Cornflower Extract and Its Active Components Alleviate Dexamethasone-Induced Muscle Wasting by Targeting Cannabinoid Receptors and Modulating Gut Microbiota
Abstract
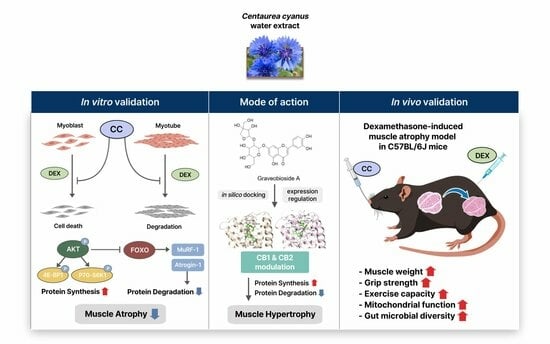
:1. Introduction
2. Materials and Methods
2.1. Plant Materials and Extraction
2.2. Isolation and Identification of CC-Derived Compounds
2.3. High-Performance Liquid Chromatography (HPLC) Profiling
2.4. Chemicals and Apparatus
2.5. C2C12 Cell Differentiation and Cytotoxicity Assay
2.6. Quantification of Myofiber
2.7. Detection of Reduced GSH Measurement
2.8. Mitotracker Assay
2.9. Determination of Cellular ATP Content
2.10. Measurement of ROS Production
2.11. Computational Docking
2.12. Mouse Model of DEX-Induced Muscle Atrophy
2.13. Grip Strength and Exercise Performance Test
2.14. Histological Analysis of Skeletal Muscle
2.15. Determination of Muscle ATP and Aconitase Contents
2.16. RNA Extraction and Quantitative Reverse Transcription–Polymerase Chain Reaction (qRT-PCR)
2.17. Western Blot Analysis
2.18. 16S rRNA Gene Sequencing in Cecum Samples
2.19. Statistical Analyses
3. Results
3.1. CC Demonstrates Protective Activity against DEX-Induced Atrophy in C2C12 Myotubes
3.2. CC Mitigates DEX-Induced Oxidative Stress and Boosts Mitochondrial Content in C2C12 Myotubes
3.3. Chemical Characterization of CC Reveals a Diverse Profile of Bioactive Phytochemicals
3.4. Multiple Compounds Isolated from CC Exhibit Anti-Atrophic, Antioxidant, and ATP-Stimulatory Properties in DEX-Treated C2C12 Myotubes
3.5. Graveobioside A Exerts Muscle-Protective Effects through Modulatory Action on Cannabinoid Receptors CB1 and CB2
3.6. CC Ameliorates DEX-Induced Muscle Atrophy in a Murine Mode
3.7. CC Enhances Exercise Capacity and Promotes Mitochondrial Function in Skeletal Muscle
3.8. CC Modulates Gut Microbial Diversity and Composition, Reversing DEX-Induced Dysbiosis
3.9. Correlation Analysis Suggests a Potential Role for Clostridium Sensu Stricto 1 in Mediating the Muscle-Protective and Performance-Enhancing Effects of CC
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
Appendix A




References
- Gonzalez-Freire, M.; Semba, R.D.; Ubaida-Mohien, C.; Fabbri, E.; Scalzo, P.; Højlund, K.; Dufresne, C.; Lyashkov, A.; Ferrucci, L. The Human Skeletal Muscle Proteome Project: A reappraisal of the current literature. J. Cachexia Sarcopenia Muscle 2017, 8, 5–18. [Google Scholar] [CrossRef] [PubMed]
- Anjanappa, M.; Corden, M.; Green, A.; Roberts, D.; Hoskin, P.; McWilliam, A.; Choudhury, A. Sarcopenia in cancer: Risking more than muscle loss. Tech. Innov. Patient Support Radiat. Oncol. 2020, 16, 50–57. [Google Scholar] [CrossRef] [PubMed]
- Larsson, L.; Degens, H.; Li, M.; Salviati, L.; Lee, Y.I.; Thompson, W.; Kirkland, J.L.; Sandri, M. Sarcopenia: Aging-Related Loss of Muscle Mass and Function. Physiol. Rev. 2019, 99, 427–511. [Google Scholar] [CrossRef] [PubMed]
- Klein, G.L. The effect of glucocorticoids on bone and muscle. Osteoporos. Sarcopenia 2015, 1, 39–45. [Google Scholar] [CrossRef] [PubMed]
- Kuo, T.; Harris, C.A.; Wang, J.C. Metabolic functions of glucocorticoid receptor in skeletal muscle. Mol. Cell. Endocrinol. 2013, 380, 79–88. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.J.; Xiao, J.J.; Liu, L.; Jiao, H.C.; Lin, H. Excessive glucocorticoid-induced muscle MuRF1 overexpression is independent of Akt/FoXO1 pathway. Biosci. Rep. 2017, 37, BSR20171056. [Google Scholar] [CrossRef] [PubMed]
- Latres, E.; Amini, A.R.; Amini, A.A.; Griffiths, J.; Martin, F.J.; Wei, Y.; Lin, H.C.; Yancopoulos, G.D.; Glass, D.J. Insulin-like Growth Factor-1 (IGF-1) Inversely Regulates Atrophy-induced Genes via the Phosphatidylinositol 3-Kinase/Akt/Mammalian Target of Rapamycin (PI3K/Akt/mTOR) Pathway. J. Biol. Chem. 2005, 280, 2737–2744. [Google Scholar] [CrossRef]
- Liu, J.; Peng, Y.; Wang, X.; Fan, Y.; Qin, C.; Shi, L.; Tang, Y.; Cao, K.; Li, H.; Long, J.; et al. Mitochondrial Dysfunction Launches Dexamethasone-Induced Skeletal Muscle Atrophy via AMPK/FOXO3 Signaling. Mol. Pharm. 2016, 13, 73–84. [Google Scholar] [CrossRef]
- Shen, S.; Liao, Q.; Liu, J.; Pan, R.; Lee, S.M.; Lin, L. Myricanol rescues dexamethasone-induced muscle dysfunction via a sirtuin 1-dependent mechanism. J. Cachexia Sarcopenia Muscle 2019, 10, 429–444. [Google Scholar] [CrossRef]
- Park, S.S.; Kwon, E.S.; Kwon, K.S. Molecular mechanisms and therapeutic interventions in sarcopenia. Osteoporos. Sarcopenia 2017, 3, 117–122. [Google Scholar] [CrossRef]
- Grosicki, G.J.; Fielding, R.A.; Lustgarten, M.S. Gut Microbiota Contribute to Age-Related Changes in Skeletal Muscle Size, Composition, and Function: Biological Basis for a Gut-Muscle Axis. Calcif. Tissue Int. 2018, 102, 433–442. [Google Scholar] [CrossRef]
- Liu, C.; Cheung, W.H.; Li, J.; Chow, S.K.; Yu, J.; Wong, S.H.; Ip, M.; Sung, J.J.Y.; Wong, R.M.Y. Understanding the gut microbiota and sarcopenia: A systematic review. J. Cachexia Sarcopenia Muscle 2021, 12, 1393–1407. [Google Scholar] [CrossRef] [PubMed]
- Mancin, L.; Wu, G.D.; Paoli, A. Gut microbiota–bile acid–skeletal muscle axis. Trends Microbiol. 2023, 31, 254–269. [Google Scholar] [CrossRef] [PubMed]
- Baek, J.S.; Shin, Y.J.; Ma, X.; Park, H.S.; Hwang, Y.H.; Kim, D.H. Bifidobacterium bifidum and Lactobacillus paracasei alleviate sarcopenia and cognitive impairment in aged mice by regulating gut microbiota-mediated AKT, NF-κB, and FOXO3a signaling pathways. Immun. Ageing 2023, 20, 56. [Google Scholar] [CrossRef]
- Lee, K.; Kim, J.; Park, S.-D.; Shim, J.-J.; Lee, J.-L. Lactobacillus plantarum HY7715 ameliorates sarcopenia by improving skeletal muscle mass and function in aged Balb/c mice. Int. J. Mol. Sci. 2021, 22, 10023. [Google Scholar] [CrossRef]
- Prado, C.M.; Orsso, C.E.; Pereira, S.L.; Atherton, P.J.; Deutz, N.E.P. Effects of β-hydroxy β-methylbutyrate (HMB) supplementation on muscle mass, function, and other outcomes in patients with cancer: A systematic review. J. Cachexia Sarcopenia Muscle 2022, 13, 1623–1641. [Google Scholar] [CrossRef]
- Bruneton, J. Pharmacognosy, Phytochemistry, Medicinal Plants; Lavoisier Publishing: Paris, France, 1995. [Google Scholar]
- Garbacki, N.; Gloaguen, V.; Damas, J.; Bodart, P.; Tits, M.; Angenot, L. Anti-inflammatory and immunological effects of Centaurea cyanus flower-heads. J. Ethnopharmacol. 1999, 68, 235–241. [Google Scholar] [CrossRef] [PubMed]
- Mirzaie, A.; Zare Karizi, S. Study of chemical composition and characteristics of Centurea cyanus extract on colon cancer cell line and analysis of apoptosis gene expression. Tehran Univ. Med. Sci. J. 2016, 74, 626–634. [Google Scholar]
- Pirvu, L.; Bubueanu, C.; Panteli, M.; Petcu, L.; Coprean, D. Centaurea cyanus L. polysaccharides and polyphenols cooperation in achieving strong rat gastric ulcer protection. Open Chem. 2015, 13, 000010151520150102. [Google Scholar] [CrossRef]
- Keyvanloo Shahrestanaki, M.; Bagheri, M.; Ghanadian, M.; Aghaei, M.; Jafari, S.M. Centaurea cyanus extracted 13-O-acetylsolstitialin A decrease Bax/Bcl-2 ratio and expression of cyclin D1/Cdk-4 to induce apoptosis and cell cycle arrest in MCF-7 and MDA-MB-231 breast cancer cell lines. J. Cell. Biochem. 2019, 120, 18309–18319. [Google Scholar] [CrossRef]
- Kim, Y.I.; Lee, H.; Nirmala, F.S.; Seo, H.-D.; Ha, T.Y.; Jung, C.H.; Ahn, J. Antioxidant Activity of Valeriana fauriei Protects against Dexamethasone-Induced Muscle Atrophy. Oxidative Med. Cell. Longev. 2022, 2022, 3645431. [Google Scholar] [CrossRef] [PubMed]
- Nair, A.B.; Jacob, S. A simple practice guide for dose conversion between animals and human. J. Basic Clin. Pharm. 2016, 7, 27–31. [Google Scholar] [CrossRef] [PubMed]
- Mula, J.; Lee, J.D.; Liu, F.; Yang, L.; Peterson, C.A. Automated image analysis of skeletal muscle fiber cross-sectional area. J. Appl. Physiol. 2013, 114, 148–155. [Google Scholar] [CrossRef]
- Yilmaz, P.; Parfrey, L.W.; Yarza, P.; Gerken, J.; Pruesse, E.; Quast, C.; Schweer, T.; Peplies, J.; Ludwig, W.; Glöckner, F.O. The SILVA and “All-species Living Tree Project (LTP)” taxonomic frameworks. Nucleic Acids Res. 2014, 42, D643–D648. [Google Scholar] [CrossRef]
- Sang, S.M.; Lapsley, K.; Jeong, W.S.; Lachance, P.A.; Ho, C.T.; Rosen, R.T. Antioxidative phenolic compounds isolated from almond skins (Prunus amygdalus batsch). J. Agric. Food Chem. 2002, 50, 2459–2463. [Google Scholar] [CrossRef] [PubMed]
- Wan, C.P.; Li, S.S.; Liu, L.; Chen, C.Y.; Fan, S.Y. Caffeoylquinic Acids from the Aerial Parts of Chrysanthemum coronarium L. Plants 2017, 6, 10. [Google Scholar] [CrossRef]
- Liu, G.Y.; Zhuang, L.W.; Song, D.D.; Lu, C.L.; Xu, X. Isolation, purification, and identification of the main phenolic compounds from leaves of celery (Apium graveolens L. var. dulce Mill./Pers.). J. Sep. Sci. 2017, 40, 472–479. [Google Scholar] [CrossRef]
- Mencherini, T.; Cau, A.; Bianco, G.; Della Loggia, R.; Aquino, R.P.; Autore, G. An extract of Apium graveolens var. dulce leaves: Structure of the major constituent, apiin, and its anti-inflammatory properties. J. Pharm. Pharmacol. 2007, 59, 891–897. [Google Scholar] [CrossRef]
- Tan, J.W.; Bednarek, P.; Liu, H.K.; Schneider, B.; Svatos, A.; Hahlbrock, K. Universally occurring phenylpropanoid and species-specific indolic metabolites in infected and uninfected Arabidopsis thaliana roots and leaves. Phytochemistry 2004, 65, 691–699. [Google Scholar] [CrossRef]
- Anh, L.T.T.; Son, N.T.; Van Tuyen, N.; Thuy, P.T.; Quan, P.M.; Ha, N.T.T.; Tra, N.T. Antioxidative and alpha-glucosidase inhibitory constituents of Polyscias guilfoylei: Experimental and computational assessments. Mol. Divers. 2022, 26, 229–243. [Google Scholar] [CrossRef]
- Ramírez-Orozco, R.E.; García-Ruiz, R.; Morales, P.; Villalón, C.M.; Villafán-Bernal, J.R.; Marichal-Cancino, B.A. Potential metabolic and behavioural roles of the putative endocannabinoid receptors GPR18, GPR55 and GPR119 in feeding. Curr. Neuropharmacol. 2019, 17, 947–960. [Google Scholar] [CrossRef] [PubMed]
- Zou, S.; Kumar, U. Cannabinoid Receptors and the Endocannabinoid System: Signaling and Function in the Central Nervous System. Int. J. Mol. Sci. 2018, 19, 833. [Google Scholar] [CrossRef] [PubMed]
- Dalle, S.; Schouten, M.; Ramaekers, M.; Koppo, K. The cannabinoid receptor 1 antagonist AM6545 stimulates the Akt-mTOR axis and in vivo muscle protein synthesis in a dexamethasone-induced muscle atrophy model. Mol. Cell. Endocrinol. 2023, 563, 111854. [Google Scholar] [CrossRef]
- O’Sullivan, S.E.; Yates, A.S.; Porter, R.K. The Peripheral Cannabinoid Receptor Type 1 (CB(1)) as a Molecular Target for Modulating Body Weight in Man. Molecules 2021, 26, 6178. [Google Scholar] [CrossRef] [PubMed]
- Ng, S.K.; Chung, D.J.; Chang, L.C.; Luo, C.K.; Jwo, S.H.; Lee, Y.H.; Lin, J.S.; Wang, C.H.; Wei, T.T. The protective effect of cannabinoids against colorectal cancer cachexia through modulation of inflammation and immune responses. Biomed. Pharmacother. 2023, 161, 114467. [Google Scholar] [CrossRef]
- Jun, L.; Robinson, M.; Geetha, T.; Broderick, T.L.; Babu, J.R. Prevalence and Mechanisms of Skeletal Muscle Atrophy in Metabolic Conditions. Int. J. Mol. Sci. 2023, 24, 2973. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Brault, J.J.; Schild, A.; Cao, P.; Sandri, M.; Schiaffino, S.; Lecker, S.H.; Goldberg, A.L. FoxO3 Coordinately Activates Protein Degradation by the Autophagic/Lysosomal and Proteasomal Pathways in Atrophying Muscle Cells. Cell Metab. 2007, 6, 472–483. [Google Scholar] [CrossRef] [PubMed]
- Goldbraikh, D.; Neufeld, D.; Eid-Mutlak, Y.; Lasry, I.; Gilda, J.E.; Parnis, A.; Cohen, S. USP1 deubiquitinates Akt to inhibit PI3K-Akt-FoxO signaling in muscle during prolonged starvation. EMBO Rep. 2020, 21, e48791. [Google Scholar] [CrossRef] [PubMed]
- Oh, H.-J.; Jin, H.; Kim, B.-Y.; Lee, O.-H.; Lee, B.-Y. A Combined Angelica gigas and Artemisia dracunculus Extract Prevents Dexamethasone-Induced Muscle Atrophy in Mice through the Akt/mTOR/FoxO3a Signaling Pathway. Cells 2022, 11, 3245. [Google Scholar] [CrossRef]
- Sandri, M.; Sandri, C.; Gilbert, A.; Skurk, C.; Calabria, E.; Picard, A.; Walsh, K.; Schiaffino, S.; Lecker, S.H.; Goldberg, A.L. Foxo transcription factors induce the atrophy-related ubiquitin ligase atrogin-1 and cause skeletal muscle atrophy. Cell 2004, 117, 399–412. [Google Scholar] [CrossRef]
- Zammit, P.S. Function of the myogenic regulatory factors Myf5, MyoD, Myogenin and MRF4 in skeletal muscle, satellite cells and regenerative myogenesis. Semin. Cell Dev. Biol. 2017, 72, 19–32. [Google Scholar] [CrossRef] [PubMed]
- Gayraud-Morel, B.; Chrétien, F.; Flamant, P.; Gomès, D.; Zammit, P.S.; Tajbakhsh, S. A role for the myogenic determination gene Myf5 in adult regenerative myogenesis. Dev. Biol. 2007, 312, 13–28. [Google Scholar] [CrossRef] [PubMed]
- Ganassi, M.; Badodi, S.; Wanders, K.; Zammit, P.S.; Hughes, S.M. Myogenin is an essential regulator of adult myofibre growth and muscle stem cell homeostasis. eLife 2020, 9, e60445. [Google Scholar] [CrossRef] [PubMed]
- Cerullo, F.; Gambassi, G.; Cesari, M. Rationale for antioxidant supplementation in sarcopenia. J. Aging Res. 2012, 2012, 316943. [Google Scholar] [CrossRef] [PubMed]
- Meister, A. Mitochondrial changes associated with glutathione deficiency. Biochim. Biophys. Acta (BBA)—Mol. Basis Dis. 1995, 1271, 35–42. [Google Scholar] [CrossRef]
- Hargreaves, M.; Spriet, L.L. Skeletal muscle energy metabolism during exercise. Nat. Metab. 2020, 2, 817–828. [Google Scholar] [CrossRef] [PubMed]
- Chistiakov, D.A.; Sobenin, I.A.; Revin, V.V.; Orekhov, A.N.; Bobryshev, Y.V. Mitochondrial aging and age-related dysfunction of mitochondria. BioMed Res. Int. 2014, 2014, 238463. [Google Scholar] [CrossRef]
- Charos, A.E.; Reed, B.D.; Raha, D.; Szekely, A.M.; Weissman, S.M.; Snyder, M. A highly integrated and complex PPARGC1A transcription factor binding network in HepG2 cells. Genome Res. 2012, 22, 1668–1679. [Google Scholar] [CrossRef]
- Alcazar Magana, A.; Kamimura, N.; Soumyanath, A.; Stevens, J.F.; Maier, C.S. Caffeoylquinic acids: Chemistry, biosynthesis, occurrence, analytical challenges, and bioactivity. Plant J. 2021, 107, 1299–1319. [Google Scholar] [CrossRef]
- Chen, X.; Yang, J.H.; Cho, S.S.; Kim, J.H.; Xu, J.; Seo, K.; Ki, S.H. 5-Caffeoylquinic acid ameliorates oxidative stress-mediated cell death via Nrf2 activation in hepatocytes. Pharm. Biol. 2020, 58, 999–1005. [Google Scholar] [CrossRef]
- Lee, H.; Kim, Y.I.; Nirmala, F.S.; Jeong, H.Y.; Seo, H.D.; Ha, T.Y.; Jung, C.H.; Ahn, J. Chrysanthemum zawadskil Herbich attenuates dexamethasone-induced muscle atrophy through the regulation of proteostasis and mitochondrial function. Biomed. Pharmacother. 2021, 136, 111226. [Google Scholar] [CrossRef]
- Dalle, S.; Schouten, M.; Meeus, G.; Slagmolen, L.; Koppo, K. Molecular networks underlying cannabinoid signaling in skeletal muscle plasticity. J. Cell. Physiol. 2022, 237, 3517–3540. [Google Scholar] [CrossRef] [PubMed]
- Iannotti, F.A.; Pagano, E.; Guardiola, O.; Adinolfi, S.; Saccone, V.; Consalvi, S.; Piscitelli, F.; Gazzerro, E.; Busetto, G.; Carrella, D.; et al. Genetic and pharmacological regulation of the endocannabinoid CB1 receptor in Duchenne muscular dystrophy. Nat. Commun. 2018, 9, 3950. [Google Scholar] [CrossRef] [PubMed]
- Ticinesi, A.; Nouvenne, A.; Cerundolo, N.; Catania, P.; Prati, B.; Tana, C.; Meschi, T. Gut Microbiota, Muscle Mass and Function in Aging: A Focus on Physical Frailty and Sarcopenia. Nutrients 2019, 11, 1633. [Google Scholar] [CrossRef] [PubMed]
- Ni, Y.; Yang, X.; Zheng, L.; Wang, Z.; Wu, L.; Jiang, J.; Yang, T.; Ma, L.; Fu, Z. Lactobacillus and Bifidobacterium Improves Physiological Function and Cognitive Ability in Aged Mice by the Regulation of Gut Microbiota. Mol. Nutr. Food Res. 2019, 63, e1900603. [Google Scholar] [CrossRef] [PubMed]
- Prokopidis, K.; Chambers, E.; Ni Lochlainn, M.; Witard, O.C. Mechanisms Linking the Gut-Muscle Axis with Muscle Protein Metabolism and Anabolic Resistance: Implications for Older Adults at Risk of Sarcopenia. Front. Physiol. 2021, 12, 770455. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.Y.; Lee, Y.; Lu, H.; Chou, C.H.; Wang, C. Analysis of gut microbiota and the effect of lauric acid against necrotic enteritis in Clostridium perfringens and Eimeria side-by-side challenge model. PLoS ONE 2019, 14, e0205784. [Google Scholar] [CrossRef] [PubMed]
- Li, C.-J.; Zhang, Z.; Zhan, P.-C.; Lv, A.-P.; Li, P.-P.; Liu, L.; Li, W.-J.; Yang, L.-L.; Zhi, X.-Y. Comparative genomic analysis and proposal of Clostridium yunnanense sp. nov., Clostridium rhizosphaerae sp. nov., and Clostridium paridis sp. nov., three novel Clostridium sensu stricto endophytes with diverse capabilities of acetic acid and ethanol production. Anaerobe 2023, 79, 102686. [Google Scholar] [CrossRef] [PubMed]
- Otsuka, R.; Zhang, S.; Furuya, K.; Tange, C.; Sala, G.; Ando, F.; Shimokata, H.; Nishita, Y.; Arai, H. Association between short-chain fatty acid intake and development of muscle strength loss among community-dwelling older Japanese adults. Exp. Gerontol. 2023, 173, 112080. [Google Scholar] [CrossRef]
- Guan, L.; Cao, Z.; Pan, Z.; Zhao, C.; Xue, M.; Yang, F.; Chen, J. Butyrate promotes C2C12 myoblast proliferation by activating ERK/MAPK pathway. Mol. Omics 2023, 19, 552–559. [Google Scholar] [CrossRef]
- Tang, G.; Du, Y.; Guan, H.; Jia, J.; Zhu, N.; Shi, Y.; Rong, S.; Yuan, W. Butyrate ameliorates skeletal muscle atrophy in diabetic nephropathy by enhancing gut barrier function and FFA2-mediated PI3K/Akt/mTOR signals. Br. J. Pharmacol. 2022, 179, 159–178. [Google Scholar] [CrossRef] [PubMed]









| Species | Target Gene | Direction | Primer Sequence (5′-3′) | Gene ID |
|---|---|---|---|---|
| Mouse | Fbxo32 | Forward | AACCCTTGGGCTTTGGGTTT | NM_026346.3 |
| Reverse | GGACTTAAGCCCGTGCAGAT | |||
| Myod1 | Forward | CATAGACTTGACAGGCCCCG | NM_010866.2 | |
| Reverse | CGGGTCCAGGTCCTCAAAAA | |||
| Myf6 | Forward | ACAGATCGTCGGAAAGCAGC | NM_008657.3 | |
| Reverse | CACTCCGCAGAATCTCCACC | |||
| Myog | Forward | AGCTATCCGGTTCCAAAGCC | NM_031189.2 | |
| Reverse | GCACAGGAGACCTTGGTCAG | |||
| Myh2 | Forward | AGCGAAGAGTAAGGCTGTCC | NM_001039545.2 | |
| Reverse | AGGCGCATGACCAAAGGTT | |||
| Ppargc1a | Forward | GTTGCCTGCATGAGTGTGTG | NM_008904.3 | |
| Reverse | CACATGTCCCAAGCCATCCA | |||
| Ucp3 | Forward | GTTTTGCGGACCTCCTCACT | NM_009464.3 | |
| Reverse | CTCTGTGCGCACCATAGTCA | |||
| Tomm20 | Forward | TGTGCGGTGTGTTGTCTGTT | NM_024214.2 | |
| Reverse | TAAGTGCCCAGAGCACAGGA | |||
| Nrf1 | Forward | CCCGTGTTCCTTTGTGGTGA | NM_001410231.1 | |
| Reverse | ATTCCATGCTCTGCTGCTGG | |||
| Tfam | Forward | GGGAATGTGGAGCGTGCTAA | NM_009360.4 | |
| Reverse | TGATAGACGAGGGGATGCGA | |||
| Gpx1 | Forward | AGTCCACCGTGTATGCCTTC | NM_001329527.1 | |
| Reverse | CCTCAGAGAGACGCGACATT | |||
| Sod1 | Forward | GGGAAGCATGGCGATGAAAG | NM_011434.2 | |
| Reverse | GCCTTCTGCTCGAAGTGGAT | |||
| Actb | Forward | CATTGCTGACAGGATGCAGAAGG | NM_007393.5 | |
| Reverse | TGCTGGAAGGTGGACAGTGAGG |
| Compounds | Docking Score (kcal mol−1) | |
|---|---|---|
| CB1 | CB2 | |
| Rimonabant (CB1 antagonist) | −13.215 | −12.170 |
| AM251 (CB1 antagonist) | −12.771 | −12.579 |
| GW-405833 (CB2 agonist) | −12.367 | −12.283 |
| CB65 (CB2 agonist) | −12.711 | −11.491 |
| Protocatechuic acid (1) | −7.006 | −8.002 |
| 3-O-caffeoylquinic acid (2) | −12.293 | −14.220 |
| 5-O-caffeoylquinic acid (3) | −11.642 | −13.024 |
| 4-O-caffeoylquinic acid (4) | −12.845 | −13.020 |
| Graveobioside A (5) | −17.714 | −19.098 |
| Apiin (6) | −14.865 | −18.306 |
| Vanillic acid (7) | −6.648 | −7.291 |
| 4-hydroxybenzoic acid (8) | −5.981 | −6.823 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nguyen, N.B.; Le, T.T.; Kang, S.W.; Cha, K.H.; Choi, S.; Youn, H.-Y.; Jung, S.H.; Kim, M. Cornflower Extract and Its Active Components Alleviate Dexamethasone-Induced Muscle Wasting by Targeting Cannabinoid Receptors and Modulating Gut Microbiota. Nutrients 2024, 16, 1130. https://doi.org/10.3390/nu16081130
Nguyen NB, Le TT, Kang SW, Cha KH, Choi S, Youn H-Y, Jung SH, Kim M. Cornflower Extract and Its Active Components Alleviate Dexamethasone-Induced Muscle Wasting by Targeting Cannabinoid Receptors and Modulating Gut Microbiota. Nutrients. 2024; 16(8):1130. https://doi.org/10.3390/nu16081130
Chicago/Turabian StyleNguyen, Ngoc Bao, Tam Thi Le, Suk Woo Kang, Kwang Hyun Cha, Sowoon Choi, Hye-Young Youn, Sang Hoon Jung, and Myungsuk Kim. 2024. "Cornflower Extract and Its Active Components Alleviate Dexamethasone-Induced Muscle Wasting by Targeting Cannabinoid Receptors and Modulating Gut Microbiota" Nutrients 16, no. 8: 1130. https://doi.org/10.3390/nu16081130