Ouabain Ameliorates Alzheimer’s Disease-Associated Neuropathology and Cognitive Impairment in FAD4T Mice
Abstract
:1. Introduction
2. Materials and Methods
2.1. Chemicals and Reagents
2.2. Bioinformatics Analysis
2.3. Assay of sTREM2 Levels in Clinical Patients
2.4. Predicting Active Ingredients and Targets of OUA
2.5. GO and KEGG Enrichment Analyses
2.6. Network Construction and Analysis
2.7. Molecular Docking of OUA with the TREM2 Protein
2.8. Animal Groups and Intervention Methods
2.9. Morris Water Maze (MWM)
2.10. Fear Conditioning Experiment
2.11. Nissl Staining
2.12. Hematoxylin–Eosin (HE) Staining
2.13. Cell Culture and Processing
2.14. Cell Viability Assay
2.15. Calcein AM/PI Detection of Apoptosis
2.16. Western Blotting
2.17. Quantitative Real-Time Fluorescent PCR (RT-PCR)
2.18. Cell Transfection Assay
2.19. Preparation Procedure for Aβ1-42 Oligomers
2.20. Statistical Analysis
3. Results
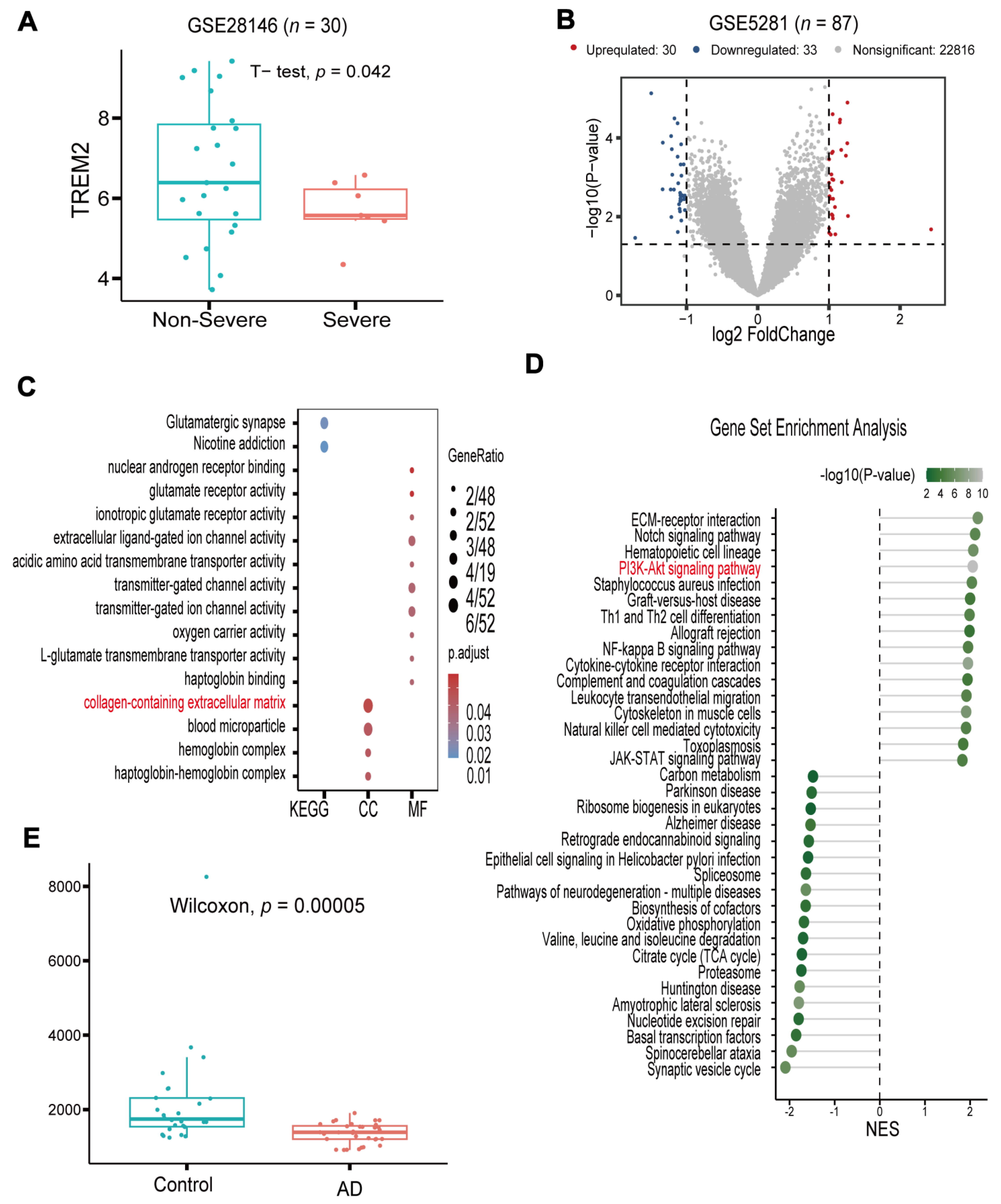
3.1. TREM2 Is a Potential Target and Has Low Expression in AD
3.2. Network Pharmacology Analyses of Potential Mechanisms of OUA Involvement in AD
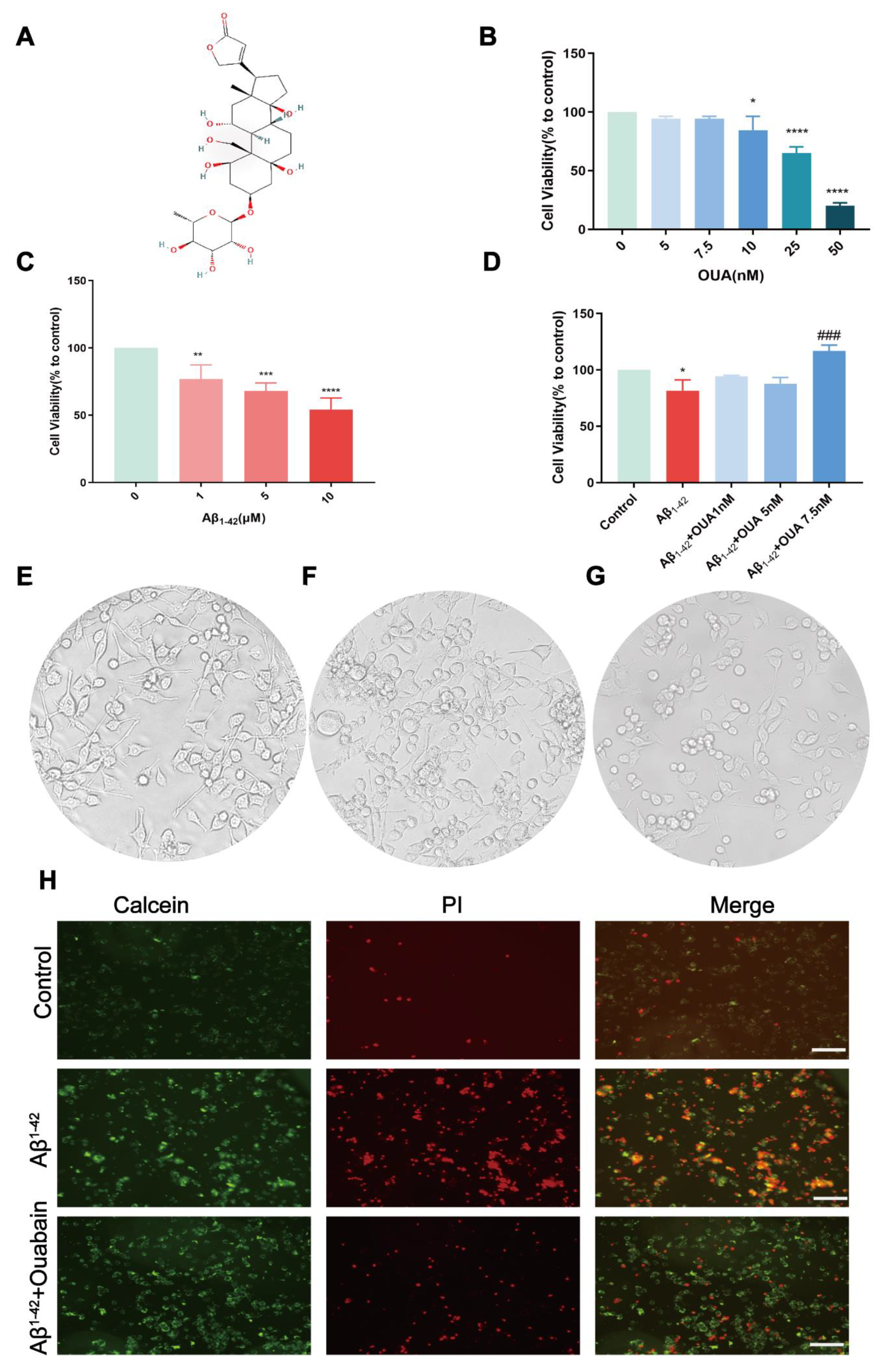
3.3. The Effect of OUA on the Aβ1-42-Induced Change in BV-2 Cell Viability
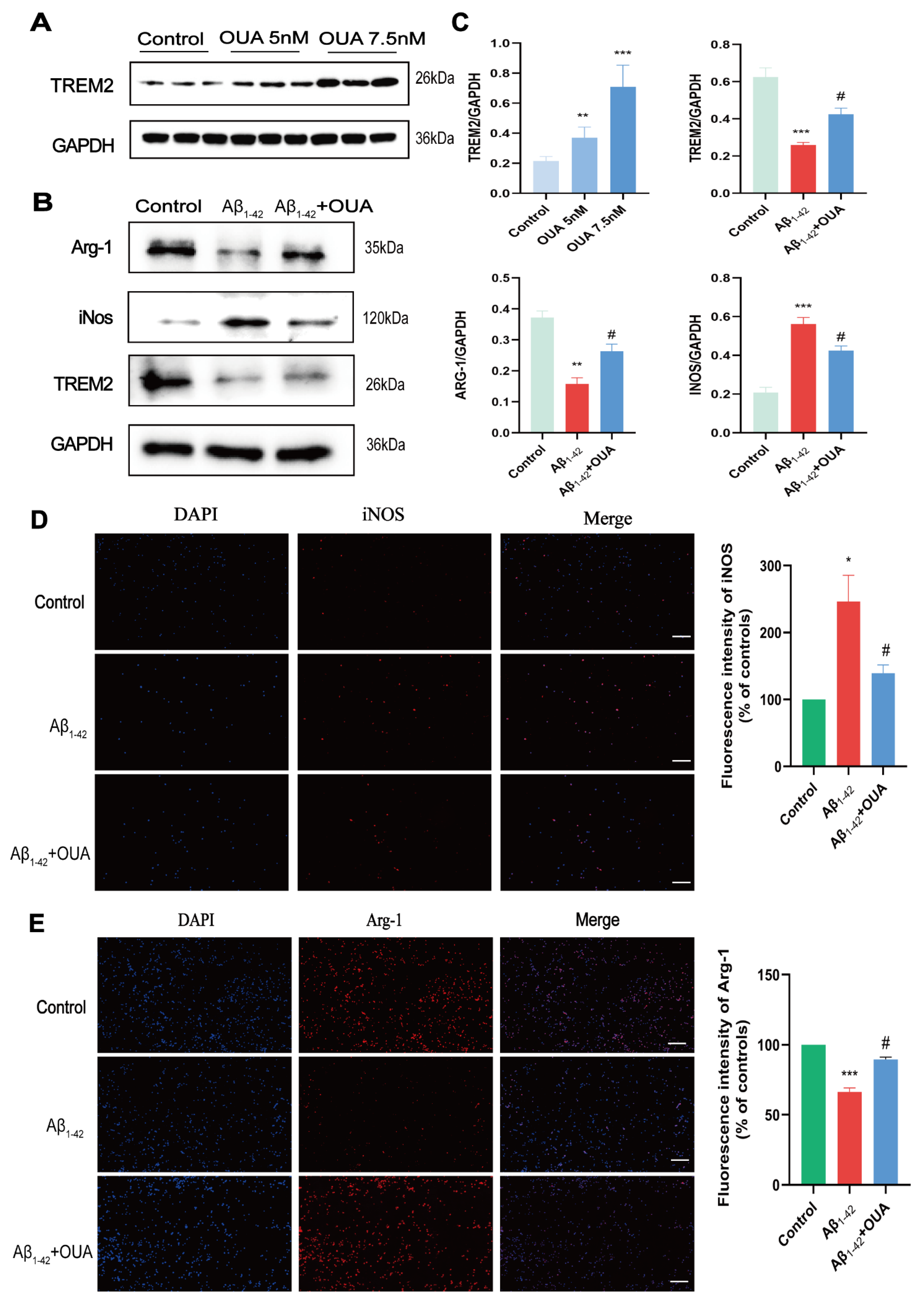
3.4. OUA Increases Aβ1-42-Induced TREM2 Expression in BV-2 Cells and Modulates Microglial Polarization
3.5. Effects of OUA on the Aβ1-42-Induced TREM2/PI3K/AKT Pathway
3.6. Effect of OUA on Aβ1-42 -Induced Inflammatory Cytokine Production in BV-2 Cells
3.7. OUA Treatment Improves Cognitive Dysfunction in FAD4T Mice
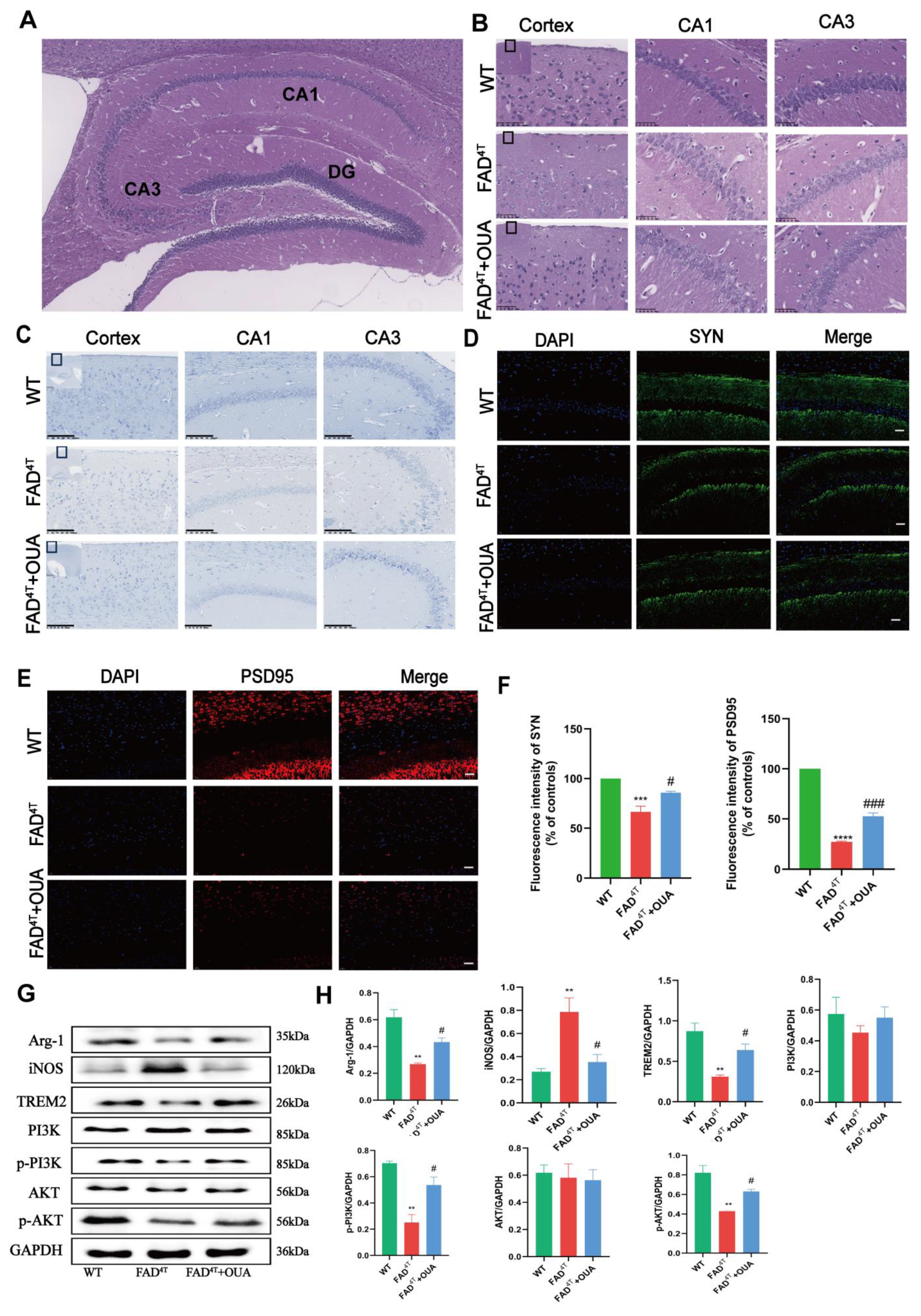
3.8. OUA Alleviates Neuronal Damage and Pathological Changes in the Hippocampal Region of FAD4T Mice
3.9. OUA Alleviates Synaptic Toxicity in the Hippocampus of FAD4T Mice
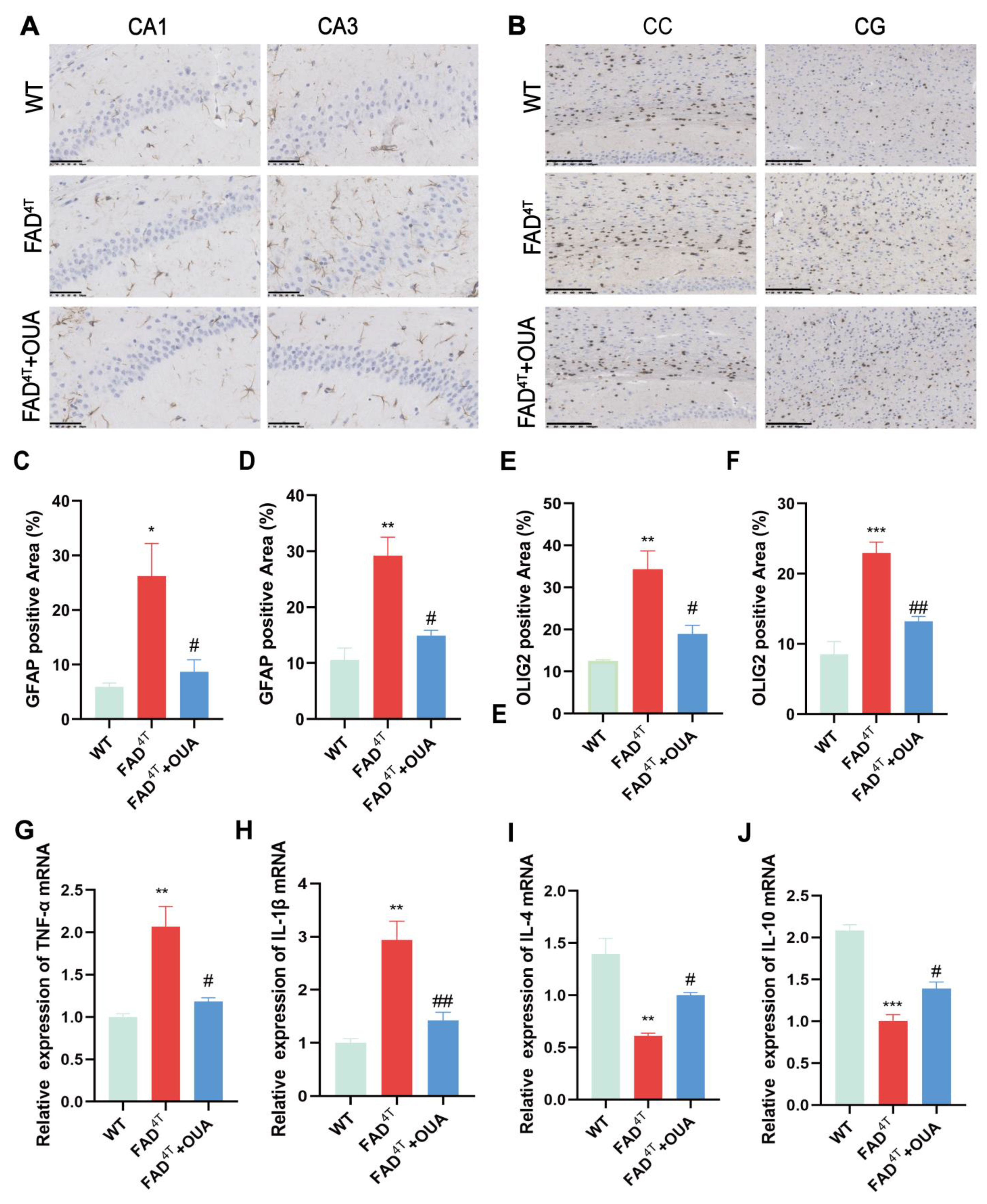
3.10. OUA Alleviates the Proliferation and Activation of Astrocytes and Oligodendrocytes in the Brains of FAD4T Mice
3.11. OUA Regulates Microglial Cell Polarization Through the TREM2/PI3K/AKT Pathway in FAD4T Mice
3.12. OUA Promotes TREM2 Expression and Inhibits Neuroinflammation in FAD4T Mice
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
References
- Ballard, C.; Gauthier, S.; Corbett, A.; Brayne, C.; Aarsland, D.; Jones, E. Alzheimer’s disease. Lancet 2011, 377, 1019–1031. [Google Scholar] [CrossRef]
- Brody, H. Alzheimer’s disease. Nature 2011, 475, S1. [Google Scholar] [CrossRef] [PubMed]
- Paouri, E.; Georgopoulos, S. Systemic and CNS Inflammation Crosstalk: Implications for Alzheimer’s Disease. Curr. Alzheimer Res. 2019, 16, 559–574. [Google Scholar] [CrossRef]
- Qiu, Z.; Zhang, H.; Xia, M.; Gu, J.; Guo, K.; Wang, H.; Miao, C. Programmed Death of Microglia in Alzheimer’s Disease: Autophagy, Ferroptosis, and Pyroptosis. J. Prev. Alzheimer’s Dis. 2023, 10, 95–103. [Google Scholar] [CrossRef]
- Borst, K.; Dumas, A.A.; Prinz, M. Microglia: Immune and non-immune functions. Immunity 2021, 54, 2194–2208. [Google Scholar] [CrossRef]
- Escoubas, C.C.; Molofsky, A.V. Microglia as integrators of brain-associated molecular patterns. Trends Immunol. 2024, 45, 358–370. [Google Scholar] [CrossRef] [PubMed]
- Orihuela, R.; McPherson, C.A.; Harry, G.J. Microglial M1/M2 polarization and metabolic states. Br. J. Pharmacol. 2016, 173, 649–665. [Google Scholar] [CrossRef] [PubMed]
- Masuda, T.; Sankowski, R.; Staszewski, O.; Prinz, M. Microglia Heterogeneity in the Single-Cell Era. Cell Rep. 2020, 30, 1271–1281. [Google Scholar] [CrossRef]
- Colonna, M.; Butovsky, O. Microglia Function in the Central Nervous System During Health and Neurodegeneration. Annu. Rev. Immunol. 2017, 35, 441–468. [Google Scholar] [CrossRef]
- Vidal-Itriago, A.; Radford, R.A.W.; Aramideh, J.A.; Maurel, C.; Scherer, N.M.; Don, E.K.; Lee, A.; Chung, R.S.; Graeber, M.B.; Morsch, M. Microglia morphophysiological diversity and its implications for the CNS. Front. Immunol. 2022, 13, 997786. [Google Scholar] [CrossRef]
- Augusto-Oliveira, M.; Arrifano, G.P.; Delage, C.I.; Tremblay, M.; Crespo-Lopez, M.E.; Verkhratsky, A. Plasticity of microglia. Biol. Rev. Camb. Philos. Soc. 2022, 97, 217–250. [Google Scholar] [CrossRef] [PubMed]
- Jansen, I.E.; Savage, J.E.; Watanabe, K.; Bryois, J.; Williams, D.M.; Steinberg, S.; Sealock, J.; Karlsson, I.K.; Hägg, S.; Athanasiu, L.; et al. Genome-wide meta-analysis identifies new loci and functional pathways influencing Alzheimer’s disease risk. Nat. Genet. 2019, 51, 404–413. [Google Scholar] [CrossRef] [PubMed]
- Qin, Q.; Teng, Z.; Liu, C.; Li, Q.; Yin, Y.; Tang, Y. TREM2, microglia, and Alzheimer’s disease. Mech. Ageing Dev. 2021, 195, 111438. [Google Scholar] [CrossRef]
- Li, J.T.; Zhang, Y. TREM2 regulates innate immunity in Alzheimer’s disease. J. Neuroinflammation 2018, 15, 107. [Google Scholar] [CrossRef]
- Abduljaleel, Z.; Al-Allaf, F.A.; Khan, W.; Athar, M.; Shahzad, N.; Taher, M.M.; Elrobh, M.; Alanazi, M.S.; El-Huneidi, W. Evidence of trem2 variant associated with triple risk of Alzheimer’s disease. PLoS ONE 2014, 9, e92648. [Google Scholar] [CrossRef] [PubMed]
- Omino, E.A.; Kokwaro, J.O. Ethnobotany of Apocynaceae species in Kenya. J. Ethnopharmacol. 1993, 40, 167–180. [Google Scholar] [CrossRef]
- Cassels, B.K. Analysis of a Maasai arrow poison. J. Ethnopharmacol. 1985, 14, 273–281. [Google Scholar] [CrossRef]
- Jaeger, H.H.; Schindler, O.; Weiss, E.; Reichstein, T. The cardenolides of Strophanthus gratus (WALL. Et HOOK.) FRANCH. Glycosides and aglycones, 265. Helv. Chim. Acta 1965, 48, 202–219. [Google Scholar] [CrossRef]
- Van Wyk, B.E. A review of commercially important African medicinal plants. J. Ethnopharmacol. 2015, 176, 118–134. [Google Scholar] [CrossRef]
- Hamlyn, J.M.; Blaustein, M.P.; Bova, S.; DuCharme, D.W.; Harris, D.W.; Mandel, F.; Mathews, W.R.; Ludens, J.H. Identification and characterization of a ouabain-like compound from human plasma. Proc. Natl. Acad. Sci. USA 1991, 88, 6259–6263. [Google Scholar] [CrossRef]
- Siman, F.D.; Silveira, E.A.; Fernandes, A.A.; Stefanon, I.; Vassallo, D.V.; Padilha, A.S. Ouabain induces nitric oxide release by a PI3K/Akt-dependent pathway in isolated aortic rings from rats with heart failure. J. Cardiovasc. Pharmacol. 2015, 65, 28–38. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.J.; Yang, J.; Lu, Z.R. New ouabain-conjugated peptide found from phage displayed peptide library. Am. J. Hypertens. 2004, 17, 619–623. [Google Scholar] [CrossRef] [PubMed]
- Tian, J.; Liu, J.; Garlid, K.D.; Shapiro, J.I.; Xie, Z. Involvement of mitogen-activated protein kinases and reactive oxygen species in the inotropic action of ouabain on cardiac myocytes. A potential role for mitochondrial K(ATP) channels. Mol. Cell. Biochem. 2003, 242, 181–187. [Google Scholar] [CrossRef]
- Lancaster, M.C.; Vegad, J.L. Suppression of the early inflammatory response in the sheep by strophanthin G. Nature 1967, 213, 840–841. [Google Scholar] [CrossRef]
- Leite, J.A.; Alves, A.K.; Galvão, J.G.; Teixeira, M.P.; Cavalcante-Silva, L.H.; Scavone, C.; Morrot, A.; Rumjanek, V.M.; Rodrigues-Mascarenhas, S. Ouabain Modulates Zymosan-Induced Peritonitis in Mice. Mediat. Inflamm. 2015, 2015, 265798. [Google Scholar] [CrossRef] [PubMed]
- Mázala-de-Oliveira, T.; de Figueiredo, C.S.; de Rezende Corrêa, G.; da Silva, M.S.; Miranda, R.L.; de Azevedo, M.A.; Cossenza, M.; Dos Santos, A.A.; Giestal-de-Araujo, E. Ouabain-Na(+)/K(+)-ATPase Signaling Regulates Retinal Neuroinflammation and ROS Production Preventing Neuronal Death by an Autophagy-Dependent Mechanism Following Optic Nerve Axotomy In Vitro. Neurochem. Res. 2022, 47, 723–738. [Google Scholar] [CrossRef]
- Sibarov, D.A.; Zhuravleva, Z.D.; Ilina, M.A.; Boikov, S.I.; Stepanenko, Y.D.; Karelina, T.V.; Antonov, S.M. Unveiling the Role of Cholesterol in Subnanomolar Ouabain Rescue of Cortical Neurons from Calcium Overload Caused by Excitotoxic Insults. Cells 2023, 12, 2011. [Google Scholar] [CrossRef]
- Xu, Y.; Kong, J.; Hu, P. Computational Drug Repurposing for Alzheimer’s Disease Using Risk Genes from GWAS and Single-Cell RNA Sequencing Studies. Front. Pharmacol. 2021, 12, 617537. [Google Scholar] [CrossRef]
- Wu, T.; Hu, E.; Xu, S.; Chen, M.; Guo, P.; Dai, Z.; Feng, T.; Zhou, L.; Tang, W.; Zhan, L.; et al. clusterProfiler 4.0: A universal enrichment tool for interpreting omics data. Innov. Camb. Mass. 2021, 2, 100141. [Google Scholar] [CrossRef]
- Zgorzynska, E. TREM2 in Alzheimer’s disease: Structure, function, therapeutic prospects, and activation challenges. Mol. Cell. Neurosci. 2024, 128, 103917. [Google Scholar] [CrossRef]
- Zhang, X.; Jiang, D.; Jiang, W.; Zhao, M.; Gan, J. Role of TLR4-Mediated PI3K/AKT/GSK-3β Signaling Pathway in Apoptosis of Rat Hepatocytes. BioMed Res. Int. 2015, 2015, 631326. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.H.; Zuo, Z.F.; Meng, L.; Yang, Q.; Lv, P.; Zhao, L.P.; Wang, X.B.; Wang, Y.F.; Huang, Y.; Fu, C.; et al. Neuroprotective effect of salidroside on hippocampal neurons in diabetic mice via PI3K/Akt/GSK-3β signaling pathway. Psychopharmacology 2023, 240, 1865–1876. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.; Liu, Y.; Xu, Q.Q.; Xian, Y.F.; Lin, Z.X. Sulforaphene Ameliorates Neuroinflammation and Hyperphosphorylated Tau Protein via Regulating the PI3K/Akt/GSK-3β Pathway in Experimental Models of Alzheimer’s Disease. Oxidative Med. Cell. Longev. 2020, 2020, 4754195. [Google Scholar] [CrossRef]
- Othman, M.Z.; Hassan, Z.; Che Has, A.T. Morris water maze: A versatile and pertinent tool for assessing spatial learning and memory. Exp. Anim. 2022, 71, 264–280. [Google Scholar] [CrossRef]
- Ghosh, S.K.; Walocha, J.A. Evolution of staining methods in neuroanatomy: Impetus for emanation of neuron doctrine during the turn of 20th century. Anat. Rec. 2024, 307, 3398–3412. [Google Scholar] [CrossRef]
- Oxford, A.E.; Stewart, E.S.; Rohn, T.T. Clinical Trials in Alzheimer’s Disease: A Hurdle in the Path of Remedy. Int. J. Alzheimer’s Dis. 2020, 2020, 5380346. [Google Scholar] [CrossRef] [PubMed]
- Cavalcante-Silva, L.H.A.; Lima, É.A.; Carvalho, D.C.M.; de Sales-Neto, J.M.; Alves, A.K.A.; Galvão, J.; da Silva, J.S.F.; Rodrigues-Mascarenhas, S. Much More than a Cardiotonic Steroid: Modulation of Inflammation by Ouabain. Front. Physiol. 2017, 8, 895. [Google Scholar] [CrossRef]
- McKenna, M.J.; Renaud, J.M.; Ørtenblad, N.; Overgaard, K. A century of exercise physiology: Effects of muscle contraction and exercise on skeletal muscle Na(+), K(+)-ATPase, Na(+) and K(+) ions, and on plasma K(+) concentration-historical developments. Eur. J. Appl. Physiol. 2024, 124, 681–751. [Google Scholar] [CrossRef] [PubMed]
- Blaustein, M.P.; Hamlyn, J.M. Sensational site: The sodium pump ouabain-binding site and its ligands. Am. J. Physiol. Cell Physiol. 2024, 326, C1120–C1177. [Google Scholar] [CrossRef]
- Trant, J.; Sanchez, G.; McDermott, J.P.; Blanco, G. Ouabain enhances renal cyst growth in a slowly progressive mouse model of autosomal dominant polycystic kidney disease. Am. J. Physiol. Ren. Physiol. 2023, 325, F857–F869. [Google Scholar] [CrossRef]
- Liu, J.; Mai, T.; Ren, H.; Chang, Y.; Li, C.; Lv, G.; Zheng, D.; Liao, X.; Yu, Y.; Zhang, F.; et al. Arrhythmia onsets triggered by acute myocardial ischemia are not mediated by lysophosphoglycerides accumulation in ventricular myocardium. Sci. Rep. 2024, 14, 9589. [Google Scholar] [CrossRef] [PubMed]
- Tejeda-Muñoz, N.; Azbazdar, Y.; Sosa, E.A.; Monka, J.; Wei, P.S.; Binder, G.; Mei, K.C.; Kurmangaliyev, Y.Z.; De Robertis, E.M. Na,K-ATPase activity promotes macropinocytosis in colon cancer via Wnt signaling. Biol. Open 2024, 13, bio060269. [Google Scholar] [CrossRef]
- Rutkoski, R.; Debarba, L.K.; Stilgenbauer, L.; Rosenthal, T.; Sadagurski, M.; Nagorny, P. Selective (α)-l-Rhamnosylation and Neuroprotective Activity Exploration of Cardiotonic Steroids. ACS Med. Chem. Lett. 2024, 15, 280–286. [Google Scholar] [CrossRef]
- Xiao, Q.; Yan, P.; Ma, X.; Liu, H.; Perez, R.; Zhu, A.; Gonzales, E.; Tripoli, D.L.; Czerniewski, L.; Ballabio, A.; et al. Neuronal-Targeted TFEB Accelerates Lysosomal Degradation of APP, Reducing Aβ Generation and Amyloid Plaque Pathogenesis. J. Neurosci. Off. J. Soc. Neurosci. 2015, 35, 12137–12151. [Google Scholar] [CrossRef]
- Battaglini, M.; Marino, A.; Montorsi, M.; Carmignani, A.; Ceccarelli, M.C.; Ciofani, G. Nanomaterials as Microglia Modulators in the Treatment of Central Nervous System Disorders. Adv. Healthc. Mater. 2024, 13, e2304180. [Google Scholar] [CrossRef] [PubMed]
- Lenz, K.M.; McCarthy, M.M. A starring role for microglia in brain sex differences. Neurosci. A Rev. J. Bringing Neurobiol. Neurol. Psychiatry 2015, 21, 306–321. [Google Scholar] [CrossRef]
- Dai, R.; Chu, T.; Zhang, M.; Wang, X.; Jourdon, A.; Wu, F.; Mariani, J.; Vaccarino, F.M.; Lee, D.; Fullard, J.F.; et al. Evaluating performance and applications of sample-wise cell deconvolution methods on human brain transcriptomic data. Sci. Adv. 2024, 10, eadh2588. [Google Scholar] [CrossRef] [PubMed]
- Silvin, A.; Ginhoux, F. Microglia heterogeneity along a spatio-temporal axis: More questions than answers. Glia 2018, 66, 2045–2057. [Google Scholar] [CrossRef] [PubMed]
- Prinz, M.; Jung, S.; Priller, J. Microglia Biology: One Century of Evolving Concepts. Cell 2019, 179, 292–311. [Google Scholar] [CrossRef]
- Tejera, D.; Heneka, M.T. Microglia in Neurodegenerative Disorders. Methods Mol. Biol. 2019, 2034, 57–67. [Google Scholar] [CrossRef]
- Wei, W.; Zhang, L.; Xin, W.; Pan, Y.; Tatenhorst, L.; Hao, Z.; Gerner, S.T.; Huber, S.; Juenemann, M.; Butz, M.; et al. TREM2 regulates microglial lipid droplet formation and represses post-ischemic brain injury. Biomed. Pharmacother. Biomed. Pharmacother. 2024, 170, 115962. [Google Scholar] [CrossRef] [PubMed]
- Xue, T.; Ji, J.; Sun, Y.; Huang, X.; Cai, Z.; Yang, J.; Guo, W.; Guo, R.; Cheng, H.; Sun, X. Sphingosine-1-phosphate, a novel TREM2 ligand, promotes microglial phagocytosis to protect against ischemic brain injury. Acta Pharm. Sinica B 2022, 12, 1885–1898. [Google Scholar] [CrossRef] [PubMed]
- Li, R.Y.; Qin, Q.; Yang, H.C.; Wang, Y.Y.; Mi, Y.X.; Yin, Y.S.; Wang, M.; Yu, C.J.; Tang, Y. TREM2 in the pathogenesis of AD: A lipid metabolism regulator and potential metabolic therapeutic target. Mol. Neurodegener. 2022, 17, 40. [Google Scholar] [CrossRef] [PubMed]
- Lee-Gosselin, A.; Jury-Garfe, N.; You, Y.; Dabin, L.; Soni, D.; Dutta, S.; Rochet, J.C.; Kim, J.; Oblak, A.L.; Lasagna-Reeves, C.A. TREM2-Deficient Microglia Attenuate Tau Spreading In Vivo. Cells 2023, 12, 1597. [Google Scholar] [CrossRef]
- Liebold, I.; Meyer, S.; Heine, M.; Kuhl, A.; Witt, J.; Eissing, L.; Fischer, A.W.; Koop, A.C.; Kluwe, J.; Wiesch, J.S.Z.; et al. TREM2 Regulates the Removal of Apoptotic Cells and Inflammatory Processes during the Progression of NAFLD. Cells 2023, 12, 341. [Google Scholar] [CrossRef]
- Li, Y.; Xu, H.; Wang, H.; Yang, K.; Luan, J.; Wang, S. TREM2: Potential therapeutic targeting of microglia for Alzheimer’s disease. Biomedecine & pharmacotherapie. 2023, 165, 115218. [Google Scholar] [CrossRef]
- Huang, W.; Huang, J.; Huang, N.; Luo, Y. The role of TREM2 in Alzheimer’s disease: From the perspective of Tau. Front. Cell Dev. Biol. 2023, 11, 1280257. [Google Scholar] [CrossRef]
- Becher, A.; Drenckhahn, A.; Pahner, I.; Ahnert-Hilger, G. The synaptophysin-synaptobrevin complex is developmentally upregulated in cultivated neurons but is absent in neuroendocrine cells. Eur. J. Cell Biol. 1999, 78, 650–656. [Google Scholar] [CrossRef]
- Flemming, A. Stroke: Can PSD95 inhibitors widen the therapeutic window? Nat. Rev. Drug Discov. 2012, 11, 272. [Google Scholar] [CrossRef]
- Dore, K.; Carrico, Z.; Alfonso, S.; Marino, M.; Koymans, K.; Kessels, H.W.; Malinow, R. PSD-95 protects synapses from β-amyloid. Cell Rep. 2021, 35, 109194. [Google Scholar] [CrossRef]
- Garcia, I.J.P.; Kinoshita, P.F.; Valadares, J.M.M.; Carvalho, L.E.D.; Cortes, V.F.; Barbosa, L.A.; Scavone, C.; Santos, H.L. Effect of Ouabain on Glutamate Transport in the Hippocampus of Rats with LPS-Induced Neuroinflammation. Biomedicines 2023, 11, 920. [Google Scholar] [CrossRef] [PubMed]
- Rajanathan, R.; Pedersen, T.M.; Guldbrandsen, H.O.; Olesen, L.F.; Thomsen, M.B.; Bøtker, H.E.; Matchkov, V.V. Augmented Ouabain-Induced Vascular Response Reduces Cardiac Efficiency in Mice with Migraine-Associated Mutation in the Na(+), K(+)-ATPase α(2)-Isoform. Biomedicines 2023, 11, 344. [Google Scholar] [CrossRef] [PubMed]



| Gene Name | Forward Primer (5′->3′) | Reverse Primer (5′->3′) |
|---|---|---|
| TREM2 | GCCTTCCTGAAGAAGCGGAA | GAGTGATGGTGACGGTTCC |
| TNF-α | CTGAACTTCGGGGTGATCGG | GGCTTGTCACTCGAATTTTGAGA |
| IL-1β | GAAATGCCACCTTTTGACAGTG | TGGATGCTCTCATCAGGACAG |
| IL-4 | GGTCTCAACCCCCAGCTAGT | GCCGATGATCTCTCTCAAGTGAT |
| IL-10 | CTTACTGACTGGCATGAGGATCA | GCAGCTCTAGGAGCATGTGG |
| GAPDH | AAGAAGGTGGTGAAGCAGG | GAAGGTGGAAGAGTGGGAGT |
| Component | Volumetric (20 μL) |
|---|---|
| cDNA | 2 μL |
| Forward primer | 0.4 μL |
| Universal miRNA qPCR Primer | 0.4 μL |
| PerfectStartTM Green qPCR SuperMix | 10 μL |
| RNAase free Water | Make up to 20 μL |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, D.; Liu, J.; Zhu, Q.; Wei, X.; Zhang, X.; Chen, Q.; Zhao, Y.; Tang, H.; Xu, W. Ouabain Ameliorates Alzheimer’s Disease-Associated Neuropathology and Cognitive Impairment in FAD4T Mice. Nutrients 2024, 16, 3558. https://doi.org/10.3390/nu16203558
Wang D, Liu J, Zhu Q, Wei X, Zhang X, Chen Q, Zhao Y, Tang H, Xu W. Ouabain Ameliorates Alzheimer’s Disease-Associated Neuropathology and Cognitive Impairment in FAD4T Mice. Nutrients. 2024; 16(20):3558. https://doi.org/10.3390/nu16203558
Chicago/Turabian StyleWang, Dan, Jiajia Liu, Qizhi Zhu, Xin Wei, Xiang Zhang, Qi Chen, Yu Zhao, Heng Tang, and Weiping Xu. 2024. "Ouabain Ameliorates Alzheimer’s Disease-Associated Neuropathology and Cognitive Impairment in FAD4T Mice" Nutrients 16, no. 20: 3558. https://doi.org/10.3390/nu16203558
APA StyleWang, D., Liu, J., Zhu, Q., Wei, X., Zhang, X., Chen, Q., Zhao, Y., Tang, H., & Xu, W. (2024). Ouabain Ameliorates Alzheimer’s Disease-Associated Neuropathology and Cognitive Impairment in FAD4T Mice. Nutrients, 16(20), 3558. https://doi.org/10.3390/nu16203558







