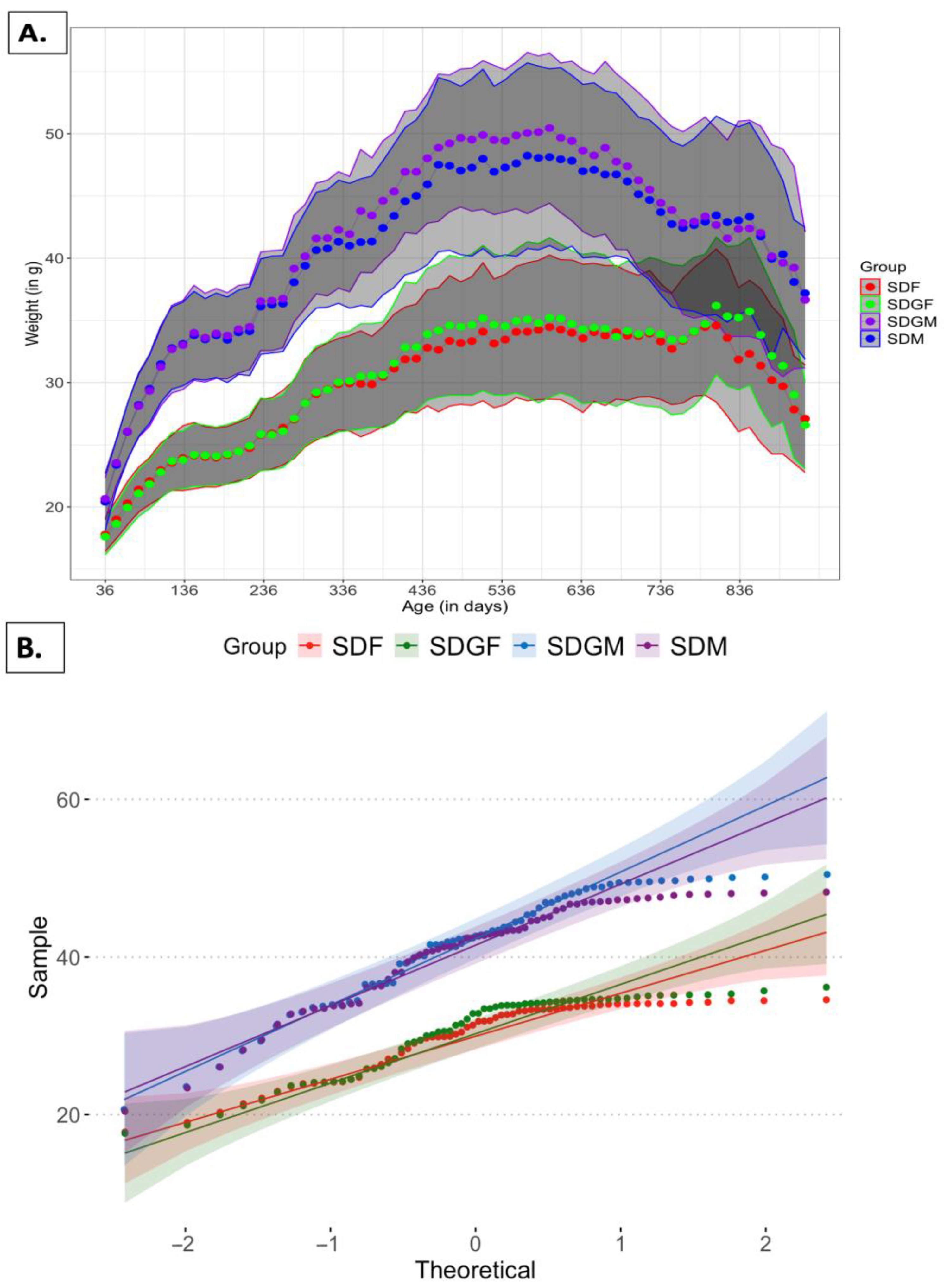
2.1. Effect of Diets on Mouse Body Weights and Tissues
2.1.1. Effect of Diets on Mouse Body Weights
2.1.2. Effect of Diets on Mouse Tissues
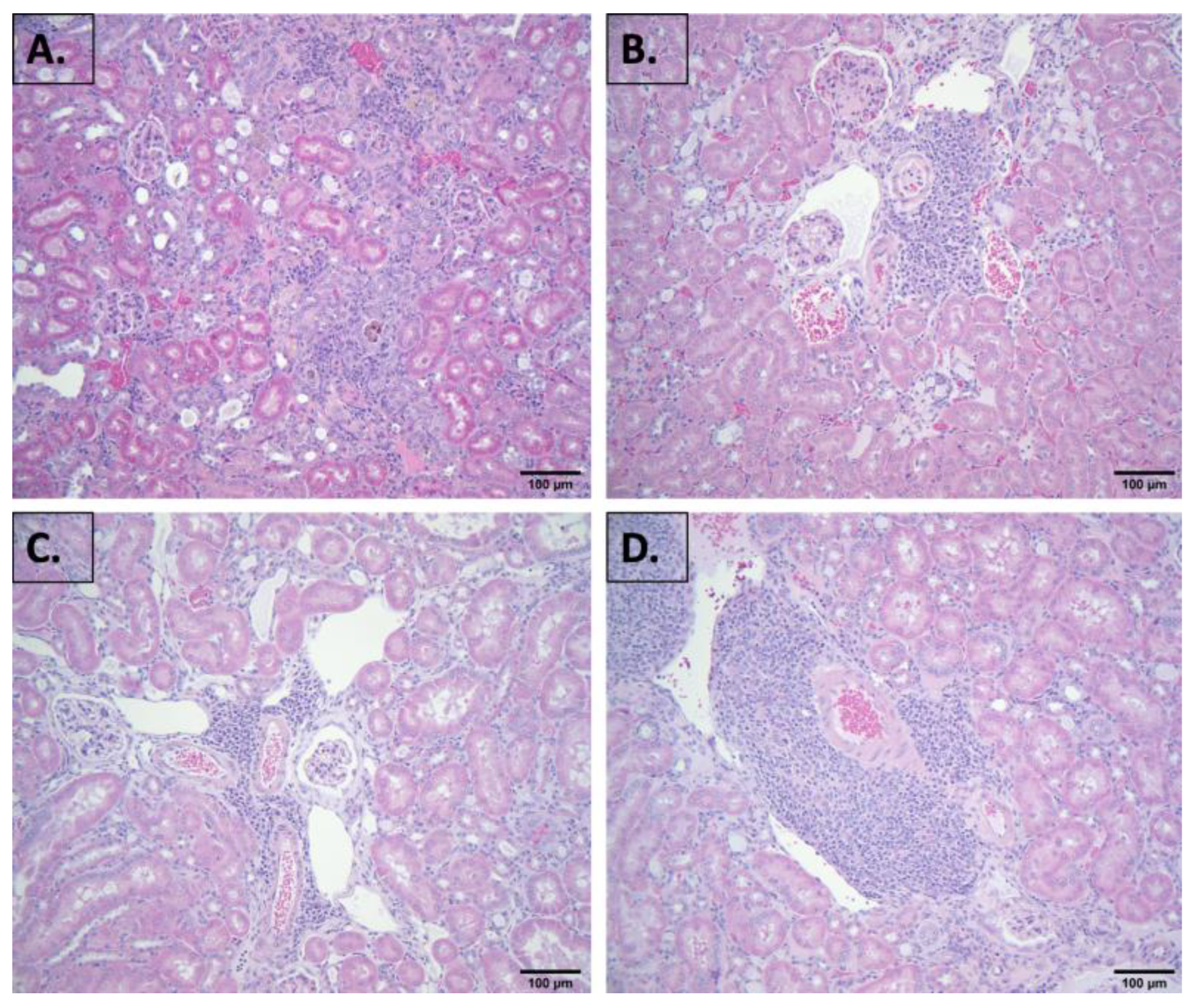
2.2. Evaluation of Blood Chemistry and Histology
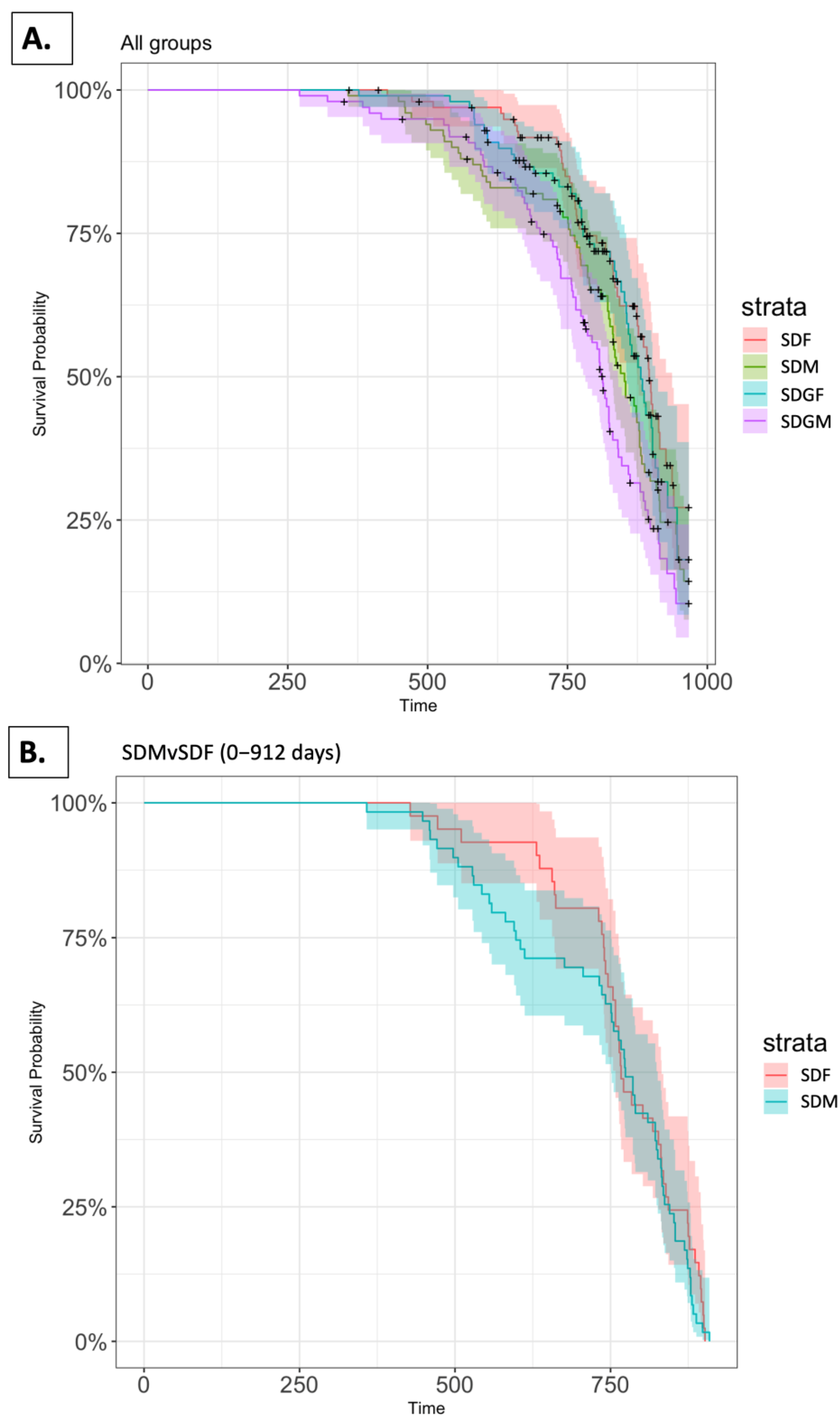
2.3. Survival Analysis
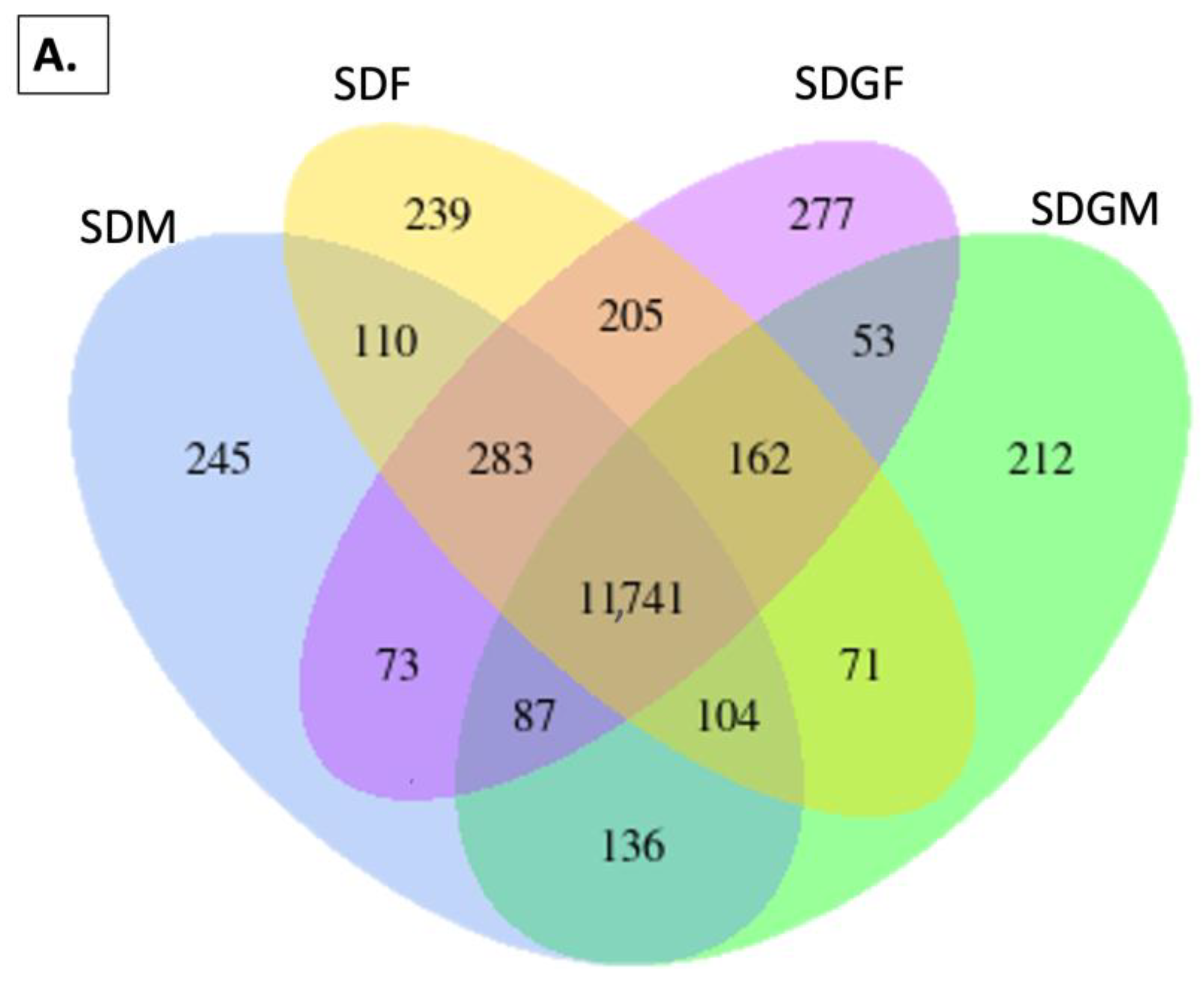
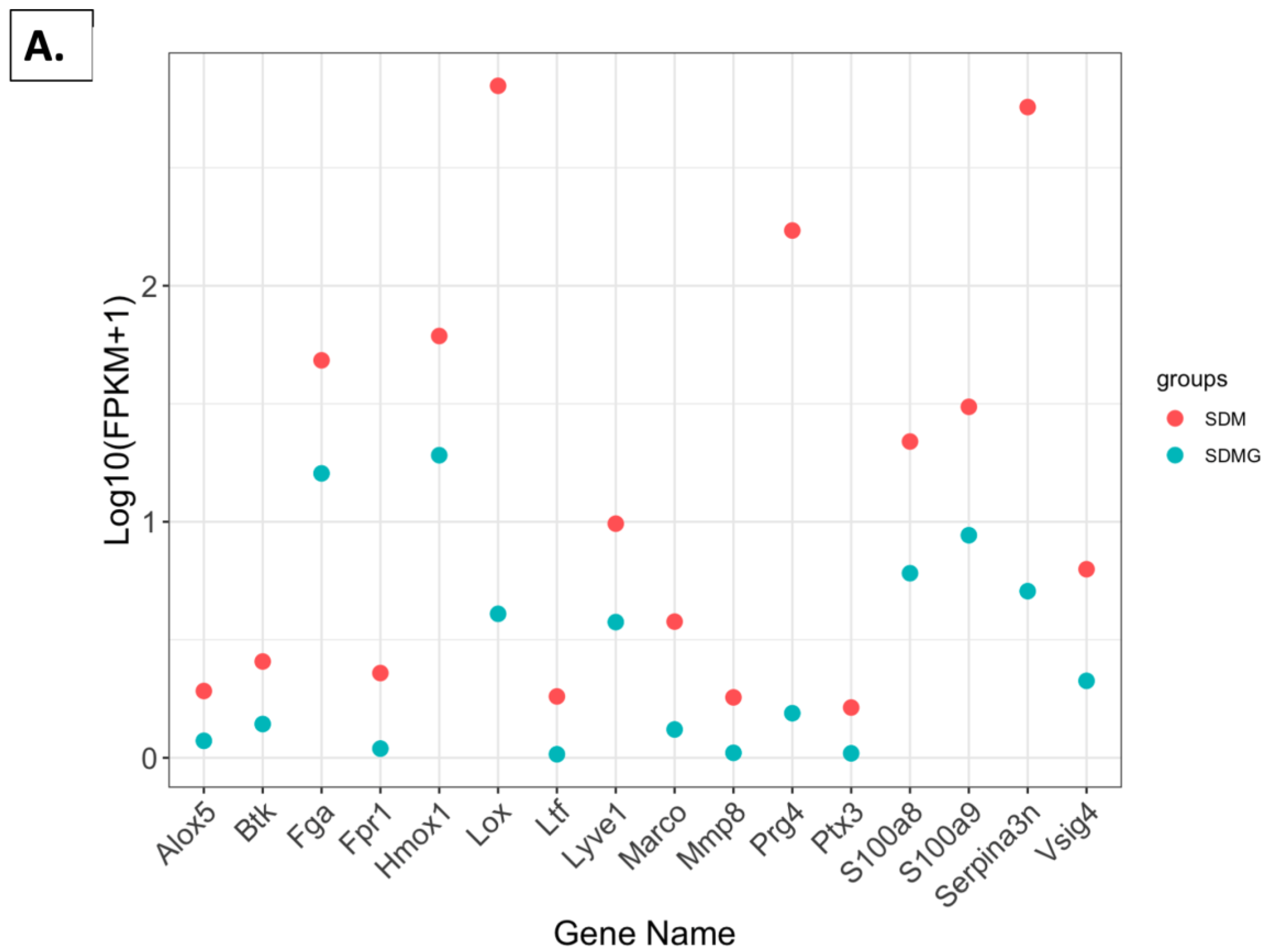
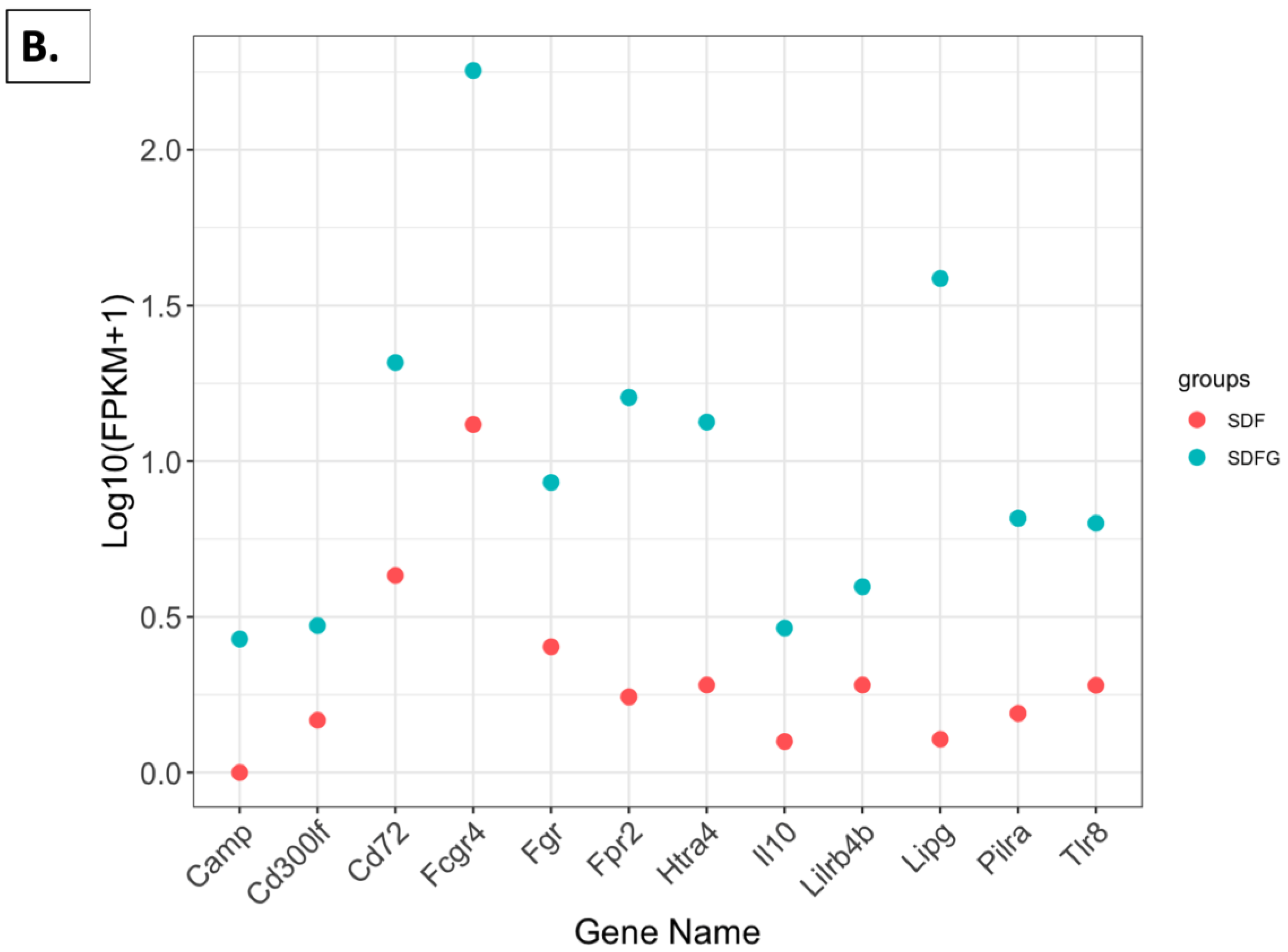
2.4. Effect of Grapes on Gene Expression in the Kidney
2.4.1. Venn Diagrams
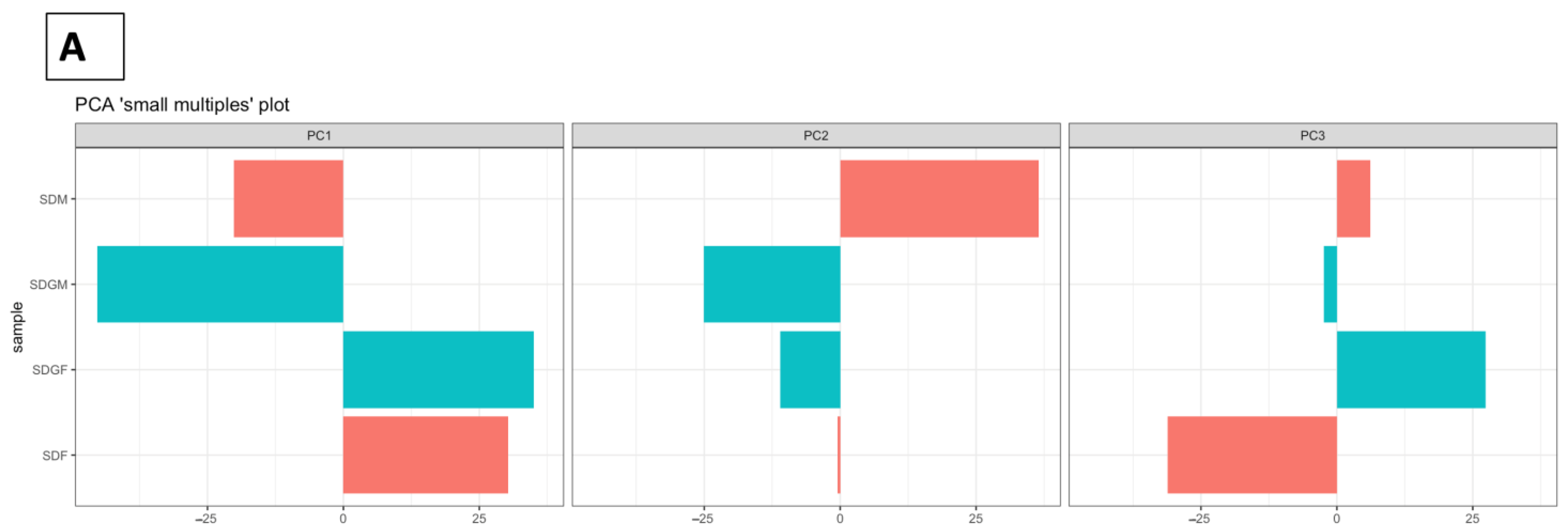
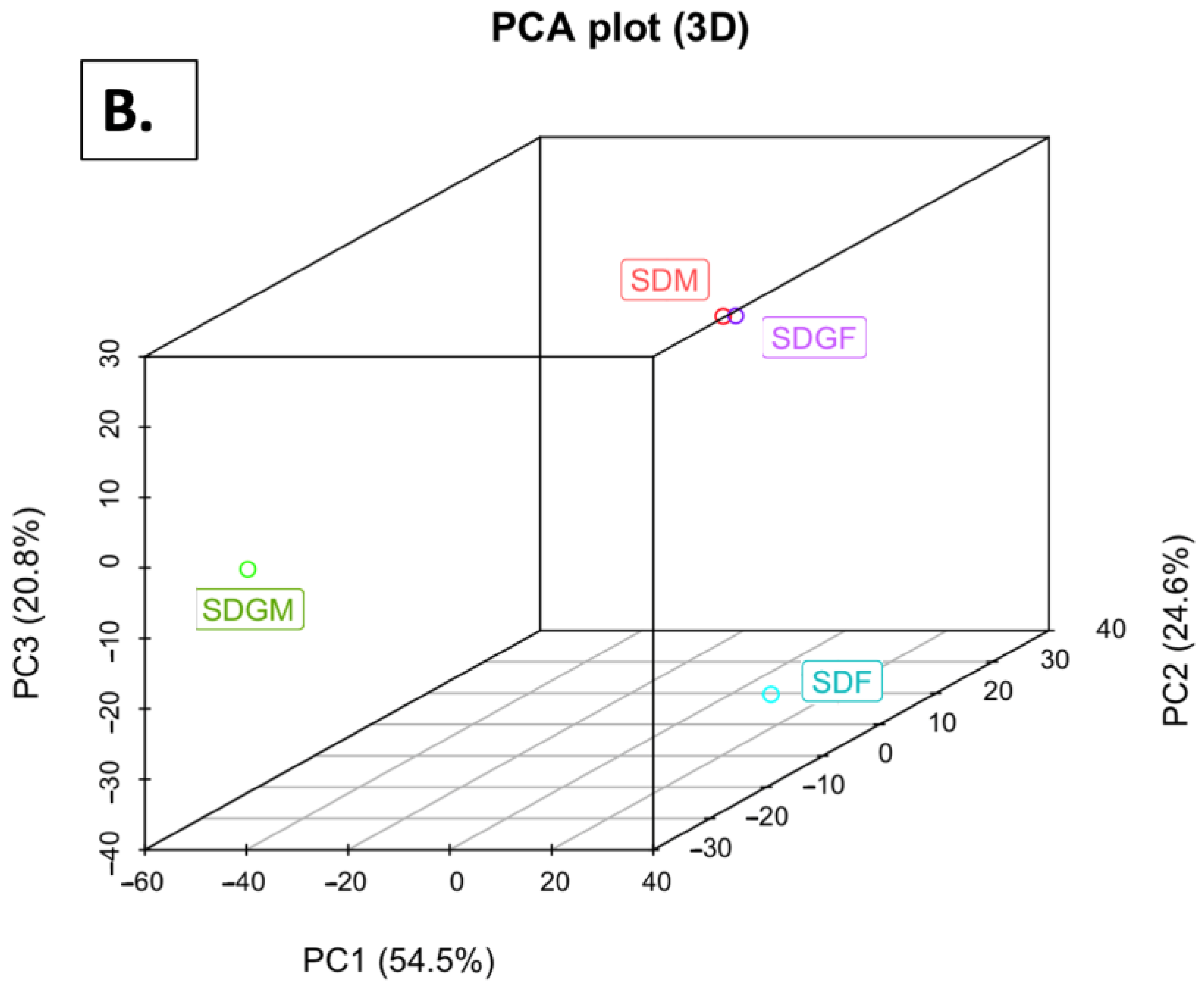
2.4.2. Principal Component Analyses
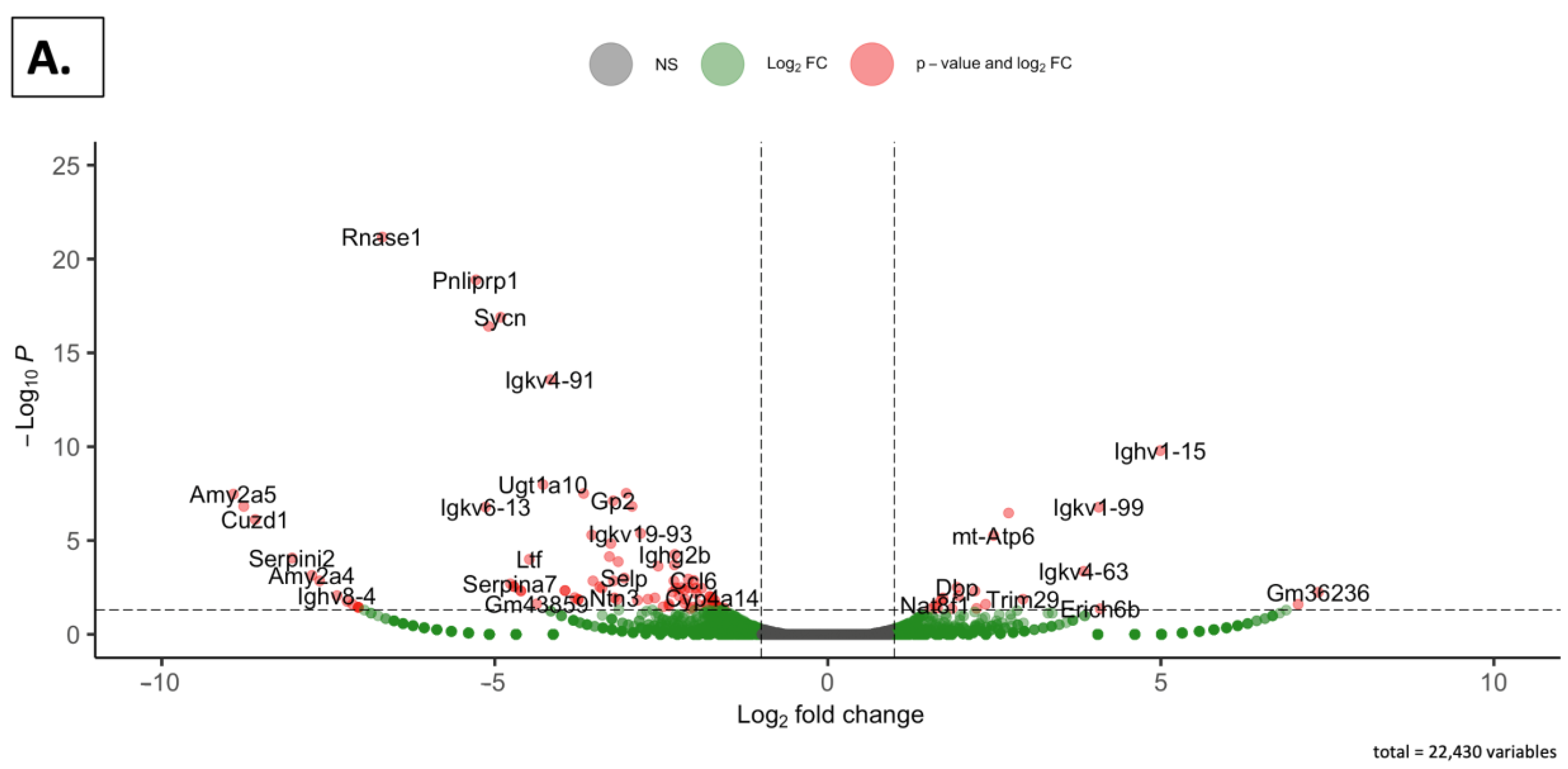
2.4.3. Volcano Plots
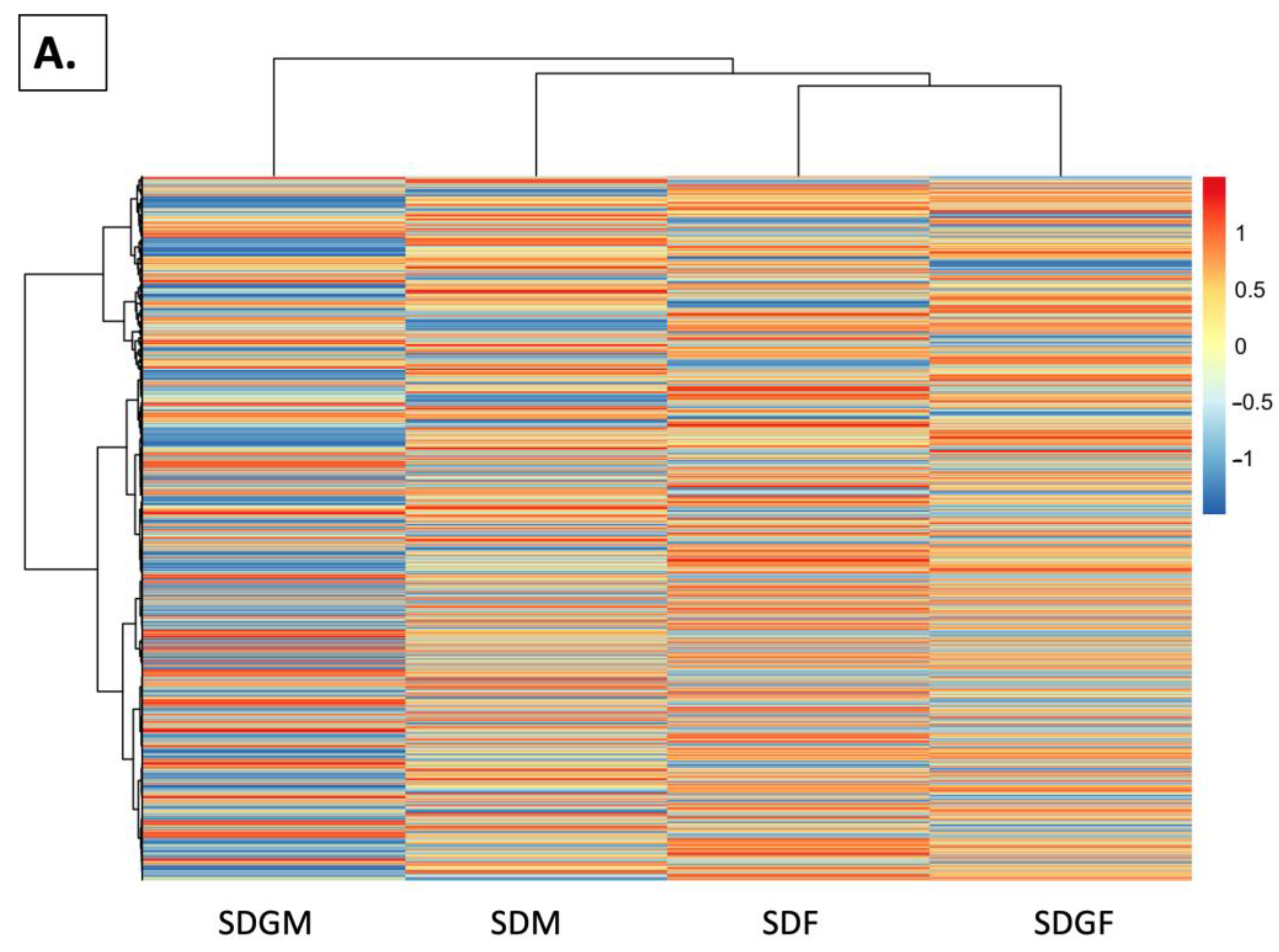
2.4.4. Heat Maps
2.4.5. FPKM Value Plots
2.4.6. FPKM Values Related to Immune Function
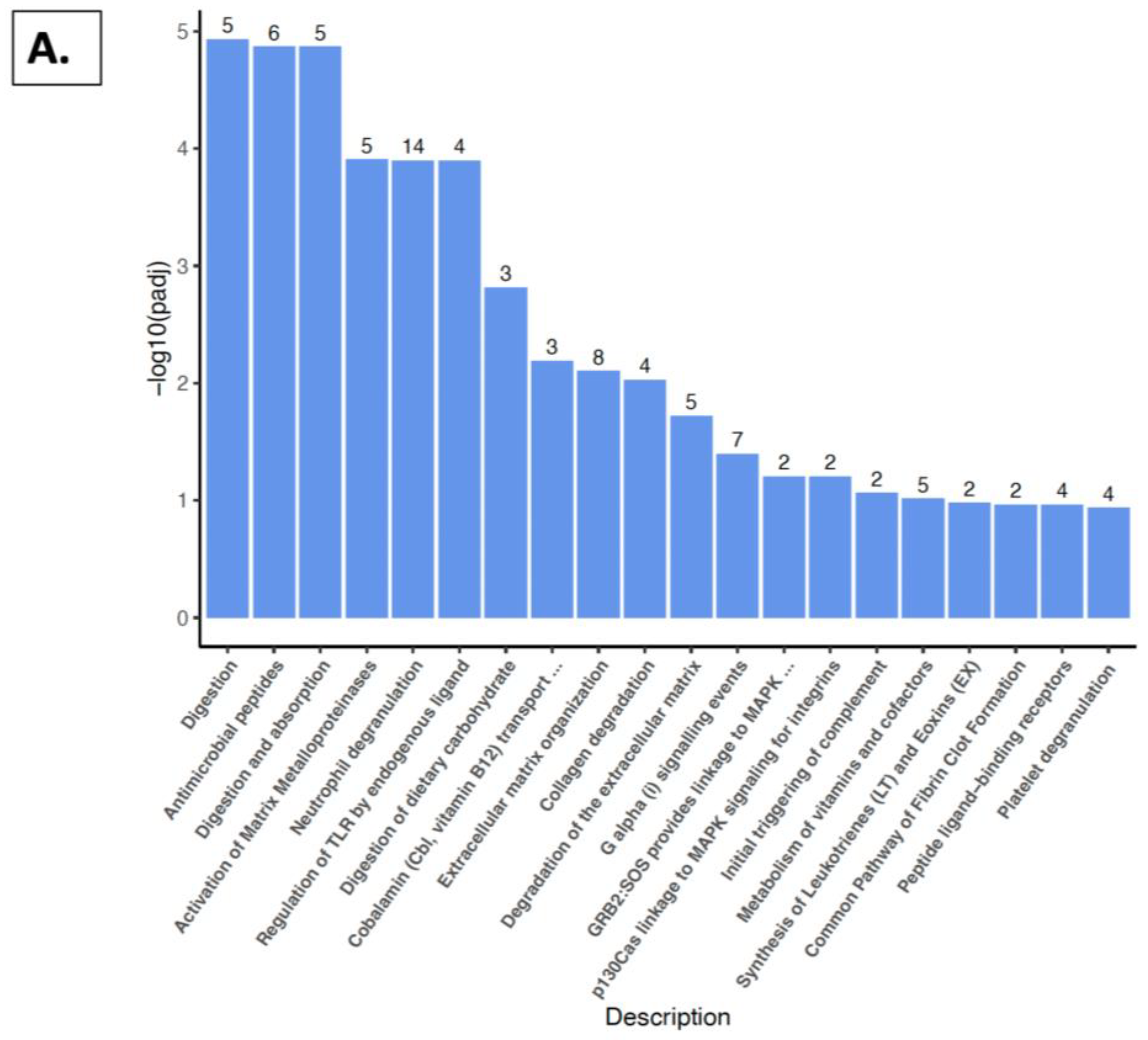
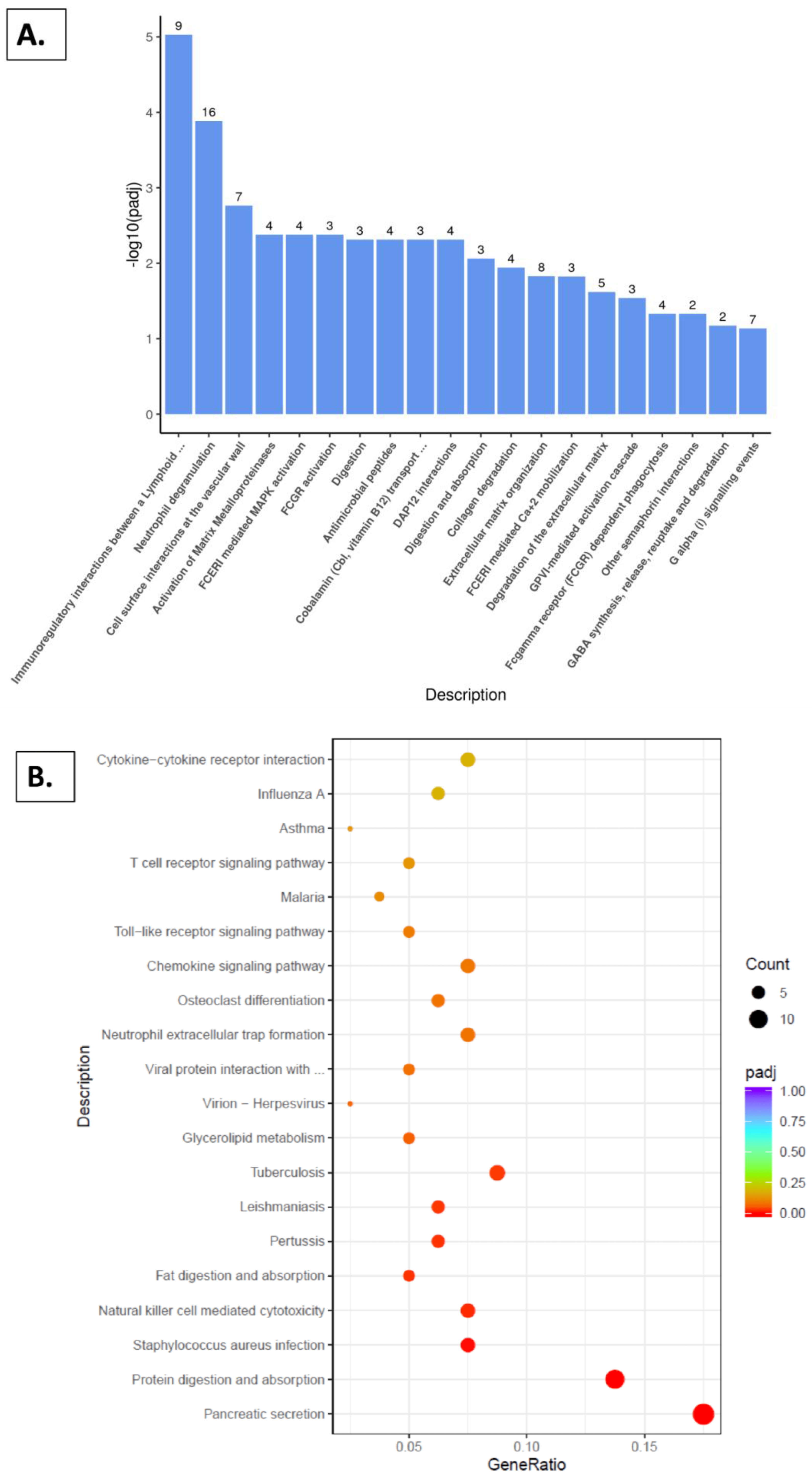
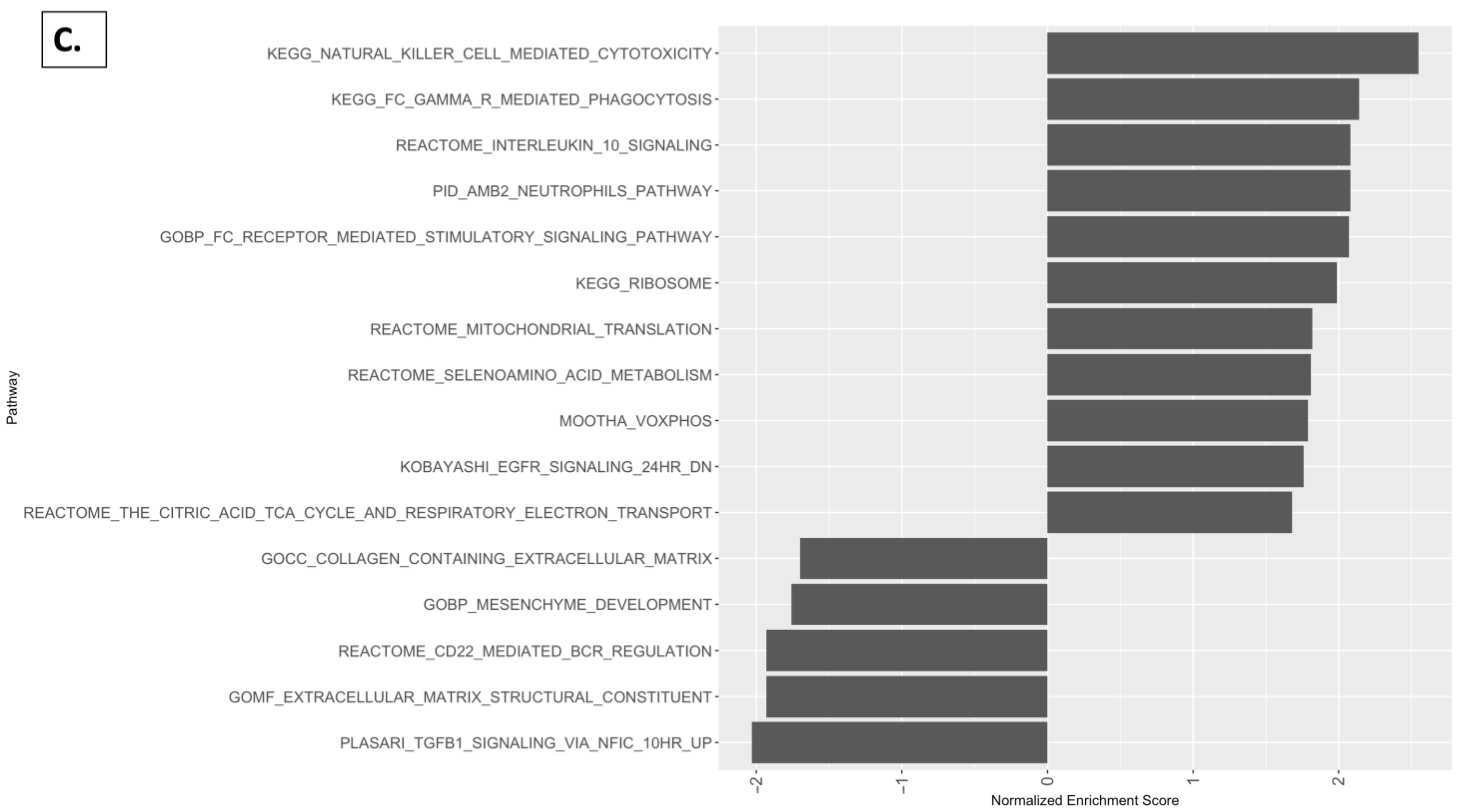
2.5. Reactome Pathway, KEGG Pathway, and GSEA Analysis with Kidney
3. Discussion
4. Materials and Methods
4.1. Animals and Diets
4.1.1. Semi-Synthetic Diets
4.1.2. Animal Protocol
4.1.3. Tissue and Serum Collection
4.2. Blood Analysis
4.3. Histopathological Examination of Kidney and Other Tissues
4.4. RNA Extraction and RNA Sequencing
4.5. Pathway and GO Term Enrichment Analyses
4.6. Heat Map Generation
4.7. Principal Component Analysis
4.8. Differential Expression Analyses
4.9. Statistical Analyses
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Wang, D.D.; Li, Y.; Bhupathiraju, S.N.; Rosner, B.A.; Sun, Q.; Giovannucci, E.L.; Rimm, E.B.; Manson, J.E.; Willett, W.C.; Stampfer, M.J.; et al. Fruit and Vegetable Intake and Mortality: Results from Two Prospective Cohort Studies of US Men and Women and a Meta-Analysis of 26 Cohort Studies. Circulation 2021, 143, 1642–1654. [Google Scholar] [CrossRef] [PubMed]
- Cragg, G.M.; Pezzuto, J.M. Natural Products as a Vital Source for the Discovery of Cancer Chemotherapeutic and Chemopreventive Agents. Med. Princ. Pract. 2016, 25, 41–59. [Google Scholar] [CrossRef] [PubMed]
- Dave, A.; Parande, F.; Park, E.J.; Pezzuto, J.M. Phytochemicals and cancer chemoprevention. J. Cancer Metastasis Treat. 2020, 6, 46. [Google Scholar] [CrossRef]
- Jang, M.; Cai, L.; Udeani, G.O.; Slowing, K.V.; Thomas, C.F.; Beecher, C.W.; Fong, H.H.; Farnsworth, N.R.; Kinghorn, A.D.; Mehta, R.G.; et al. Cancer Chemopreventive Activity of Resveratrol, a Natural Product Derived from Grapes. Science 1997, 275, 218–220. [Google Scholar] [CrossRef] [PubMed]
- Park, E.-J.; Pezzuto, J.M. The Pharmacology of Resveratrol in Animals and Humans. Biochim. Biophys. Acta BBA Mol. Basis Dis. 2015, 1852, 1071–1113. [Google Scholar] [CrossRef] [PubMed]
- Pezzuto, J.M. Grapes and Human Health: A Perspective. J. Agric. Food Chem. 2008, 56, 6777–6784. [Google Scholar] [CrossRef] [PubMed]
- Pezzuto, J.M. (Ed.) Grapes and Health; Springer International Publishing: Cham, Switzerland, 2016; p. 235. [Google Scholar]
- Yang, J.; Kurnia, P.; Henning, S.M.; Lee, R.; Huang, J.; Garcia, M.C.; Surampudi, V.; Heber, D.; Li, Z. Effect of Standardized Grape Powder Consumption on the Gut Microbiome of Healthy Subjects: A Pilot Study. Nutrients 2021, 13, 3965. [Google Scholar] [CrossRef] [PubMed]
- Dave, A.; Beyoğlu, D.; Park, E.-J.; Idle, J.R.; Pezzuto, J.M. Influence of Grape Consumption on the Human Microbiome. Sci. Rep. 2023, 13, 7706. [Google Scholar] [CrossRef] [PubMed]
- Swindell, W.R. Genes and Gene Expression Modules Associated with Caloric Restriction and Aging in the Laboratory Mouse. BMC Genom. 2009, 10, 585. [Google Scholar] [CrossRef]
- Dave, A.; Park, E.-J.; Pezzuto, J.M. Multi-Organ Nutrigenomic Effects of Dietary Grapes in a Mouse Model. Antioxidants 2023, 12, 1821. [Google Scholar] [CrossRef]
- Parande, F.; Dave, A.; Park, E.-J.; McAllister, C.; Pezzuto, J.M. Effect of Dietary Grapes on Female C57BL6/J Mice Consuming a High-Fat Diet: Behavioral and Genetic Changes. Antioxidants 2022, 11, 414. [Google Scholar] [CrossRef] [PubMed]
- Dave, A.; Park, E.-J.; Kumar, A.; Parande, F.; Beyoğlu, D.; Idle, J.R.; Pezzuto, J.M. Consumption of Grapes Modulates Gene Expression, Reduces Non-Alcoholic Fatty Liver Disease, and Extends Longevity in Female C57BL/6J Mice Provided with a High-Fat Western-Pattern Diet. Foods 2022, 11, 1984. [Google Scholar] [CrossRef] [PubMed]
- Beyoğlu, D.; Park, E.-J.; Quiñones-Lombraña, A.; Dave, A.; Parande, F.; Pezzuto, J.M.; Idle, J.R. Addition of Grapes to Both a Standard and a High-Fat Western Pattern Diet Modifies Hepatic and Urinary Metabolite Profiles in the Mouse. Food Funct. 2022, 13, 8489–8499. [Google Scholar] [CrossRef] [PubMed]
- Nair, A.B.; Jacob, S. A Simple Practice Guide for Dose Conversion between Animals and Human. J. Basic Clin. Pharm. 2016, 7, 27–31. [Google Scholar] [CrossRef] [PubMed]
- California Table Grape Commission. Guidelines for Use of California Table Grape Powder in Health Research Studies; California Table Grape Commission: Fresno, CA, USA, 2021. [Google Scholar]
- Russell, E. Lifespan and Aging Patterns; McGraw-Hill Book: New York, NY, USA, 1966. [Google Scholar]
- Bou Sleiman, M.; Roy, S.; Gao, A.W.; Sadler, M.C.; von Alvensleben, G.V.G.; Li, H.; Sen, S.; Harrison, D.E.; Nelson, J.F.; Strong, R.; et al. Sex- and Age-Dependent Genetics of Longevity in a Heterogeneous Mouse Population. Science 2022, 377, eabo3191. [Google Scholar] [CrossRef] [PubMed]
- Flurkey, K.; Currer, J.M.; Harrison, D.E. Mouse Models in Aging Research. In The Mouse in Biomedical Research; Academic Press: Cambridge, MA, USA, 2007; Volume 3, pp. 637–672. [Google Scholar]
- Sellers, R.S.; Mortan, D.; Michael, B.; Roome, N.; Johnson, J.K.; Yano, B.L.; Perry, R.; Schafer, K. Society of Toxicologic Pathology Position Paper: Organ Weight Recommendations for Toxicology Studies. Toxicol. Pathol. 2007, 35, 751–755. [Google Scholar] [CrossRef] [PubMed]
- The Jackson Laboratory. Organ Weight Summary. Available online: https://www.jax.org/de/-/media/jaxweb/files/jax-mice-and-services/b6j-data-summary.xlsx (accessed on 15 April 2022).
- Brisco, M.A.; Coca, S.G.; Chen, J.; Owens, A.T.; McCauley, B.D.; Kimmel, S.E.; Testani, J.M. Blood Urea Nitrogen/Creatinine Ratio Identifies a High-Risk but Potentially Reversible Form of Renal Dysfunction in Patients with Decompensated Heart Failure. Circ. Heart Fail. 2013, 6, 233–239. [Google Scholar] [CrossRef] [PubMed]
- Montford, J.R.; Bauer, C.; Dobrinskikh, E.; Hopp, K.; Levi, M.; Weiser-Evans, M.; Nemenoff, R.; Furgeson, S.B. Inhibition of 5-Lipoxygenase Decreases Renal Fibrosis and Progression of Chronic Kidney Disease. Am. J. Physiol. Renal. Physiol. 2019, 316, F732–F742. [Google Scholar] [CrossRef]
- Ekpenyong-Akiba, A.E.; Poblocka, M.; Althubiti, M.; Rada, M.; Jurk, D.; Germano, S.; Kocsis-Fodor, G.; Shi, Y.; Canales, J.J.; Macip, S. Amelioration of Age-related Brain Function Decline by Bruton’s Tyrosine Kinase Inhibition. Aging Cell 2020, 19, e13079. [Google Scholar] [CrossRef]
- Sivalingam, V.; Patel, B.K. Familial Mutations in Fibrinogen Aα (FGA) Chain Identified in Renal Amyloidosis Increase in Vitro Amyloidogenicity of FGA Fragment. Biochimie 2016, 127, 44–49. [Google Scholar] [CrossRef]
- Zhang, J.; Ding, T.; Tang, D.; Wang, J.; Huang, P. Formyl Peptide Receptor 1 Promotes Podocyte Injury through Regulation of Mitogen-Activated Protein Kinase Pathways. Exp. Biol. Med. 2022, 247, 87–96. [Google Scholar] [CrossRef] [PubMed]
- Lever, J.M.; Boddu, R.; George, J.F.; Agarwal, A. Heme Oxygenase-1 in Kidney Health and Disease. Antioxid. Redox Signal. 2016, 25, 165–183. [Google Scholar] [CrossRef]
- Zhang, X.; Zhou, W.; Niu, Y.; Zhu, S.; Zhang, Y.; Li, X.; Yu, C. Lysyl Oxidase Promotes Renal Fibrosis via Accelerating Collagen Cross-link Driving by Β-arrestin/ERK/STAT3 Pathway. FASEB J. 2022, 36, e22427. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Zhu, J.; Zhao, B.; Nie, D.; Wang, W.; Qi, Y.; Chen, L.; Li, B.; Chen, B. LTF Induces Senescence and Degeneration in the Meniscus via the NF-κB Signaling Pathway: A Study Based on Integrated Bioinformatics Analysis and Experimental Validation. Front. Mol. Biosci. 2023, 10, 1134253. [Google Scholar] [CrossRef]
- Zarjou, A.; Black, L.M.; Bolisetty, S.; Traylor, A.M.; Bowhay, S.A.; Zhang, M.-Z.; Harris, R.C.; Agarwal, A. Dynamic Signature of Lymphangiogenesis during Acute Kidney Injury and Chronic Kidney Disease. Lab. Investig. 2019, 99, 1376–1388. [Google Scholar] [CrossRef]
- Yamada, C.; Beron-Pelusso, C.; Algazzaz, N.; Heidari, A.; Luz, D.; Rawas-Qalaji, M.; Toderas, I.; Mascarenhas, A.K.; Kawai, T.; Movila, A. Age-dependent Effect between MARCO and TLR4 on PMMA Particle Phagocytosis by Macrophages. J. Cell Mol. Med. 2019, 23, 5827–5831. [Google Scholar] [CrossRef] [PubMed]
- Tammaro, A.; Florquin, S.; Brok, M.; Claessen, N.; Butter, L.M.; Teske, G.J.D.; de Boer, O.J.; Vogl, T.; Leemans, J.C.; Dessing, M.C. S100A8/A9 Promotes Parenchymal Damage and Renal Fibrosis in Obstructive Nephropathy. Clin. Exp. Immunol. 2018, 193, 361–375. [Google Scholar] [CrossRef] [PubMed]
- Sánchez-Navarro, A.; Murillo-de-Ozores, A.R.; Pérez-Villalva, R.; Linares, N.; Carbajal-Contreras, H.; Flores, M.E.; Gamba, G.; Castañeda-Bueno, M.; Bobadilla, N.A. Transient Response of serpinA3 during Cellular Stress. FASEB J. 2022, 36, e22190. [Google Scholar] [CrossRef]
- Cha, D.R.; Han, S.Y.; Ko, Y.-J.; Cha, J.J. #915 Role of VSIG4 in Renal Aging Process. Nephrol. Dial. Transplant. 2024, 39, gfae069-1363-915. [Google Scholar] [CrossRef]
- Evans, F.; Alí-Ruiz, D.; Rego, N.; Negro-Demontel, M.L.; Lago, N.; Cawen, F.A.; Pannunzio, B.; Sanchez-Molina, P.; Reyes, L.; Paolino, A.; et al. CD300f Immune Receptor Contributes to Healthy Aging by Regulating Inflammaging, Metabolism, and Cognitive Decline. Cell Rep. 2023, 42, 113269. [Google Scholar] [CrossRef]
- Lee, C.; Han, J.; Jung, Y. Formyl Peptide Receptor 2 Is an Emerging Modulator of Inflammation in the Liver. Exp. Mol. Med. 2023, 55, 325–332. [Google Scholar] [CrossRef]
- Clausen, T.; Southan, C.; Ehrmann, M. The HtrA Family of Proteases: Implications for Protein Composition and Cell Fate. Mol. Cell 2002, 10, 443–455. [Google Scholar] [CrossRef]
- Wei, W.; Zhao, Y.; Zhang, Y.; Jin, H.; Shou, S. The Role of IL-10 in Kidney Disease. Int. Immunopharmacol. 2022, 108, 108917. [Google Scholar] [CrossRef] [PubMed]
- Cella, M.; Döhring, C.; Samaridis, J.; Dessing, M.; Brockhaus, M.; Lanzavecchia, A.; Colonna, M. A Novel Inhibitory Receptor (ILT3) Expressed on Monocytes, Macrophages, and Dendritic Cells Involved in Antigen Processing. J. Exp. Med. 1997, 185, 1743–1751. [Google Scholar] [CrossRef] [PubMed]
- Jang, H.R.; Gandolfo, M.T.; Ko, G.J.; Satpute, S.R.; Racusen, L.; Rabb, H. B Cells Limit Repair after Ischemic Acute Kidney Injury. J. Am. Soc. Nephrol. 2010, 21, 654–665. [Google Scholar] [CrossRef] [PubMed]
- Cheung, M.D.; Erman, E.N.; Moore, K.H.; Lever, J.M.; Li, Z.; LaFontaine, J.R.; Ghajar-Rahimi, G.; Liu, S.; Yang, Z.; Karim, R.; et al. Resident Macrophage Subpopulations Occupy Distinct Microenvironments in the Kidney. JCI Insight 2022, 7, e161078. [Google Scholar] [CrossRef] [PubMed]
- Orlofsky, A.; Lin, E.Y.; Prystowsky, M.B. Selective Induction of the Beta Chemokine C10 by IL-4 in Mouse Macrophages. J. Immunol. 1994, 152, 5084–5091. [Google Scholar] [CrossRef]
- Lee, J.; Lee, Y.; Kim, K.-H.; Kim, D.-K.; Joo, K.-W.; Shin, S.-J.; Kim, Y.-S.; Yang, S.-H. Chemokine (C-C Motif) Ligand 8 and Tubulo-Interstitial Injury in Chronic Kidney Disease. Cells 2022, 11, 658. [Google Scholar] [CrossRef] [PubMed]
- Greenman, R.; Segal-Salto, M.; Barashi, N.; Hay, O.; Katav, A.; Levi, O.; Vaknin, I.; Aricha, R.; Aharoni, S.; Snir, T.; et al. CCL24 Regulates Biliary Inflammation and Fibrosis in Primary Sclerosing Cholangitis. JCI Insight 2023, 8, e162270. [Google Scholar] [CrossRef]
- Bhargava, P.; Schnellmann, R.G. Mitochondrial Energetics in the Kidney. Nat. Rev. Nephrol. 2017, 13, 629–646. [Google Scholar] [CrossRef]
- Wang, S.N.; Lapage, J.; Hirschberg, R. Glomerular Ultrafiltration and Apical Tubular Action of IGF-I, TGF-Beta, and HGF in Nephrotic Syndrome. Kidney Int. 1999, 56, 1247–1251. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Boadle, R.A.; Harris, D.C. Toxicity of Holotransferrin but Not Albumin in Proximal Tubule Cells in Primary Culture. J. Am. Soc. Nephrol. 1998, 9, 77–84. [Google Scholar] [CrossRef]
- Bülow, R.D.; Boor, P. Extracellular Matrix in Kidney Fibrosis: More Than Just a Scaffold. J. Histochem. Cytochem. 2019, 67, 643–661. [Google Scholar] [CrossRef] [PubMed]
- Järveläinen, H.; Sainio, A.; Koulu, M.; Wight, T.N.; Penttinen, R. Extracellular Matrix Molecules: Potential Targets in Pharmacotherapy. Pharmacol. Rev. 2009, 61, 198–223. [Google Scholar] [CrossRef] [PubMed]
- Schaefer, L.; Iozzo, R.V. Biological Functions of the Small Leucine-Rich Proteoglycans: From Genetics to Signal Transduction. J. Biol. Chem. 2008, 283, 21305–21309. [Google Scholar] [CrossRef] [PubMed]
- Frantz, C.; Stewart, K.M.; Weaver, V.M. The Extracellular Matrix at a Glance. J. Cell Sci. 2010, 123, 4195–4200. [Google Scholar] [CrossRef] [PubMed]
- Schaefer, L.; Schaefer, R.M. Proteoglycans: From Structural Compounds to Signaling Molecules. Cell Tissue Res. 2010, 339, 237–246. [Google Scholar] [CrossRef] [PubMed]
- Genovese, F.; Manresa, A.A.; Leeming, D.J.; Karsdal, M.A.; Boor, P. The Extracellular Matrix in the Kidney: A Source of Novel Non-Invasive Biomarkers of Kidney Fibrosis? Fibrogenesis Tissue Repair 2014, 7, 4. [Google Scholar] [CrossRef] [PubMed]
- Schroder, K.; Tschopp, J. The Inflammasomes. Cell 2010, 140, 821–832. [Google Scholar] [CrossRef] [PubMed]
- Nathan, C.; Ding, A. Nonresolving Inflammation. Cell 2010, 140, 871–882. [Google Scholar] [CrossRef]
- Sharma, A.K.; Mauer, S.M.; Kim, Y.; Michael, A.F. Interstitial Fibrosis in Obstructive Nephropathy. Kidney Int. 1993, 44, 774–788. [Google Scholar] [CrossRef] [PubMed]
- Rozario, T.; DeSimone, D.W. The Extracellular Matrix in Development and Morphogenesis: A Dynamic View. Dev. Biol. 2010, 341, 126–140. [Google Scholar] [CrossRef] [PubMed]
- Verma, S.K.; Garikipati, V.N.S.; Krishnamurthy, P.; Schumacher, S.M.; Grisanti, L.A.; Cimini, M.; Cheng, Z.; Khan, M.; Yue, Y.; Benedict, C.; et al. Interleukin-10 Inhibits Bone Marrow Fibroblast Progenitor Cell-Mediated Cardiac Fibrosis in Pressure-Overloaded Myocardium. Circulation 2017, 136, 940–953. [Google Scholar] [CrossRef] [PubMed]
- Jin, Y.; Liu, R.; Xie, J.; Xiong, H.; He, J.C.; Chen, N. Interleukin-10 Deficiency Aggravates Kidney Inflammation and Fibrosis in the Unilateral Ureteral Obstruction Mouse Model. Lab. Investig. 2013, 93, 801–811. [Google Scholar] [CrossRef] [PubMed]
- Sureshbabu, A.; Muhsin, S.A.; Choi, M.E. TGF-β Signaling in the Kidney: Profibrotic and Protective Effects. Am. J. Physiol. Renal. Physiol. 2016, 310, F596–F606. [Google Scholar] [CrossRef] [PubMed]
- Cabała, S.; Ożgo, M.; Herosimczyk, A. The Kidney-Gut Axis as a Novel Target for Nutritional Intervention to Counteract Chronic Kidney Disease Progression. Metabolites 2024, 14, 78. [Google Scholar] [CrossRef] [PubMed]
- van Breemen, R.B.; Wright, B.; Li, Y.; Nosal, D.; Burton, T. Standardized Grape Powder for Basic and Clinical Research. In Grapes and Health; Springer International Publishing: Cham, Switzerland, 2016; ISBN 978-3-319-28993-9. [Google Scholar]
- Joshi, T.; Patel, I.; Kumar, A.; Donovan, V.; Levenson, A.S. Grape Powder Supplementation Attenuates Prostate Neoplasia Associated with Pten Haploinsufficiency in Mice Fed High-Fat Diet. Mol. Nutr. Food Res. 2020, 64, e2000326. [Google Scholar] [CrossRef] [PubMed]
- Yu, G.; Wang, L.-G.; Han, Y.; He, Q.-Y. clusterProfiler: An R Package for Comparing Biological Themes among Gene Clusters. OMICS 2012, 16, 284–287. [Google Scholar] [CrossRef]
- Anders, S.; Huber, W. Differential Expression Analysis for Sequence Count Data. Genome Biol. 2010, 11, R106. [Google Scholar] [CrossRef]
- Chambers, L.M.; Hastie, T.J. (Eds.) Statistical Models in S, 1st ed.; Routledge: London, UK, 2017; ISBN 978-1-351-41422-7. [Google Scholar]
- Kassambara, A. rstatix: Pipe-Friendly Framework for Basic Statistical Tests. R Package Version 0.7.2. 2023. Available online: https://CRAN.R-project.org/package=rstatix (accessed on 11 July 2024).






| Weight or Length | SDM (n = 5) | SDGM (n = 5) | p-Value * | SDF (n = 5) | SDGF (n = 5) | p-Value * |
|---|---|---|---|---|---|---|
| Body weight (BW, g) | 38.1 ± 3.0 | 44.7 ± 6.2 | 0.062 | 25.3 ± 4.4 | 29.3 ± 5.6 | 0.23 |
| Liver (g) | 2.86 ± 0.48 | 2.73 ± 0.62 | 0.73 | 1.76 ± 0.89 | 1.64 ± 0.34 | 0.75 |
| Liver/BW ratio | 0.075 ± 0.011 | 0.062 ± 0.017 | 0.18 | 0.067 ± 0.031 | 0.059 ± 0.013 | 0.44 |
| Colon length (cm) | 9.1 ± 0.8 | 9.5 ± 0.7 | 0.46 | 8.9 ± 1.4 | 8.9 ± 1.0 | 1.00 |
| Colon (g) | 0.222 ± 0.032 | 0.201 ± 0.051 | 0.62 | 0.204 ± 0.058 | 0.248 ± 0.042 | 0.20 |
| Colon/BW ratio | 0.006 ± 0.001 | 0.005 ± 0.001 | 0.12 | 0.008 ± 0.002 | 0.009 ± 0.001 | 0.55 |
| Kidney (g) | 0.522 ± 0.046 | 0.525 ± 0.037 | 0.93 | 0.494 ± 0.110 | 0.413 ± 0.095 | 0.25 |
| Kidney/BW ratio | 0.014 ± 0.001 | 0.012 ± 0.001 | 0.01 | 0.020 ± 0.045 | 0.014 ± 0.002 | 0.03 |
| Prostate or ovary | 0.328 ± 0.170 | 0.545 ± 0.290 | 0.19 | 0.0698 ± 0.053 | 0.0838 ± 0.068 | 0.73 |
| Prostate/BW ratio | 0.010 ± 0.005 | 0.0010 ± 0.004 | 0.91 | NA ** | NA ** | NA ** |
| Ovary/BW ratio | NA ** | NA ** | NA ** | 0.003 ± 0.002 | 0.003 ± 0.003 | 0.76 |
| Hip muscle (g) | 0.53 ± 0.13 | 0.63 ± 0.15 | 0.27 | 0.46 ± 0.16 | 0.46 ± 0.68 | 0.92 |
| Hip muscle/BW ratio | 0.013 ± 0.003 | 0.014 ± 0.004 | 0.85 | 0.050 ± 0.071 | 0.046 ± 0.067 | 0.93 |
| Male | Female | |||||||
|---|---|---|---|---|---|---|---|---|
| Substance | SDM (n = 5) | SDGM (n = 5) | p-Value | Reference * | SDF (n = 5) | SDGF (n = 4) | p-Value | Reference * |
| ALB (g/dL) | 3.46 ± 0.57 | 3.88 ± 0.30 | 0.18 | 2.8–3.8 (3.2) | 3.44 ± 0.84 | 3.30 ± 0.68 | 0.79 | 2.4–4.3 (3.4) |
| ALP (U/L) | 85.2 ± 42.3 | 91.6 ± 27.1 | 0.78 | 111–275 (195) | 35.0 ± 16.4 | 28.0 ± 12.3 | 0.50 | 105–370 (95) |
| ALT (U/L) | 241.8 ± 157.1 | 121.0 ± 56.9 | 0.14 | 28–129 (68) | 51.2 ± 25.7 | 47.2 ± 17.8 | 0.80 | 27–195 (57) |
| AMY (U/L) | 1016 ± 95 | 1057 ± 242 | 0.73 | NA ** | 1613 ± 263 | 1396 ± 185 | 0.21 | NA ** |
| TBIL (mg/dL) | 0.32 ± 0.045 | 0.32 ± 0.045 | 1.00 | 0.2–0.6 (0.3) | 0.32 ± 0.45 | 0.375 ± 0.15 | 0.46 | 0.2–0.6 (0.3) |
| BUN (mg/dL) | 42.6 ± 25.2 | 24.2 ± 9.1 | 0.16 | 7–28 (14) | 55.0 ± 51.6 | 20.2 ± 5.4 | 0.23 | 5–26 (14) |
| Ca (mg/dL) | 10.9 ± 0.31 | 11.0 ± 0.48 | 0.76 | 9.7–12.5 (11.0) | 10.9 ± 0.78 | 10.7 ± 0.24 | 0.58 | 9.7–12.3 (11.1) |
| PHOS (mg/dL) | 7.6 ± 4.0 | 9.4 ± 1.5 | 0.37 | 7.9–14.5 (11.1) | 12.1 ± 4.7 | 9.0 ± 1.5 | 0.24 | 7.3–13.5 (10.5) |
| CRE (mg/dL) | 0.34 ± 0.17 | 0.38 ± 0.18 | 0.72 | 0.2–0.5 (0.3) | 0.36 ± 0.21 | 0.50 ± 0.29 | 0.43 | 0.2–0.5 (0.3) |
| GLU (mg/dL) | 153.2 ± 48.9 | 195.0 ± 43.2 | 0.19 | 172–372 (259) | 88.2 ± 47.2 | 124 ± 41.0 | 0.27 | 177–348 (240) |
| Na (mmol/L) | 159.8 ± 2.5 | 158.0 ± 3.6 | 0.39 | 145–176 (158) | 162.2 ± 5.6 | 155.0 ± 1.4 | 0.043 | 147–181 (159) |
| K (mmol/L) | 6.88 ± 0.41 | 7.06 ± 0.46 | 0.53 | 7.6–11.2 (9.4) | 7.24 ± 0.78 | 6.73 ± 0.30 | 0.25 | 7.3–11.8 (8.8) |
| TPR (g/dL) | 5.86 ± 0.56 | 6.12 ± 0.23 | 0.36 | 4.8–7.0 (5.6) | 5.76 ± 0.56 | 5.43 ± 1.3 | 0.62 | 4.8–7.2 (5.7) |
| GLOB (g/dL) | 2.42 ± 0.26 | 2.20 ± 0.39 | 0.32 | NA ** | 2.30 ± 0.49 | 2.15 ± 0.77 | 0.73 | NA ** |
| Gene Name | Padj. | Log2 (FoldChange) | SDGF_fpkm | SDF_fpkm | Sex |
|---|---|---|---|---|---|
| Cxcl13 | <0.001 | 4.013 | 195.847 | 12.281 | Female |
| Ccl4 | <0.001 | 4.017 | 17.779 | 1.103 | Female |
| Ccl3 | 0.001 | 2.877 | 4.646 | 0.634 | Female |
| Il10 | 0.004 | 2.876 | 1.910 | 0.259 | Female |
| Cxcl13 | <0.001 | −2.940 | 4.815 | 35.901 | Male |
| Ccl6 | 0.001 | −2.013 | 16.040 | 62.831 | Male |
| Ccl8 | 0.021 | −1.714 | 22.995 | 73.282 | Male |
| Ccl24 | 0.050 | −2.635 | 0.285 | 1.750 | Male |
| Standard Diet (TD.160157) 3 | Standard Diet with 5% Grape Powder (TD.160158) 3 | |
|---|---|---|
| Formula (g/kg) | ||
| Casein | 195 | 192.9 |
| DL-Methionine | 3.0 | 3.0 |
| Sucrose | 191.1 | 191.0 |
| Dextrose, anhydrous | 66.45 | 44.3 |
| Fructose | 66.45 | 44.3 |
| Corn starch | 235.03 | 232.88 |
| Maltodextrin | 100 | 100.0 |
| Anhydrous milk fat 1 | 30 | 29.85 |
| Soybean oil | 10 | 10 |
| Cellulose | 50 | 50 |
| Mineral mix, AIN-76 (170915) | 35 | 35 |
| Potassium citrate, monohydrate | 4.03 | 2.69 |
| Calcium carbonate | 4.0 | 4.0 |
| Vitamin mix, Teklad (40060) | 10.0 | 10.0 |
| Ethoxyquin, antioxidant | 0.04 | 0.04 |
| Grape powder, freeze-dried 2 | 0 | 50 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dave, A.; Park, E.-J.; Kofsky, P.; Dufresne, A.; Chakraborty, S.; Pezzuto, J.M. Long-Term Dietary Consumption of Grapes Affects Kidney Health in C57BL/6J Mice. Nutrients 2024, 16, 2309. https://doi.org/10.3390/nu16142309
Dave A, Park E-J, Kofsky P, Dufresne A, Chakraborty S, Pezzuto JM. Long-Term Dietary Consumption of Grapes Affects Kidney Health in C57BL/6J Mice. Nutrients. 2024; 16(14):2309. https://doi.org/10.3390/nu16142309
Chicago/Turabian StyleDave, Asim, Eun-Jung Park, Paulette Kofsky, Alexandre Dufresne, Soma Chakraborty, and John M. Pezzuto. 2024. "Long-Term Dietary Consumption of Grapes Affects Kidney Health in C57BL/6J Mice" Nutrients 16, no. 14: 2309. https://doi.org/10.3390/nu16142309
APA StyleDave, A., Park, E.-J., Kofsky, P., Dufresne, A., Chakraborty, S., & Pezzuto, J. M. (2024). Long-Term Dietary Consumption of Grapes Affects Kidney Health in C57BL/6J Mice. Nutrients, 16(14), 2309. https://doi.org/10.3390/nu16142309






