Effect of a Novel Food Rich in Miraculin on the Intestinal Microbiome of Malnourished Patients with Cancer and Dysgeusia
Abstract
:1. Introduction
2. Materials and Methods
2.1. Statement of Ethical Principles
2.2. Participants and Experimental Design
2.3. Sequencing of Biological Samples
2.3.1. Extraction of DNA
2.3.2. 16S rRNA Gene Sequencing and Taxonomic Assignment
2.4. Plasma Cytokines and Biochemical Parameters
2.5. Dietary Pattern Assessment
2.6. Short-Chain Fatty Acid Determination by Gas Chromatography/Mass Spectrometry
2.7. Electrical Taste Perception
2.8. Statistical Analysis
3. Results
3.1. Phylum Level
3.2. Genus Level
3.3. Species Level
3.4. Short-Chain Fatty Acids
3.5. Rivera-Pinto Test for Microbiome Balance
3.6. Analysis of the Relationships Among the Intestinal Microbiome, Nutritional Status, Electrical Taste Perception Inflammatory Cytokines, and Plasma Short-Chain Fatty Acids
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Brown, J.S.; Amend, S.R.; Austin, R.H.; Gatenby, R.A.; Hammarlund, E.U.; Pienta, K.J. Updating the Definition of Cancer. Mol. Cancer Res. 2023, 21, 1142–1147. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization (WHO). Cancer. Available online: https://www.who.int/health-topics/cancer#tab=tab_1 (accessed on 14 June 2024).
- FitzGerald, T.J.; Bishop-Jodoin, M.; Laurie, F.; Sacher, A.; Aghababian, R.V.; Dickson, E. Treatment Toxicity: Radiation. In Oncologic Emergency Medicine: Principles and Practice; Todd, K.H., Thomas, J.C.R., Eds.; Springer International Publishing: Cham, Switzerland, 2016; pp. 407–419. [Google Scholar] [CrossRef]
- Arends, J.; Baracos, V.; Bertz, H.; Bozzetti, F.; Calder, P.C.; Deutz, N.E.P.; Erickson, N.; Laviano, A.; Lisanti, M.P.; Lobo, D.N.; et al. ESPEN expert group recommendations for action against cancer-related malnutrition. Clin. Nutr. 2017, 36, 1187–1196. [Google Scholar] [CrossRef] [PubMed]
- Amezaga, J.; Alfaro, B.; Rios, Y.; Larraioz, A.; Ugartemendia, G.; Urruticoechea, A.; Tueros, I. Assessing taste and smell alterations in cancer patients undergoing chemotherapy according to treatment. Support. Care Cancer 2018, 26, 4077–4086. [Google Scholar] [CrossRef] [PubMed]
- Denda, Y.; Niikura, N.; Satoh-Kuriwada, S.; Yokoyama, K.; Terao, M.; Morioka, T.; Tsuda, B.; Okamura, T.; Ota, Y.; Tokuda, Y.; et al. Taste alterations in patients with breast cancer following chemotherapy: A cohort study. Breast Cancer 2020, 27, 954–962. [Google Scholar] [CrossRef]
- Fark, T.; Hummel, C.; Hahner, A.; Nin, T.; Hummel, T. Characteristics of taste disorders. Eur. Arch. Otorhinolaryngol. 2013, 270, 1855–1860. [Google Scholar] [CrossRef]
- Campagna, S.; Gonella, S.; Sperlinga, R.; Giuliano, P.L.; Marchese, R.; Pedersini, R.; Berchialla, P.; Dimonte, V. Prevalence, Severity, and Self-Reported Characteristics of Taste Alterations in Patients Receiving Chemotherapy. Oncol. Nurs. Forum 2018, 45, 342–353. [Google Scholar] [CrossRef]
- Spotten, L.E.; Corish, C.A.; Lorton, C.M.; Ui Dhuibhir, P.M.; O’Donoghue, N.C.; O’Connor, B.; Walsh, T.D. Subjective and objective taste and smell changes in cancer. Ann. Oncol. 2017, 28, 969–984. [Google Scholar] [CrossRef]
- Bleumer, T.; Abel, J.; Bohmerle, W.; Schroder, S.; Yap, S.A.; Schaeper, N.D.E.; Hummel, T.; Stintzing, S.; Stephan, L.U.; Pelzer, U. Smell and Taste Alterations in Patients Receiving Curative or Palliative Chemotherapy-The CONKO 021-ChemTox Trial. Cancers 2024, 16, 2495. [Google Scholar] [CrossRef]
- Hovan, A.J.; Williams, P.M.; Stevenson-Moore, P.; Wahlin, Y.B.; Ohrn, K.E.; Elting, L.S.; Spijkervet, F.K.; Brennan, M.T.; Dysgeusia Section, Oral Care Study Group, Multinational Association of Supportive Care in Cancer (MASCC)/International Society of Oral Oncology (ISOO). A systematic review of dysgeusia induced by cancer therapies. Support. Care Cancer 2010, 18, 1081–1087. [Google Scholar] [CrossRef]
- Erkurt, E.; Erkisi, M.; Tunali, C. Supportive treatment in weight-losing cancer patients due to the additive adverse effects of radiation treatment and/or chemotherapy. J. Exp. Clin. Cancer Res. CR 2000, 19, 431–439. [Google Scholar] [PubMed]
- Whitcroft, K.L.; Altundag, A.; Balungwe, P.; Boscolo-Rizzo, P.; Douglas, R.; Enecilla, M.L.B.; Fjaeldstad, A.W.; Fornazieri, M.A.; Frasnelli, J.; Gane, S.; et al. Position paper on olfactory dysfunction: 2023. Rhinology 2023, 61, 1–108. [Google Scholar] [CrossRef]
- Togni, L.; Mascitti, M.; Vignini, A.; Alia, S.; Sartini, D.; Barlattani, A.; Emanuelli, M.; Santarelli, A. Treatment-related dysgeusia in oral and oropharyngeal cancer: A comprehensive review. Nutrients 2021, 13, 3325. [Google Scholar] [CrossRef] [PubMed]
- Wilken, M.K.; Satiroff, B.A. Pilot study of “miracle fruit” to improve food palatability for patients receiving chemotherapy. Clin. J. Oncol. Nurs. 2012, 16, E173–E177. [Google Scholar] [CrossRef]
- Solemdal, K.; Sandvik, L.; Willumsen, T.; Mowe, M.; Hummel, T. The impact of oral health on taste ability in acutely hospitalized elderly. PLoS ONE 2012, 7, e36557. [Google Scholar] [CrossRef]
- Merkonidis, C.; Grosse, F.; Ninh, T.; Hummel, C.; Haehner, A.; Hummel, T. Characteristics of chemosensory disorders—Results from a survey. Eur. Arch. Otorhinolaryngol. 2015, 272, 1403–1416. [Google Scholar] [CrossRef]
- Dominiak, H.S.; Hasselsteen, S.D.; Nielsen, S.W.; Andersen, J.R.; Herrstedt, J. Prevention of taste alterations in patients with cancer receiving Paclitaxel-or Oxaliplatin-based chemotherapy—A pilot trial of cannabidiol. Nutrients 2023, 15, 3014. [Google Scholar] [CrossRef] [PubMed]
- Epstein, J.B.; de Andrade e Silva, S.M.; Epstein, G.L.; Leal, J.H.S.; Barasch, A.; Smutzer, G. Taste disorders following cancer treatment: Report of a case series. Support. Care Cancer 2019, 27, 4587–4595. [Google Scholar] [CrossRef] [PubMed]
- Heckmann, S.M.; Hujoel, P.; Habiger, S.; Friess, W.; Wichmann, M.; Heckmann, J.G.; Hummel, T. Zinc gluconate in the treatment of dysgeusia—A randomized clinical trial. J. Dent. Res. 2005, 84, 35–38. [Google Scholar] [CrossRef] [PubMed]
- Riquelme, E.; Zhang, Y.; Zhang, L.; Montiel, M.; Zoltan, M.; Dong, W.; Quesada, P.; Sahin, I.; Chandra, V.; San Lucas, A. Tumor microbiome diversity and composition influence pancreatic cancer outcomes. Cell 2019, 178, 795–806.e12. [Google Scholar] [CrossRef]
- Pushalkar, S.; Hundeyin, M.; Daley, D.; Zambirinis, C.P.; Kurz, E.; Mishra, A.; Mohan, N.; Aykut, B.; Usyk, M.; Torres, L.E. The pancreatic cancer microbiome promotes oncogenesis by induction of innate and adaptive immune suppression. Cancer Discov. 2018, 8, 403–416. [Google Scholar] [CrossRef]
- de Vos, W.M.; de Vos, E.A. Role of the intestinal microbiome in health and disease: From correlation to causation. Nutr. Rev. 2012, 70, S45–S56. [Google Scholar] [CrossRef] [PubMed]
- Lozupone, C.A.; Stombaugh, J.I.; Gordon, J.I.; Jansson, J.K.; Knight, R. Diversity, stability and resilience of the human gut microbiota. Nature 2012, 489, 220–230. [Google Scholar] [CrossRef]
- Grenham, S.; Clarke, G.; Cryan, J.F.; Dinan, T.G. Brain-gut-microbe communication in health and disease. Front. Physiol. 2011, 2, 94. [Google Scholar] [CrossRef]
- Enaud, R.; Vandenborght, L.E.; Coron, N.; Bazin, T.; Prevel, R.; Schaeverbeke, T.; Berger, P.; Fayon, M.; Lamireau, T.; Delhaes, L. The Mycobiome: A Neglected Component in the Microbiota-Gut-Brain Axis. Microorganisms 2018, 6, 22. [Google Scholar] [CrossRef] [PubMed]
- Goralczyk-Binkowska, A.; Szmajda-Krygier, D.; Kozlowska, E. The Microbiota-Gut-Brain Axis in Psychiatric Disorders. Int. J. Mol. Sci. 2022, 23, 11245. [Google Scholar] [CrossRef] [PubMed]
- Carding, S.; Verbeke, K.; Vipond, D.T.; Corfe, B.M.; Owen, L.J. Dysbiosis of the gut microbiota in disease. Microb. Ecol. Health Dis. 2015, 26, 26191. [Google Scholar] [CrossRef] [PubMed]
- Virtue, A.T.; McCright, S.J.; Wright, J.M.; Jimenez, M.T.; Mowel, W.K.; Kotzin, J.J.; Joannas, L.; Basavappa, M.G.; Spencer, S.P.; Clark, M.L. The gut microbiota regulates white adipose tissue inflammation and obesity via a family of microRNAs. Sci. Transl. Med. 2019, 11, eaav1892. [Google Scholar] [CrossRef] [PubMed]
- Koh, A.; Molinaro, A.; Ståhlman, M.; Khan, M.T.; Schmidt, C.; Mannerås-Holm, L.; Wu, H.; Carreras, A.; Jeong, H.; Olofsson, L.E. Microbially produced imidazole propionate impairs insulin signaling through mTORC1. Cell 2018, 175, 947–961.e17. [Google Scholar] [CrossRef]
- Rekdal, V.; Bess, E.; Bisanz, J.; Turnbaugh, P.; Balskus, E. Discovery and inhibition of an interspecies gut bacterial pathway for Levodopa metabolism. Science 2019, 364, eaau6323. [Google Scholar] [CrossRef] [PubMed]
- Jin, C.; Lagoudas, G.K.; Zhao, C.; Bullman, S.; Bhutkar, A.; Hu, B.; Ameh, S.; Sandel, D.; Liang, X.S.; Mazzilli, S. Commensal microbiota promote lung cancer development via γδ T cells. Cell 2019, 176, 998–1013.e16. [Google Scholar] [CrossRef]
- El Tekle, G.; Garrett, W.S. Bacteria in cancer initiation, promotion and progression. Nat. Rev. Cancer 2023, 23, 600–618. [Google Scholar] [CrossRef]
- Plummer, M.; de Martel, C.; Vignat, J.; Ferlay, J.; Bray, F.; Franceschi, S. Global burden of cancers attributable to infections in 2012: A synthetic analysis. Lancet Glob. Health 2016, 4, e609–e616. [Google Scholar] [CrossRef] [PubMed]
- de Martel, C.; Georges, D.; Bray, F.; Ferlay, J.; Clifford, G.M. Global burden of cancer attributable to infections in 2018: A worldwide incidence analysis. Lancet Glob. Health 2020, 8, e180–e190. [Google Scholar] [CrossRef]
- Cullin, N.; Azevedo Antunes, C.; Straussman, R.; Stein-Thoeringer, C.K.; Elinav, E. Microbiome and cancer. Cancer Cell 2021, 39, 1317–1341. [Google Scholar] [CrossRef] [PubMed]
- Hou, K.; Wu, Z.X.; Chen, X.Y.; Wang, J.Q.; Zhang, D.; Xiao, C.; Zhu, D.; Koya, J.B.; Wei, L.; Li, J.; et al. Microbiota in health and diseases. Signal Transduct. Target. Ther. 2022, 7, 135. [Google Scholar] [CrossRef]
- Hekmatshoar, Y.; Saadat, Y.R.; Khatibi, S.M.H.; Ozkan, T.; Vahed, F.Z.; Nariman-Saleh-Fam, Z.; Gargari, B.P.; Sunguroglu, A.; Vahed, S.Z. The impact of tumor and gut microbiotas on cancer therapy: Beneficial or detrimental? Life Sci. 2019, 233, 116680. [Google Scholar] [CrossRef]
- Gomez de Cedron, M.; Wagner, S.; Reguero, M.; Menendez-Rey, A.; de Molina, A.R. Miracle Berry as a Potential Supplement in the Control of Metabolic Risk Factors in Cancer. Antioxidants 2020, 9, 1282. [Google Scholar] [CrossRef] [PubMed]
- Soares, H.; Cusnir, M.; Schwartz, M.; Pizzolato, J.; Lutzky, J.; Campbell, R.; Beaumont, J.; Eton, D.; Stonick, S.; Lilenbaum, R. Treatment of taste alterations in chemotherapy patients using the “miracle fruit”: Preliminary analysis of a pilot study. J. Clin. Oncol. 2010, 28, e19523. [Google Scholar] [CrossRef]
- Osabor, V.; Etiuma, R.; Ntinya, M. Chemical profile of leaves and roots of miracle fruit (Synsepalum dulcificum). Am. Chem. Sci. J. 2016, 12, 1–8. [Google Scholar] [CrossRef]
- He, Z.; Tan, J.S.; Abbasiliasi, S.; Lai, O.M.; Tam, Y.J.; Ariff, A.B. Phytochemicals, nutritionals and antioxidant properties of miracle fruit Synsepalum dulcificum. Ind. Crops Prod. 2016, 86, 87–94. [Google Scholar] [CrossRef]
- Ma, F.Y.; Zhang, X.M.; Li, Y.; Zhang, M.; Tu, X.H.; Du, L.Q. Identification of phenolics from miracle berry (Synsepalum dulcificum) leaf extract and its antiangiogenesis and anticancer activities. Front. Nutr. 2022, 9, 970019. [Google Scholar] [CrossRef]
- Swamy, K.B.; Hadi, S.A.; Sekaran, M.; Pichika, M.R. The clinical effects of Synsepalum dulcificum: A review. J. Med. Food 2014, 17, 1165–1169. [Google Scholar] [CrossRef] [PubMed]
- Wagner, S.; Gomez de Cedron, M.; Navarro Del Hierro, J.; Martin-Hernandez, D.; Siles, M.L.N.; Santoyo, S.; Jaime, L.; Martin, D.; Fornari, T.; Ramirez de Molina, A. Biological Activities of Miracle Berry Supercritical Extracts as Metabolic Regulators in Chronic Diseases. Int. J. Mol. Sci. 2023, 24, 6957. [Google Scholar] [CrossRef]
- Afzal, S.; Raju, A.V.; Raju, C.S.; Chong, G.; Wuii, Z.; Eseyin, O.A. Evaluation of the antimicrobial and anticancer properties of the fruits of Synsepalum dulcificum (Sapotaceae). Trop. J. Pharm. Res. 2021, 20, 1925–1930. [Google Scholar] [CrossRef]
- Zollapi, N.N.H.; Taib, R.; Zakaria, N.; Khayat, M.E. Identification of Dipeptidyl-Peptidase 4 (DPP-4) Inhibitors from Miracle Berry Fruit (Synsepalum dulcificum) Extract. J. Biochem. Microbiol. Biotechnol. 2023, 11, 42–47. [Google Scholar] [CrossRef]
- Misaka, T. Molecular mechanisms of the action of miraculin, a taste-modifying protein. Semin. Cell Dev. Biol. 2013, 24, 222–225. [Google Scholar] [CrossRef] [PubMed]
- López-Plaza, B.; Gil, Á.; Menéndez-Rey, A.; Bensadon-Naeder, L.; Hummel, T.; Feliú-Batlle, J.; Palma-Milla, S. Effect of Regular Consumption of a Miraculin-Based Food Supplement on Taste Perception and Nutritional Status in Malnourished Cancer Patients: A Triple-Blind, Randomized, Placebo-Controlled Clinical Trial-CLINMIR Pilot Protocol. Nutrients 2023, 15, 4639. [Google Scholar] [CrossRef] [PubMed]
- Plaza-Diaz, J.; Ruiz-Ojeda, F.J.; Lopez-Plaza, B.; Brandimonte-Hernandez, M.; Alvarez-Mercado, A.I.; Arcos-Castellanos, L.; Feliu-Batlle, J.; Hummel, T.; Palma-Milla, S.; Gil, A. Effect of a Novel Food Rich in Miraculin on the Oral Microbiome of Malnourished Oncologic Patients with Dysgeusia. Cancers 2024, 16, 3414. [Google Scholar] [CrossRef]
- López-Plaza, B.; Álvarez-Mercado, A.I.; Arcos-Castellanos, L.; Plaza-Diaz, J.; Ruiz-Ojeda, F.J.; Brandimonte-Hernández, M.; Feliú-Batlle, J.; Hummel, T.; Gil, Á.; Palma-Milla, S. Efficacy and Safety of Habitual Consumption of a Food Supplement Containing Miraculin in Malnourished Cancer Patients: The CLINMIR Pilot Study. Nutrients 2024, 16, 1905. [Google Scholar] [CrossRef] [PubMed]
- EFSA Panel on Nutrition, N.F.; Allergens, F.; Turck, D.; Castenmiller, J.; De Henauw, S.; Hirsch-Ernst, K.I.; Kearney, J.; Maciuk, A.; Mangelsdorf, I.; McArdle, H.J.; et al. Safety of dried fruits of Synsepalum dulcificum as a novel food pursuant to Regulation (EU) 2015/2283. EFSA J. 2021, 19, e06600. [Google Scholar] [CrossRef] [PubMed]
- Tafazoli, S.; Vo, T.D.; Roberts, A.; Rodriguez, C.; Viñas, R.; Madonna, M.E.; Chiang, Y.-H.; Noronha, J.W.; Holguin, J.C.; Ryder, J.A. Safety assessment of miraculin using in silico and in vitro digestibility analyses. Food Chem. Toxicol. 2019, 133, 110762. [Google Scholar] [CrossRef] [PubMed]
- Fernández-Sánchez, H.; Guzmán-Facundo, F.R.; Herrera-Medina, D.; Sidani, S. Importancia del estudio piloto en un proyecto de intervención. Index Enfermería 2023, 32, e12860. [Google Scholar] [CrossRef]
- Lancaster, G.A.; Dodd, S.; Williamson, P.R. Design and analysis of pilot studies: Recommendations for good practice. J. Eval. Clin. Pract. 2004, 10, 307–312. [Google Scholar] [CrossRef] [PubMed]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef]
- Bokulich, N.A.; Kaehler, B.D.; Rideout, J.R.; Dillon, M.; Bolyen, E.; Knight, R.; Huttley, G.A.; Gregory Caporaso, J. Optimizing taxonomic classification of marker-gene amplicon sequences with QIIME 2’s q2-feature-classifier plugin. Microbiome 2018, 6, 90. [Google Scholar] [CrossRef] [PubMed]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glockner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2013, 41, D590–D596. [Google Scholar] [CrossRef]
- Dixon, P. VEGAN, a package of R functions for community ecology. J. Veg. Sci. 2003, 14, 927–930. [Google Scholar] [CrossRef]
- Alvarez-Mercado, A.I.; Lopez-Plaza, B.; Plaza-Diaz, J.; Arcos-Castellano, L.; Ruiz-Ojeda, F.J.; Brandimonte-Hernandez, M.; Feliu-Batlle, J.; Hummel, T.; Palma-Milla, S.; Gil, A. The Regular Consumption of a Food Supplement Containing Miraculin Can Contribute to Reducing Biomarkers of Inflammation and Cachexia in Malnourished Patients with Cancer and Taste Disorders: The CLINMIR Pilot Study. medRxiv 2024. [Google Scholar] [CrossRef]
- Frank, M.E. Electrogustometry: A simple way to test taste. In Smell and Taste in Health and Disease; Raven Press: New York, NY, USA, 1991. [Google Scholar]
- Barry, M.A.; Gatenby, J.C.; Zeiger, J.D.; Gore, J.C. Hemispheric dominance of cortical activity evoked by focal electrogustatory stimuli. Chem. Senses 2001, 26, 471–482. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Graves, B.; Rosseel, Y.; Merkle, E.C. Computation and application of generalized linear mixed model derivatives using lme4. Psychometrika 2022, 87, 1173–1193. [Google Scholar] [CrossRef]
- Wei, T.S.V.; Levy, M.; Xie, Y.; Jin, Y.; Zemla, J.; Freidank, M.; Cai, J.; Protivinsky, T. Package ‘Corrplot’. 2022. Available online: https://github.com/taiyun/corrplot (accessed on 5 December 2024).
- Benjamini, Y.; Hochberg, Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. Ser. B 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Rivera-Pinto, J.; Egozcue, J.J.; Pawlowsky-Glahn, V.; Paredes, R.; Noguera-Julian, M.; Calle, M.L. Balances: A New Perspective for Microbiome Analysis. mSystems 2018, 3, e00053-18. [Google Scholar] [CrossRef] [PubMed]
- Halyard, M.Y. Taste and smell alterations in cancer patients-real problems with few solutions. J. Support. Oncol. 2009, 7, 68–69. [Google Scholar] [PubMed]
- Hes, C.; Desilets, A.; Tonneau, M.; El Ouarzadi, O.; De Figueiredo Sousa, M.; Bahig, H.; Filion, E.; Nguyen-Tan, P.F.; Christopoulos, A.; Benlaifaoui, M.; et al. Gut microbiome predicts gastrointestinal toxicity outcomes from chemoradiation therapy in patients with head and neck squamous cell carcinoma. Oral Oncol. 2024, 148, 106623. [Google Scholar] [CrossRef] [PubMed]
- Ma, C.; Han, M.; Heinrich, B.; Fu, Q.; Zhang, Q.; Sandhu, M.; Agdashian, D.; Terabe, M.; Berzofsky, J.A.; Fako, V. Gut microbiome–mediated bile acid metabolism regulates liver cancer via NKT cells. Science 2018, 360, eaan5931. [Google Scholar] [CrossRef] [PubMed]
- Herrera-Quintana, L.; Vazquez-Lorente, H.; Silva, R.; Olivares-Arancibia, J.; Reyes-Amigo, T.; Pires, B.R.B.; Plaza-Diaz, J. The Role of the Microbiome and of Radiotherapy-Derived Metabolites in Breast Cancer. Cancers 2024, 16, 3671. [Google Scholar] [CrossRef] [PubMed]
- Luck, R.; Deppenmeier, U. Genetic tools for the redirection of the central carbon flow towards the production of lactate in the human gut bacterium Phocaeicola (Bacteroides) vulgatus. Appl. Microbiol. Biotechnol. 2022, 106, 1211–1225. [Google Scholar] [CrossRef] [PubMed]
- Nabavi, S.F.; Bilotto, S.; Russo, G.L.; Orhan, I.E.; Habtemariam, S.; Daglia, M.; Devi, K.P.; Loizzo, M.R.; Tundis, R.; Nabavi, S.M. Omega-3 polyunsaturated fatty acids and cancer: Lessons learned from clinical trials. Cancer Metastasis Rev. 2015, 34, 359–380. [Google Scholar] [CrossRef]
- Watson, H.; Mitra, S.; Croden, F.C.; Taylor, M.; Wood, H.M.; Perry, S.L.; Spencer, J.A.; Quirke, P.; Toogood, G.J.; Lawton, C.L.; et al. A randomised trial of the effect of omega-3 polyunsaturated fatty acid supplements on the human intestinal microbiota. Gut 2018, 67, 1974–1983. [Google Scholar] [CrossRef] [PubMed]
- Olsson, L.M.; Boulund, F.; Nilsson, S.; Khan, M.T.; Gummesson, A.; Fagerberg, L.; Engstrand, L.; Perkins, R.; Uhlen, M.; Bergström, G. Dynamics of the normal gut microbiota: A longitudinal one-year population study in Sweden. Cell Host Microbe 2022, 30, 726–739. [Google Scholar] [CrossRef]
- Tudela, H.; Claus, S.P.; Saleh, M. Next generation microbiome research: Identification of keystone species in the metabolic regulation of host-gut microbiota interplay. Front. Cell Dev. Biol. 2021, 9, 719072. [Google Scholar] [CrossRef]
- Chaput, N.; Lepage, P.; Coutzac, C.; Soularue, E.; Le Roux, K.; Monot, C.; Boselli, L.; Routier, E.; Cassard, L.; Collins, M. Baseline gut microbiota predicts clinical response and colitis in metastatic melanoma patients treated with ipilimumab. Ann. Oncol. 2017, 28, 1368–1379. [Google Scholar] [CrossRef] [PubMed]
- Gopalakrishnan, V.; Spencer, C.N.; Nezi, L.; Reuben, A.; Andrews, M.C.; Karpinets, T.V.; Prieto, P.; Vicente, D.; Hoffman, K.; Wei, S.C. Gut microbiome modulates response to anti–PD-1 immunotherapy in melanoma patients. Science 2018, 359, 97–103. [Google Scholar] [CrossRef] [PubMed]
- Coutzac, C.; Jouniaux, J.-M.; Paci, A.; Schmidt, J.; Mallardo, D.; Seck, A.; Asvatourian, V.; Cassard, L.; Saulnier, P.; Lacroix, L. Systemic short chain fatty acids limit antitumor effect of CTLA-4 blockade in hosts with cancer. Nat. Commun. 2020, 11, 2168. [Google Scholar] [CrossRef] [PubMed]
- Limeta, A.; Ji, B.; Levin, M.; Gatto, F.; Nielsen, J. Meta-analysis of the gut microbiota in predicting response to cancer immunotherapy in metastatic melanoma. JCI Insight 2020, 5, e140940. [Google Scholar] [CrossRef] [PubMed]
- Spencer, C.N.; McQuade, J.L.; Gopalakrishnan, V.; McCulloch, J.A.; Vetizou, M.; Cogdill, A.P.; Khan, M.A.W.; Zhang, X.; White, M.G.; Peterson, C.B. Dietary fiber and probiotics influence the gut microbiome and melanoma immunotherapy response. Science 2021, 374, 1632–1640. [Google Scholar] [CrossRef] [PubMed]
- Lili, L.; Ye, J. Characterization of gut microbiota in patients with primary hepatocellular carcinoma received immune checkpoint inhibitors: A Chinese population-based study. Medicine 2020, 99, e21788. [Google Scholar] [CrossRef] [PubMed]
- Newsome, R.C.; Gharaibeh, R.Z.; Pierce, C.M.; da Silva, W.V.; Paul, S.; Hogue, S.R.; Yu, Q.; Antonia, S.; Conejo-Garcia, J.R.; Robinson, L.A. Interaction of bacterial genera associated with therapeutic response to immune checkpoint PD-1 blockade in a United States cohort. Genome Med. 2022, 14, 35. [Google Scholar] [CrossRef]
- Sakamoto, M.; Ikeyama, N.; Toyoda, A.; Murakami, T.; Mori, H.; Morohoshi, S.; Kunihiro, T.; Iino, T.; Ohkuma, M. Coprobacter secundus subsp. similis subsp. nov. and Solibaculum mannosilyticum gen. nov., sp. nov., isolated from human feces. Microbiol. Immunol. 2021, 65, 245–256. [Google Scholar]
- Riveros Escalona, M.A.; Poloni, J.F.; Krause, M.J.; Dorn, M. Meta-analyses of host metagenomes from colorectal cancer patients reveal strong relationship between colorectal cancer-associated species. Mol. Omics 2023, 19, 429–444. [Google Scholar] [CrossRef]
- Zuo, W.; Michail, S.; Sun, F. Metagenomic analyses of multiple gut datasets revealed the association of phage signatures in colorectal cancer. Front. Cell. Infect. Microbiol. 2022, 12, 918010. [Google Scholar] [CrossRef] [PubMed]
- Arthur, J.C.; Perez-Chanona, E.; Mühlbauer, M.; Tomkovich, S.; Uronis, J.M.; Fan, T.-J.; Campbell, B.J.; Abujamel, T.; Dogan, B.; Rogers, A.B. Intestinal inflammation targets cancer-inducing activity of the microbiota. Science 2012, 338, 120–123. [Google Scholar] [CrossRef]
- Heshiki, Y.; Vazquez-Uribe, R.; Li, J.; Ni, Y.; Quainoo, S.; Imamovic, L.; Li, J.; Sørensen, M.; Chow, B.K.; Weiss, G.J. Predictable modulation of cancer treatment outcomes by the gut microbiota. Microbiome 2020, 8, 28. [Google Scholar] [CrossRef]
- Thomas, A.M.; Manghi, P.; Asnicar, F.; Pasolli, E.; Armanini, F.; Zolfo, M.; Beghini, F.; Manara, S.; Karcher, N.; Pozzi, C. Metagenomic analysis of colorectal cancer datasets identifies cross-cohort microbial diagnostic signatures and a link with choline degradation. Nat. Med. 2019, 25, 667–678. [Google Scholar] [CrossRef] [PubMed]
- Ulger Toprak, N.; Yagci, A.; Gulluoglu, B.; Akin, M.; Demirkalem, P.; Celenk, T.; Soyletir, G. A possible role of Bacteroides fragilis enterotoxin in the aetiology of colorectal cancer. Clin. Microbiol. Infect. 2006, 12, 782–786. [Google Scholar] [CrossRef] [PubMed]
- Wirbel, J.; Pyl, P.T.; Kartal, E.; Zych, K.; Kashani, A.; Milanese, A.; Fleck, J.S.; Voigt, A.Y.; Palleja, A.; Ponnudurai, R. Meta-analysis of fecal metagenomes reveals global microbial signatures that are specific for colorectal cancer. Nat. Med. 2019, 25, 679–689. [Google Scholar] [CrossRef] [PubMed]
- Zeller, G.; Tap, J.; Voigt, A.Y.; Sunagawa, S.; Kultima, J.R.; Costea, P.I.; Amiot, A.; Böhm, J.; Brunetti, F.; Habermann, N. Potential of fecal microbiota for early-stage detection of colorectal cancer. Mol. Syst. Biol. 2014, 10, 766. [Google Scholar] [CrossRef] [PubMed]
- Briscoe, L.; Balliu, B.; Sankararaman, S.; Halperin, E.; Garud, N.R. Evaluating supervised and unsupervised background noise correction in human gut microbiome data. PLoS Comput. Biol. 2022, 18, e1009838. [Google Scholar] [CrossRef] [PubMed]
- Yachida, S.; Mizutani, S.; Shiroma, H.; Shiba, S.; Nakajima, T.; Sakamoto, T.; Watanabe, H.; Masuda, K.; Nishimoto, Y.; Kubo, M. Metagenomic and metabolomic analyses reveal distinct stage-specific phenotypes of the gut microbiota in colorectal cancer. Nat. Med. 2019, 25, 968–976. [Google Scholar] [CrossRef] [PubMed]
- Sankar, S.A.; Lagier, J.-C.; Pontarotti, P.; Raoult, D.; Fournier, P.-E. The human gut microbiome, a taxonomic conundrum. Syst. Appl. Microbiol. 2015, 38, 276–286. [Google Scholar] [CrossRef]
- Abdulamir, A.S.; Hafidh, R.R.; Mahdi, L.K.; Al-jeboori, T.; Abubaker, F. Investigation into the controversial association of Streptococcus gallolyticus with colorectal cancer and adenoma. BMC Cancer 2009, 9, 403. [Google Scholar] [CrossRef] [PubMed]
- Boleij, A.; Hechenbleikner, E.M.; Goodwin, A.C.; Badani, R.; Stein, E.M.; Lazarev, M.G.; Ellis, B.; Carroll, K.C.; Albesiano, E.; Wick, E.C. The Bacteroides fragilis toxin gene is prevalent in the colon mucosa of colorectal cancer patients. Clin. Infect. Dis. 2015, 60, 208–215. [Google Scholar] [CrossRef] [PubMed]
- Sears, C.L.; Geis, A.L.; Housseau, F. Bacteroides fragilis subverts mucosal biology: From symbiont to colon carcinogenesis. J. Clin. Investig. 2014, 124, 4166–4172. [Google Scholar] [CrossRef] [PubMed]
- McCoy, A.N.; Araújo-Pérez, F.; Azcarate-Peril, A.; Yeh, J.J.; Sandler, R.S.; Keku, T.O. Fusobacterium is associated with colorectal adenomas. PLoS ONE 2013, 8, e53653. [Google Scholar] [CrossRef] [PubMed]
- Kostic, A.D.; Chun, E.; Robertson, L.; Glickman, J.N.; Gallini, C.A.; Michaud, M.; Clancy, T.E.; Chung, D.C.; Lochhead, P.; Hold, G.L. Fusobacterium nucleatum potentiates intestinal tumorigenesis and modulates the tumor-immune microenvironment. Cell Host Microbe 2013, 14, 207–215. [Google Scholar] [CrossRef] [PubMed]
- Nosho, K.; Sukawa, Y.; Adachi, Y.; Ito, M.; Mitsuhashi, K.; Kurihara, H.; Kanno, S.; Yamamoto, I.; Ishigami, K.; Igarashi, H. Association of Fusobacterium nucleatum with immunity and molecular alterations in colorectal cancer. World J. Gastroenterol. 2016, 22, 557. [Google Scholar] [CrossRef]
- Loftus, M.; Hassouneh, S.A.-D.; Yooseph, S. Bacterial community structure alterations within the colorectal cancer gut microbiome. BMC Microbiol. 2021, 21, 98. [Google Scholar] [CrossRef]
- Garutti, M.; Noto, C.; Pasto, B.; Cucciniello, L.; Alajmo, M.; Casirati, A.; Pedrazzoli, P.; Caccialanza, R.; Puglisi, F. Nutritional Management of Oncological Symptoms: A Comprehensive Review. Nutrients 2023, 15, 5068. [Google Scholar] [CrossRef]
- Rinninella, E.; Raoul, P.; Cintoni, M.; Franceschi, F.; Miggiano, G.A.D.; Gasbarrini, A.; Mele, M.C. What is the Healthy Gut Microbiota Composition? A Changing Ecosystem across Age, Environment, Diet, and Diseases. Microorganisms 2019, 7, 14. [Google Scholar] [CrossRef]
- Dunnack, H.J.; Judge, M.P.; Cong, X.; Salner, A.; Duffy, V.B.; Xu, W. An Integrative Review of the Role of the Oral and Gut Microbiome in Oral Health Symptomatology During Cancer Therapy. Oncol. Nurs. Forum 2021, 48, 317–331. [Google Scholar] [CrossRef]
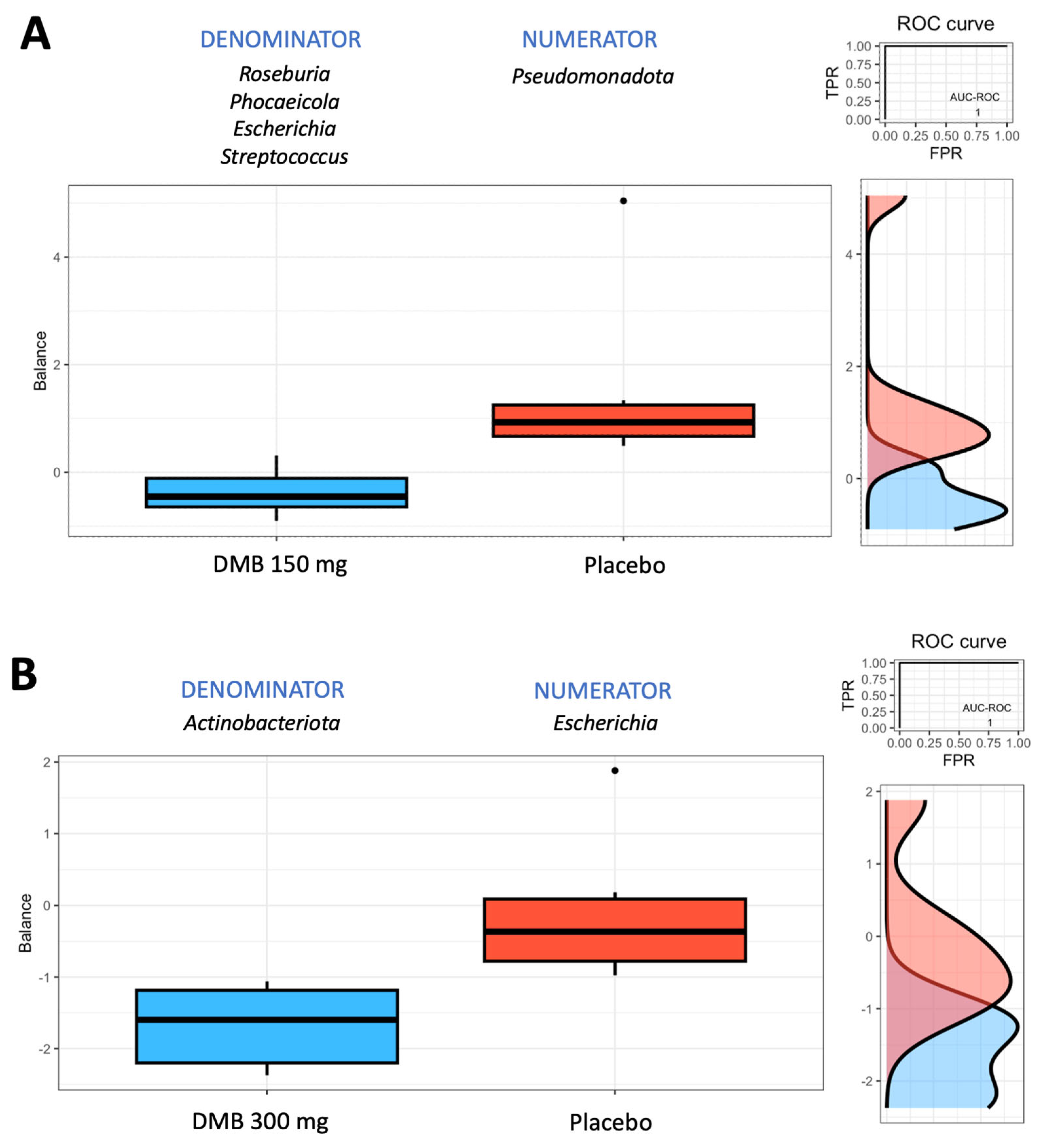
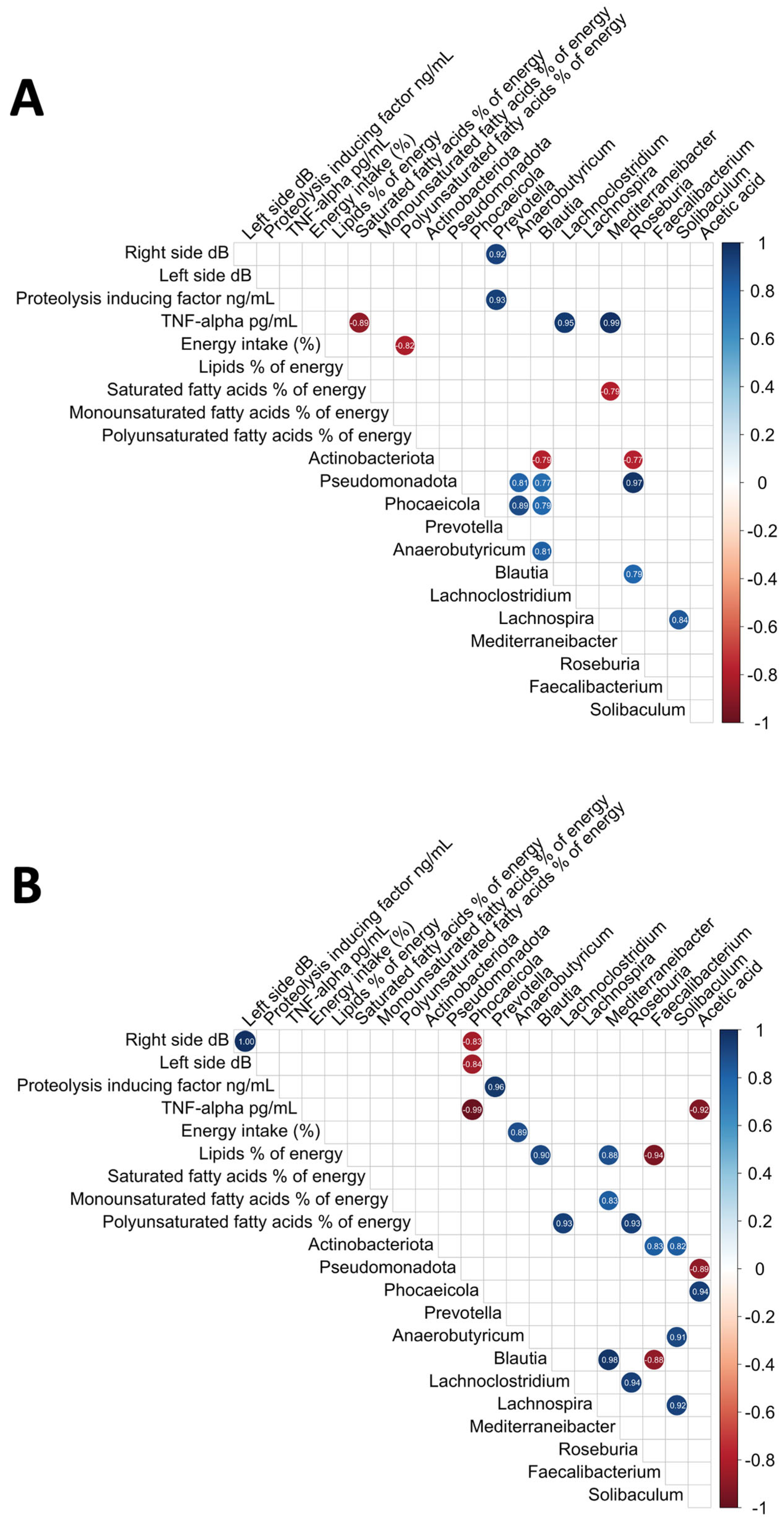
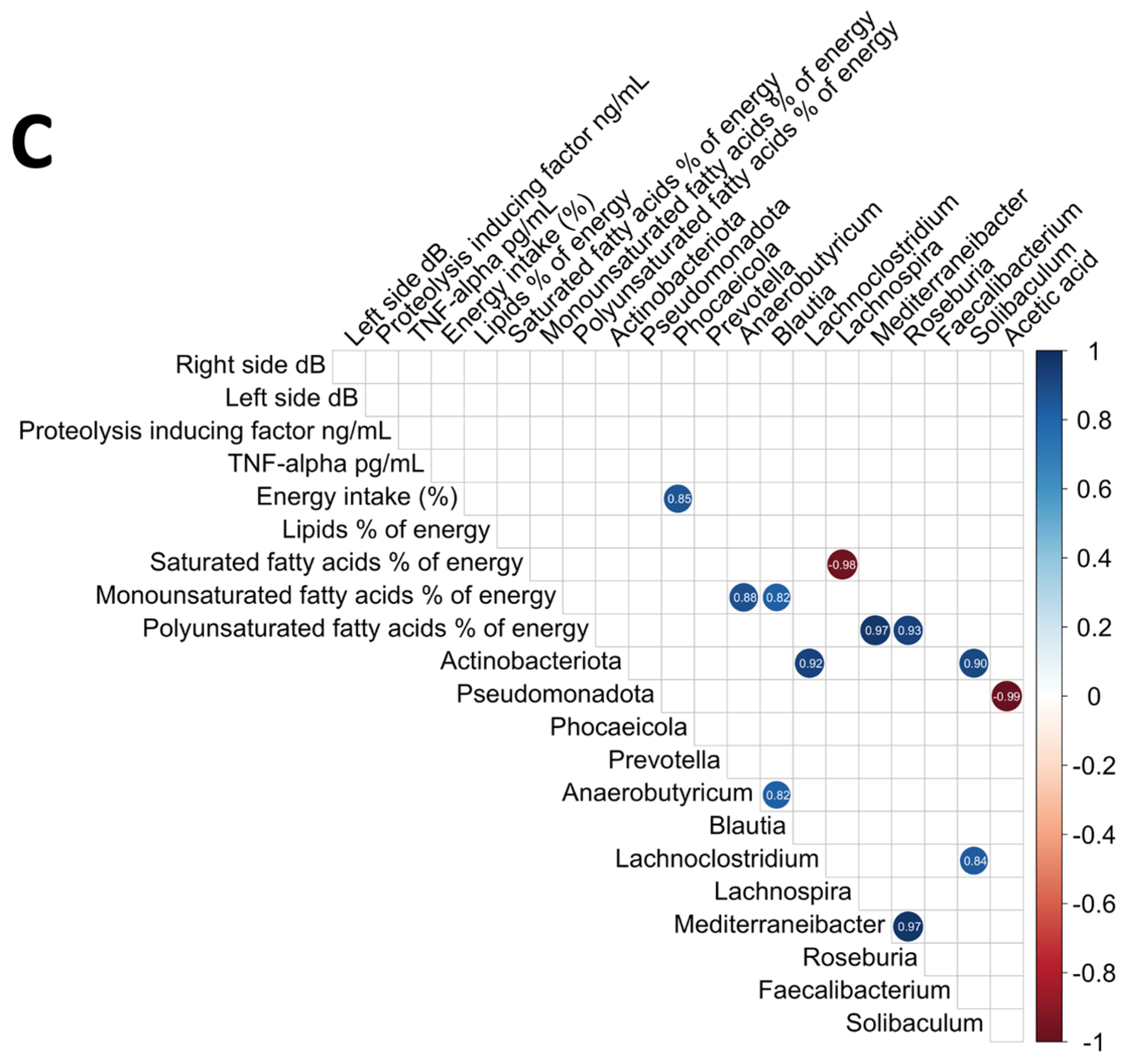
| Phylum | DMB 150 mg | DMB 300 mg | Placebo | p-Value | |||||
|---|---|---|---|---|---|---|---|---|---|
| Baseline | 3 Months | Baseline | 3 Months | Baseline | 3 Months | Treatment (T) | Time (t) | T × t | |
| Actinobacteriota | 0.3 (0.1–0.7) | 0.4 (0.1–0.4) | 0.2 (0.03–0.7) | 0.2 (0.07–0.5) | 0.3 (0.01–0.8) | 0.1 (0.03–0.6) | 0.717 | 0.357 | 0.468 |
| Bacillota | 78.1 (70.6–94.1) | 88.6 (76–95.1) | 75.7 (49.3–86) | 74.7 (67.7–80.6) | 69.1 (13.1–88.1) | 73.7 (50.3–91.1) | 0.062 | 0.224 | 0.598 |
| Bacteroidota | 14.6 (1.4–24.5) | 8.2 (1.9–20.7) | 8.9 (3.1–41.4) | 16 (3–26.4) | 10.2 (0.008–18.1) | 6.7 (0.1–32.4) | 0.444 | 0.674 | 0.888 |
| Pseudomonadota | 3.8 (2.2–7.9) | 2 (1.2–12) | 7.2 (4.6–24) | 8.1 (3.4–21.5) | 16.8 (2.7–86.8) | 15.9 (4.2–31.4) | 0.043 * | 0.253 | 0.366 |
| Tenericutes | 0.2 (0.01–0.3) | 0.06 (0.01–0.2) | 0.09 (0.01–0.2) | 0.03 (0.008–0.2) | 0.1 (0.006–0.4) | 0.06 (0.01–0.2) | 0.542 | 0.092 | 0.703 |
| Synergistetes | 0.1 (0.008–0.3) | 0.08 (0.01–0.2) | 0.08 (0.01–1.3) | 0.2 (0.02–0.5) | 0.2 (0.05–0.2) | 0.1 (0.01–0.2) | 0.282 | 0.397 | 0.876 |
| Verrucomicrobiota | 0.1 (0.006–0.9) | 0.1 (0.01–1.5) | 0.03 (0.007–1.3) | 0.05 (0.01–0.2) | 0.08 (0.07–1.2) | 0.04 (0.01–2.4) | 0.616 | 0.466 | 0.388 |
| Shannon | 3.3 (2.5–3.6) | 3.3 (3.1–3.8) | 3.3 (2.3–3.7) | 3.3 (2.6–3.4) | 3.3 (1.5–3.3) | 3.0 (2.1–3.8) | 0.150 | 0.879 | 0.745 |
| Simpson | 0.9 (0.8–0.9) | 0.9 (0.9–1.0) | 0.9 (0.8–0.9) | 0.9 (0.9–0.9) | 0.9 (0.5–0.9) | 0.9 (0.8–1.0) | 0.135 | 0.825 | 0.579 |
| Chao1 | 435.2 (308.3–548.2) | 388.7 (315.0–540.0) | 404.7 (255.1–684.8) | 422.6 (261.0–491.2) | 394.7 (264.3–514.2) | 415.6 (272.1–547.0) | 0.367 | 0.139 | 0.202 |
| Genus | DMB 150 mg | DMB 300 mg | Placebo | p-Value | |||||
|---|---|---|---|---|---|---|---|---|---|
| Baseline | 3 Months | Baseline | 3 Months | Baseline | 3 Months | Treatment (T) | Time (t) | T × t | |
| Faecalibacterium | 17.5 (5.7–34.1) | 13.4 (12–24.2) | 19.7 (4.6–28.8) | 17.2 (4.6–28.8) | 11.4 (0.01–35) | 12.2 (0.05–46.3) | 0.614 | 0.707 | 0.670 |
| Prevotella | 0.04 (0.01–10.8) | 0.4 (0.01–15.8) | 1.7 (0.02–30.9) | 5.6 (0.1–23.8) | 3.9 (0.01–15.5) | 3.7 (0.1–27.9) | 0.669 | 0.936 | 0.722 |
| Blautia | 4.2 (2.4–8.4) | 4.9 (2.3–7.8) | 4.7 (0.5–10.1) | 3.9 (2.3–7.5) | 2.9 (0.02–9.0) | 2.9 (0.02–4.4) | 0.382 | 0.236 | 0.828 |
| Anaerobutyricum | 4.0 (1.0–7.8) | 2.4 (1.6–5.4) | 2.6 (0.2–6.5) | 2.3 (1.2–5.3) | 1.9 (0.5–2.9) | 3 (0.01–4.2) | 0.369 | 0.848 | 0.126 |
| Dysosmobacter | 3.3 (0.8–4) | 3.7 (0.7–6.3) | 1.5 (0.5–7.1) | 2.9 (0.3–9.3) | 2.6 (1.3–3.8) | 2.6 (0.02–6.2) | 0.841 | 0.299 | 0.859 |
| Vescimonas | 3.3 (0.2–10.6) | 3.3 (1.1–18.8) | 2.6 (0.1–8.6) | 1.3 (0.03–8.4) | 6.0 (0.5–17.4) | 2.8 (0.01–12.6) | 0.450 | 0.977 | 0.130 |
| Roseburia | 2.8 (0.7–21.4) | 1.7 (0.9–23.4) | 2.4 (1.2–13.2) | 3.6 (0.4–11.6) | 1.6 (1.4–4.9) | 3.2 (1.6–12.4) | 0.830 | 0.506 | 0.547 |
| Sulcia | 2.8 (2.8–2.8) | 1.1 (1.1–1.1) | 0 (0–0) | 0 (0–0) | 0.3 (0.3–0.3) | 0 (0–0) | 1.0 | 1.0 | 1.0 |
| Bacteroides | 2.5 (0.4–9.4) | 1.3 (0.7–4.8) | 1.9 (0.8–14) | 2.2 (0.6–8) | 0.9 (0.3–15.5) | 0.7 (0.02–7.7) | 0.915 | 0.131 | 0.957 |
| Lachnospira | 2.1 (1.3–4) | 2.5 (0.5–3.4) | 2.5 (0.4–3.4) | 1.3 (0.3–3.8) | 1.1 (0.6–10.8) | 1.8 (0.6–6.4) | 0.576 | 0.459 | 0.874 |
| Clostridium | 1.8 (1.2–2.4) | 1.9 (0.9–4.3) | 2.1 (0.2–4.1) | 1.3 (0.9–16.2) | 1.2 (0.9–3.1) | 1.4 (0.7–40.1) | 0.614 | 0.182 | 0.513 |
| Coprococcus | 1.5 (1.0–3.6) | 2.2 (0.8–2.8) | 0.9 (0.06–5.0) | 1.1 (0.2–2.2) | 1.0 (0.2–2.7) | 1.9 (0.5–2.9) | 0.558 | 0.630 | 0.590 |
| Blattabacterium | 1.5 (1.5–1.5) | 0.5 (0.5–0.5) | 0 (0–0) | 0 (0–0) | 0.2 (0.2–0.2) | 0 (0–0) | 1.0 | 1.0 | 1.0 |
| Phascolarctobacterium | 1.4 (0.2–3) | 1.9 (1.4–10.8) | 2 (1.1–2.6) | 1.6 (0.08–6.7) | 0.03 (0.007–8.4) | 0.01 (0.006–9.2) | 0.966 | 0.206 | 0.586 |
| Mediterraneibacter | 1.3 (0.5–3.3) | 0.8 (0.4–2.5) | 1.6 (0.3–9) | 1.5 (0.4–4.5) | 0.7 (0.5–1.5) | 0.8 (0.03–4.8) | 0.379 | 0.514 | 0.350 |
| Dorea | 1.2 (0.9–2.9) | 1.3 (0.8–2.8) | 2.3 (0.3–4.3) | 1.0 (0.07–10.2) | 0.8 (0.06–1.9) | 1.0 (0.09–1.4) | 0.214 | 0.628 | 0.668 |
| Phocaeicola | 1.2 (0.3–5.2) | 0.9 (0.09–2.8) | 2.7 (0.3–4.0) | 2.2 (0.6–4.2) | 0.5 (0.3–0.7) | 0.4 (0.4–0.5) | 0.036 * | 0.430 | 0.619 |
| Ruminococcus | 1.1 (0.4–22.9) | 0.9 (0.7–19.2) | 0.8 (0.1–5.3) | 0.8 (0.3–1) | 1.7 (0.2–5.3) | 1.2 (0.008–3.2) | 0.491 | 0.055 | 0.841 |
| Solibaculum | 1.1 (0.4–10.8) | 3.4 * (0.8–7.8) | 1.0 (0.07–6.7) | 1.0 (0.08–6.4) | 5.8 (0.03–8.0) | 1.4 * (0.02–3.0) | 0.782 | 0.172 | 0.046 * |
| Herbinix | 1.0 (0.3–2.8) | 1.6 (0.2–2.4) | 0.6 (0.2–1.9) | 0.8 (0.1–1.5) | 1.1 (0.5–4.6) | 1.5 (0.8–3.3) | 0.403 | 0.740 | 0.685 |
| Lachnoclostridium | 0.9 (0.5–2.3) | 0.6 (0.4–3.0) | 0.8 (0.2–1.9) | 0.6 (0.3–3.5) | 0.6 (0.01–0.8) | 0.6 (0.2–2.8) | 0.487 | 0.103 | 0.934 |
| Anaerostipes | 0.9 (0.3–6.7) | 1.0 (0.4–7.2) | 0.8 (0.3–4.0) | 0.8 (0.3–1.2) | 0.5 (0.2–6.0) | 1.0 (0.01–5.7) | 0.801 | 0.617 | 0.806 |
| Escherichia | 0.8 (0.2–2.7) | 0.3 (0.1–2.9) | 1.3 (0.1–14.0) | 1.7 (0.2–5.0) | 1.1 (0.3–19.3) | 3.4 (0.3–9.2) | 0.012 * | 0.291 | 0.756 |
| Species | DMB 150 mg | DMB 300 mg | Placebo | p-Value | |||||
|---|---|---|---|---|---|---|---|---|---|
| Baseline | 3 Months | Baseline | 3 Months | Baseline | 3 Months | Treatment (T) | Time (t) | T × t | |
| Bacteroides caccae | 0.4 (0.04–0.9) | 0.2 (0.01–1.0) | 0.2 (0.07–1.1) | 0.08 (0.04–0.3) | 0.03 (0.02–0.4) | 0.02 (0.02–1.0) | 0.985 | 0.197 | 0.084 |
| Bacteroides stercoris | 0.2 (0.03–0.4) | 0.2 (0.01–1.0) | 0.2 (0.01–1.5) | 0.5 (0.05–0.8) | 0.1 (0.08–0.1) | 0.3 (0.03–0.5) | 0.587 | 0.608 | 0.713 |
| Bacteroides thetaiotaomicron | 0.5 (0.06–1.0) | 0.3 (0.03–0.8) | 0.07 (0.04–0.8) | 0.1 (0.06–0.5) | 0.1 (0.04–5.3) | 0.2 (0.09–2.1) | 0.591 | 0.197 | 0.475 |
| Bacteroides uniformis | 0.5 (0.03–2.7) | 0.4 (0.08–2.1) | 0.3 (0.03–1.9) | 0.6 (0.03–2.1) | 0.2 (0.06–0.4) | 0.2 (0.1–0.5) | 0.552 | 0.554 | 0.655 |
| Bacteroides sp. PHL 2737 | 1.5 (0.8–2.2) | 0.3 * (0.3–0.3) | 2.1 (0.2–4.0) | 0.4 * (0.2–2.2) | 0.4 (0.3–0.4) | 0.2 (0.2–0.2) | 0.469 | <0.001 * | <0.001 * |
| Anaerobutyricum hallii | 4.0 (1.0–7.8) | 2.5 (1.6–5.4) | 2.6 (0.2–6.6) | 2.3 (1.2–5.4) | 1.9 (0.5–2.9) | 3.0 (0.01–4.3) | 0.366 | 0.853 | 0.128 |
| Blautia argi | 0.3 (0.2–0.9) | 0.2 (0.2–0.5) | 0.3 (0.04–0.9) | 0.2 (0.1–0.4) | 0.2 (0.1–0.3) | 0.2 (0.1–0.3) | 0.136 | 0.287 | 0.688 |
| Blautia liquoris | 1.0 (0.6–5.8) | 1.0 (0.5–3.9) | 1.0 (0.2–1.6) | 1.2 (0.1–1.7) | 0.4 (0.2–3.0) | 0.9 (0.6–2.9) | 0.624 | 0.606 | 0.364 |
| Blautia massiliensis | 0.6 (0.3–2.5) | 0.9 (0.4–2.5) | 1.4 (0.05–3.8) | 1.0 (0.2–2.3) | 0.5 (0.01–1.2) | 0.4 (0.1–0.7) | 0.194 | 0.265 | 0.426 |
| Blautia obeum | 0.4 (0.04–1.1) | 0.07 (0.05–0.9) | 0.1 (0.08–2.0) | 0.04 (0.03–0.3) | 0.1 (0.02–0.5) | 0.1 (0.06–0.3) | 0.620 | 0.269 | 0.521 |
| Blautia pseudococcoides | 0.1 (0.09–0.2) | 0.2 (0.1–0.3) | 0.2 (0.04–0.3) | 0.1 (0.08–0.2) | 0.1 (0.07–0.2) | 0.09 (0.05–0.2) | 0.199 | 0.945 | 0.801 |
| Blautia sp. SC05B48 | 1.0 (0.4–1.9) | 0.8 (0.4–2.9) | 1.1 (0.1–3.8) | 1.0 (0.5–4.4) | 1.2 (0.5–8.3) | 0.8 (0.3–2.9) | 0.720 | 0.283 | 0.278 |
| Lachnospira eligens | 2.1 (1.3–4.0) | 2.5 (0.6–3.4) | 2.5 (0.4–3.4) | 1.4 (0.3–3.8) | 1.1 (0.6–10.8) | 1.8 (0.6–6.5) | 0.576 | 0.462 | 0.873 |
| Roseburia hominis | 2.5 (0.6–19.3) | 1.7 (0.8–17.3) | 2.0 (0.2–4.8) | 2.3 (0.1–6.9) | 1.4 (1.1–3.9) | 2.7 (1.3–4.2) | 0.561 | 0.975 | 0.356 |
| Roseburia intestinalis | 0.3 (0.07–2.0) | 0.5 (0.1–5.5) | 0.6 (0.03–8.8) | 1.2 (0.2–4.5) | 0.3 (0.1 -1.0) | 0.5 (0.3–5.8) | 0.533 | 0.421 | 0.494 |
| Roseburia sp. NSJ-69 | 0.01 (0.007–0.3) | 0.04 (0.02–0.8) | 0.1 (0.01–0.3) | 0.1 (0.01–0.5) | 0.03 (0.02–0.09) | 0.1 (0.08–2.4) | 0.423 | 0.172 | 0.659 |
| Faecalibacterium prausnitzii | 17.7 (5.8–34.4) | 13.9 (12.1–24.6) | 19.8 (4.6–28.9) | 17.3 (4.6–29.0) | 11.5 (0.01–35.7) | 12.3 (0.07–46.7) | 0.620 | 0.712 | 0.678 |
| Vescimonas coprocola | 2.3 (0.06–7.7) | 1.6 (0.5–9.6) | 1.6 (0.1–2.6) | 0.9 (0.03–3.3) | 3.9 (0.5–13.0) | 2.2 * (0.02–8.9) | 0.245 | 0.889 | 0.049 * |
| Vescimonas fastidiosa | 1.1 (0.2–9.0) | 2.5 (0.6–9.5) | 1.0 (0.02–6.4) | 0.7 (0.04–6.1) | 2.2 (1.9–4.6) | 1.3 (0.01–3.8) | 0.719 | 0.881 | 0.563 |
| Dysosmobacter marseille | 0.5 (0.3–1.0) | 0.8 (0.2–2.2) | 0.2 (0.03–1.5) | 0.4 (0.03–1.3) | 0.8 (0.5–0.9) | 0.9 (0.4–1.1) | 0.421 | 0.405 | 0.416 |
| Dysosmobacter welbionis | 2.5 (0.6–3.4) | 3.1 (0.5–4.6) | 1.3 (0.5–7.1) | 2.3 (0.3–9.2) | 1.8 (0.6–2.9) | 1.9 (0.3–5.2) | 0.897 | 0.207 | 0.988 |
| Short-Chain Fatty Acids | DMB 150 mg | DMB 300 mg | Placebo | p-Value | |||||
|---|---|---|---|---|---|---|---|---|---|
| Baseline | 3 Months | Baseline | 3 Months | Baseline | 3 Months | Treatment (T) | Time (t) | T × t | |
| Acetic acid (µmol/L) | 12.8 ± 6.4 | 26.7 ± 6.7 * | 36.6 ± 7.0 | 31.7 ± 7.3 | 25.0 ± 7.8 | 15.0 ± 8.2 * | 0.082 | 0.032 * | 0.027 * |
| Propionic acid (µmol/L) | 0.8 ± 0.6 | 2.2 ± 0.9 | 1.5 ± 0.6 | 0.6 ± 1.0 | 1.9 ± 0.7 | 2.6 ± 1.1 | 0.357 | 0.420 | 0.559 |
| Isobutyric acid (µmol/L) | 0.3 ± 0.1 | 0.4 ± 0.1 | 0.3 ± 0.1 | 0.3 ± 0.1 | 0.4 ± 0.1 | 0.4 ± 0.1 | 0.993 | 0.893 | 0.955 |
| Butyric acid (µmol/L) | 0.9 ± 0.3 | 1.3 ± 0.3 | 0.9 ± 0.3 | 0.8 ± 0.4 | 1.1 ± 0.3 | 1.3 ± 0.4 | 0.698 | 0.414 | 0.591 |
| Isovaleric acid (µmol/L) | 0.2 ± 0.1 | 0.3 ± 0.1 | 0.2 ± 0.1 | 0.3 ± 0.1 | 0.2 ± 0.1 | 0.4 ± 0.1 | 0.865 | 0.991 | 0.603 |
| Valeric acid (µmol/L) | 1.0 ± 0.8 | 2.2 ± 0.9 | 1.0 ± 1.5 | 0.7 ± 1.0 | 1.0 ± 1.9 | 2.7 ± 1.1 | 0.573 | 0.559 | 0.878 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Plaza-Diaz, J.; Brandimonte-Hernández, M.; López-Plaza, B.; Ruiz-Ojeda, F.J.; Álvarez-Mercado, A.I.; Arcos-Castellanos, L.; Feliú-Batlle, J.; Hummel, T.; Palma-Milla, S.; Gil, A. Effect of a Novel Food Rich in Miraculin on the Intestinal Microbiome of Malnourished Patients with Cancer and Dysgeusia. Nutrients 2025, 17, 246. https://doi.org/10.3390/nu17020246
Plaza-Diaz J, Brandimonte-Hernández M, López-Plaza B, Ruiz-Ojeda FJ, Álvarez-Mercado AI, Arcos-Castellanos L, Feliú-Batlle J, Hummel T, Palma-Milla S, Gil A. Effect of a Novel Food Rich in Miraculin on the Intestinal Microbiome of Malnourished Patients with Cancer and Dysgeusia. Nutrients. 2025; 17(2):246. https://doi.org/10.3390/nu17020246
Chicago/Turabian StylePlaza-Diaz, Julio, Marco Brandimonte-Hernández, Bricia López-Plaza, Francisco Javier Ruiz-Ojeda, Ana Isabel Álvarez-Mercado, Lucía Arcos-Castellanos, Jaime Feliú-Batlle, Thomas Hummel, Samara Palma-Milla, and Angel Gil. 2025. "Effect of a Novel Food Rich in Miraculin on the Intestinal Microbiome of Malnourished Patients with Cancer and Dysgeusia" Nutrients 17, no. 2: 246. https://doi.org/10.3390/nu17020246
APA StylePlaza-Diaz, J., Brandimonte-Hernández, M., López-Plaza, B., Ruiz-Ojeda, F. J., Álvarez-Mercado, A. I., Arcos-Castellanos, L., Feliú-Batlle, J., Hummel, T., Palma-Milla, S., & Gil, A. (2025). Effect of a Novel Food Rich in Miraculin on the Intestinal Microbiome of Malnourished Patients with Cancer and Dysgeusia. Nutrients, 17(2), 246. https://doi.org/10.3390/nu17020246








