Diet Impacts on Gene Expression in Healthy Colon Tissue: Insights from the BarcUVa-Seq Study
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Population and Design
2.2. Dietary and Lifestyle Data Collection
2.3. RNA Processing and Quality Control
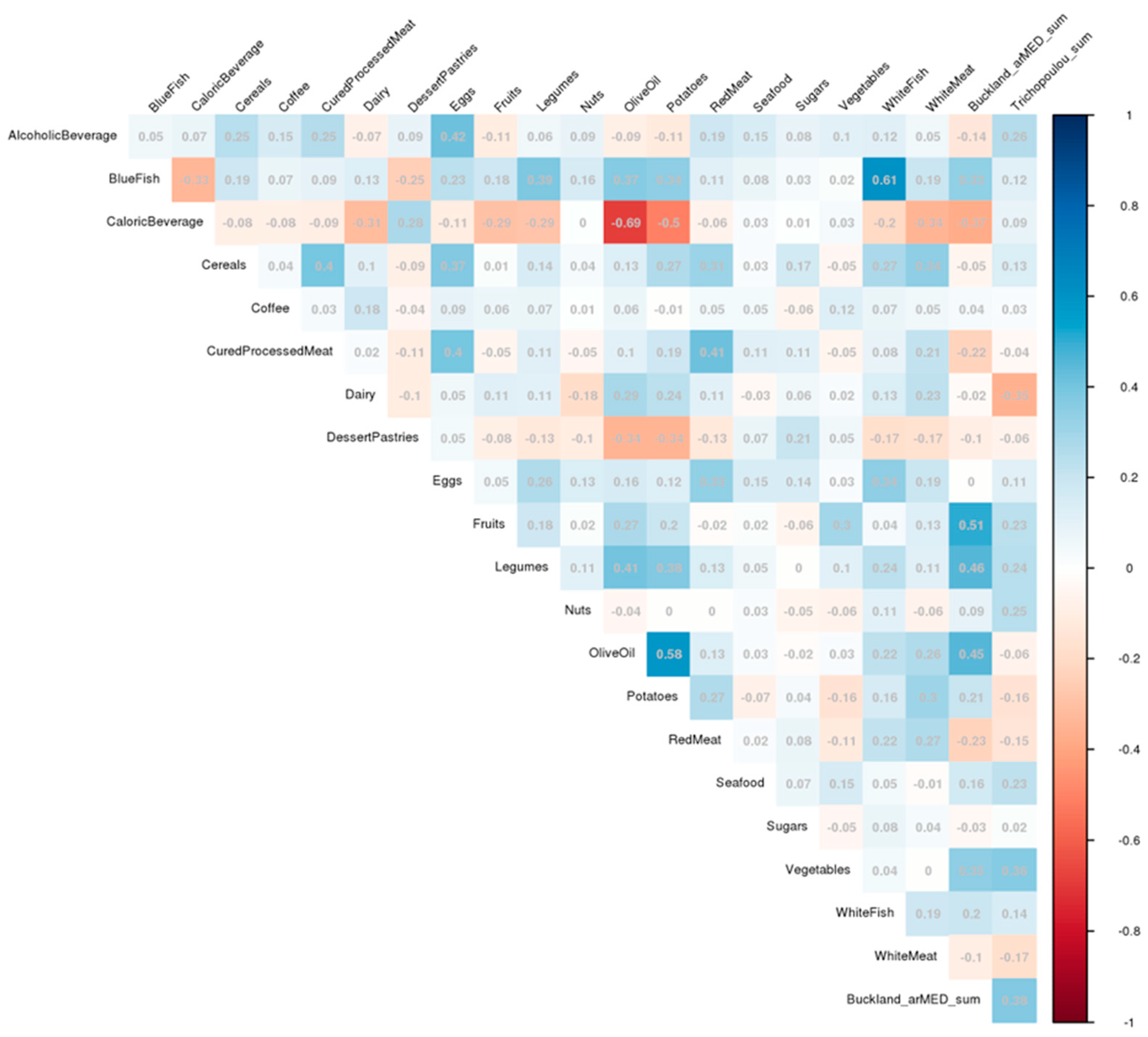
2.4. Dietary Variable and Pattern Assessment
2.5. Differential Expression Analysis
2.6. Functional Analysis of Food Group Expression Profiles
3. Results
3.1. Study Population
3.2. Differential Expression Analysis
3.3. Functional Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wang, R.; Li, Z.; Liu, S.; Zhang, D. Global, Regional, and National Burden of 10 Digestive Diseases in 204 Countries and Territories from 1990 to 2019. Front. Public Health 2023, 11, 1061453. [Google Scholar] [CrossRef] [PubMed]
- Keum, N.; Giovannucci, E. Global Burden of Colorectal Cancer: Emerging Trends, Risk Factors and Prevention Strategies. Nat. Rev. Gastroenterol. Hepatol. 2019, 16, 713–732. [Google Scholar] [CrossRef] [PubMed]
- Carreras-Torres, R.; Ibanez-Sanz, G.; Obon-Santacana, M.; Duell, E.J.; Moreno, V. Identifying Environmental Risk Factors for Inflammatory Bowel Diseases: A Mendelian Randomization Study. Sci. Rep. 2020, 10, 19273. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Xin, H.; Zhao, M.; Bi, C.; Xiao, Y.; Li, Y.; Qin, C. Global Research Trends on the Relationship between IBD and CRC: A Bibliometric Analysis from 2000 to 2023. J. Health Popul. Nutr. 2024, 43, 83. [Google Scholar] [CrossRef]
- Gravina, A.G.; Pellegrino, R.; Durante, T.; Palladino, G.; D’Onofrio, R.; Mammone, S.; Arboretto, G.; Auletta, S.; Imperio, G.; Ventura, A.; et al. Inflammatory Bowel Diseases Patients Suffer from Significant Low Levels and Barriers to Physical Activity: The “BE-FIT-IBD” Study. World J. Gastroenterol. 2023, 29, 5668–5682. [Google Scholar] [CrossRef]
- World Cancer Research Fund/American Institute for Cancer Research. Continuous Update Project Expert Report 2018. In Recommendations and Public Health and Policy Implications; CUP: London, UK, 2018. [Google Scholar]
- Guo, A.; Ludvigsson, J.; Brantsæter, A.L.; Klingberg, S.; Östensson, M.; Størdal, K.; Mårild, K. Early-Life Diet and Risk of Inflammatory Bowel Disease: A Pooled Study in Two Scandinavian Birth Cohorts. Gut 2024, 73, 590–600. [Google Scholar] [CrossRef]
- Fernandez-Villa, T.; Alvarez-Alvarez, L.; Rubin-Garcia, M.; Obon-Santacana, M.; Moreno, V. The Role of Dietary Patterns in Colorectal Cancer: A 2019 Update. Expert Rev. Gastroenterol. Hepatol. 2020, 14, 281–290. [Google Scholar] [CrossRef]
- Peters, V.; Bolte, L.; Schuttert, E.M.; Andreu-Sánchez, S.; Dijkstra, G.; Weersma, R.K.; Campmans-Kuijpers, M.J.E. Western and Carnivorous Dietary Patterns Are Associated with Greater Likelihood of IBD Development in a Large Prospective Population-Based Cohort. J. Crohn’s Colitis 2022, 16, 931–939. [Google Scholar] [CrossRef]
- Mena, M.-P.; Sacanella, E.; Vazquez-Agell, M.; Morales, M.; Fitó, M.; Escoda, R.; Serrano-Martínez, M.; Salas-Salvadó, J.; Benages, N.; Casas, R.; et al. Inhibition of Circulating Immune Cell Activation: A Molecular Antiinflammatory Effect of the Mediterranean Diet. Am. J. Clin. Nutr. 2009, 89, 248–256. [Google Scholar] [CrossRef]
- Barbaresko, J.; Koch, M.; Schulze, M.B.; Nöthlings, U. Dietary Pattern Analysis and Biomarkers of Low-Grade Inflammation: A Systematic Literature Review. Nutr. Rev. 2013, 71, 511–527. [Google Scholar] [CrossRef]
- Nasir, A.; Bullo, M.M.H.; Ahmed, Z.; Imtiaz, A.; Yaqoob, E.; Jadoon, M.; Ahmed, H.; Afreen, A.; Yaqoob, S. Nutrigenomics: Epigenetics and Cancer Prevention: A Comprehensive Review. Crit. Rev. Food Sci. Nutr. 2020, 60, 1375–1387. [Google Scholar] [CrossRef] [PubMed]
- Mastrogamvraki, N.; Zaravinos, A. Signatures of Co-Deregulated Genes and Their Transcriptional Regulators in Colorectal Cancer. NPJ Syst. Biol. Appl. 2020, 6, 23. [Google Scholar] [CrossRef]
- Slattery, M.L.; Pellatt, D.F.; Mullany, L.E.; Wolff, R.K.; Herrick, J.S. Gene Expression in Colon Cancer: A Focus on Tumor Site and Molecular Phenotype. Genes Chromosomes Cancer 2015, 54, 527–541. [Google Scholar] [CrossRef]
- Diez-Obrero, V.; Moratalla-Navarro, F.; Ibanez-Sanz, G.; Guardiola, J.; Rodriguez-Moranta, F.; Obon-Santacana, M.; Diez-Villanueva, A.; Dampier, C.H.; Devall, M.; Carreras-Torres, R.; et al. Transcriptome-Wide Association Study for Inflammatory Bowel Disease Reveals Novel Candidate Susceptibility Genes in Specific Colon Subsites and Tissue Categories. J. Crohn’s Colitis 2022, 16, 275–285. [Google Scholar] [CrossRef]
- Reynés, B.; Palou, M.; Palou, A. Gene Expression Modulation of Lipid and Central Energetic Metabolism Related Genes by High-Fat Diet Intake in the Main Homeostatic Tissues. Food Funct. 2017, 8, 629–650. [Google Scholar] [CrossRef] [PubMed]
- Bouchard-Mercier, A.; Paradis, A.-M.; Rudkowska, I.; Lemieux, S.; Couture, P.; Vohl, M.-C. Associations between Dietary Patterns and Gene Expression Profiles of Healthy Men and Women: A Cross-Sectional Study. Nutr. J. 2013, 12, 24. [Google Scholar] [CrossRef]
- Christensen, J.J.; Ulven, S.M.; Thoresen, M.; Westerman, K.; Holven, K.B.; Andersen, L.F. Associations between Dietary Patterns and Gene Expression Pattern in Peripheral Blood Mononuclear Cells: A Cross-Sectional Study. Nutr. Metab. Cardiovasc. Dis. NMCD 2020, 30, 2111–2122. [Google Scholar] [CrossRef] [PubMed]
- de Mello, V.D.F.; Kolehmanien, M.; Schwab, U.; Pulkkinen, L.; Uusitupa, M. Gene Expression of Peripheral Blood Mononuclear Cells as a Tool in Dietary Intervention Studies: What Do We Know so Far? Mol. Nutr. Food Res. 2012, 56, 1160–1172. [Google Scholar] [CrossRef]
- Milanski, M.; Degasperi, G.; Coope, A.; Morari, J.; Denis, R.; Cintra, D.E.; Tsukumo, D.M.L.; Anhe, G.; Amaral, M.E.; Takahashi, H.K.; et al. Saturated Fatty Acids Produce an Inflammatory Response Predominantly through the Activation of TLR4 Signaling in Hypothalamus: Implications for the Pathogenesis of Obesity. J. Neurosci. 2009, 29, 359–370. [Google Scholar] [CrossRef]
- Zhang, D.; Jin, W.; Wu, R.; Li, J.; Park, S.-A.; Tu, E.; Zanvit, P.; Xu, J.; Liu, O.; Cain, A.; et al. High Glucose Intake Exacerbates Autoimmunity through Reactive-Oxygen-Species-Mediated TGF-β Cytokine Activation. Immunity 2019, 51, 671–681.e5. [Google Scholar] [CrossRef]
- Jawhara, S. How Do Polyphenol-Rich Foods Prevent Oxidative Stress and Maintain Gut Health? Microorganisms 2024, 12, 1570. [Google Scholar] [CrossRef] [PubMed]
- Bjørklund, G.; Chirumbolo, S. Role of Oxidative Stress and Antioxidants in Daily Nutrition and Human Health. Nutrition 2017, 33, 311–321. [Google Scholar] [CrossRef] [PubMed]
- Nogal, A.; Valdes, A.M.; Menni, C. The Role of Short-Chain Fatty Acids in the Interplay between Gut Microbiota and Diet in Cardio-Metabolic Health. Gut Microbes 2021, 13, e1897212. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Mantrana, I.; Selma-Royo, M.; Alcantara, C.; Collado, M.C. Shifts on Gut Microbiota Associated to Mediterranean Diet Adherence and Specific Dietary Intakes on General Adult Population. Front. Microbiol. 2018, 9, 890. [Google Scholar] [CrossRef] [PubMed]
- Costantini, L.; Molinari, R.; Farinon, B.; Merendino, N. Impact of Omega-3 Fatty Acids on the Gut Microbiota. Int. J. Mol. Sci. 2017, 18, 2645. [Google Scholar] [CrossRef]
- Slattery, M.L.; Pellatt, D.F.; Mullany, L.E.; Wolff, R.K. Differential Gene Expression in Colon Tissue Associated With Diet, Lifestyle, and Related Oxidative Stress. PLoS ONE 2015, 10, e0134406. [Google Scholar] [CrossRef]
- Slattery, M.L.; Pellatt, D.F.; Wolff, R.K.; Lundgreen, A. Genes, Environment and Gene Expression in Colon Tissue: A Pathway Approach to Determining Functionality. Int. J. Mol. Epidemiol. Genet. 2016, 7, 45–57. [Google Scholar]
- Pellatt, A.J.; Slattery, M.L.; Mullany, L.E.; Wolff, R.K.; Pellatt, D.F. Dietary Intake Alters Gene Expression in Colon Tissue: Possible Underlying Mechanism for the Influence of Diet on Disease. Pharmacogenetics Genom. 2016, 26, 294–306. [Google Scholar] [CrossRef]
- Obon-Santacana, M.; Mas-Lloret, J.; Bars-Cortina, D.; Criado-Mesas, L.; Carreras-Torres, R.; Diez-Villanueva, A.; Moratalla-Navarro, F.; Guino, E.; Ibanez-Sanz, G.; Rodriguez-Alonso, L.; et al. Meta-Analysis and Validation of a Colorectal Cancer Risk Prediction Model Using Deep Sequenced Fecal Metagenomes. Cancers 2022, 14, 4214. [Google Scholar] [CrossRef]
- Obon-Santacana, M.; Diez-Villanueva, A.; Alonso, M.H.; Ibanez-Sanz, G.; Guino, E.; Lopez, A.; Rodriguez-Alonso, L.; Mata, A.; Garcia-Rodriguez, A.; Palomo, A.G.; et al. Polygenic Risk Score across Distinct Colorectal Cancer Screening Outcomes: From Premalignant Polyps to Colorectal Cancer. BMC Med. 2021, 19, 261. [Google Scholar] [CrossRef]
- Peris, M.; Espinàs, J.A.; Muñoz, L.; Navarro, M.; Binefa, G.; Borràs, J.M. Catalan Colorectal Cancer Screening Pilot Programme Group Lessons Learnt from a Population-Based Pilot Programme for Colorectal Cancer Screening in Catalonia (Spain). J. Med. Screen 2007, 14, 81–86. [Google Scholar] [CrossRef] [PubMed]
- Binefa, G.; Garcia, M.; Milà, N.; Fernández, E.; Rodríguez-Moranta, F.; Gonzalo, N.; Benito, L.; Clopés, A.; Guardiola, J.; Moreno, V. Colorectal Cancer Screening Programme in Spain: Results of Key Performance Indicators After Five Rounds (2000–2012). Sci. Rep. 2016, 6, 19532. [Google Scholar] [CrossRef] [PubMed]
- Atkin, W.S.; Valori, R.; Kuipers, E.J.; Hoff, G.; Senore, C.; Segnan, N.; Jover, R.; Schmiegel, W.; Lambert, R.; Pox, C.; et al. European Guidelines for Quality Assurance in Colorectal Cancer Screening and Diagnosis. First Edition--Colonoscopic Surveillance Following Adenoma Removal. Endoscopy 2012, 44 (Suppl. S3), SE151–SE163. [Google Scholar] [CrossRef]
- Diez-Obrero, V.; Dampier, C.H.; Moratalla-Navarro, F.; Devall, M.; Plummer, S.J.; Diez-Villanueva, A.; Peters, U.; Bien, S.; Huyghe, J.R.; Kundaje, A.; et al. Genetic Effects on Transcriptome Profiles in Colon Epithelium Provide Functional Insights for Genetic Risk Loci. Cell. Mol. Gastroenterol. Hepatol. 2021, 12, 181–197. [Google Scholar] [CrossRef] [PubMed]
- García-Closas, R.; García-Closas, M.; Kogevinas, M.; Malats, N.; Silverman, D.; Serra, C.; Tardón, A.; Carrato, A.; Castaño-Vinyals, G.; Dosemeci, M.; et al. Food, Nutrient and Heterocyclic Amine Intake and the Risk of Bladder Cancer. Eur. J. Cancer 2007, 43, 1731–1740. [Google Scholar] [CrossRef] [PubMed]
- Farran, A.; Zamora, R.; Cervera, P. Tablas de Composicion de Alimentos Del CESNID; McGraw-Hill Interamericana: Madrid, Spain, 2004. [Google Scholar]
- Willett, W. Nutritional Epidemiology; Oxford University Press: Oxford, UK, 2012; ISBN 0-19-024084-9. [Google Scholar]
- Trichopoulou, A.; Costacou, T.; Bamia, C.; Trichopoulos, D. Adherence to a Mediterranean Diet and Survival in a Greek Population. N. Engl. J. Med. 2003, 348, 2599–2608. [Google Scholar] [CrossRef]
- Buckland, G.; Travier, N.; Cottet, V.; Gonzalez, C.A.; Lujan-Barroso, L.; Agudo, A.; Trichopoulou, A.; Lagiou, P.; Trichopoulos, D.; Peeters, P.H.; et al. Adherence to the Mediterranean Diet and Risk of Breast Cancer in the European Prospective Investigation into Cancer and Nutrition Cohort Study. Int.J.Cancer 2013, 132, 2918–2927. [Google Scholar] [CrossRef]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor Package for Differential Expression Analysis of Digital Gene Expression Data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef]
- R Core Team. The R Project for Statistical Computing. Available online: https://www.r-project.org/ (accessed on 25 October 2022).
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P.; et al. STRING V11: Protein-Protein Association Networks with Increased Coverage, Supporting Functional Discovery in Genome-Wide Experimental Datasets. Nucleic Acids Res. 2019, 47, D607–D613. [Google Scholar] [CrossRef]
- Csardi, G.; Nepusz, T. The Igraph Software Package for Complex Network Research. InterJournal Complex Syst. 2005, 1695, 1–9. [Google Scholar]
- Yu, G.; He, Q.-Y. ReactomePA: An R/Bioconductor Package for Reactome Pathway Analysis and Visualization. Mol. BioSyst. 2016, 12, 477–479. [Google Scholar] [CrossRef]
- Das, U.N. Essential Fatty Acids and Their Metabolites as Modulators of Stem Cell Biology with Reference to Inflammation, Cancer, and Metastasis. Cancer Metastasis Rev. 2011, 30, 311–324. [Google Scholar] [CrossRef] [PubMed]
- Khan, I.; Hussain, M.; Jiang, B.; Zheng, L.; Pan, Y.; Hu, J.; Khan, A.; Ashraf, A.; Zou, X. Omega-3 Long-Chain Polyunsaturated Fatty Acids: Metabolism and Health Implications. Prog. Lipid Res. 2023, 92, 101255. [Google Scholar] [CrossRef]
- Ferrucci, L.; Cherubini, A.; Bandinelli, S.; Bartali, B.; Corsi, A.; Lauretani, F.; Martin, A.; Andres-Lacueva, C.; Senin, U.; Guralnik, J.M. Relationship of Plasma Polyunsaturated Fatty Acids to Circulating Inflammatory Markers. J. Clin. Endocrinol. Metab. 2006, 91, 439–446. [Google Scholar] [CrossRef]
- Yamaguchi, A.; Botta, E.; Holinstat, M. Eicosanoids in Inflammation in the Blood and the Vessel. Front. Pharmacol. 2022, 13, 997403. [Google Scholar] [CrossRef]
- Park, H.G.; Han, S.I.; Oh, S.Y.; Kang, H.S. Cellular Responses to Mild Heat Stress. Cell. Mol. Life Sci. Cmls 2005, 62, 10–23. [Google Scholar] [CrossRef] [PubMed]
- Jakobsen, C.H.; Størvold, G.L.; Bremseth, H.; Follestad, T.; Sand, K.; Mack, M.; Olsen, K.S.; Lundemo, A.G.; Iversen, J.G.; Krokan, H.E.; et al. DHA Induces ER Stress and Growth Arrest in Human Colon Cancer Cells: Associations with Cholesterol and Calcium Homeostasis. J. Lipid Res. 2008, 49, 2089–2100. [Google Scholar] [CrossRef]
- Wei, Z.; Li, D.; Zhu, L.; Yang, L.; Chen, C.; Bai, C.; Li, G. Omega 3 Polyunsaturated Fatty Acids Inhibit Cell Proliferation by Regulating Cell Cycle in Fad3b Transgenic Mouse Embryonic Stem Cells. Lipids Health Dis. 2018, 17, 210. [Google Scholar] [CrossRef] [PubMed]
- Bordoni, A.; Astolfi, A.; Morandi, L.; Pession, A.; Danesi, F.; Di Nunzio, M.; Franzoni, M.; Biagi, P.; Pession, A. N−3 PUFAs Modulate Global Gene Expression Profile in Cultured Rat Cardiomyocytes. Implications in Cardiac Hypertrophy and Heart Failure. FEBS Lett. 2007, 581, 923–929. [Google Scholar] [CrossRef]
- Vedin, I.; Cederholm, T.; Freund-Levi, Y.; Basun, H.; Garlind, A.; Irving, G.F.; Eriksdotter-Jönhagen, M.; Wahlund, L.-O.; Dahlman, I.; Palmblad, J. Effects of DHA-Rich n-3 Fatty Acid Supplementation on Gene Expression in Blood Mononuclear Leukocytes: The OmegAD Study. PLoS ONE 2012, 7, e35425. [Google Scholar] [CrossRef]
- Durkin, L.A.; Childs, C.E.; Calder, P.C. Omega-3 Polyunsaturated Fatty Acids and the Intestinal Epithelium—A Review. Foods 2021, 10, 199. [Google Scholar] [CrossRef] [PubMed]
- Tsermpini, E.E.; Plemenitaš Ilješ, A.; Dolžan, V. Alcohol-Induced Oxidative Stress and the Role of Antioxidants in Alcohol Use Disorder: A Systematic Review. Antioxidants 2022, 11, 1374. [Google Scholar] [CrossRef] [PubMed]
- Wu, D.; Cederbaum, A.I. Alcohol, Oxidative Stress, and Free Radical Damage. Alcohol Res. Health 2003, 27, 277–284. [Google Scholar] [PubMed]
- Moraes, L.; Dries, S.S.; Seibert, B.S.; Linden, R.; Perassolo, M.S. Evaluation of Oxidative Stress Markers in Ethanol Users. Braz. J. Med. Biol. Res. 2023, 56, e12465. [Google Scholar] [CrossRef]
- Simon, L.; Souza-Smith, F.M.; Molina, P.E. Alcohol-Associated Tissue Injury: Current Views on Pathophysiological Mechanisms. Annu. Rev. Physiol. 2022, 84, 87–112. [Google Scholar] [CrossRef]
- Anderson, B.O.; Berdzuli, N.; Ilbawi, A.; Kestel, D.; Kluge, H.P.; Krech, R.; Mikkelsen, B.; Neufeld, M.; Poznyak, V.; Rekve, D.; et al. Health and Cancer Risks Associated with Low Levels of Alcohol Consumption. Lancet Public Health 2023, 8, e6–e7. [Google Scholar] [CrossRef]
- Atkinson, F.S.; Brand-Miller, J.C.; Foster-Powell, K.; Buyken, A.E.; Goletzke, J. International Tables of Glycemic Index and Glycemic Load Values 2021: A Systematic Review. Am. J. Clin. Nutr. 2021, 114, 1625–1632. [Google Scholar] [CrossRef]
- Robertson, T.M.; Alzaabi, A.Z.; Robertson, M.D.; Fielding, B.A. Starchy Carbohydrates in a Healthy Diet: The Role of the Humble Potato. Nutrients 2018, 10, 1764. [Google Scholar] [CrossRef]
- Gombart, A.F.; Pierre, A.; Maggini, S. A Review of Micronutrients and the Immune System–Working in Harmony to Reduce the Risk of Infection. Nutrients 2020, 12, 236. [Google Scholar] [CrossRef]
- Brazil, J.C.; Parkos, C.A. Finding the Sweet Spot: Glycosylation Mediated Regulation of Intestinal Inflammation. Mucosal Immunol. 2022, 15, 211–222. [Google Scholar] [CrossRef]
- Kim, T.; Xie, Y.; Li, Q.; Artegoitia, V.M.; Lebrilla, C.B.; Keim, N.L.; Adams, S.H.; Krishnan, S. Diet Affects Glycosylation of Serum Proteins in Women at Risk for Cardiometabolic Disease. Eur. J. Nutr. 2021, 60, 3727–3741. [Google Scholar] [CrossRef] [PubMed]
- Pongracz, T.; Mayboroda, O.A.; Wuhrer, M. The Human Blood N-Glycome: Unraveling Disease Glycosylation Patterns. JACS Au 2024, 4, 1696–1708. [Google Scholar] [CrossRef] [PubMed]
- Reily, C.; Stewart, T.J.; Renfrow, M.B.; Novak, J. Glycosylation in Health and Disease. Nat. Rev. Nephrol. 2019, 15, 346–366. [Google Scholar] [CrossRef]
- Food-Based Dietary Guidelines Recommendations for Potatoes|Knowledge for Policy. Available online: https://knowledge4policy.ec.europa.eu/health-promotion-knowledge-gateway/food-based-dietary-guidelines-europe-table-2_en (accessed on 12 June 2024).

| All Participants | Female | Male | ||
|---|---|---|---|---|
| Mean (sd)/N (%) | Mean (sd)/N (%) | Mean (sd)/N (%) | p-Value * | |
| Sex | 277 (63.5) | 159 (36.5) | ||
| Age | 59.8 (7.0) | 60.0 (7.1) | 59.5 (6.7) | 0.35 |
| Location | ||||
| Ascending | 139 (31.9) | 88 (63.3) | 51 (36.7) | 0.97 |
| Transverse | 135 (30.9) | 85 (63.0) | 50 (37.0) | 0.97 |
| Descending | 162 (37.2) | 104 (64.2) | 58 (35.8) | |
| Batch | ||||
| 1 | 196 (45.0) | 126 (64.3) | 70 (35.7) | 0.90 |
| 2 | 94 (21.6) | 61 (64.9) | 33 (35.1) | 0.90 0.13 |
| 3 | 46 (10.6) | 27 (58.7) | 19 (41.3) | |
| 4 | 100 (22.9) | 63 (63.0) | 37 (37.0) | |
| BMI (kg/m2) | 27.5 (4.2) | 27.3 (4.6) | 27.7 (3.6) | |
| Energy intake (kcal/day) | 1909.2 (439.6) | 1754.5 (344.5) | 2176.1 (458.5) | 2.2 × 10−16 |
| Potatoes (g/day) | 86.2 (39.1) | 82.8 (37.1) | 91.9 (41.8) | 0.00062 |
| Potatoes (RM, continuous) | 6.0 (1.4) | 6.0 (1.5) | 6.1 (1.3) | 0.13 |
| Q1 | 69 (≤5.88) | 40 (≤5.85) | ||
| Q2 | 69 (5.89–6.36) | 40 (5.86–6.46) | ||
| Q3 | 69 (6.40–6.70) | 40 (6.46–6.94) | ||
| Q4 | 69 (>6.71) | 40 (>6.94) | ||
| Olive oil (g/day) | 32.7 (10.3) | 33.3 (9.9) | 31.8 (11.0) | 0.14 |
| Olive oil (RM) | 4.8 (1.0) | 4.9 (0.9) | 4.8 (1.1) | 0.33 |
| Alcoholic beverages (g/day) | 73.1 (156.1) | 26.3 (78.3) | 154.6 (214.1) | 2.2 × 10−16 |
| Alcoholic beverages (RM) | 1.2 (4.9) | −0.7 (4.1) | 4.4 (4.4) | 2.2 × 10−16 |
| Non-consumers | 191 (≤−3.23) | 36 (≤−1.89) | ||
| Below-median consumers | 42 (2.72–4.91) | 63 (−0.57–6.85) | ||
| Above-median consumers | 43 (>4.96) | 61 (>6.88) | ||
| Blue fish (g/day) | 16.7 (8.0) | 16.4 (8.0) | 17.2 (7.9) | 1.3 × 10−8 |
| Blue fish (RM) | 3.7 (1.4) | 3.7 (1.3) | 3.8 (1.5) | 0.0043 |
| Q1 | 69 (≤3.86) | 40 (≤3.86) | ||
| Q2 | 69 (3.87–4.01) | 40 (3.87–4.12) | ||
| Q3 | 69 (4.02–4.15) | 40 (4.13–4.30) | ||
| Q4 | 69 (>4.15) | 40 (>4.30) | ||
| Caloric beverages (g/day) | 32.2 (97.1) | 18.8 (69.3) | 55.6 (129.2) | 0.00017 |
| Caloric beverages (RM) | −0.8 (4.2) | −1.4 (3.7) | 0.3 (4.8) | 0.11 |
| Top 5 Food Groups | |||||
|---|---|---|---|---|---|
| Potatoes | Caloric Beverages | Olive Oil | Blue Fish | Alcoholic Beverages | |
| DEG | n | n | n | n | n |
| Upregulated | 1242 | 1093 | 618 | 214 | 186 |
| Downregulated | 1344 | 693 | 369 | 324 | 216 |
| Total | 2586 | 1786 | 987 | 538 | 402 |
| DEG adjusted by food groups (sensitivity analysis) | |||||
| Upregulated | 142 | 5 | 2 | - | - |
| Downregulated | 84 | 5 | 3 | - | - |
| Total | 226 | 10 | 5 | - | - |
| Food Group | Reactome ID | Description | GeneRatio | BgRatio | p-Value | Genes |
|---|---|---|---|---|---|---|
| Blue fish | R-HSA-69620 | Cell cycle checkpoints | 30/260 | 293/10554 | 2.89 × 10−11 | CENPE/ERCC6L/SGO2/CENPF/CLSPN/KNL1/SGO1/DBF4/BRCA1/MCM6/RMI1/SKA2/PSMD12/BARD1/PPP2R5E/ORC5/PSME3/BRCC3/KIF2A/SEM1/PSME4/RAD50/RANBP2/YWHAB/CKAP5/MAPRE1/SEH1L/AHCTF1/NUP160/COP1 |
| R-HSA-2470946 | Cohesin loading onto Chromatin | 6/260 | 10/10554 | 4.08 × 10−8 | MAU2/SMC3/RAD21/PDS5B/NIPBL/PDS5A | |
| R-HSA-6811442 | Intra-Golgi and retrograde Golgi-to-ER traffic | 19/260 | 200/10554 | 4.72 × 10−7 | CENPE/KIF11/KIF20B/KIF15/PLA2G6/GOLGA4/KIF21A/KIF2A/KLC4/KIF5B/RINT1/TRIP11/GOLGA5/GCC2/TMF1/ARFGAP2/GOSR1/MAN1A2/DCTN1 | |
| R-HSA-72203 | Processing of capped Intron-containing pre-mRNA | 19/260 | 243/10554 | 8.68 × 10−6 | SNRPF/SNRPD1/SMNDC1/THOC7/RANBP2/CWC22/THOC5/CSTF3/PRPF40A/DHX16/CPSF2/SUGP1/SEH1L/CD2BP2/POLR2D/NUP160/NUP153/HNRNPU/RBM17 | |
| R-HSA-199992 | Trans-Golgi network vesicle budding | 10/260 | 72/10554 | 9.68 × 10−6 | TFRC/ARRB1/AP1G2/FTL/CPD/AP4M1/CLTC/AP3B1/PIK3C2A/GOLGB1 | |
| R-HSA-3371556 | Cellular response to heat stress | 11/260 | 88/10554 | 9.81 × 10−6 | HSPA4L/HSPA5/HSPH1/PTGES3/DNAJC2/RANBP2/HSPA4/HDAC6/SEH1L/NUP160/NUP153 | |
| R-HSA-5693568 | Resolution of D-loop structures through Holliday junction intermediates | 7/260 | 33/10554 | 1.25 × 10−5 | BRCA2/BRCA1/RMI1/BARD1/RAD50/PALB2/SPIDR | |
| R-HSA-983169 | Class I MHC mediated antigen processing and presentation | 24/260 | 371/10554 | 1.48 × 10−5 | HSPA5/PDIA3/PSMD12/KLHL21/LRSAM1/UBE2K/SEC24A/KCTD6/PSME3/FBXO31/ASB13/SEM1/RNF123/PSME4/TRIM11/IKBKB/ATG7/RNF220/CUL2/FBXO11/UBA5/UBE3A/CUL1/TRIP12 | |
| R-HSA-5663202 | Diseases of signal transduction | 23/260 | 378/10554 | 5.85 × 10−5 | HDAC5/PSMD12/ARRB1/ERBB3/ERBB2/ARRB2/PPP2R5E/PSME3/KRAS/SEM1/PSME4/YWHAB/TRIM24/ADAM10/ATG7/PTPN11/HDAC6/HDAC2/AP3B1/POLR2D/APC/CUL1/RASA1 | |
| Alcoholic beverages | R-HSA-163200 | Respiratory electron transport, ATP synthesis by chemiosmotic coupling, and heat production by uncoupling proteins. | 13/177 | 123/10554 | 1.32 × 10−7 | NDUFB6/COX6C/NDUFC1/NDUFAB1/NDUFA1/UQCR11/NDUFAF4/ATP5PD/UQCRB/ATP5F1E/NDUFS6/UQCRQ/NDUFB5 |
| R-HSA-983695 | Antigen activates B cell receptor (BCR), leading to generation of second messengers | 7/177 | 32/10554 | 7.83 × 10−7 | CD22/CD19/CD79B/BLK/CD79A/PLCG2/PTPN6 | |
| R-HSA-5368286 | Mitochondrial translation initiation | 9/177 | 87/10554 | 1.43 × 10−5 | MRPL33/MRPS36/MRPL13/MRPL15/CHCHD1/MRPS35/MRPS22/MTFMT/MRPL19 | |
| R-HSA-6811442 | Intra-Golgi and retrograde Golgi-to-ER traffic | 11/177 | 200/10554 | 5.40 × 10−4 | CYTH2/GOLGA4/ACTR10/BET1L/MAN2A1/DCTN1/VPS54/STX5/CYTH1/TMF1/VPS52 | |
| Potatoes * | R-HSA-72203 | Processing of capped Intron-containing pre-mRNA | 11/131 | 243/10554 | 2.14 × 10−4 | POLR2L/LSM7/AAAS/PPIL4/GTF2F2/THOC5/POLDIP3/RANBP2/DHX38/PRPF40A/EFTUD2 |
| R-HSA-9020558 | Interleukin-2 signaling | 3/131 | 12/10554 | 3.79 × 10−4 | IL2RG/PTK2B/SHC1 | |
| R-HSA-381038 | XBP1(S) activates chaperone genes | 5/131 | 57/10554 | 6.81 × 10−4 | CTDSP2/PREB/DCTN1/SHC1/GOSR2 | |
| R-HSA-446203 | Asparagine N-linked glycosylation | 10/131 | 302/10554 | 4.33 × 10−3 | UGGT2/MPDU1/PREB/RNF5/ARFGAP2/GMPPA/COG4/B4GALT3/DCTN1/GOSR2 | |
| R-HSA-3108232 | SUMO E3 ligases SUMOylate target proteins | 7/131 | 181/10554 | 7.29 × 10−3 | HIC1/AAAS/SMC6/SMC3/RANBP2/DAXX/TOPORS |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Obón-Santacana, M.; Moratalla-Navarro, F.; Guinó, E.; Carreras-Torres, R.; Díez-Obrero, V.; Bars-Cortina, D.; Ibáñez-Sanz, G.; Rodríguez-Alonso, L.; Mata, A.; García-Rodríguez, A.; et al. Diet Impacts on Gene Expression in Healthy Colon Tissue: Insights from the BarcUVa-Seq Study. Nutrients 2024, 16, 3131. https://doi.org/10.3390/nu16183131
Obón-Santacana M, Moratalla-Navarro F, Guinó E, Carreras-Torres R, Díez-Obrero V, Bars-Cortina D, Ibáñez-Sanz G, Rodríguez-Alonso L, Mata A, García-Rodríguez A, et al. Diet Impacts on Gene Expression in Healthy Colon Tissue: Insights from the BarcUVa-Seq Study. Nutrients. 2024; 16(18):3131. https://doi.org/10.3390/nu16183131
Chicago/Turabian StyleObón-Santacana, Mireia, Ferran Moratalla-Navarro, Elisabet Guinó, Robert Carreras-Torres, Virginia Díez-Obrero, David Bars-Cortina, Gemma Ibáñez-Sanz, Lorena Rodríguez-Alonso, Alfredo Mata, Ana García-Rodríguez, and et al. 2024. "Diet Impacts on Gene Expression in Healthy Colon Tissue: Insights from the BarcUVa-Seq Study" Nutrients 16, no. 18: 3131. https://doi.org/10.3390/nu16183131
APA StyleObón-Santacana, M., Moratalla-Navarro, F., Guinó, E., Carreras-Torres, R., Díez-Obrero, V., Bars-Cortina, D., Ibáñez-Sanz, G., Rodríguez-Alonso, L., Mata, A., García-Rodríguez, A., Devall, M., Casey, G., Li, L., & Moreno, V. (2024). Diet Impacts on Gene Expression in Healthy Colon Tissue: Insights from the BarcUVa-Seq Study. Nutrients, 16(18), 3131. https://doi.org/10.3390/nu16183131






