Hesperetin Alleviated Experimental Colitis via Regulating Ferroptosis and Gut Microbiota
Abstract
1. Introduction
2. Methods
2.1. Reagents and Instruments
2.2. Animal Experiments
2.3. Histopathological Analysis
2.4. Immunohistochemical (IHC) Staining
2.5. 16S rRNA Gene Sequencing
2.6. ELISA
2.7. Network Pharmacology
2.7.1. Screening UC-HT Target Genes (UCHTTGs)
2.7.2. Protein–Protein Intersection (PPI) Network Establishment and Core Targets Identification
2.7.3. The Gene Ontology (GO) and KEGG Enrichment Analysis
2.7.4. Molecular Docking between HT and Its Target Proteins
2.8. Real-Time Quantitative Polymerase Chain Reaction (RT-qPCR)
2.9. Western Blot Analysis
2.10. Transmission Electron Microscope
2.11. Malondialdehyde (MDA) and Superoxide Dismutase (SOD) Examination
2.12. Cell Culture and Treatment
2.13. Cell Viability
2.14. Intracellular Reactive Oxygen Species (ROS), Lipid Peroxidation (LPD), and Mitochondrial Membrane Potential (MMP)
2.15. Data Analysis
3. Results
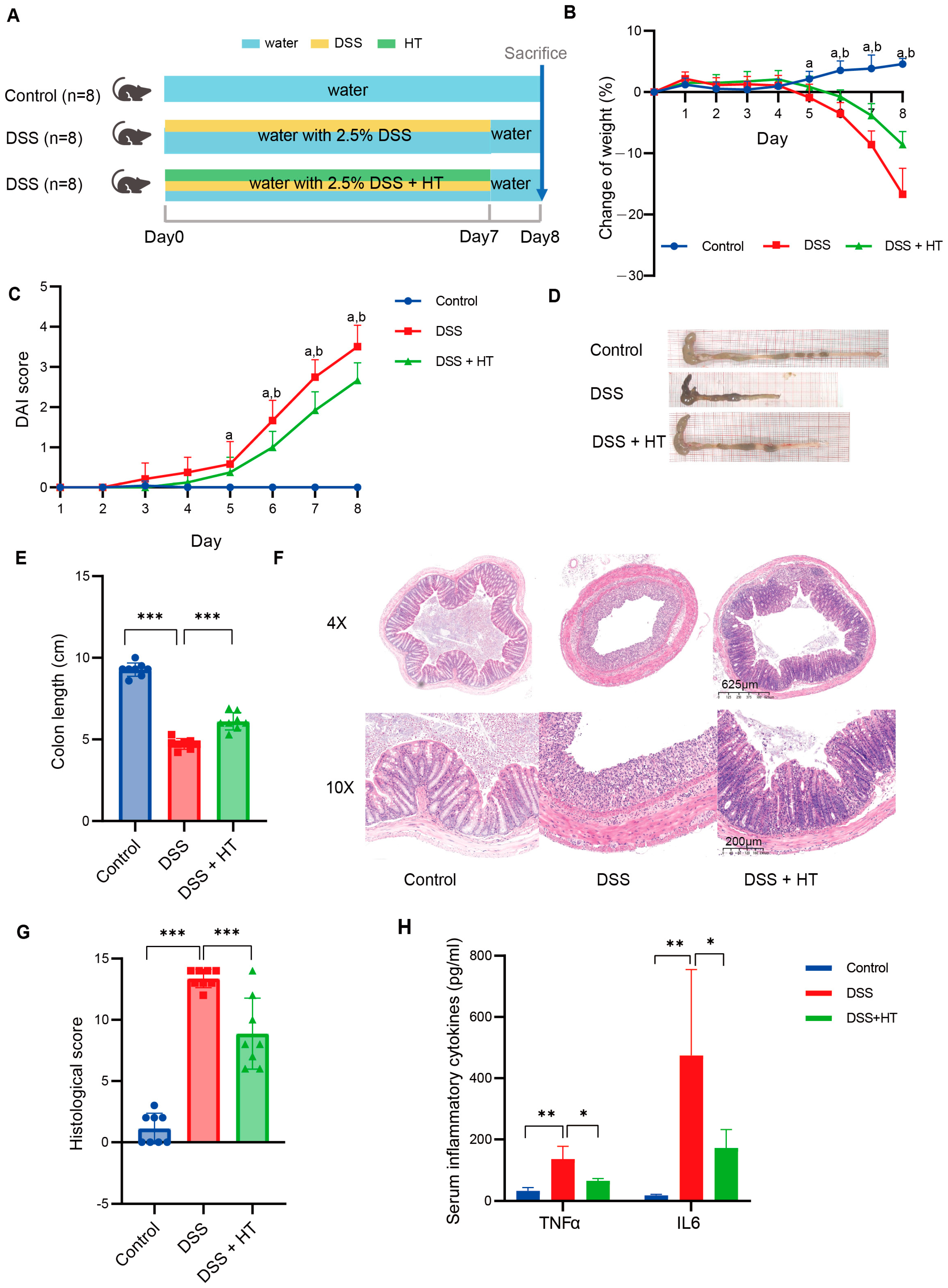
3.1. Hesperetin Alleviated DSS-Induced Colitis in Mice
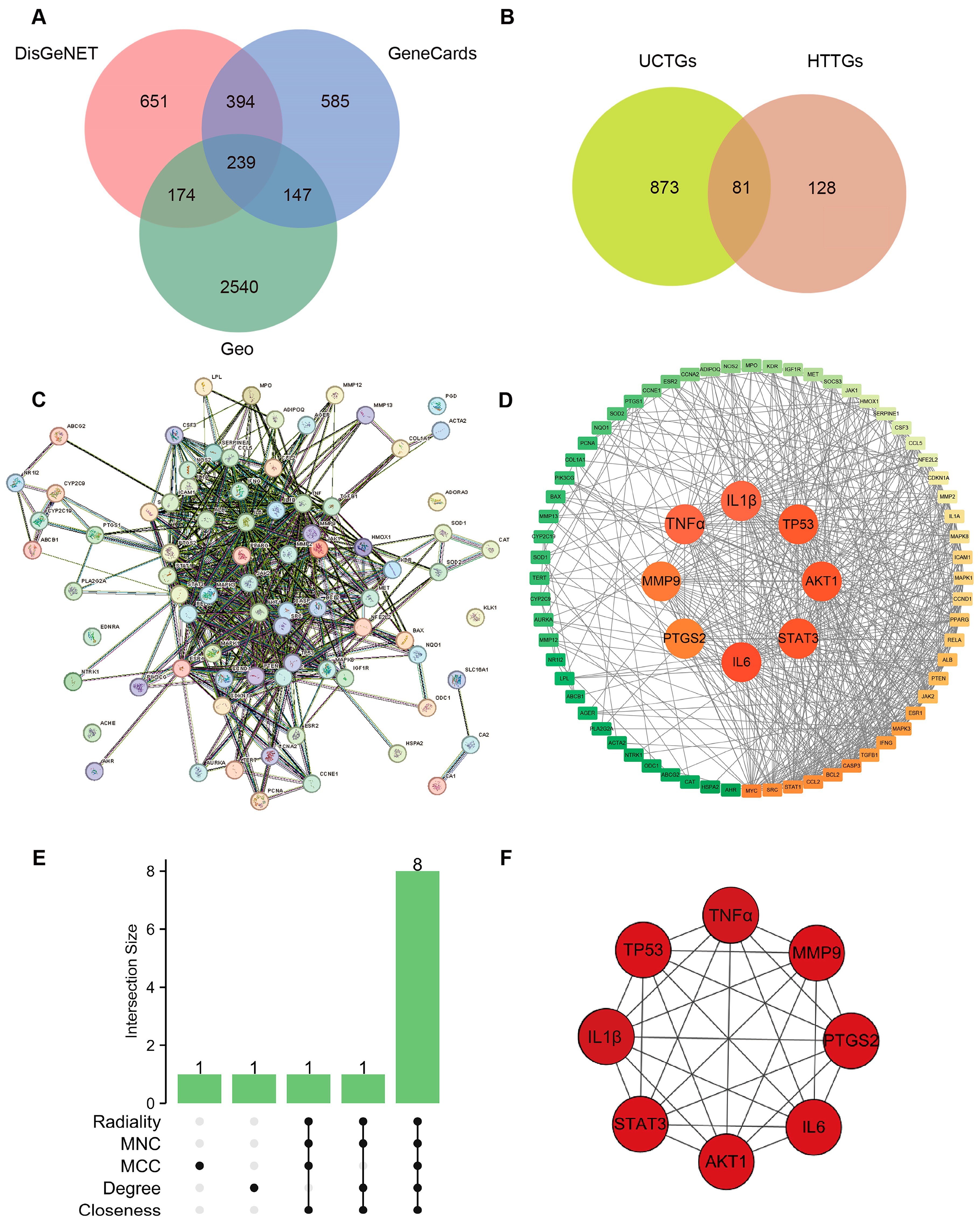
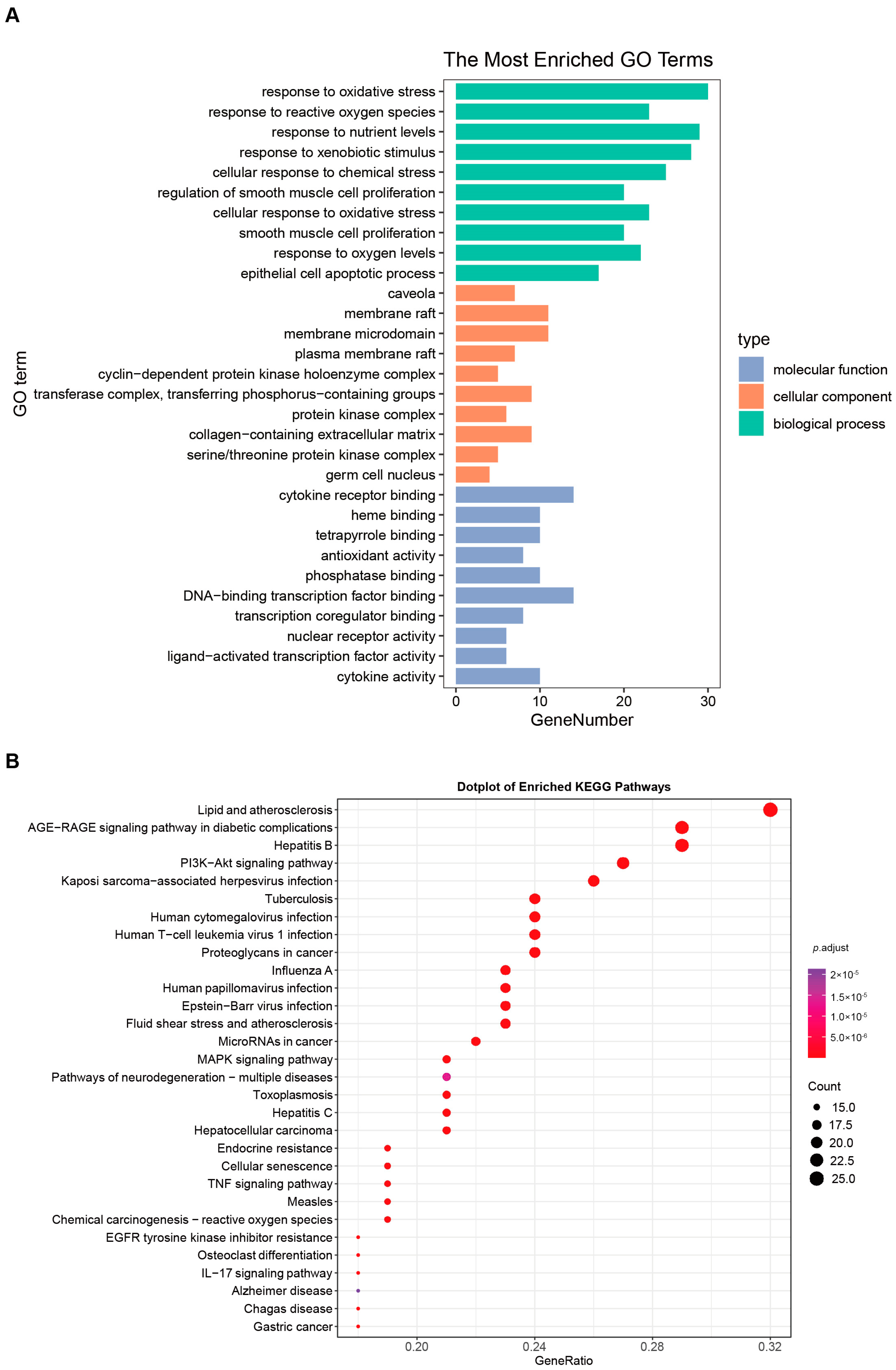
3.2. Combination of Bioinformatics, Pharmacology Network and Molecular Docking Revealed the Potential Targets of HT in UC
3.3. Verification of Target Genes of HT In Vivo
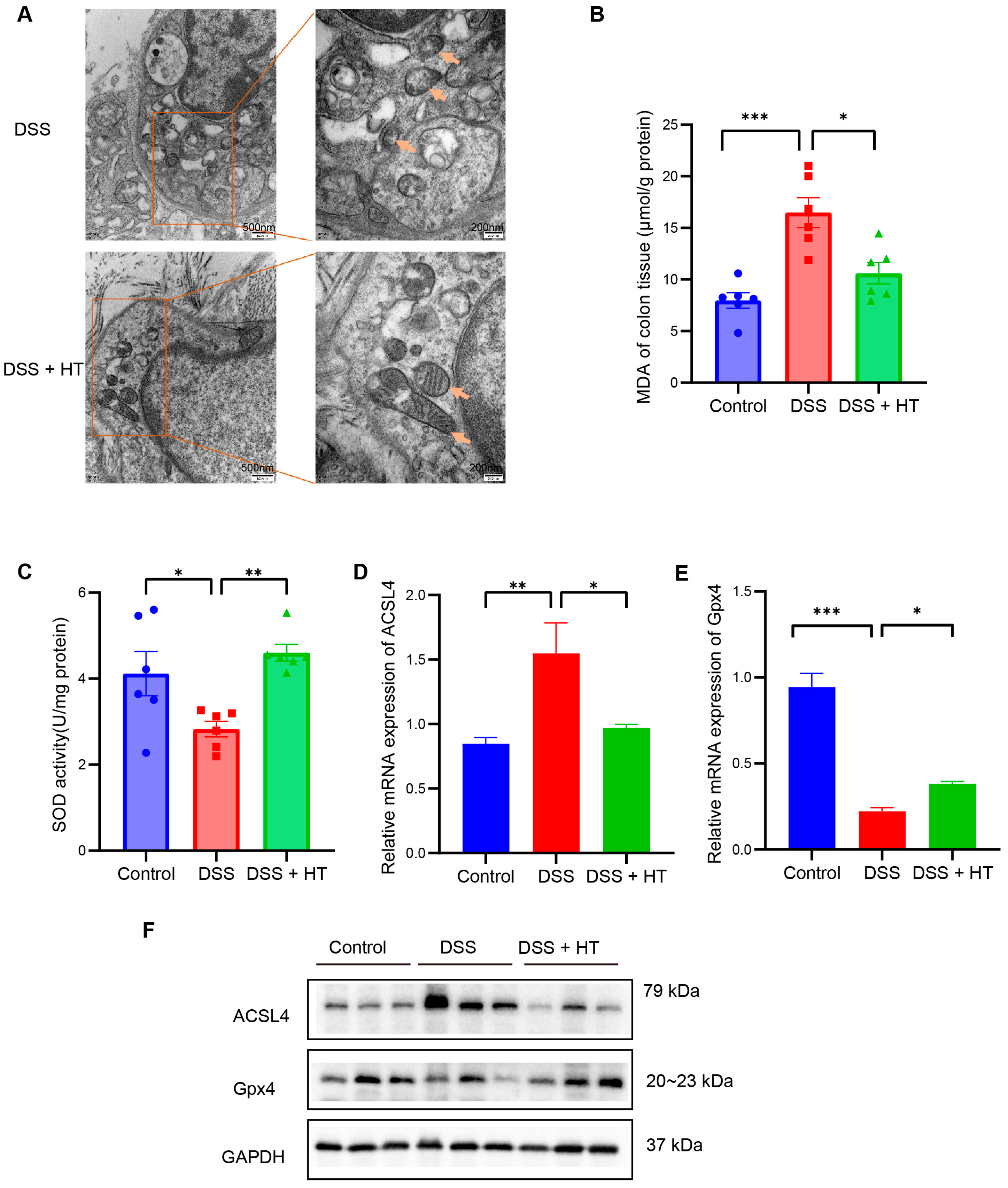
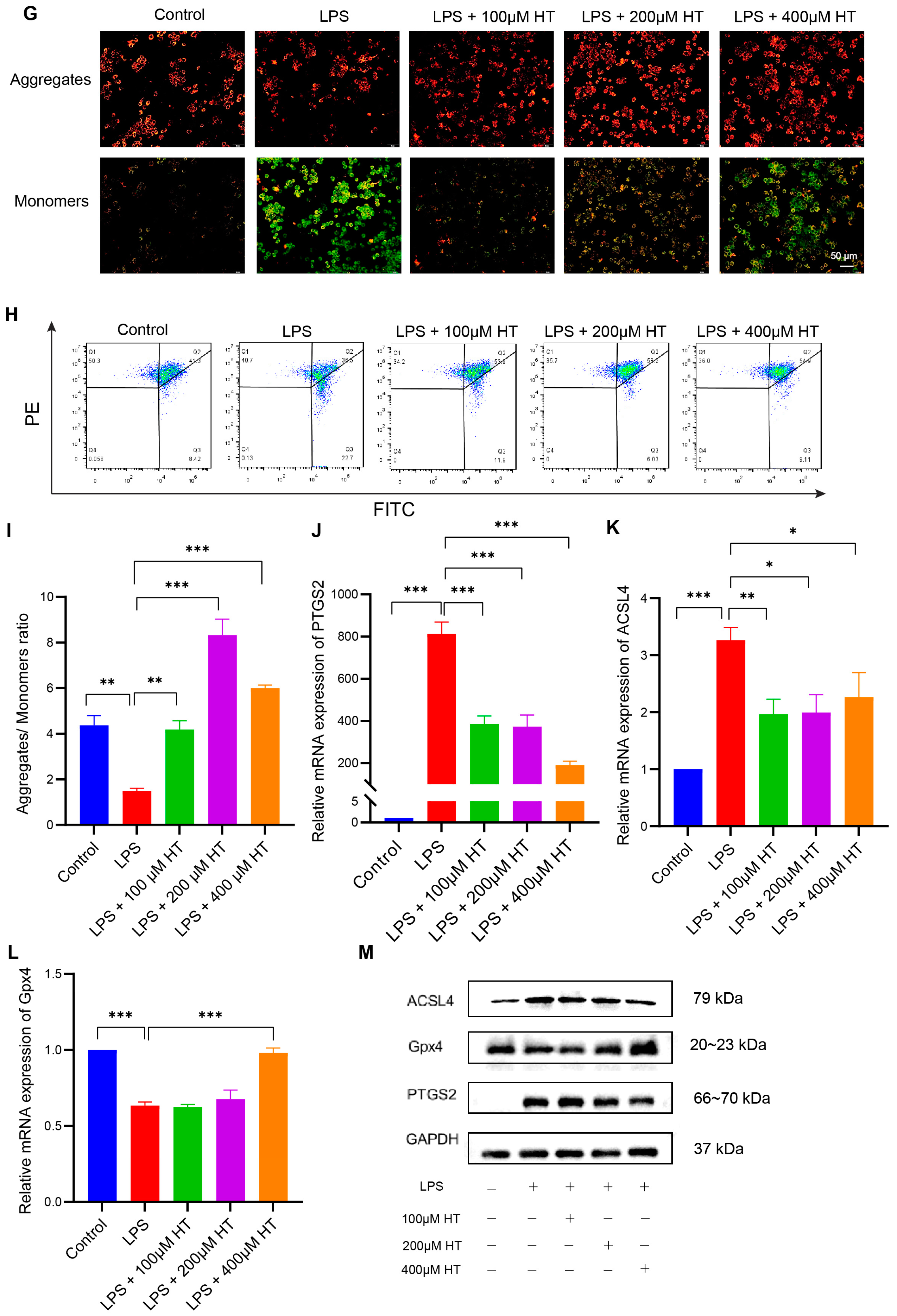
3.4. HT Suppressed Ferroptosis in Colitis In Vivo
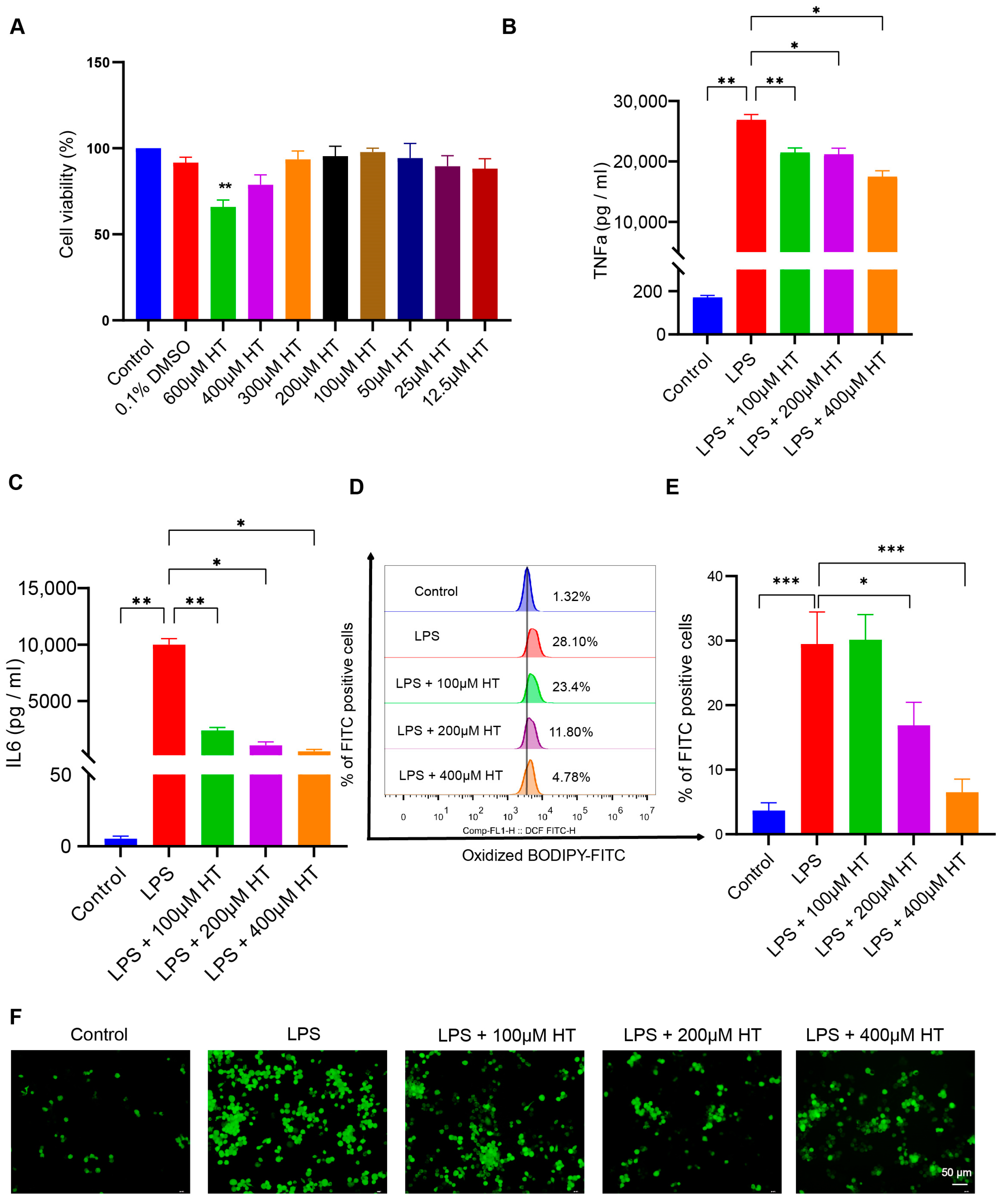
3.5. HT Suppressed Ferroptosis in LPS-Induced RAW264.7 Cells
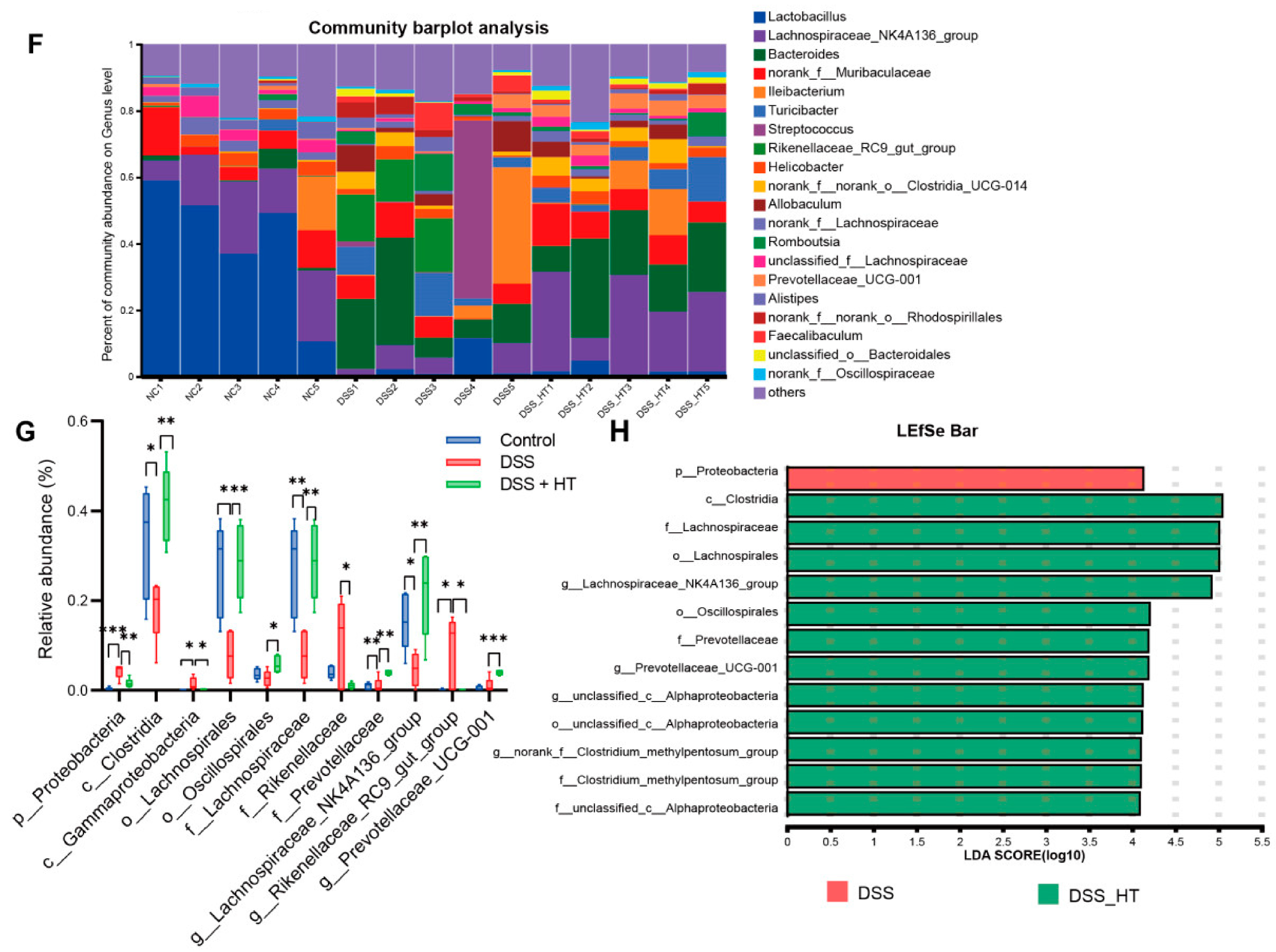
3.6. HT Regulated the Gut Microbiota in DSS-Induced Colitis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Kaplan, G.G.; Windsor, J.W. The four epidemiological stages in the global evolution of inflammatory bowel disease. Nat. Rev. Gastroenterol. Hepatol. 2021, 18, 56–66. [Google Scholar] [CrossRef] [PubMed]
- Lautenschlager, S.A.; Barry, M.P.; Rogler, G.; Biedermann, L.; Schreiner, P.; Siebenhüner, A.R.; Abdelrahman, K.; Ademi, G.; Aepli, P.; Thomas, A.; et al. Lifestyle factors associated with inflammatory bowel disease: Data from the Swiss IBD cohort study. BMC Gastroenterol. 2023, 23, 71. [Google Scholar] [CrossRef] [PubMed]
- The Lancet Gastroenterology, H. The economic burden of inflammatory bowel disease. Lancet Gastroenterol. Hepatol. 2023, 8, 391. [Google Scholar] [CrossRef] [PubMed]
- Matsuoka, K.; Yamazaki, H.; Nagahori, M.; Kobayashi, T.; Omori, T.; Mikami, Y.; Fujii, T.; Shinzaki, S.; Saruta, M.; Matsuura, M.; et al. Association of ulcerative colitis symptom severity and proctocolectomy with multidimensional patient-reported outcomes: A cross-sectional study. J. Gastroenterol. 2023, 58, 751–765. [Google Scholar] [CrossRef] [PubMed]
- Bian, F.; Yan, D.; Wu, X.; Yang, C. A Biological Perspective of TLR8 Signaling in Host Defense and Inflammation. Infect. Microbes Dis. 2023, 5, 44–55. [Google Scholar] [CrossRef]
- Li, Y.; Ma, M.; Wang, X.; Li, J.; Fang, Z.; Li, J.; Yang, B.; Lu, Y.; Xu, X.; Li, Y. Celecoxib alleviates the DSS-induced ulcerative colitis in mice by enhancing intestinal barrier function, inhibiting ferroptosis and suppressing apoptosis. Immunopharmacol. Immunotoxicol. 2024, 46, 240–254. [Google Scholar] [CrossRef] [PubMed]
- Juyal, G.; Sood, A.; Midha, V.; Singh, A.; Singh, D.; Mahajan, R.; Verma, V.; Bhatnagar, R.; Joshi, M.C. Enrichment of Lactic Acid-Producing Bacteria in the Fecal Microbiota of Patients with Ulcerative Colitis in North India. Adv. Gut Microbiome Res. 2023, 2023, 7333511. [Google Scholar] [CrossRef]
- Joshi, M.; Pandey, M.; Ved, A. Assessment of Ameliorative Effect of Myrica esculenta in a DSS-Induced Murine Model against Ulcerative Colitis. Adv. Gut Microbiome Res. 2024, 2024, 6616549. [Google Scholar] [CrossRef]
- Martins, A.L.; Galhardi Gasparini, R.; Sassaki, L.Y.; Saad-Hossne, R.; Ritter, A.M.V.; Barreto, T.B.; Marcolino, T.; Santos, C.Y. Intestinal complications in Brazilian patients with ulcerative colitis treated with conventional therapy between 2011 and 2020. World J. Gastroenterol. 2023, 29, 1330–1343. [Google Scholar] [CrossRef]
- Lindsay, J.O.; Picker, N.; Kromer, D.; Smyth, M.; Patel, H. The incidence of remission and indicators of inadequate response to advanced therapy in patients with ulcerative colitis: Results from medical charts in the United Kingdom. Curr. Med. Res. Opin. 2023, 39, 681–689. [Google Scholar] [CrossRef]
- Kurti, Z.; Gonczi, L.; Lakatos, L.; Golovics, P.A.; Pandur, T.; David, G.; Erdelyi, Z.; Szita, I.; Lakatos, P.L. Epidemiology, Treatment Strategy, Natural Disease Course and Surgical Outcomes of Patients with Ulcerative Colitis in Western Hungary—A Population-based Study Between 2007 and 2018: Data from the Veszprem County Cohort. J. Crohn’s Colitis 2023, 17, 352–360. [Google Scholar] [CrossRef] [PubMed]
- Bokemeyer, B.; Picker, N.; Wilke, T.; Rosin, L.; Patel, H. Inadequate Response, Treatment Patterns, Health Care Utilization, and Associated Costs in Patients With Ulcerative Colitis: Retrospective Cohort Study Based on German Claims Data. Inflamm. Bowel Dis. 2022, 28, 1647–1657. [Google Scholar] [CrossRef] [PubMed]
- Garg, A.; Garg, S.; Zaneveld, L.J.D.; Singla, A.K. Chemistry and pharmacology of the Citrus bioflavonoid hesperidin. Phytother. Res. 2001, 15, 655–669. [Google Scholar] [CrossRef] [PubMed]
- Nectoux, A.M.; Abe, C.; Huang, S.-W.; Ohno, N.; Tabata, J.; Miyata, Y.; Tanaka, K.; Tanaka, T.; Yamamura, H.; Matsui, T. Absorption and Metabolic Behavior of Hesperidin (Rutinosylated Hesperetin) after Single Oral Administration to Sprague-Dawley Rats. J. Agric. Food Chem. 2019, 67, 9812–9819. [Google Scholar] [CrossRef] [PubMed]
- Hassan, A.K.; El-Kalaawy, A.M.; El-Twab, S.M.A.; Alblihed, M.A.; Ahmed, O.M. Hesperetin and Capecitabine Abate 1,2 Dimethylhydrazine-Induced Colon Carcinogenesis in Wistar Rats via Suppressing Oxidative Stress and Enhancing Antioxidant, Anti-Inflammatory and Apoptotic Actions. Life 2023, 13, 984. [Google Scholar] [CrossRef] [PubMed]
- Lu, Q.; Lai, Y.; Zhang, H.; Ren, K.; Liu, W.; An, Y.; Yao, J.; Fan, H. Hesperetin Inhibits TGF-β1-Induced Migration and Invasion of Triple Negative Breast Cancer MDA-MB-231 Cells via Suppressing Fyn/Paxillin/RhoA Pathway. Integr. Cancer Ther. 2022, 21, 15347354221086900. [Google Scholar] [CrossRef] [PubMed]
- Liu, P.; Chen, J.; Qi, J.; Liu, M.; Zhang, M.; Xue, Y.; Li, L.; Liu, Y.; Shi, J.; Zhang, Y.; et al. Hesperetin ameliorates ischemia/hypoxia-induced myocardium injury via inhibition of oxidative stress, apoptosis, and regulation of Ca2+ homeostasis. Phytother. Res. 2023, 37, 1787–1805. [Google Scholar] [CrossRef] [PubMed]
- Lai, M.C.; Liu, W.Y.; Liou, S.-S.; Liu, I.-M. The Citrus Flavonoid Hesperetin Encounters Diabetes-Mediated Alzheimer-Type Neuropathologic Changes through Relieving Advanced Glycation End-Products Inducing Endoplasmic Reticulum Stress. Nutrients 2022, 14, 745. [Google Scholar] [CrossRef] [PubMed]
- Cafaro, D.; Sturiale, A.; Fortuna, V.; Sinicropi, M.S.; Naldini, G. Clinical effect of the bergamot-derived gel (Benebeo gel®) associated with mesalazine for the treatment of anitis and proctitis. Trends Gen. Pract. 2018, 1, 1–3. [Google Scholar]
- Polat, F.R.; Karaboğa, I. Immunohistochemical examination of anti-inflammatory and anti-apoptotic effects of hesperetin on trinitrobenzene sulfonic acid induced colitis in rats. Biotech. Histochem. 2019, 94, 151–158. [Google Scholar] [CrossRef]
- He, W.; Li, Y.; Liu, M.; Yu, H.; Chen, Q.; Chen, Y.; Ruan, J.; Ding, Z.; Zhang, Y.; Wang, T. Citrus aurantium L. and Its Flavonoids Regulate TNBS-Induced Inflammatory Bowel Disease through Anti-Inflammation and Suppressing Isolated Jejunum Contraction. Int. J. Mol. Sci. 2018, 19, 3057. [Google Scholar] [CrossRef] [PubMed]
- Ferlazzo, N.; Visalli, G.; Smeriglio, A.; Cirmi, S.; Lombardo, G.E.; Campiglia, P.; Di Pietro, A.; Navarra, M. Flavonoid Fraction of Orange and Bergamot Juices Protect Human Lung Epithelial Cells from Hydrogen Peroxide-Induced Oxidative Stress. Evid.-Based Complement. Alternat. Med. 2015, 2015, 957031. [Google Scholar] [CrossRef] [PubMed]
- Filocamo, A.; Bisignano, C.; Ferlazzo, N.; Cirmi, S.; Mandalari, G.; Navarra, M. In vitro effect of bergamot (Citrus bergamia) juice against cagA-positive and-negative clinical isolates of Helicobacter pylori. BMC Complement. Altern. Med. 2015, 15, 256. [Google Scholar] [CrossRef] [PubMed]
- Dixon, S.J.; Lemberg, K.M.; Lamprecht, M.R.; Skouta, R.; Zaitsev, E.M.; Gleason, C.E.; Patel, D.N.; Bauer, A.J.; Cantley, A.M.; Yang, W.S.; et al. Ferroptosis: An iron-dependent form of nonapoptotic cell death. Cell 2012, 149, 1060–1072. [Google Scholar] [CrossRef] [PubMed]
- Dong, S.; Lu, Y.; Peng, G.; Li, J.; Li, W.; Li, M.; Wang, H.; Liu, L.; Zhao, Q. Furin inhibits epithelial cell injury and alleviates experimental colitis by activating the Nrf2-Gpx4 signaling pathway. Dig. Liver Dis. 2021, 53, 1276–1285. [Google Scholar] [CrossRef] [PubMed]
- Xu, M.; Tao, J.; Yang, Y.; Tan, S.; Liu, H.; Jiang, J.; Zheng, F.; Wu, B. Ferroptosis involves in intestinal epithelial cell death in ulcerative colitis. Cell Death Dis. 2020, 11, 86. [Google Scholar] [CrossRef] [PubMed]
- Ni, J.; Zhang, L.; Feng, G.; Bao, W.; Wang, Y.; Huang, Y.; Chen, T.; Chen, J.; Cao, X.; You, K.; et al. Vanillic acid restores homeostasis of intestinal epithelium in colitis through inhibiting CA9/STIM1-mediated ferroptosis. Pharmacol. Res. 2024, 202, 107128. [Google Scholar] [CrossRef] [PubMed]
- Basha, O.M.; Hafez, R.A.; Salem, S.M.; Anis, R.H.; Hanafy, A.S. Impact of gut Microbiome alteration in Ulcerative Colitis patients on disease severity and outcome. Clin. Exp. Med. 2023, 23, 1763–1772. [Google Scholar] [CrossRef] [PubMed]
- Khorsand, B.; Asadzadeh Aghdaei, H.; Nazemalhosseini-Mojarad, E.; Nadalian, B.; Nadalian, B.; Houri, H. Overrepresentation of Enterobacteriaceae and Escherichia coli is the major gut microbiome signature in Crohn’s disease and ulcerative colitis; a comprehensive metagenomic analysis of IBDMDB datasets. Front. Cell. Infect. Microbiol. 2022, 12, 1015890. [Google Scholar] [CrossRef]
- Hsia, K.; Zhao, N.; Chung, M.; Algarrahi, K.; Kouhsari, L.M.; Fu, M.; Chen, H.; Singh, S.; Michaud, D.S.; Jangi, S. Alterations in the Fungal Microbiome in Ulcerative Colitis. Inflamm. Bowel Dis. 2023, 29, 1613–1621. [Google Scholar] [CrossRef]
- Hassan-Zahraee, M.; Ye, Z.; Xi, L.; Dushin, E.; Lee, J.; Romatowski, J.; Leszczyszyn, J.; Danese, S.; Sandborn, W.J.; Banfield, C.; et al. Baseline serum and stool microbiome biomarkers predict clinical efficacy and tissue molecular response after ritlecitinib induction therapy in ulcerative colitis. J. Crohn’s Colitis 2023, jjad213. [Google Scholar] [CrossRef]
- Lee, S.H.; Seo, D.; Lee, K.-H.; Park, S.-J.; Park, S.; Kim, H.; Kim, T.; Joo, I.H.; Park, J.-M.; Kang, Y.-H.; et al. Biometabolites of Citrus unshiu Peel Enhance Intestinal Permeability and Alter Gut Commensal Bacteria. Nutrients 2023, 15, 319. [Google Scholar] [CrossRef]
- Sang, L.X.; Chang, B.; Dai, C.; Gao, N.; Liu, W.-X.; Jiang, M. Heat-killed VSL#3 ameliorates dextran sulfate sodium (DSS)-induced acute experimental colitis in rats. Int. J. Mol. Sci. 2013, 15, 15–28. [Google Scholar] [CrossRef]
- Tang, X.; Li, X.; Wang, Y.; Zhang, Z.; Deng, A.; Wang, W.; Zhang, H.; Qin, H.; Wu, L. Butyric Acid Increases the Therapeutic Effect of EHLJ7 on Ulcerative Colitis by Inhibiting JAK2/STAT3/SOCS1 Signaling Pathway. Front. Pharmacol. 2019, 10, 1553. [Google Scholar] [CrossRef]
- Ren, Y.; Yu, G.; Shi, C.; Liu, L.; Guo, Q.; Han, C.; Zhang, D.; Zhang, L.; Liu, B.; Gao, H.; et al. Majorbio Cloud: A one-stop, comprehensive bioinformatic platform for multiomics analyses. iMeta 2022, 1, e12. [Google Scholar] [CrossRef]
- Xu, H.Y.; Zhang, Y.Q.; Liu, Z.-M.; Chen, T.; Lv, C.-Y.; Tang, S.-H.; Zhang, X.-B.; Zhang, W.; Li, Z.-Y.; Zhou, R.-R.; et al. ETCM: An encyclopaedia of traditional Chinese medicine. Nucleic Acids Res. 2019, 47, D976–D982. [Google Scholar] [CrossRef]
- Davis, A.P.; Wiegers, T.C.; Wiegers, J.; Wyatt, B.; Johnson, R.J.; Sciaky, D.; Barkalow, F.; Strong, M.; Planchart, A.; Mattingly, C.J. CTD Tetramers: A new online tool that computationally links curated chemicals, genes, phenotypes, and diseases to inform molecular mechanisms for environmental health. Toxicol. Sci. 2023, 195, 155–168. [Google Scholar] [CrossRef]
- Daina, A.; Michielin, O.; Zoete, V. SwissTargetPrediction: Updated data and new features for efficient prediction of protein targets of small molecules. Nucleic Acids Res. 2019, 47, W357–W364. [Google Scholar] [CrossRef]
- Kim, S.; Chen, J.; Cheng, T.; Gindulyte, A.; He, J.; He, S.; Li, Q.; Shoemaker, B.A.; Thiessen, P.A.; Yu, B.; et al. PubChem 2023 update. Nucleic Acids Res. 2023, 51, D1373–D1380. [Google Scholar] [CrossRef]
- Piñero, J.; Saüch, J.; Sanz, F.; Furlong, L.I. The DisGeNET cytoscape app: Exploring and visualizing disease genomics data. Comput. Struct. Biotechnol. J. 2021, 19, 2960–2967. [Google Scholar] [CrossRef]
- Safran, M.; Rosen, N.; Twik, M.; BarShir, R.; Stein, T.I.; Dahary, D.; Fishilevich, S.; Lancet, D. The GeneCards Suite. In Practical Guide to Life Science Databases; Abugessaisa, I., Kasukawa, T., Eds.; Springer Nature: Singapore, 2021; pp. 27–56. [Google Scholar]
- Clough, E.; Barrett, T. The Gene Expression Omnibus Database. Methods Mol. Biol. 2016, 1418, 93–110. [Google Scholar]
- Bardou, P.; Mariette, J.; Escudié, F.; Djemiel, C.; Klopp, C. jvenn: An interactive Venn diagram viewer. BMC Bioinform. 2014, 15, 293. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Kirsch, R.; Koutrouli, M.; Nastou, K.; Mehryary, F.; Hachilif, R.; Gable, A.L.; Fang, T.; Doncheva, N.T.; Pyysalo, S.; et al. The STRING database in 2023: Protein-protein association networks and functional enrichment analyses for any sequenced genome of interest. Nucleic Acids Res. 2023, 51, D638–D646. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Qiu, J.; Chen, J.; Deng, G.; Yuan, S. Identification of MiR-223 Associated with Diagnosis in Ectopic Pregnancy. Int. J. Gen. Med. 2023, 16, 2693–2705. [Google Scholar] [CrossRef]
- Berman, H.M.; Westbrook, J.; Feng, Z.; Gilliland, G.; Bhat, T.N.; Weissig, H.; Shindyalov, I.N.; Bourne, P.E. The Protein Data Bank. Nucleic Acids Res. 2000, 28, 235–242. [Google Scholar] [CrossRef]
- O’Boyle, N.M.; Banck, M.; James, C.A.; Morley, C.; Vandermeersch, T.; Hutchison, G.R. Open Babel: An open chemical toolbox. J. Cheminform. 2011, 3, 33. [Google Scholar] [CrossRef]
- Liu, Y.; Yang, X.; Gan, J.; Chen, S.; Xiao, Z.X.; Cao, Y. CB-Dock2: Improved protein-ligand blind docking by integrating cavity detection, docking and homologous template fitting. Nucleic Acids Res. 2022, 50, W159–W164. [Google Scholar] [CrossRef]
- Fang, T.; Liu, L.; Liu, W. Exploring the mechanism of fraxetin against acute myeloid leukemia through cell experiments and network pharmacology. BMC Complement. Med. Ther 2024, 24, 226. [Google Scholar] [CrossRef]
- Yamada, N.; Karasawa, T.; Kimura, H.; Watanabe, S.; Komada, T.; Kamata, R.; Sampilvanjil, A.; Ito, J.; Nakagawa, K.; Kuwata, H.; et al. Ferroptosis driven by radical oxidation of n-6 polyunsaturated fatty acids mediates acetaminophen-induced acute liver failure. Cell Death Dis. 2020, 11, 144. [Google Scholar] [CrossRef]
- Xu, Z.; Li, J.; Zhou, K.; Wang, K.; Hu, H.; Hu, Y.; Gao, Y.; Luo, Z.; Huang, J. Exocarpium Citri Grandis ameliorates LPS-induced acute lung injury by suppressing inflammation, NLRP3 inflammasome, and ferroptosis. J. Ethnopharmacol. 2024, 329, 118162. [Google Scholar] [CrossRef]
- Elhennawy, M.G.; Abdelaleem, E.A.; Zaki, A.A.; Mohamed, W.R. Cinnamaldehyde and hesperetin attenuate TNBS-induced ulcerative colitis in rats through modulation of the JAk2/STAT3/SOCS3 pathway. J. Biochem. Mol. Toxicol. 2021, 35, e22730. [Google Scholar] [CrossRef]
- He, W.; Liu, M.; Li, Y.; Yu, H.; Wang, D.; Chen, Q.; Chen, Y.; Zhang, Y.; Wang, T. Flavonoids from Citrus aurantium ameliorate TNBS-induced ulcerative colitis through protecting colonic mucus layer integrity. Eur. J. Pharmacol. 2019, 857, 172456. [Google Scholar] [CrossRef]
- Zhang, X.; Ma, Y.; Lv, G.; Wang, H. Ferroptosis as a therapeutic target for inflammation-related intestinal diseases. Front. Pharmacol. 2023, 14, 1095366. [Google Scholar] [CrossRef]
- Yu, X.; Liu, Z.; Yu, Y.; Qian, C.; Lin, Y.; Jin, S.; Wu, L.; Li, S. Hesperetin promotes diabetic wound healing by inhibiting ferroptosis through the activation of SIRT3. Phytother. Res. 2024, 38, 1478–1493. [Google Scholar] [CrossRef]
- Živanović, N.; Lesjak, M.; Simin, N.; Srai, S.K. Beyond Mortality: Exploring the Influence of Plant Phenolics on Modulating Ferroptosis—A Systematic Review. Antioxidants 2024, 13, 334. [Google Scholar] [CrossRef]
- Ye, Y.; Liu, L.; Feng, Z.; Liu, Y.; Miao, J.; Wei, X.; Li, H.; Yang, J.; Cao, X.; Zhao, J. The ERK-cPLA2-ACSL4 axis mediating M2 macrophages ferroptosis impedes mucosal healing in ulcerative colitis. Free Radic. Biol. Med. 2024, 214, 219–235. [Google Scholar] [CrossRef]
- Wu, Y.T.; Zhong, L.S.; Huang, C.; Guo, Y.-Y.; Jin, F.-J.; Hu, Y.-Z.; Zhao, Z.-B.; Ren, Z.; Wang, Y.-F. β-Caryophyllene Acts as a Ferroptosis Inhibitor to Ameliorate Experimental Colitis. Int. J. Mol. Sci. 2022, 23, 16055. [Google Scholar] [CrossRef]
- Wang, C.; Bai, J.; Chen, X.; Song, J.; Zhang, Y.; Wang, H.; Suo, H. Gut microbiome-based strategies for host health and disease. Crit. Rev. Food Sci. Nutr. 2023, 64, 6834–6849. [Google Scholar] [CrossRef]
- Michail, S.; Durbin, M.; Turner, D.; Griffiths, A.M.; Mack, D.R.; Hyams, J.; Leleiko, N.; Kenche, H.; Stolfi, A.; Wine, E. Alterations in the gut microbiome of children with severe ulcerative colitis. Inflamm. Bowel Dis. 2012, 18, 1799–1808. [Google Scholar] [CrossRef]
- Han, X.; Zang, D.; Liu, D.; Chen, J. The multifaceted roles of common gut microbiota in immune checkpoint inhibitor-mediated colitis: From mechanism to clinical application. Front. Immunol. 2022, 13, 988849. [Google Scholar] [CrossRef] [PubMed]
- Unno, T.; Hisada, T.; Takahashi, S. Hesperetin Modifies the Composition of Fecal Microbiota and Increases Cecal Levels of Short-Chain Fatty Acids in Rats. J. Agric. Food Chem. 2015, 63, 7952–7957. [Google Scholar] [CrossRef] [PubMed]
- Ran, X.; Hu, G.; Guo, W.; Li, K.; Wang, X.; Liu, J.; Fu, S. Hesperetin regulates the intestinal flora and inhibits the TLR4/NF-κB signaling axis to protect the blood-milk barrier and prevent mastitis. Life Sci. 2024, 342, 122533. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Gu, Y.; Chu, Q.; Wang, X.; Ding, Y.; Qin, X.; Liu, T.; Wang, S.; Liu, X.; Wang, B.; et al. Gut microbiota and metabolites as predictors of biologics response in inflammatory bowel disease: A comprehensive systematic review. Microbiol. Res. 2024, 282, 127660. [Google Scholar] [CrossRef]
- Vesci, L.; Tundo, G.; Soldi, S.; Galletti, S.; Stoppoloni, D.; Bernardini, R.; Modolea, A.B.; Luberto, L.; Marra, E.; Giorgi, F.; et al. A Novel Lactobacillus brevis Fermented with a Vegetable Substrate (AL0035) Counteracts TNBS-Induced Colitis by Modulating the Gut Microbiota Composition and Intestinal Barrier. Nutrients 2024, 16, 937. [Google Scholar] [CrossRef] [PubMed]
- Yan, C.; Huang, S.H.; Ding, H.-F.; Kwek, E.; Liu, J.-H.; Chen, Z.-X.; Ma, K.Y.; Chen, Z.-Y. Adverse effect of oxidized cholesterol exposure on colitis is mediated by modulation of gut microbiota. J. Hazard. Mater. 2023, 459, 132057. [Google Scholar] [CrossRef] [PubMed]
- Nong, K.; Qin, X.; Liu, Z.; Wang, Z.; Wu, Y.; Zhang, B.; Chen, W.; Fang, X.; Liu, Y.; Wang, X.; et al. Potential effects and mechanism of flavonoids extract of Callicarpa nudiflora Hook on DSS-induced colitis in mice. Phytomedicine 2024, 128, 155523. [Google Scholar] [CrossRef] [PubMed]
- Dong, D.; Su, T.; Chen, W.; Wang, D.; Xue, Y.; Lu, Q.; Jiang, C.; Ni, Q.; Mao, E.; Peng, Y. Clostridioides difficile aggravates dextran sulfate solution (DSS)-induced colitis by shaping the gut microbiota and promoting neutrophil recruitment. Gut Microbes 2023, 15, 2192478. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Yu, D.; Ge, C.; Luo, X.; Du, L.; Zhang, X.; Hui, C. Combined effects of microplastics and chlortetracycline on the intestinal barrier, gut microbiota, and antibiotic resistome of Muscovy ducks (Cairina moschata). Sci. Total Environ. 2023, 887, 164050. [Google Scholar] [CrossRef]
- Zhong, Y.; Xiao, Q.; Xiao, F.-J.; Zhao, X.; Zhang, W.-Y.; Xia, Y.-J.; Wang, L.-S. Ginsenoside Rg1 Alleviates Ulcerative Colitis in Obese Mice by Regulating the Gut Microbiota-Lipid Metabolism-Th1/Th2/Th17 Cells Axis. J. Agric. Food Chem. 2023, 71, 20073–20091. [Google Scholar] [CrossRef]
- Wang, H.; Sun, Y.; Xiao, F.-J.; Zhao, X.; Zhang, W.-Y.; Xia, Y.-J.; Wang, L.-S. Mesenchymal Stem Cells Ameliorate DSS-Induced Experimental Colitis by Modulating the Gut Microbiota and MUC-1 Pathway. J. Inflamm. Res. 2023, 16, 2023–2039. [Google Scholar] [CrossRef] [PubMed]



| Database | Website |
|---|---|
| ETCM | http://www.tcmip.cn/ETCM/ |
| CTD | https://ctdbase.org/ |
| STP | http://swisstargetprediction.ch/ |
| Pubchem | https://pubchem.ncbi.nlm.nih.gov/ |
| DisGeNET | https://www.disgenet.org/ |
| GeneCards | https://www.genecards.org/ |
| GEO | https://www.ncbi.nlm.nih.gov/geo/ |
| CB-DOCK2 | https://cadd.labshare.cn/cb-dock2/php/index.php |
| jvenn | https://jvenn.toulouse.inrae.fr/app/example.html |
| STRING | https://cn.string-db.org/ |
| PDB | https://www.rcsb.org/ |
| Xiantao Academic Tools | https://www.xiantaozi.com/products |
| Gene Symbol | Forward(F)/Reverse(R) | Sequence |
|---|---|---|
| TNFα | F | CAGGCGGTGCCTATGTCTC |
| R | CGATCACCCCGAAGTTCAGTAG | |
| IL6 | F | GAGGATACCACTCCCAACAGACC |
| R | AAGTGCATCATCGTTGTTCATACA | |
| IL1β | F | GCCACCTTTTGACAGTGATGAG |
| R | ATGTGCTGCTGCGAGATTTG | |
| STAT3 | F | AGAACCTCCAGGACGACTTTG |
| R | TCACAATGCTTCTCCGCATCT | |
| PTGS2 | F | TTCCAATCCATGTCAAAACCGT |
| R | AGTCCGGGTACAGTCACACTT | |
| MMP9 | F | GGACCCGAAGCGGACATTG |
| R | CGTCGTCGAAATGGGCATCT | |
| AKT1 | F | ATGAACGACGTAGCCATTGTG |
| R | TTGTAGCCAATAAAGGTGCCAT | |
| Gpx4 | F | TGTGCATCCCGCGATGATT |
| R | CCCTGTACTTATCCAGGCAGA | |
| TP53 | F | CCCCTGTCATCTTTTGTCCCT |
| R | AGCTGGCAGAATAGCTTATTGAG | |
| R | GGACCAAAGACCTCCAGAATG | |
| GAPDH | F | AGGTCGGTGTGAACGGATTTG |
| R | TGTAGACCATGTAGTTGAGGTCA |
| PDB ID | Protein Name | Vina Score(kcal/mol) | Cavity Volume | Center | Docking Size | ||||
|---|---|---|---|---|---|---|---|---|---|
| x | y | z | x | y | z | ||||
| 7JRA | TNFα | −8.5 | 1364 | −16 | −3 | −25 | 21 | 21 | 21 |
| 5R7W | IL1β | −7.5 | 189 | 40 | 9 | 57 | 22 | 22 | 22 |
| 1AIE | TP53 | −6 | 13 | 8 | 17 | 9 | 22 | 22 | 22 |
| 7NH5 | AKT1 | −7.7 | 246 | 13 | −2 | 4 | 22 | 22 | 22 |
| 6NJS | STAT3 | −7.3 | 892 | 5 | 32 | 20 | 22 | 22 | 22 |
| 7NXZ | IL6 | −6.3 | 116 | −4 | −12 | 2 | 22 | 22 | 22 |
| 5F19 | PTGS2 | −8.6 | 1234 | 32 | 37 | 18 | 22 | 22 | 32 |
| 6ESM | MMP9 | −10.3 | 350 | 1 | 48 | 23 | 22 | 22 | 22 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, J.; Yao, Y.; Yao, T.; Shi, Q.; Zeng, Y.; Li, L. Hesperetin Alleviated Experimental Colitis via Regulating Ferroptosis and Gut Microbiota. Nutrients 2024, 16, 2343. https://doi.org/10.3390/nu16142343
Wang J, Yao Y, Yao T, Shi Q, Zeng Y, Li L. Hesperetin Alleviated Experimental Colitis via Regulating Ferroptosis and Gut Microbiota. Nutrients. 2024; 16(14):2343. https://doi.org/10.3390/nu16142343
Chicago/Turabian StyleWang, Jinzhi, Yuanyuan Yao, Ting Yao, Qingmiao Shi, Yifan Zeng, and Lanjuan Li. 2024. "Hesperetin Alleviated Experimental Colitis via Regulating Ferroptosis and Gut Microbiota" Nutrients 16, no. 14: 2343. https://doi.org/10.3390/nu16142343
APA StyleWang, J., Yao, Y., Yao, T., Shi, Q., Zeng, Y., & Li, L. (2024). Hesperetin Alleviated Experimental Colitis via Regulating Ferroptosis and Gut Microbiota. Nutrients, 16(14), 2343. https://doi.org/10.3390/nu16142343






