The Impact of Fermented Milk Products Containing Bifidobacterium longum BB536 on the Gut Environment: A Randomized Double-Blind Placebo-Controlled Trial
Highlights
- Consumption of fermented milk containing BB536 increases the relative abundance of Faecalibacterium.
- Consumption of fermented milk containing BB536 affects the concentrations of Tryptophan metabolites.
- The number of viable BB536 bacteria in the gut is positively correlated with the abundance of Faecalibacterium and Tryptophan metabolites.
- The alpha diversity of fecal microbiota affects the survival rate of BB536.
Abstract
:1. Introduction
2. Materials and Methods
2.1. Clinical Trial Design
2.2. Bacterial Strains and Culture Conditions
2.3. 16S rRNA Gene Analysis
2.4. Faecalibacterium-Specific Quantitative PCR Analysis
2.5. Metabolomic Analysis of Fecal Metabolites
2.5.1. Chemicals and Reagents
2.5.2. Extraction of Metabolites from Fecal Samples
2.5.3. Liquid Chromatography–Tandem Mass Spectrometry (LC-MS/MS) Analysis
2.5.4. HPLC Analysis
2.6. Statistical Analysis
2.7. Data Availability
3. Results
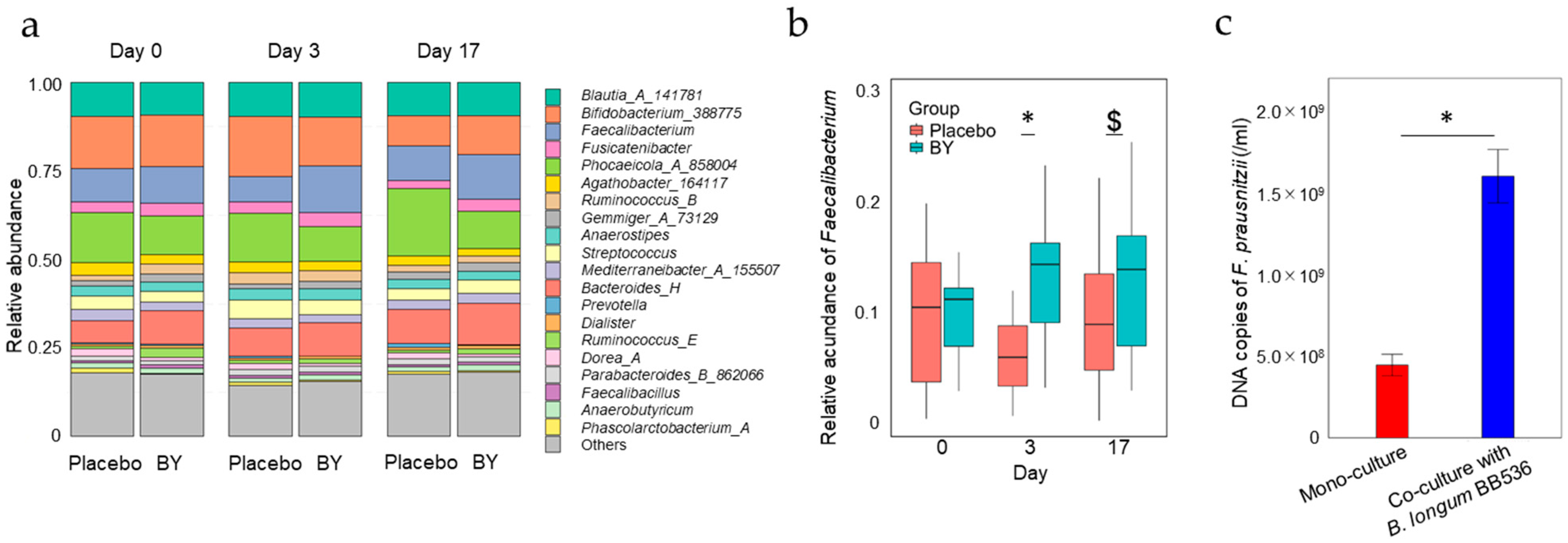
3.1. Differences in Fecal Microbiota Composition Between the BY and Placebo Groups
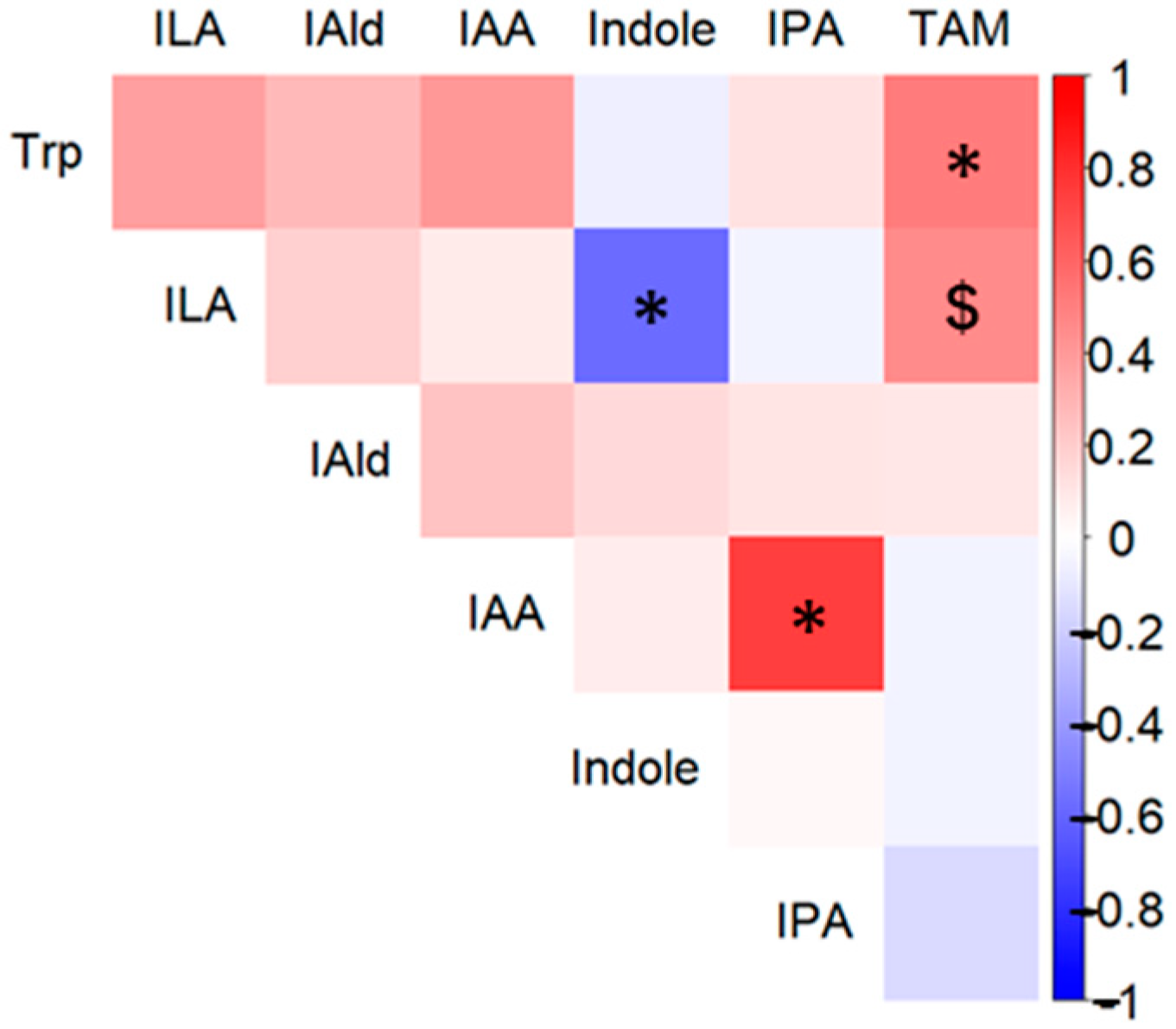
3.2. Differences in Fecal Metabolite Abundance Between the BY and Placebo Groups
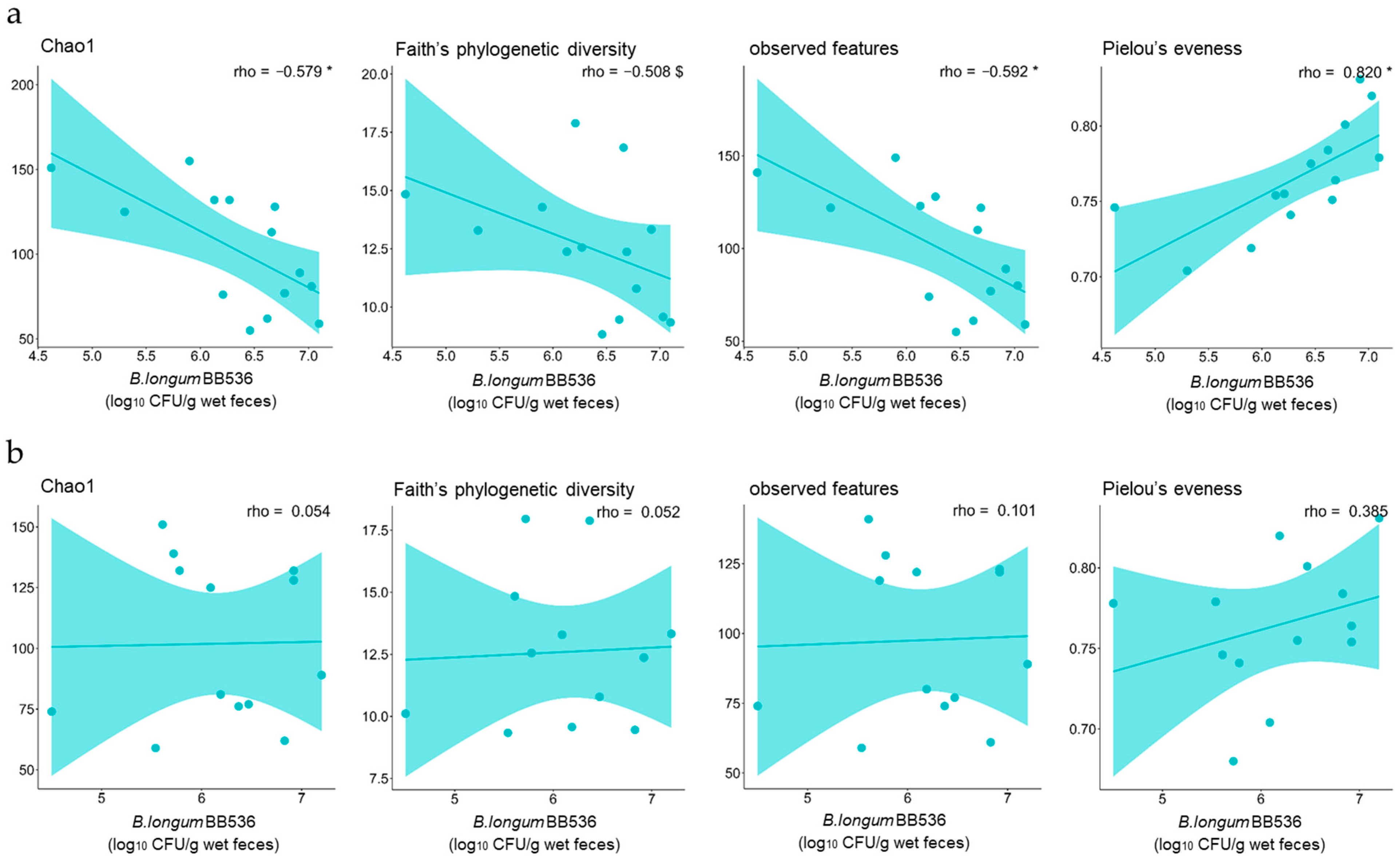
3.3. Correlations Between the Number of B. longum BB536 Bacteria That Survived Digestion and the Diversity of the Fecal Microbiota
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Daniel, N.; Lécuyer, E.; Chassaing, B. Host/microbiota interactions in health and diseases-Time for mucosal microbiology! Mucosal Immunol. 2021, 14, 1006–1016. [Google Scholar] [CrossRef] [PubMed]
- Di Tommaso, N.; Gasbarrini, A.; Ponziani, F.R. Intestinal barrier in human health and disease. Int. J. Environ. Res. Public Health 2021, 18, 12836. [Google Scholar] [CrossRef] [PubMed]
- Bohan, R.; Tianyu, X.; Tiantian, Z.; Ruonan, F.; Hongtao, H.; Qiong, W.; Chao, S. Gut microbiota: A potential manipulator for host adipose tissue and energy metabolism. J. Nutr. Biochem. 2019, 64, 206–217. [Google Scholar] [CrossRef]
- Yoo, J.Y.; Groer, M.; Dutra, S.V.O.; Sarkar, A.; McSkimming, D.I. Gut microbiota and immune system interactions. Microorganisms 2020, 8, 1587. [Google Scholar] [CrossRef] [PubMed]
- Margolis, K.G.; Cryan, J.F.; Mayer, E.A. The microbiota-gut-brain axis: From motility to mood. Gastroenterology 2022, 160, 1486–1501. [Google Scholar] [CrossRef]
- Levy, M.; Kolodziejczyk, A.A.; Thaiss, C.A.; Elinav, E. Dysbiosis and the immune system. Nat. Rev. Immunol. 2017, 17, 219–232. [Google Scholar] [CrossRef]
- Markowiak, P.; Śliżewska, K. Effects of probiotics, prebiotics, and synbiotics on human health. Nutrients 2017, 9, 1021. [Google Scholar] [CrossRef]
- Wong, C.B.; Odamaki, T.; Xiao, J.-Z. Insights into the reason of Human-Residential Bifidobacteria (HRB) being the natural inhabitants of the human gut and their potential health-promoting benefits. FEMS Microbiol. Rev. 2020, 44, 369–385. [Google Scholar] [CrossRef]
- Nakamura, Y.; Suzuki, S.; Murakami, S.; Nishimoto, Y.; Higashi, K.; Watarai, N.; Umetsu, J.; Ishii, C.; Ito, Y.; Mori, Y.; et al. Integrated gut microbiome and metabolome analyses identified fecal biomarkers for bowel movement regulation by Bifidobacterium longum BB536 supplementation: A RCT. Comput. Struct. Biotechnol. J. 2022, 20, 5847–5858. [Google Scholar] [CrossRef]
- Odamaki, T.; Xiao, J.-Z.; Iwabuchi, N.; Sakamoto, M.; Takahashi, N.; Kondo, S.; Miyaji, K.; Iwatsuki, K.; Togashi, H.; Enomoto, T.; et al. Influence of Bifidobacterium longum BB536 intake on faecal microbiota in individuals with Japanese cedar pollinosis during the pollen season. J. Med. Microbiol. 2007, 56, 1301–1308. [Google Scholar] [CrossRef]
- Li, Y.; Arai, S.; Kato, K.; Iwabuchi, S.; Iwabuchi, N.; Muto, N.; Motobayashi, H.; Ebihara, S.; Tanaka, M.; Hashimoto, S. The potential immunomodulatory effect of Bifidobacterium longum subsp. longum BB536 on healthy adults through plasmacytoid dendritic cell activation in the peripheral blood. Nutrients 2023, 16, 42. [Google Scholar] [CrossRef] [PubMed]
- Sato, S.; Arai, S.; Kato, K.; Yoshida, K.; Iwabuchi, N.; Sagami, T.; Tanaka, M. Effects of Bifidobacterium longum BB536 and Bifidobacterium breve MCC1274 on body composition in normal and overweight adults in randomized placebo-controlled study. Nutrients 2024, 16, 815. [Google Scholar] [CrossRef] [PubMed]
- Yong, C.C.; Sakurai, T.; Kaneko, H.; Horigome, A.; Mitsuyama, E.; Nakajima, A.; Katoh, T.; Sakanaka, M.; Abe, T.; Xiao, J.-Z.; et al. Human gut-associated Bifidobacterium species salvage exogenous indole, a uremic toxin precursor, to synthesize indole-3-lactic acid via tryptophan. Gut Microbes 2024, 16, 2347728. [Google Scholar] [CrossRef]
- Starke, S.; Harris, D.M.M.; Zimmermann, J.; Schuchardt, S.; Oumari, M.; Frank, D.; Bang, C.; Rosenstiel, P.; Schreiber, S.; Frey, N.; et al. Amino acid auxotrophies in human gut bacteria are linked to higher microbiome diversity and long-term stability. ISME J. 2023, 17, 2370–2380. [Google Scholar] [CrossRef]
- Sun, M.; Ma, N.; He, T.; Johnston, L.J.; Ma, X. Tryptophan (Trp) modulates gut homeostasis via aryl hydrocarbon receptor (AhR). Crit. Rev. Food Sci. Nutr. 2019, 60, 1760–1768. [Google Scholar] [CrossRef] [PubMed]
- Illés, P.; Krasulová, K.; Vyhlídalová, B.; Poulíková, K.; Marcalíková, A.; Pečinková, P.; Sirotová, N.; Vrzal, R.; Mani, S.; Dvořák, Z. Indole microbial intestinal metabolites expand the repertoire of ligands and agonists of the human pregnane X receptor. Toxicol. Lett. 2020, 334, 87–93. [Google Scholar] [CrossRef]
- Sakurai, T.; Horigome, A.; Odamaki, T.; Shimizu, T.; Xiao, J.-Z. Production of hydroxycarboxylic acid receptor 3 (HCA3) ligands by Bifidobacterium. Microorganisms 2021, 9, 2397. [Google Scholar] [CrossRef]
- Sen, A.; Kimura, M.; Ejima, R.; Arai, S.; Mitsuyama, E.; Kaneko, H.; Mishima, R.; Muto, N.; Hiraku, A.; Kato, K.; et al. Assessing probiotic viability in the gastrointestinal tract in randomized placebo controlled trial: A combined approach of molecular biology and novel cultivation techniques. Benef. Microbes, 2024; submitted. [Google Scholar]
- Martín, R.; Rios-Covian, D.; Huillet, E.; Auger, S.; Khazaal, S.; Bermúdez-Humarán, L.G.; Sokol, H.; Chatel, J.-M.; Langella, P. Faecalibacterium: A bacterial genus with promising human health applications. FEMS Microbiol. Rev. 2023, 47, 1–18. [Google Scholar] [CrossRef]
- McCann, J.R.; Rawls, J.F. Essential amino acid metabolites as chemical mediators of host-microbe interaction in the gut. Annu. Rev. Microbiol. 2023, 77, 479–497. [Google Scholar] [CrossRef]
- Murakami, R.; Hashikura, N.; Yoshida, K.; Xiao, J.-Z.; Odamaki, T. Growth-promoting effect of alginate on Faecalibacterium prausnitzii through cross-feeding with Bacteroides. Food Res. Int. 2021, 144, 110326. [Google Scholar] [CrossRef]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef] [PubMed]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef] [PubMed]
- Sánchez, B.; Delgado, S.; Blanco-Míguez, A.; Lourenço, A.; Gueimonde, M.; Margolles, A. Probiotics, gut microbiota, and their influence on host health and disease. Mol. Nutr. Food Res. 2016, 61, 1600240. [Google Scholar] [CrossRef] [PubMed]
- Galdeano, C.M.; Cazorla, S.I.; Dumit, J.M.L.; Vélez, E.; Perdigón, G. Beneficial effects of probiotic consumption on the immune system. Ann. Nutr. Metab. 2019, 74, 115–124. [Google Scholar] [CrossRef]
- Wieërs, G.; Belkhir, L.; Enaud, R.; Leclercq, S.; de Foy, J.-M.P.; Dequenne, I.; de Timary, P.; Cani, P.D. How probiotics affect the microbiota. Front. Cell. Infect. Microbiol. 2020, 9, 454. [Google Scholar] [CrossRef]
- Markowiak-Kopeć, P.; Śliżewska, K. The effect of probiotics on the production of short-chain fatty acids by human intestinal microbiome. Nutrients 2020, 12, 1107. [Google Scholar] [CrossRef]
- Lau, A.S.Y.; Yanagisawa, N.; Hor, Y.Y.; Lew, L.C.; Ong, J.S.; Chuah, L.O.; Lee, Y.Y.; Choi, S.B.; Rashid, F.; Wahid, N.; et al. Bifidobacterium longum BB536 alleviated upper respiratory illnesses and modulated gut microbiota profiles in Malaysian pre-school children. Benef. Microbes 2018, 9, 61–70. [Google Scholar] [CrossRef]
- Ramirez-Farias, C.; Slezak, K.; Fuller, Z.; Duncan, A.; Holtrop, G.; Louis, P. Effect of inulin on the human gut microbiota: Stimulation of Bifidobacterium adolescentis and Faecalibacterium prausnitzii. Br. J. Nutr. 2008, 101, 541–550. [Google Scholar] [CrossRef]
- Louis, P.; Young, P.; Holtrop, G.; Flint, H.J. Diversity of human colonic butyrate-producing bacteria revealed by analysis of the butyryl-CoA:acetate CoA-transferase gene. Environ. Microbiol. 2010, 12, 304–314. [Google Scholar] [CrossRef]
- Zhou, L.; Zhang, M.; Wang, Y.; Dorfman, R.G.; Liu, H.; Yu, T.; Chen, X.; Tang, D.; Xu, L.; Yin, Y.; et al. Faecalibacterium prausnitzii produces butyrate to maintain Th17/Treg balance and to ameliorate colorectal colitis by inhibiting histone deacetylase 1. Inflamm. Bowel Dis. 2018, 24, 1926–1940. [Google Scholar] [CrossRef]
- Zhuang, M.; Shang, W.; Ma, Q.; Strappe, P.; Zhou, Z. Abundance of probiotics and butyrate-production microbiome manages constipation via short-chain fatty acids production and hormones secretion. Mol. Nutr. Food Res. 2019, 63, 1801187. [Google Scholar] [CrossRef]
- Zagato, E.; Pozzi, C.; Bertocchi, A.; Schioppa, T.; Saccheri, F.; Guglietta, S.; Fosso, B.; Melocchi, L.; Nizzoli, G.; Troisi, J.; et al. Endogenous murine microbiota member Faecalibaculum rodentium and its human homologue protect from intestinal tumour growth. Nat. Microbiol. 2020, 5, 511–524. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Li, G.; Zhao, W.; Wang, X.; He, J.; Zhou, L.; Zhang, X.; An, P.; Liu, Y.; Zhang, C.; et al. Efficacy of Bifidobacterium animalis subsp. lactis BL-99 in the treatment of functional dyspepsia: A randomized placebo-controlled clinical trial. Nat. Commun. 2024, 15, 227. [Google Scholar] [CrossRef]
- Touchefeu, Y.; Duchalais, E.; des Varannes, S.B.; Alameddine, J.; Mirallie, E.; Matysiak-Budnik, T.; Le Bastard, Q.; Javaudin, F.; Rimbert, M.; Jotereau, F.; et al. Concomitant decrease of double-positive lymphocyte population CD4CD8αα and Faecalibacterium prausnitzii in patients with colorectal cancer. Eur. J. Gastroenterol. Hepatol. 2020, 32, 149–156. [Google Scholar] [CrossRef]
- Gupta, S.K.; Vyavahare, S.; Blanes, I.L.D.; Berger, F.; Isales, C.; Fulzele, S. Microbiota-derived tryptophan metabolism: Impacts on health, aging, and disease. Exp. Gerontol. 2023, 183, 112319. [Google Scholar] [CrossRef]
- Laursen, M.F.; Sakanaka, M.; von Burg, N.; Mörbe, U.; Andersen, D.; Moll, J.M.; Pekmez, C.T.; Rivollier, A.; Michaelsen, K.F.; Mølgaard, C.; et al. Bifidobacterium species associated with breastfeeding produce aromatic lactic acids in the infant gut. Nat. Microbiol. 2021, 6, 1367–1382. [Google Scholar] [CrossRef] [PubMed]
- Henrick, B.M.; Rodriguez, L.; Lakshmikanth, T.; Pou, C.; Henckel, E.; Arzoomand, A.; Olin, A.; Wang, J.; Mikes, J.; Tan, Z.; et al. Bifidobacteria-mediated immune system imprinting early in life. Cell 2021, 184, 3884–3898.e11. [Google Scholar] [CrossRef] [PubMed]
- Sakurai, T.; Odamaki, T.; Xiao, J.-Z. Production of indole-3-lactic acid by Bifidobacterium strains isolated fromhuman infants. Microorganisms 2019, 7, 340. [Google Scholar] [CrossRef] [PubMed]
- Lamas, B.; Richard, M.L.; Leducq, V.; Pham, H.-P.; Michel, M.-L.; Da Costa, G.; Bridonneau, C.; Jegou, S.; Hoffmann, T.W.; Natividad, J.M.; et al. CARD9 impacts colitis by altering gut microbiota metabolism of tryptophan into aryl hydrocarbon receptor ligands. Nat. Med. 2016, 22, 598–605. [Google Scholar] [CrossRef]
- Hendrikx, T.; Duan, Y.; Wang, Y.; Oh, J.-H.; Alexander, L.M.; Huang, W.; Stärkel, P.; Ho, S.B.; Gao, B.; Fiehn, O.; et al. Bacteria engineered to produce IL-22 in intestine induce expression of REG3G to reduce ethanol-induced liver disease in mice. Gut 2019, 68, 1504–1515. [Google Scholar] [CrossRef]
- Zelante, T.; Iannitti, R.G.; Cunha, C.; De Luca, A.; Giovannini, G.; Pieraccini, G.; Zecchi, R.; D’Angelo, C.; Massi-Benedetti, C.; Fallarino, F.; et al. Tryptophan catabolites from microbiota engage aryl hydrocarbon receptor and balance mucosal reactivity via interleukin-22. Immunity 2013, 39, 372–385. [Google Scholar] [CrossRef] [PubMed]
- Scott, S.A.; Fu, J.; Chang, P.V. Microbial tryptophan metabolites regulate gut barrier function via the aryl hydrocarbon receptor. Proc. Natl. Acad. Sci. USA 2020, 117, 19376–19387. [Google Scholar] [CrossRef] [PubMed]
- Bröer, S. Intestinal amino acid transport and metabolic health. Annu. Rev. Nutr. 2023, 43, 73–99. [Google Scholar] [CrossRef]
- Xue, C.; Li, G.; Zheng, Q.; Gu, X.; Shi, Q.; Su, Y.; Chu, Q.; Yuan, X.; Bao, Z.; Lu, J.; et al. Tryptophan metabolism in health and disease. Cell Metab. 2023, 35, 1304–1326. [Google Scholar] [CrossRef]
- Siska, P.J.; Jiao, J.; Matos, C.; Singer, K.; Berger, R.S.; Dettmer, K.; Oefner, P.J.; Cully, M.D.; Wang, Z.; QuinnIii, W.J.; et al. Kynurenine induces T cell fat catabolism and has limited suppressive effects in vivo. eBioMedicine 2021, 74, 103734. [Google Scholar] [CrossRef]
- Wieckiewicz, M.; Martynowicz, H.; Lavigne, G.; Lobbezoo, F.; Kato, T.; Winocur, E.; Wezgowiec, J.; Danel, D.; Wojakowska, A.; Mazur, G.; et al. An exploratory study on the association between serotonin and sleep breathing disorders. Sci. Rep. 2023, 13, 11800. [Google Scholar] [CrossRef]
- Walden, K.E.; Moon, J.M.; Hagele, A.M.; Allen, L.E.; Gaige, C.J.; Krieger, J.M.; Jager, R.; Mumford, P.W.; Pane, M.; Kerksick, C.M. A randomized controlled trial to examine the impact of a multi-strain probiotic on self-reported indicators of depression, anxiety, mood, and associated biomarkers. Front. Nutr. 2023, 10, 1219313. [Google Scholar] [CrossRef] [PubMed]
- Pinto-Sanchez, M.I.; Hall, G.B.; Ghajar, K.; Nardelli, A.; Bolino, C.; Lau, J.T.; Martin, F.-P.; Cominetti, O.; Welsh, C.; Rieder, A.; et al. Probiotic Bifidobacterium longum NCC3001 reduces depression scores and alters brain activity: A pilot study in patients with irritable bowel syndrome. Gastroenterology 2017, 153, 448–459.e8. [Google Scholar] [CrossRef]
- Su, X.; Gao, Y.; Yang, R. Gut microbiota-derived tryptophan metabolites maintain gut and systemic homeostasis. Cells 2022, 11, 2296. [Google Scholar] [CrossRef]
- Koper, J.E.; Troise, A.D.; Loonen, L.M.; Vitaglione, P.; Capuano, E.; Fogliano, V.; Wells, J.M. Tryptophan supplementation increases the production of microbial-derived AhR agonists in an in vitro simulator of intestinal microbial ecosystem. J. Agric. Food Chem. 2022, 70, 3958–3968. [Google Scholar] [CrossRef]
- Tian, P.; Chen, Y.; Zhu, H.; Wang, L.; Qian, X.; Zou, R.; Zhao, J.; Zhang, H.; Qian, L.; Wang, Q.; et al. Bifidobacterium breve CCFM1025 attenuates major depression disorder via regulating gut microbiome and tryptophan metabolism: A randomized clinical trial. Brain Behav. Immun. 2022, 100, 233–241. [Google Scholar] [CrossRef] [PubMed]
- Sharma, V.; Rodionov, D.A.; Leyn, S.A.; Tran, D.; Iablokov, S.N.; Ding, H.; Peterson, D.A.; Osterman, A.L.; Peterson, S.N. B-vitamin sharing promotes stability of gut microbial communities. Front. Microbiol. 2019, 10, 1485. [Google Scholar] [CrossRef] [PubMed]
- van Elsas, J.D.; Chiurazzi, M.; Mallon, C.A.; Elhottova, D.; Kristufek, V.; Salles, J.F. Microbial diversity determines the invasion of soil by a bacterial pathogen. Proc. Natl. Acad. Sci. USA 2012, 109, 1159–1164. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ejima, R.; Mishima, R.; Sen, A.; Yamaguchi, K.; Mitsuyama, E.; Kaneko, H.; Kimura, M.; Arai, S.; Muto, N.; Hiraku, A.; et al. The Impact of Fermented Milk Products Containing Bifidobacterium longum BB536 on the Gut Environment: A Randomized Double-Blind Placebo-Controlled Trial. Nutrients 2024, 16, 3580. https://doi.org/10.3390/nu16213580
Ejima R, Mishima R, Sen A, Yamaguchi K, Mitsuyama E, Kaneko H, Kimura M, Arai S, Muto N, Hiraku A, et al. The Impact of Fermented Milk Products Containing Bifidobacterium longum BB536 on the Gut Environment: A Randomized Double-Blind Placebo-Controlled Trial. Nutrients. 2024; 16(21):3580. https://doi.org/10.3390/nu16213580
Chicago/Turabian StyleEjima, Ryuta, Riko Mishima, Akira Sen, Kana Yamaguchi, Eri Mitsuyama, Hiroki Kaneko, Madoka Kimura, Satoshi Arai, Natsumi Muto, Akari Hiraku, and et al. 2024. "The Impact of Fermented Milk Products Containing Bifidobacterium longum BB536 on the Gut Environment: A Randomized Double-Blind Placebo-Controlled Trial" Nutrients 16, no. 21: 3580. https://doi.org/10.3390/nu16213580
APA StyleEjima, R., Mishima, R., Sen, A., Yamaguchi, K., Mitsuyama, E., Kaneko, H., Kimura, M., Arai, S., Muto, N., Hiraku, A., Kato, K., Kuwano, Y., Maruyama, H., Nakamura, M., Iwabuchi, N., Nakano, M., Odamaki, T., & Tanaka, M. (2024). The Impact of Fermented Milk Products Containing Bifidobacterium longum BB536 on the Gut Environment: A Randomized Double-Blind Placebo-Controlled Trial. Nutrients, 16(21), 3580. https://doi.org/10.3390/nu16213580





