TAS1R2/TAS1R3 Single-Nucleotide Polymorphisms Affect Sweet Taste Receptor Activation by Sweeteners: The SWEET Project
Abstract
:1. Introduction
2. Materials and Methods
2.1. The SWEET Project
2.2. Sweeteners
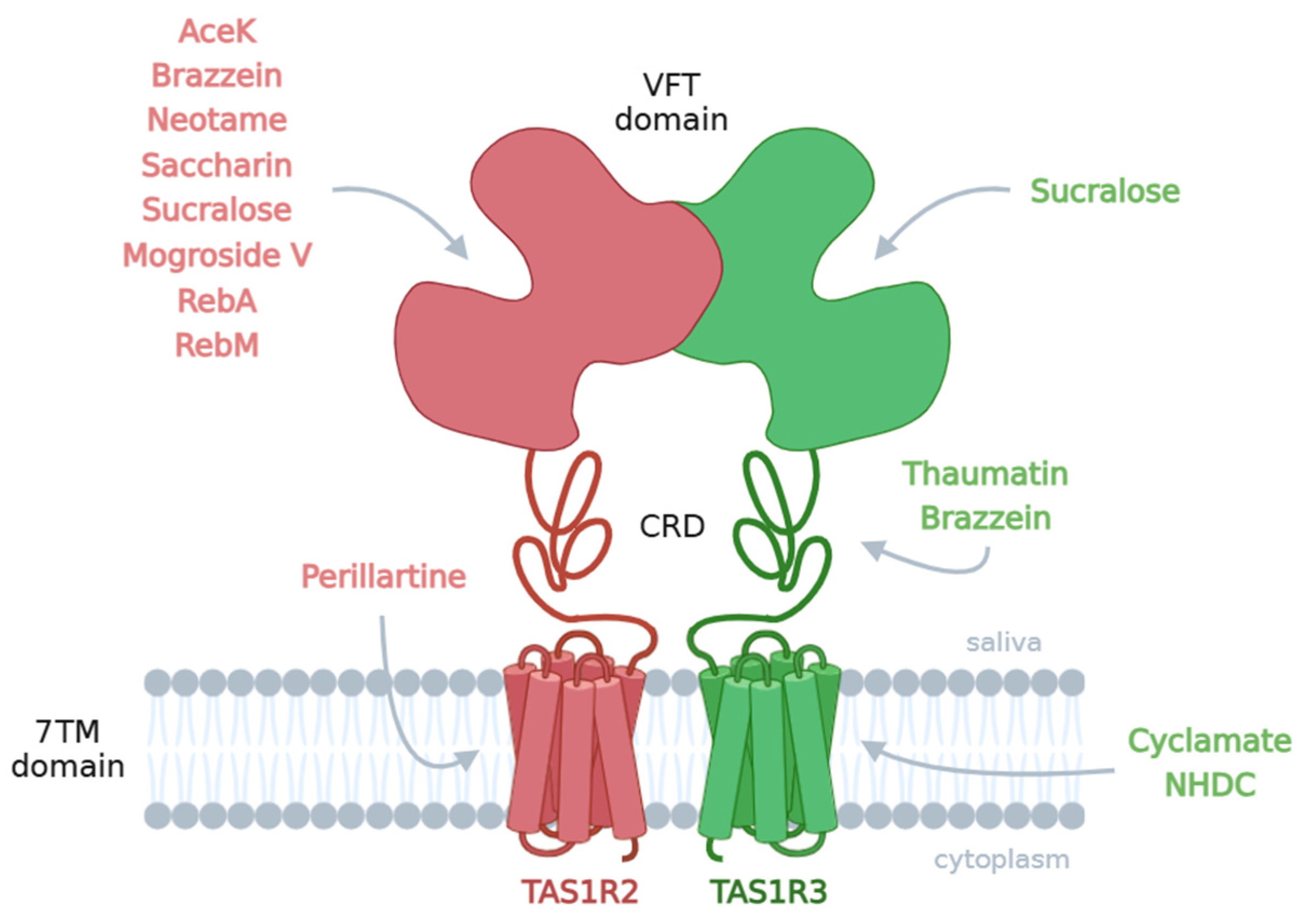
2.3. In Vitro Cell-Based Assays Studying Human TAS1R2- and TAS1R3-SNPs
2.3.1. Selection and Expression of TAS1R2- and TAS1R3-SNPs
2.3.2. Calcium Mobilisation Assay
2.4. In Vitro Cell-Based Assays for Studying Mouse TAS1R3
2.5. Statistical Analysis
2.6. In Vivo SNP Genotyping Assays
2.6.1. Subjects
2.6.2. Blood Sample Polymorphism Analysis
3. Results
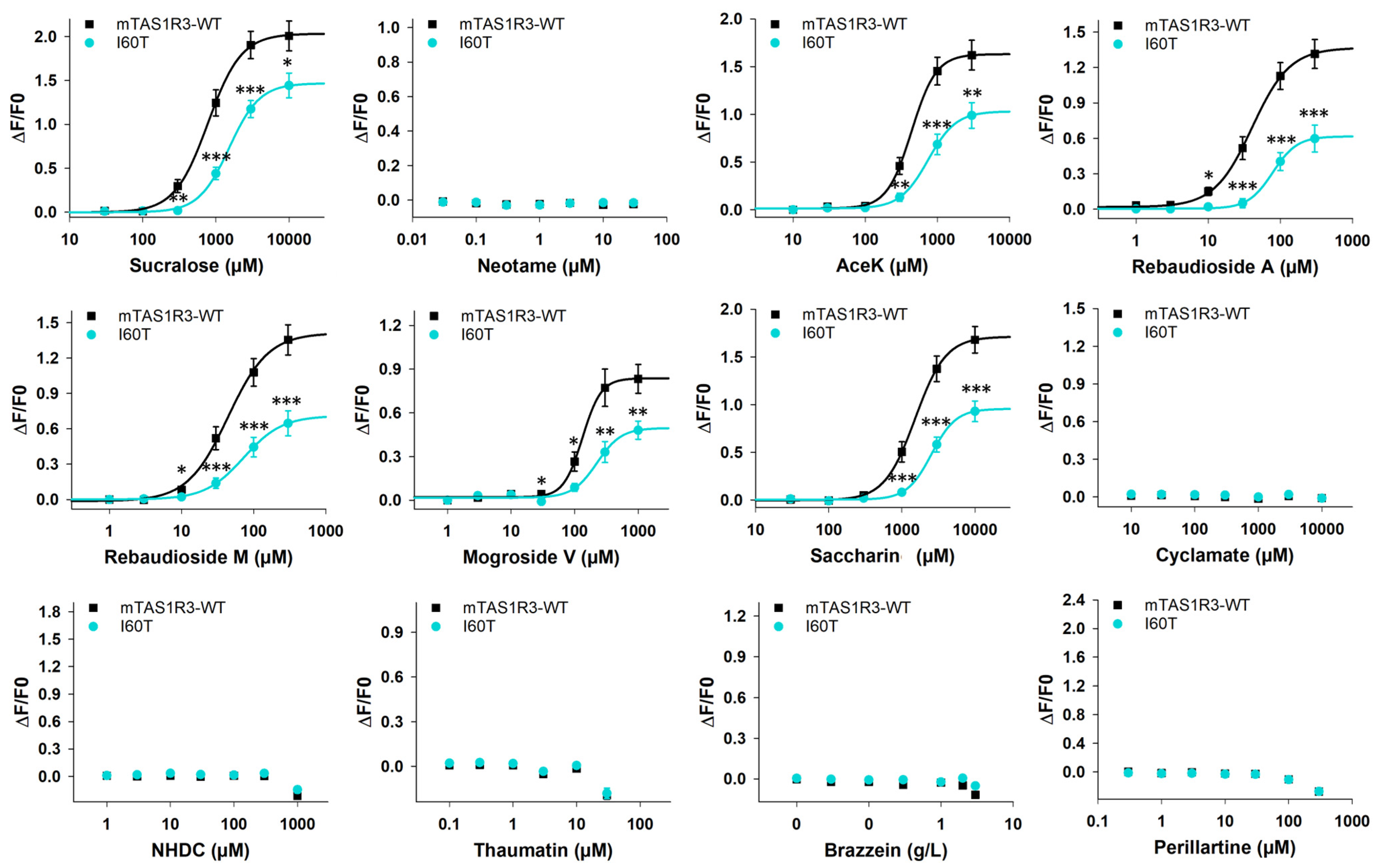
3.1. Response of Mouse TAS1R3-I60T Variant to Sweeteners
3.2. Response of Human TAS1R2 and TAS1R3 Variants to Sweeteners
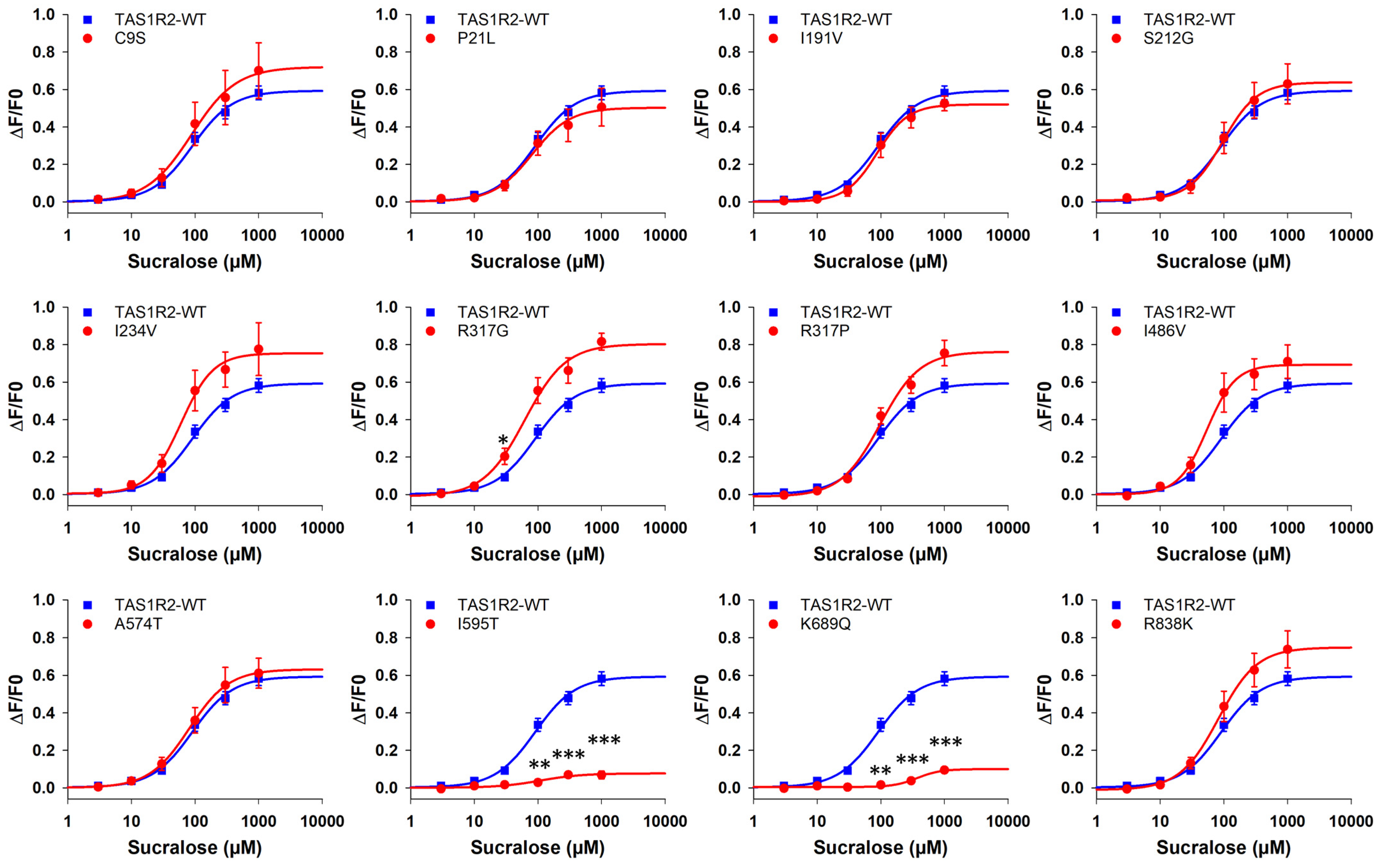
3.2.1. TAS1R2 Variants
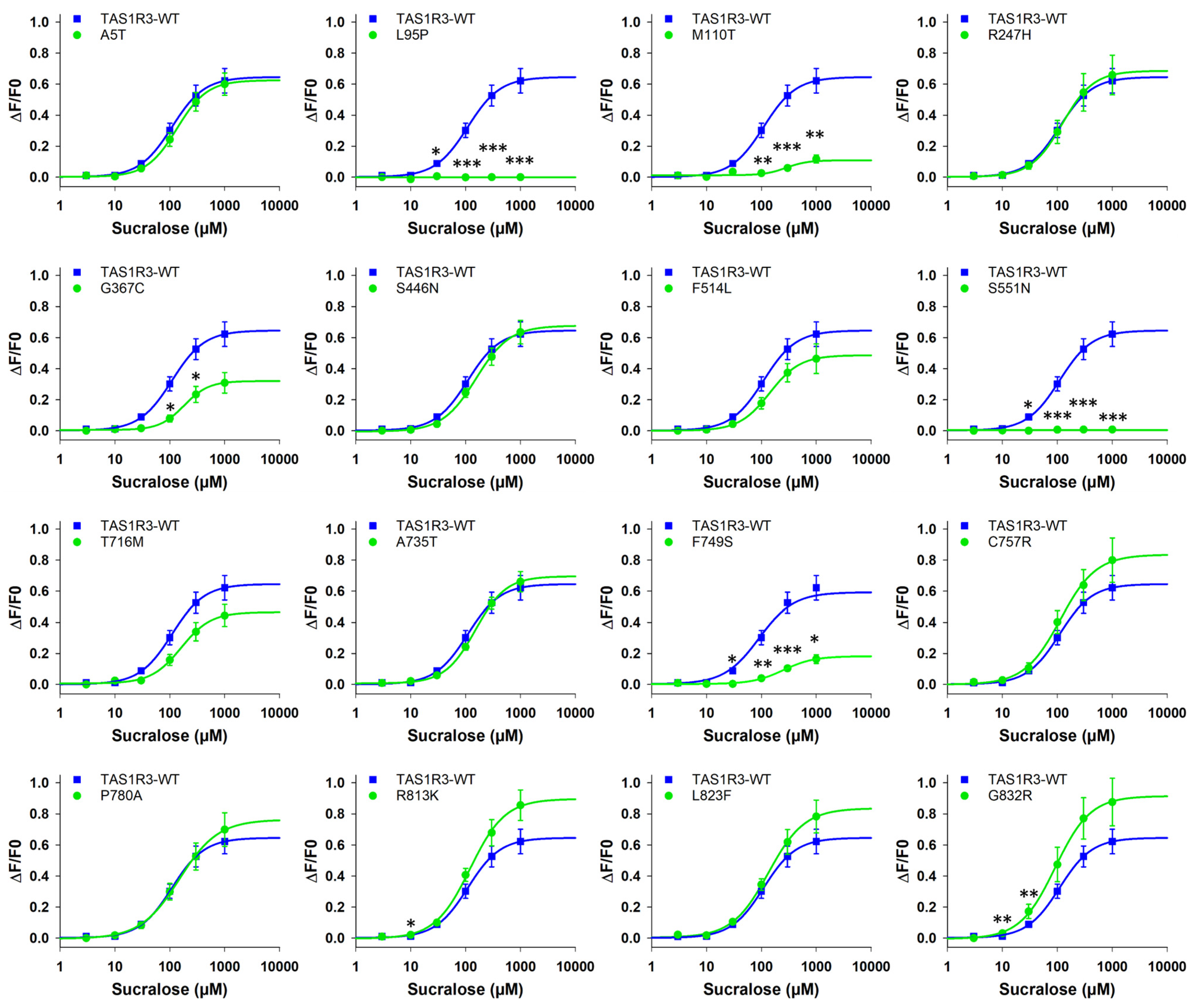
3.2.2. TAS1R3 Variants
3.3. Results of TAS1R2/TAS1R3 SNP Sequencing in an Obese Population
4. Discussion
4.1. Is There a Clear Link Between Certain Phenotypes (Higher Sugar Intake and/or Higher Detection Threshold) and the Cellular Response of Associated SNPs?
4.2. Possible/Probable SNPs of TAS1R2/TAS1R3 Associated with a Loss of Function of the Sweet Taste Receptor
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Briand, L.; Salles, C. Taste Perception and Integration. In Flavor from Food to Behaviors, Wellbeing and Health; Etiévant, P., Guichard, E., Salles, C., Voilley, A., Eds.; Elsevier Ltd.: Duxford, UK, 2016; pp. 101–119. ISBN 978-0-08-100295-7. [Google Scholar]
- Belloir, C.; Neiers, F.; Briand, L. Sweeteners and Sweetness Enhancers. Curr. Opin. Clin. Nutr. Metab. Care 2017, 20, 279. [Google Scholar] [CrossRef] [PubMed]
- Malik, V.S.; Popkin, B.M.; Bray, G.A.; Després, J.-P.; Willett, W.C.; Hu, F.B. Sugar-Sweetened Beverages and Risk of Metabolic Syndrome and Type 2 Diabetes: A Meta-Analysis. Diabetes Care 2010, 33, 2477–2483. [Google Scholar] [CrossRef] [PubMed]
- Peres, M.A.; Sheiham, A.; Liu, P.; Demarco, F.F.; Silva, A.E.R.; Assunção, M.C.; Menezes, A.M.; Barros, F.C.; Peres, K.G. Sugar Consumption and Changes in Dental Caries from Childhood to Adolescence. J. Dent. Res. 2016, 95, 388–394. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. Diet, Nutrition, and the Prevention of Chronic Diseases: Report of a Joint WHO/FAO Expert Consultation; World Health Organization: Geneva, Switzerland, 2003; ISBN 978-92-4-120916-8. [Google Scholar]
- Li, X.; Staszewski, L.; Xu, H.; Durick, K.; Zoller, M.; Adler, E. Human Receptors for Sweet and Umami Taste. Proc. Natl. Acad. Sci. USA 2002, 99, 4692–4696. [Google Scholar] [CrossRef]
- Nelson, G.; Hoon, M.A.; Chandrashekar, J.; Zhang, Y.; Ryba, N.J.P.; Zuker, C.S. Mammalian Sweet Taste Receptors. Cell 2001, 106, 381–390. [Google Scholar] [CrossRef]
- Nelson, G.; Chandrashekar, J.; Hoon, M.A.; Feng, L.; Zhao, G.; Ryba, N.J.P.; Zuker, C.S. An Amino-Acid Taste Receptor. Nature 2002, 416, 199–202. [Google Scholar] [CrossRef]
- Jiang, P.; Ji, Q.; Liu, Z.; Snyder, L.A.; Benard, L.M.J.; Margolskee, R.F.; Max, M. The Cysteine-Rich Region of T1R3 Determines Responses to Intensely Sweet Proteins. J. Biol. Chem. 2004, 279, 45068–45075. [Google Scholar] [CrossRef]
- Assadi-Porter, F.M.; Maillet, E.L.; Radek, J.T.; Quijada, J.; Markley, J.L.; Max, M. Key Amino Acid Residues Involved in Multi-Point Binding Interactions between Brazzein, a Sweet Protein, and the T1R2–T1R3 Human Sweet Receptor. J. Mol. Biol. 2010, 398, 584–599. [Google Scholar] [CrossRef]
- Masuda, T.; Taguchi, W.; Sano, A.; Ohta, K.; Kitabatake, N.; Tani, F. Five Amino Acid Residues in Cysteine-Rich Domain of Human T1R3 Were Involved in the Response for Sweet-Tasting Protein, Thaumatin. Biochimie 2013, 95, 1502–1505. [Google Scholar] [CrossRef]
- Nie, Y.; Vigues, S.; Hobbs, J.R.; Conn, G.L.; Munger, S.D. Distinct Contributions of T1R2 and T1R3 Taste Receptor Subunits to the Detection of Sweet Stimuli. Curr. Biol. 2005, 15, 1948–1952. [Google Scholar] [CrossRef]
- Zhang, F.; Klebansky, B.; Fine, R.M.; Liu, H.; Xu, H.; Servant, G.; Zoller, M.; Tachdjian, C.; Li, X. Molecular Mechanism of the Sweet Taste Enhancers. Proc. Natl. Acad. Sci. USA 2010, 107, 4752–4757. [Google Scholar] [CrossRef] [PubMed]
- Behrens, M.; Meyerhof, W.; Hellfritsch, C.; Hofmann, T. Sweet and Umami Taste: Natural Products, Their Chemosensory Targets, and Beyond. Angew. Chem. Int. Ed. 2011, 50, 2220–2242. [Google Scholar] [CrossRef]
- DuBois, G.E. Molecular Mechanism of Sweetness Sensation. Physiol. Behav. 2016, 164, 453–463. [Google Scholar] [CrossRef] [PubMed]
- Laffitte, A.; Belloir, C.; Neiers, F.; Briand, L. Functional Characterization of the Venus Flytrap Domain of the Human TAS1R2 Sweet Taste Receptor. Int. J. Mol. Sci. 2022, 23, 9216. [Google Scholar] [CrossRef] [PubMed]
- Belloir, C.; Jeannin, M.; Karolkowski, A.; Scott, C.; Briand, L. A Receptor-Based Assay to Study the Sweet and Bitter Tastes of Sweeteners and Binary Sweet Blends: The SWEET Project. Chem. Senses 2024, 49, bjae041. [Google Scholar] [CrossRef]
- Jiang, P.; Cui, M.; Ji, Q.; Snyder, L.; Liu, Z.; Benard, L.; Margolskee, R.F.; Osman, R.; Max, M. Molecular Mechanisms of Sweet Receptor Function. Chem. Senses 2005, 30, i17–i18. [Google Scholar] [CrossRef]
- Xu, H.; Staszewski, L.; Tang, H.; Adler, E.; Zoller, M.; Li, X. Different Functional Roles of T1R Subunits in the Heteromeric Taste Receptors. Proc. Natl. Acad. Sci. USA 2004, 101, 14258–14263. [Google Scholar] [CrossRef]
- Liu, B.; Ha, M.; Meng, X.-Y.; Kaur, T.; Khaleduzzaman, M.; Zhang, Z.; Jiang, P.; Li, X.; Cui, M. Molecular Mechanism of Species-Dependent Sweet Taste toward Artificial Sweeteners. J. Neurosci. 2011, 31, 11070–11076. [Google Scholar] [CrossRef]
- Maillet, E.L.; Cui, M.; Jiang, P.; Mezei, M.; Hecht, E.; Quijada, J.; Margolskee, R.F.; Osman, R.; Max, M. Characterization of the Binding Site of Aspartame in the Human Sweet Taste Receptor. Chem. Senses 2015, 40, 577–586. [Google Scholar] [CrossRef]
- Masuda, K.; Koizumi, A.; Nakajima, K.; Tanaka, T.; Abe, K.; Misaka, T.; Ishiguro, M. Characterization of the Modes of Binding between Human Sweet Taste Receptor and Low-Molecular-Weight Sweet Compounds. PLoS ONE 2012, 7, e35380. [Google Scholar] [CrossRef]
- Jiang, P.; Cui, M.; Zhao, B.; Snyder, L.A.; Benard, L.M.J.; Osman, R.; Max, M.; Margolskee, R.F. Identification of the Cyclamate Interaction Site within the Transmembrane Domain of the Human Sweet Taste Receptor Subunit T1R3. J. Biol. Chem. 2005, 280, 34296–34305. [Google Scholar] [CrossRef] [PubMed]
- Winnig, M.; Bufe, B.; Kratochwil, N.A.; Slack, J.P.; Meyerhof, W. The Binding Site for Neohesperidin Dihydrochalcone at the Human Sweet Taste Receptor. BMC Struct. Biol. 2007, 7, 66. [Google Scholar] [CrossRef] [PubMed]
- Jiang, P.; Cui, M.; Zhao, B.; Liu, Z.; Snyder, L.A.; Benard, L.M.J.; Osman, R.; Margolskee, R.F.; Max, M. Lactisole Interacts with the Transmembrane Domains of Human T1R3 to Inhibit Sweet Taste. J. Biol. Chem. 2005, 280, 15238–15246. [Google Scholar] [CrossRef] [PubMed]
- Morini, G.; Bassoli, A.; Temussi, P.A. From Small Sweeteners to Sweet Proteins: Anatomy of the Binding Sites of the Human T1R2_T1R3 Receptor. J. Med. Chem. 2005, 48, 5520–5529. [Google Scholar] [CrossRef]
- Ohta, K.; Masuda, T.; Tani, F.; Kitabatake, N. Introduction of a Negative Charge at Arg82 in Thaumatin Abolished Responses to Human T1R2–T1R3 Sweet Receptors. Biochem. Biophys. Res. Commun. 2011, 413, 41–45. [Google Scholar] [CrossRef]
- Choi, J.-H.; Lee, J.; Yang, S.; Kim, J. Genetic Variations in Taste Perception Modify Alcohol Drinking Behavior in Koreans. Appetite 2017, 113, 178–186. [Google Scholar] [CrossRef]
- Dotson, C.D.; Babich, J.; Steinle, N.I. Genetic Predisposition and Taste Preference: Impact on Food Intake and Risk of Chronic Disease. Curr. Nutr. Rep. 2012, 1, 175–183. [Google Scholar] [CrossRef]
- Lee, R.J.; Hariri, B.M.; McMahon, D.B.; Chen, B.; Doghramji, L.; Adappa, N.D.; Palmer, J.N.; Kennedy, D.W.; Jiang, P.; Margolskee, R.F.; et al. Bacterial D-Amino Acids Suppress Sinonasal Innate Immunity through Sweet Taste Receptors in Solitary Chemosensory Cells. Sci. Signal. 2017, 10, eaam7703. [Google Scholar] [CrossRef]
- Lee, R.J.; Cohen, N.A. Taste Receptors in Innate Immunity. Cell. Mol. Life Sci. 2015, 72, 217–236. [Google Scholar] [CrossRef]
- Mfuna Endam, L.; Filali-Mouhim, A.; Boisvert, P.; Boulet, L.-P.; Bossé, Y.; Desrosiers, M. Genetic Variations in Taste Receptors Are Associated with Chronic Rhinosinusitis: A Replication Study. Int. Forum Allergy Rhinol. 2014, 4, 200–206. [Google Scholar] [CrossRef]
- Tarragon, E.; Moreno, J.J. Role of Endocannabinoids on Sweet Taste Perception, Food Preference, and Obesity-Related Disorders. Chem. Senses 2018, 43, 3–16. [Google Scholar] [CrossRef]
- Triantafillou, V.; Workman, A.D.; Kohanski, M.A.; Cohen, N.A. Taste Receptor Polymorphisms and Immune Response: A Review of Receptor Genotypic-Phenotypic Variations and Their Relevance to Chronic Rhinosinusitis. Front. Cell. Infect. Microbiol. 2018, 8, 64. [Google Scholar] [CrossRef]
- Barham, H.P.; Taha, M.A.; Broyles, S.T.; Stevenson, M.M.; Zito, B.A.; Hall, C.A. Association Between Bitter Taste Receptor Phenotype and Clinical Outcomes Among Patients with COVID-19. JAMA Netw. Open 2021, 4, e2111410. [Google Scholar] [CrossRef]
- Santin, A.; Spedicati, B.; Pecori, A.; Nardone, G.G.; Concas, M.P.; Piatti, G.; Menini, A.; Tirelli, G.; Boscolo-Rizzo, P.; Girotto, G. The Bittersweet Symphony of COVID-19: Associations between TAS1Rs and TAS2R38 Genetic Variations and COVID-19 Symptoms. Life 2024, 14, 219. [Google Scholar] [CrossRef]
- Lin, C.; Civantos, A.M.; Arnold, M.; Stevens, E.M.; Cowart, B.J.; Colquitt, L.R.; Mansfield, C.; Kennedy, D.W.; Brooks, S.G.; Workman, A.D.; et al. Divergent Bitter and Sweet Taste Perception Intensity in Chronic Rhinosinusitis Patients. Int. Forum Allergy Rhinol. 2021, 11, 857–865. [Google Scholar] [CrossRef]
- Liszt, K.I.; Wang, Q.; Farhadipour, M.; Segers, A.; Thijs, T.; Nys, L.; Deleus, E.; der Schueren, B.V.; Gerner, C.; Neuditschko, B.; et al. Human Intestinal Bitter Taste Receptors Regulate Innate Immune Responses and Metabolic Regulators in Obesity. J. Clin. Investig. 2022, 132, e144828. [Google Scholar] [CrossRef]
- Laffitte, A.; Neiers, F.; Briand, L. Functional Roles of the Sweet Taste Receptor in Oral and Extraoral Tissues. Curr. Opin. Clin. Nutr. Metab. Care 2014, 17, 379. [Google Scholar] [CrossRef]
- Chamoun, E.; Liu, A.S.; Duizer, L.M.; Feng, Z.; Darlington, G.; Duncan, A.M.; Haines, J.; Ma, D.W.L. Single Nucleotide Polymorphisms in Sweet, Fat, Umami, Salt, Bitter and Sour Taste Receptor Genes Are Associated with Gustatory Function and Taste Preferences in Young Adults. Nutr. Res. 2021, 85, 40–46. [Google Scholar] [CrossRef]
- Garcia-Bailo, B.; Toguri, C.; Eny, K.M.; El-Sohemy, A. Genetic Variation in Taste and Its Influence on Food Selection. OMICS 2009, 13, 69–80. [Google Scholar] [CrossRef]
- Bufe, B.; Breslin, P.A.S.; Kuhn, C.; Reed, D.R.; Tharp, C.D.; Slack, J.P.; Kim, U.-K.; Drayna, D.; Meyerhof, W. The Molecular Basis of Individual Differences in Phenylthiocarbamide and Propylthiouracil Bitterness Perception. Curr. Biol. 2005, 15, 322–327. [Google Scholar] [CrossRef]
- Raliou, M.; Grauso, M.; Hoffmann, B.; Schlegel–Le-Poupon, C.; Nespoulous, C.; Débat, H.; Belloir, C.; Wiencis, A.; Sigoillot, M.; Preet Bano, S.; et al. Human Genetic Polymorphisms in T1R1 and T1R3 Taste Receptor Subunits Affect Their Function. Chem. Senses 2011, 36, 527–537. [Google Scholar] [CrossRef]
- Shigemura, N.; Shirosaki, S.; Ohkuri, T.; Sanematsu, K.; Islam, A.S.; Ogiwara, Y.; Kawai, M.; Yoshida, R.; Ninomiya, Y. Variation in Umami Perception and in Candidate Genes for the Umami Receptor in Mice and Humans. Am. J. Clin. Nutr. 2009, 90, 764S–769S. [Google Scholar] [CrossRef]
- Shigemura, N.; Shirosaki, S.; Sanematsu, K.; Yoshida, R.; Ninomiya, Y. Genetic and Molecular Basis of Individual Differences in Human Umami Taste Perception. PLoS ONE 2009, 4, e6717. [Google Scholar] [CrossRef]
- Fushan, A.A.; Simons, C.T.; Slack, J.P.; Manichaikul, A.; Drayna, D. Allelic Polymorphism within the TAS1R3 Promoter Is Associated with Human Taste Sensitivity to Sucrose. Curr. Biol. 2009, 19, 1288–1293. [Google Scholar] [CrossRef]
- Mainland, J.D.; Matsunami, H. Taste Perception: How Sweet It Is (To Be Transcribed by You). Curr. Biol. 2009, 19, R655–R656. [Google Scholar] [CrossRef]
- Kim, U.; Wooding, S.; Riaz, N.; Jorde, L.B.; Drayna, D. Variation in the Human TAS1R Taste Receptor Genes. Chem. Senses 2006, 31, 599–611. [Google Scholar] [CrossRef]
- Eny, K.M.; Wolever, T.M.; Corey, P.N.; El-Sohemy, A. Genetic Variation in TAS1R2 (Ile191Val) Is Associated with Consumption of Sugars in Overweight and Obese Individuals in 2 Distinct Populations. Am. J. Clin. Nutr. 2010, 92, 1501–1510. [Google Scholar] [CrossRef]
- Pioltine, M.B.; De Melo, M.E.; Santos, A.S.; Machado, A.D.; Fernandes, A.E.; Fujiwara, C.T.; Cercato, C.; Mancini, M.C. Genetic Variations in Sweet Taste Receptor Gene Are Related to Chocolate Powder and Dietary Fiber Intake in Obese Children and Adolescents. J. Pers. Med. 2018, 8, 7. [Google Scholar] [CrossRef]
- Dias, A.G.; Eny, K.M.; Cockburn, M.; Chiu, W.; Nielsen, D.E.; Duizer, L.; El-Sohemy, A. Variation in the TAS1R2 Gene, Sweet Taste Perception and Intake of Sugars. J. Nutr. Nutr. 2015, 8, 81–90. [Google Scholar] [CrossRef]
- Han, P.; Keast, R.S.J.; Roura, E. Salivary Leptin and TAS1R2/TAS1R3 Polymorphisms Are Related to Sweet Taste Sensitivity and Carbohydrate Intake from a Buffet Meal in Healthy Young Adults. Br. J. Nutr. 2017, 118, 763–770. [Google Scholar] [CrossRef]
- Liang, Y.; Yao, J.; Qiu, R.; Chen, A.; Huang, H.; Lin, H.; Yu, L. The Rs35874116 Single Nucleotide Polymorphism Increases Sweet Intake and the Risk of Severe Early Childhood Caries: A Case–Control Study. BMC Oral Health 2022, 22, 471. [Google Scholar] [CrossRef]
- Melis, M.; Mastinu, M.; Naciri, L.C.; Muroni, P.; Tomassini Barbarossa, I. Associations between Sweet Taste Sensitivity and Polymorphisms (SNPs) in the TAS1R2 and TAS1R3 Genes, Gender, PROP Taster Status, and Density of Fungiform Papillae in a Genetically Homogeneous Sardinian Cohort. Nutrients 2022, 14, 4903. [Google Scholar] [CrossRef]
- Poirier, N.; Roudnitzky, N.; Brockhoff, A.; Belloir, C.; Maison, M.; Thomas-Danguin, T.; Meyerhof, W.; Briand, L. Efficient Production and Characterization of the Sweet-Tasting Brazzein Secreted by the Yeast Pichia pastoris. J. Agric. Food Chem. 2012, 60, 9807–9814. [Google Scholar] [CrossRef]
- Kim, S.-K.; Chen, Y.; Abrol, R.; Goddard, W.A.; Guthrie, B. Activation Mechanism of the G Protein-Coupled Sweet Receptor Heterodimer with Sweeteners and Allosteric Agonists. Proc. Natl. Acad. Sci. USA 2017, 114, 2568–2573. [Google Scholar] [CrossRef]
- Cai, C.; Jiang, H.; Li, L.; Liu, T.; Song, X.; Liu, B. Characterization of the Sweet Taste Receptor Tas1r2 from an Old World Monkey Species Rhesus Monkey and Species-Dependent Activation of the Monomeric Receptor by an Intense Sweetener Perillartine. PLoS ONE 2016, 11, e0160079. [Google Scholar] [CrossRef]
- Choi, J.-H.; Lee, J.; Choi, I.J.; Kim, Y.-W.; Ryu, K.W.; Kim, J. Variations in TAS1R Taste Receptor Gene Family Modify Food Intake and Gastric Cancer Risk in a Korean Population. Mol. Nutr. Food Res. 2016, 60, 2433–2445. [Google Scholar] [CrossRef]
- Reed, D.R.; Tanaka, T.; McDaniel, A.H. Diverse Tastes: Genetics of Sweet and Bitter Perception. Physiol. Behav. 2006, 88, 215–226. [Google Scholar] [CrossRef]
- Dotson, C.D.; Zhang, L.; Xu, H.; Shin, Y.-K.; Vigues, S.; Ott, S.H.; Elson, A.E.T.; Choi, H.J.; Shaw, H.; Egan, J.M.; et al. Bitter Taste Receptors Influence Glucose Homeostasis. PLoS ONE 2008, 3, e3974. [Google Scholar] [CrossRef]
- Melo, S.V.; Agnes, G.; Vitolo, M.R.; Mattevi, V.S.; Campagnolo, P.D.B.; Almeida, S. Evaluation of the Association between the TAS1R2 and TAS1R3 Variants and Food Intake and Nutritional Status in Children. Genet. Mol. Biol. 2017, 40, 415–420. [Google Scholar] [CrossRef]
- Ramos-Lopez, O.; Panduro, A.; Martinez-Lopez, E.; Roman, S. Sweet Taste Receptor TAS1R2 Polymorphism (Val191Val) Is Associated with a Higher Carbohydrate Intake and Hypertriglyceridemia among the Population of West Mexico. Nutrients 2016, 8, 101. [Google Scholar] [CrossRef]
- Chen, Q.-Y.; Alarcon, S.; Tharp, A.; Ahmed, O.M.; Estrella, N.L.; Greene, T.A.; Rucker, J.; Breslin, P.A. Perceptual Variation in Umami Taste and Polymorphisms in TAS1R Taste Receptor Genes123. Am. J. Clin. Nutr. 2009, 90, 770S–779S. [Google Scholar] [CrossRef]
- Belloir, C.; Gautier, A.; Karolkowski, A.; Delompré, T.; Jeannin, M.; Moitrier, L.; Neiers, F.; Briand, L. Optimized Vector for Functional Expression of the Human Bitter Taste Receptor TAS2R14 in HEK293 Cells. Protein Expr. Purif. 2025, 227, 106643. [Google Scholar] [CrossRef]
- Belloir, C.; Brulé, M.; Tornier, L.; Neiers, F.; Briand, L. Biophysical and Functional Characterization of the Human TAS1R2 Sweet Taste Receptor Overexpressed in a HEK293S Inducible Cell Line. Sci. Rep. 2021, 11, 22238. [Google Scholar] [CrossRef]
- Slack, J.P.; Brockhoff, A.; Batram, C.; Menzel, S.; Sonnabend, C.; Born, S.; Galindo, M.M.; Kohl, S.; Thalmann, S.; Ostopovici-Halip, L.; et al. Modulation of Bitter Taste Perception by a Small Molecule hTAS2R Antagonist. Curr. Biol. 2010, 20, 1104–1109. [Google Scholar] [CrossRef]
- Ueda, T.; Ugawa, S.; Yamamura, H.; Imaizumi, Y.; Shimada, S. Functional Interaction between T2R Taste Receptors and G-Protein α Subunits Expressed in Taste Receptor Cells. J. Neurosci. 2003, 23, 7376–7380. [Google Scholar] [CrossRef]
- Gibbons, C.; O’Hara, B.; O’Connor, D.; Hardman, C.; Wilton, M.; Harrold, J.A.; Almiron-Roig, E.; Navas-Carretero, S.; Hodgkins, C.E.; Nazare, J.A.; et al. Acute and Repeated Impact of Sweeteners and Sweetness Enhancers in Solid and Semi-Solid Foods on Appetite: Protocol for a Multicentre, Cross-over, RCT in People with Overweight/Obesity—The SWEET Project. BMJ Open 2022, 12, e063903. [Google Scholar] [CrossRef]
- Almiron-Roig, E.; Navas-Carretero, S.; Castelnuovo, G.; Kjølbæk, L.; Romo-Hualde, A.; Normand, M.; Maloney, N.; Hardman, C.A.; Hodgkins, C.E.; Moshoyiannis, H.; et al. Impact of Acute Consumption of Beverages Containing Plant-Based or Alternative Sweetener Blends on Postprandial Appetite, Food Intake, Metabolism, and Gastro-Intestinal Symptoms: Results of the SWEET Beverages Trial. Appetite 2023, 184, 106515. [Google Scholar] [CrossRef]
- Gnirke, A.; Melnikov, A.; Maguire, J.; Rogov, P.; LeProust, E.M.; Brockman, W.; Fennell, T.; Giannoukos, G.; Fisher, S.; Russ, C.; et al. Solution Hybrid Selection with Ultra-Long Oligonucleotides for Massively Parallel Targeted Sequencing. Nat. Biotechnol. 2009, 27, 182–189. [Google Scholar] [CrossRef]
- Adzhubei, I.A.; Schmidt, S.; Peshkin, L.; Ramensky, V.E.; Gerasimova, A.; Bork, P.; Kondrashov, A.S.; Sunyaev, S.R. A Method and Server for Predicting Damaging Missense Mutations. Nat. Methods 2010, 7, 248–249. [Google Scholar] [CrossRef]
- Bachmanov, A.A.; Tordoff, M.G.; Beauchamp, G.K. Sweetener Preference of C57BL/6ByJ and 129P3/J Mice. Chem. Senses 2001, 26, 905–913. [Google Scholar] [CrossRef]
- Reed, D.R.; Li, S.; Li, X.; Huang, L.; Tordoff, M.G.; Starling-Roney, R.; Taniguchi, K.; West, D.B.; Ohmen, J.D.; Beauchamp, G.K.; et al. Polymorphisms in the Taste Receptor Gene (Tas1r3) Region Are Associated with Saccharin Preference in 30 Mouse Strains. J. Neurosci. 2004, 24, 938–946. [Google Scholar] [CrossRef]
- Methven, L.; Ellis, L.; Kavaliauskaite, G. Investigating Perception and Liking of Non-Nutritive Sweeteners in Individuals Representing Different Taste Receptor Genotypes. In Proceedings of the 15th Weurman Flavour Research Symposium, Graz, Austria, 18–22 September 2017; University and Reading: Reading, UK, 2018; pp. 193–198. [Google Scholar]
- Koc, G.; Soyocak, A.; Andac-Ozturk, S. TAS1R2 Rs35874116 and TRPM5 Rs886277 Polymorphisms Are Not Related with Risk of Obesity. Int. J. Clin. Pract. 2021, 75, e14562. [Google Scholar] [CrossRef]
- Smith, N.J.; Grant, J.N.; Moon, J.I.; So, S.S.; Finch, A.M. Critically Evaluating Sweet Taste Receptor Expression and Signaling through a Molecular Pharmacology Lens. FEBS J. 2021, 288, 2660–2672. [Google Scholar] [CrossRef]
- Hwang, L.-D.; Lin, C.; Gharahkhani, P.; Cuellar-Partida, G.; Ong, J.-S.; An, J.; Gordon, S.D.; Zhu, G.; MacGregor, S.; Lawlor, D.A.; et al. New Insight into Human Sweet Taste: A Genome-Wide Association Study of the Perception and Intake of Sweet Substances. Am. J. Clin. Nutr. 2019, 109, 1724–1737. [Google Scholar] [CrossRef]
- Diószegi, J.; Mohammad Kurshed, A.A.; Pikó, P.; Kósa, Z.; Sándor, J.; Ádány, R. Association of Single Nucleotide Polymorphisms with Taste and Food Preferences of the Hungarian General and Roma Populations. Appetite 2021, 164, 105270. [Google Scholar] [CrossRef]
- Fernández-Carrión, R.; Sorlí, J.V.; Coltell, O.; Pascual, E.C.; Ortega-Azorín, C.; Barragán, R.; Giménez-Alba, I.M.; Alvarez-Sala, A.; Fitó, M.; Ordovas, J.M.; et al. Sweet Taste Preference: Relationships with Other Tastes, Liking for Sugary Foods and Exploratory Genome-Wide Association Analysis in Subjects with Metabolic Syndrome. Biomedicines 2022, 10, 79. [Google Scholar] [CrossRef]
- Frayling, T.M.; Beaumont, R.N.; Jones, S.E.; Yaghootkar, H.; Tuke, M.A.; Ruth, K.S.; Casanova, F.; West, B.; Locke, J.; Sharp, S.; et al. A Common Allele in FGF21 Associated with Sugar Intake Is Associated with Body Shape, Lower Total Body-Fat Percentage, and Higher Blood Pressure. Cell Rep. 2018, 23, 327–336. [Google Scholar] [CrossRef]
- Graham, C.A.-M.; Spedicati, B.; Pelliccione, G.; Gasparini, P.; Concas, M.P. Regulator of G-Protein Signalling 9: A New Candidate Gene for Sweet Food Liking? Foods 2023, 12, 1739. [Google Scholar] [CrossRef]
- Kawafune, K.; Hachiya, T.; Nogawa, S.; Takahashi, S.; Jia, H.; Saito, K.; Kato, H. Strong Association between the 12q24 Locus and Sweet Taste Preference in the Japanese Population Revealed by Genome-Wide Meta-Analysis. J. Hum. Genet. 2020, 65, 939–947. [Google Scholar] [CrossRef]
- Kogelman, L.J.A.; Zhernakova, D.V.; Westra, H.-J.; Cirera, S.; Fredholm, M.; Franke, L.; Kadarmideen, H.N. An Integrative Systems Genetics Approach Reveals Potential Causal Genes and Pathways Related to Obesity. Genome Med. 2015, 7, 105. [Google Scholar] [CrossRef]
- Priego, T.; Sánchez, J.; Picó, C.; Ahrens, W.; De Henauw, S.; Kourides, Y.; Lissner, L.; Molnár, D.; Moreno, L.A.; Russo, P.; et al. TAS1R3 and UCN2 Transcript Levels in Blood Cells Are Associated with Sugary and Fatty Food Consumption in Children. J. Clin. Endocrinol. Metab. 2015, 100, 3556–3564. [Google Scholar] [CrossRef] [PubMed]
- Keskitalo, K.; Knaapila, A.; Kallela, M.; Palotie, A.; Wessman, M.; Sammalisto, S.; Peltonen, L.; Tuorila, H.; Perola, M. Sweet Taste Preferences Are Partly Genetically Determined: Identification of a Trait Locus on Chromosome 161. Am. J. Clin. Nutr. 2007, 86, 55–63. [Google Scholar] [CrossRef] [PubMed]
- Stevens, H.; Piluso, F.; Gasparini, P.; Mavrommatis, Y.; Pilic, L.; Graham, C.A.-M.; Concas, M.P. The Genetics of Sweet Taste: Perception, Feeding Behaviours, and Health. Proceedings 2024, 91, 342. [Google Scholar] [CrossRef]
- Bahauddin, A.R.; Shaari, N.; Shariff, Z.; Karim, R. Association Between TAS1R2 Gene Polymorphism (Rs12033832) and Sweet Taste Perception Amongst Malay Obese and Non- Obese Subjects. Malays. J. Med. Health Sci. 2020, 16, 2636–9346. [Google Scholar]
| Gene | SNP Reference Sequence | Allele a | mRNA Position | Amino Acid Encoded | Position in Protein | Domain Within Sweet Taste Receptor b | Variant Name Studied | Minor Allele Frequency c | References |
|---|---|---|---|---|---|---|---|---|---|
| TAS1R2 | rs9701796 | G C | 27 | Ser (S) Cys (C) | 9 | Signal peptide | C9S | 0.202 | [48,49,50,51,58,59,60] |
| rs72953144 | G A | 63 | Pro (P) Leu (L) | 21 | VFT | P21L | 0.022 | [48] | |
| rs35874116 | T C | 572 | Ile (I) Val (V) | 191 | VFT | I191V | 0.267 | [29,48,49,50,51,58,61,62] | |
| rs74604275 | T C | 634 | Ser (S) Gly (G) | 212 | VFT | S212G | 0.004 | [48] | |
| rs139655863 | T C | 700 | Ile (I) Val (V) | 234 | VFT | I234V | 0.005 | [48] | |
| rs34545913 rs34447754 | C>G G>C | 951 | Arg (R) Pro (P) Gly (G) | 317 | VFT | R317P R317G | 0.014 0.238 | [48] | |
| rs28374389 | T C | 1457 | Ile (I) Val (V) | 486 | CRD | I486V | 0.126 | [48,59,60] | |
| rs6662276 | C A | 1721 | Ala (A) Thr (T) | 574 | TM1 | A574T | 0.091 | [48,58,59,60] | |
| rs41273167 | A G | 1785 | Ile (I) Thr (T) | 595 | ICL1 | I595T | 0.012 | ||
| rs1212135598 | C T | 2065 | Lys (K) Gln (Q) | 689 | TM4 | K689Q | <0.001 | [48] | |
| rs9988418 | C T | 2514 | Arg (R) Lys (K) | 838 | C-terminal | R838K | 0.058 | [48,59] | |
| TAS1R3 | rs76755863 | G A | 13 | Ala (A) Thr (T) | 5 | Signal peptide | A5T | 0.019 | [48,63] |
| rs776847816 | T C | 284 | Leu (L) Pro (P) | 95 | VFT | L95P | <0.001 | [48] | |
| rs746577143 | T C | 329 | Met (M) Thr (T) | 110 | VFT | M110T | <0.001 | [45] | |
| rs111615792 | G A | 740 | Arg (R) His (H) | 247 | VFT | R247H | 0.067 | [48,63] | |
| rs147600530 | G T | 1099 | Gly (G) Cys (C) | 367 | VFT | G367C | 0.016 | [48] | |
| rs141949092 | G A | 1338 | Ser (S) Asn (N) | 446 | VFT | S446N | <0.001 | [58] | |
| rs200580453 | C G | 1542 | Phe (F) Leu (L) | 514 | VFT | F514L | <0.001 | [58] | |
| rs1425170639 | C A | 1652 | Ser (S) Asn (N) | 551 | VFT | S551N | <0.001 | [45] | |
| rs147441599 | C A | 2148 | Thr (T) Met (M) | 716 | ECL2 | T716M | <0.001 | [58] | |
| rs112507608 | G A | 2203 | Ala (A) Thr (T) | 735 | TM5 | A735T | 0.060 | [48] | |
| rs79148073 | T C | 2246 | Phe (F) Ser (S) | 749 | TM5 | F749S | <0.001 | [43] | |
| rs307377 | T C | 2269 | Cys (C) Arg (R) | 757 | ICL3 | C757R | 0.048 | [29,44,48,59,60,63] | |
| rs199779671 | C G | 2340 | Pro (P) Ala (A) | 780 | TM6 | P780A | <0.001 | [58] | |
| rs34810828 | G A | 2439 | Lys (K) Arg (R) | 813 | TM7 | R813K | 0.006 | [44,45] | |
| rs12030797 | C A | 2469 | Phe (F) Leu (L) | 823 | TM7 | L823F | <0.001 | [44,59,60] | |
| rs35913253 | G A | 2496 | Arg (R) Gly (G) | 832 | C-terminal | G832R | <0.001 | [44] |
| Food Products | ||||
|---|---|---|---|---|
| Centres | Beverages | Biscuits | Cereal, Yogurt and Chocolate | Total/ Centre |
| CRNH | - | 24 | - | 24 |
| UNAV | 23 | - | 27 | 50 |
| UCPH | 21 | - | 8 | 29 |
| UNILIV | - | - | 30 | 30 |
| UNILEEDS | - | 29 | - | 29 |
| Total/product | 44 | 53 | 65 | 162 |
| mTAS1R3-WT | mTAS1R3-I60T | |||||
|---|---|---|---|---|---|---|
| Sweeteners | EC50 (µM) | Max ΔF/F0 | EC50 (µM) | Max ΔF/F0 | R1 | R2 |
| Sucralose | 778 ± 22 | 2.03 ± 0.02 | 1520 ± 56 | 1.47 ± 0.03 | 2.0 | 0.7 |
| Neotame | - | - | - | - | - | - |
| Acesulfame K | 439 ± 10 | 1.63 ± 0.02 | 744 ± 19 | 1.03 ± 0.02 | 1.7 | 0.6 |
| Rebaudioside A | 41 ± 2 | 1.37 ± 0.02 | 78 ± 3 | 0.62 ± 0.01 | 1.9 | 0.5 |
| Rebaudioside M | 44 ± 4 | 1.41 ± 0.05 | 73 ± 2 | 0.71 ± 0.01 | 1.7 | 0.5 |
| Mogroside V | 133 ± 6 | 0.84 ± 0.02 | 225 ± 25 | 0.50 ± 0.03 | 1.7 | 0.5 |
| Saccharin | 1521 ± 17 | 1.71 ± 0.01 | 2539 ± 61 | 0.96 ± 0.02 | 1.7 | 0.6 |
| Cyclamate | - | - | - | - | - | - |
| NHDC | - | - | - | - | - | - |
| Thaumatin | - | - | - | - | - | - |
| Brazzein * | - | - | - | - | - | - |
| Perillartine | - | - | - | - | - | - |
| Sucralose | Neotame | Acesulfame K | ||||||||||
| Variant | EC50 (µM) | R1 | Max ΔF/F0 | R2 | EC50 (µM) | R1 | Max ΔF/F0 | R2 | EC50 (µM) | R1 | Max ΔF/F0 | R2 |
| WT | 90 ± 11 | 1.0 | 0.59 ± 0.03 | 1.0 | 0.84 ± 0.13 | 1.0 | 0.56 ± 0.03 | 1.0 | 705 ± 7 | 1.0 | 0.31 ± 0.01 | 1.0 |
| C9S | 88 ± 15 | 1.0 | 0.72 ± 0.05 | 1.2 | 0.85 ± 0.16 | 1.0 | 0.66 ± 0.04 | 1.2 | 466 ± 54 | 0.7 | 0.32 ± 0.01 | 1.0 |
| P21L | 80 ± 13 | 0.9 | 0.50 ± 0.03 | 0.8 | 0.67 ± 0.16 | 0.8 | 0.53 ± 0.04 | 0.9 | 374 ± 11 | 0.5 | 0.24 ± 0.01 | 0.8 |
| I191V | 87 ± 7 | 1.0 | 0.52 ± 0.02 | 0.9 | 0.84 ± 0.08 | 1.0 | 0.64 ± 0.02 | 1.1 | 492 ± 30 | 0.7 | 0.27 ± 0.01 | 0.9 |
| S212G | 96 ± 6 | 1.1 | 0.64 ± 0.02 | 1.1 | 0.99 ± 0.05 | 1.2 | 0.63 ± 0.01 | 1.1 | 438 ± 25 | 0.6 | 0.28 ± 0.01 | 0.9 |
| I234V | 60 ± 7 | 0.7 | 0.75 ± 0.03 | 1.3 | 0.80 ± 0.08 | 1.0 | 0.66 ± 0.02 | 1.2 | 510 ± 63 | 0.7 | 0.41 ± 0.02 | 1.3 |
| R317G | 62 ± 11 | 0.7 | 0.80 ± 0.05 | 1.4 | 0.73 ± 0.13 | 0.9 | 0.56 ± 0.03 | 1.0 | 465 ± 55 | 0.7 | 0.46 ± 0.02 | 1.5 |
| R317P | 98 ± 19 | 1.1 | 0.76 ± 0.06 | 1.3 | 0.86 ± 0.16 | 1.0 | 0.63 ± 0.04 | 1.1 | 685 ± 41 | 1.0 | 0.42 ± 0.01 | 1.4 |
| I486V | 54 ± 5 | 0.6 | 0.69 ± 0.02 | 1.2 | 0.71 ± 0.06 | 0.8 | 0.61 ± 0.01 | 1.1 | 778 ± 108 | 1.1 | 0.46 ± 0.03 | 1.5 |
| A574T | 81 ± 2 | 0.9 | 0.63 ± 0.01 | 1.1 | 0.73 ± 0.03 | 0.9 | 0.60 ± 0.01 | 1.1 | 658 ± 65 | 0.9 | 0.34 ± 0.02 | 1.1 |
| I595T | 108 ± 5 | 1.2 | 0.08 ± 0.02 | 0.1 | 1.27 ± 0.18 | 1.5 | 0.09 ± 0.01 | 0.2 | 300 ± 139 | 0.4 | 0.02 ± 0.01 | 0.1 |
| K689Q | 307 ± nd | 3.4 | 0.08 ± 0.01 | 0.1 | 3.58 ± 0.46 | 4.3 | 0.12 ± 0.01 | 0.2 | 3000 ± ns | 4.3 | 0.02 ± nd | 0.1 |
| R838K | 82 ± 6 | 0.9 | 0.75 ± 0.02 | 1.3 | 0.66 ± 0.05 | 0.8 | 0.75 ± 0.02 | 1.3 | 484 ± 47 | 0.7 | 0.42 ± 0.02 | 1.4 |
| Rebaudioside A | Rebaudioside M | Mogroside V | ||||||||||
| Variant | EC50 (µM) | R1 | Max ΔF/F0 | R2 | EC50 (µM) | R1 | Max ΔF/F0 | R2 | EC50 (µM) | R1 | Max ΔF/F0 | R2 |
| WT | 22.8 ± 1.0 | 1.0 | 1.06 ± 0.02 | 1.0 | 16.9 ± 0.5 | 1.0 | 1.12 ± 0.01 | 1.0 | 17.3 ± 1.2 | 1.0 | 1.28 ± 0.03 | 1.0 |
| C9S | 26.4 ± 0.5 | 1.2 | 1.18 ± 0.01 | 1.1 | 23.7 ± 1.6 | 1.4 | 1.18 ± 0.03 | 1.1 | 18.6 ± 1.6 | 1.1 | 1.32 ± 0.04 | 1.0 |
| P21L | 25.3 ± 0.5 | 1.1 | 1.09 ± 0.01 | 1.0 | 19.5 ± 0.9 | 1.2 | 0.99 ± 0.02 | 0.9 | 19.9 ± 1.1 | 1.2 | 1.18 ± 0.02 | 0.9 |
| I191V | 28.2 ± 1.8 | 1.2 | 1.19 ± 0.03 | 1.1 | 21.0 ± 0.4 | 1.2 | 1.10 ± 0.01 | 1.0 | 19.2 ± 1.7 | 1.1 | 1.33 ± 0.04 | 1.0 |
| S212G | 25.5 ± 0.6 | 1.1 | 1.16 ± 0.01 | 1.1 | 23.2 ± 1.1 | 1.4 | 1.14 ± 0.02 | 1.0 | 21.2 ± 2.1 | 1.2 | 1.32 ± 0.05 | 1.0 |
| I234V | 28.4 ± 3.8 | 1.2 | 1.06 ± 0.06 | 1.0 | 18.4 ± 0.6 | 1.1 | 1.14 ± 0.01 | 1.0 | 19.4 ± 1.6 | 1.1 | 1.28 ± 0.04 | 1.0 |
| R317G | 27.2 ± 1.0 | 1.2 | 1.07 ± 0.02 | 1.0 | 20.4 ± 0.4 | 1.2 | 1.23 ± 0.01 | 1.1 | 18.4 ± 1.0 | 1.1 | 1.34 ± 0.03 | 1.0 |
| R317P | 37.2 ± 3.8 | 1.6 | 1.15 ± 0.05 | 1.1 | 26.9 ± 0.5 | 1.6 | 1.19 ± 0.01 | 1.1 | 27.9 ± 2.5 | 1.6 | 1.51 ± 0.05 | 1.2 |
| I486V | 22.7 ± 1.1 | 1.0 | 1.06 ± 0.02 | 1.0 | 17.6 ± 0.6 | 1.0 | 1.20 ± 0.01 | 1.1 | 20.1 ± 0.9 | 1.2 | 1.39 ± 0.02 | 1.1 |
| A574T | 19.8 ± 1.3 | 0.9 | 0.98 ± 0.02 | 0.9 | 16.3 ± 0.5 | 1.0 | 1.08 ± 0.01 | 1.0 | 16.7 ± 3.1 | 1.0 | 1.25 ± 0.08 | 1.0 |
| I595T | 27.6 ± 2.0 | 1.2 | 0.16 ± 0.01 | 0.2 | 35.8 ± 4.0 | 2.1 | 0.23 ± 0.01 | 0.2 | 31.4 ± 6.0 | 1.8 | 0.27 ± 0.03 | 0.2 |
| K689Q | 77.0 ± 5.7 | 3.4 | 0.29 ± 0.01 | 0.3 | 75.8 ± 2.3 | 4.5 | 0.36 ± 0.01 | 0.3 | 57.9 ± 0.4 | 3.3 | 0.54 ± 0.01 | 0.4 |
| R838K | 23.0 ± 1.4 | 1.0 | 1.09 ± 0.03 | 1.0 | 18.2 ± 1.1 | 1.1 | 1.20 ± 0.01 | 1.1 | 17.1 ± 0.9 | 1.0 | 1.31 ± 0.02 | 1.0 |
| Saccharin | Cyclamate | NHDC | ||||||||||
| Variant | EC50 (µM) | R1 | Max ΔF/F0 | R2 | EC50 (µM) | R1 | Max ΔF/F0 | R2 | EC50 (µM) | R1 | Max ΔF/F0 | R2 |
| WT | 274.4 ± 4.1 | 1.0 | 0.64 ± 0.01 | 1.0 | 1889 ± 46 | 1.0 | 0.87 ± 0.01 | 1.0 | 76.7 ± 4.4 | 1.0 | 0.71 ± 0.02 | 1.0 |
| C9S | 370.6 ± 38.3 | 1.3 | 0.55 ± 0.03 | 0.9 | 1806 ± 55 | 1.0 | 0.86 ± 0.01 | 1.0 | 99.0 ± 5.2 | 1.3 | 0.83 ± 0.02 | 1.2 |
| P21L | 352.6 ± 32.5 | 1.3 | 0.46 ± 0.03 | 0.7 | 2114 ± 55 | 1.1 | 0.86 ± 0.01 | 1.0 | 100.1 ± 5.1 | 1.3 | 0.81 ± 0.02 | 1.1 |
| I191V | 285.0 ± 2.5 | 1.0 | 0.37 ± 0.01 | 0.6 | 2017 ± 31 | 1.1 | 0.88 ± 0.01 | 1.0 | 108.5 ± 4.6 | 1.4 | 0.73 ± 0.01 | 1.0 |
| S212G | 304.6 ± 9.3 | 1.1 | 0.51 ± 0.01 | 0.8 | 1767 ± 44 | 0.9 | 0.92 ± 0.01 | 1.1 | 90.7 ± 8.1 | 1.2 | 0.84 ± 0.03 | 1.2 |
| I234V | 289.0 ± 14.4 | 1.0 | 0.66 ± 0.02 | 1.0 | 2250 ± 42 | 1.2 | 1.07 ± 0.01 | 1.2 | 94.7 ± 6.8 | 1.2 | 0.80 ± 0.02 | 1.1 |
| R317G | 224.5 ± 6.1 | 0.8 | 0.84 ± 0.01 | 1.3 | 2161 ± 44 | 1.1 | 1.15 ± 0.01 | 1.3 | 97.1 ± 5.0 | 1.3 | 0.85 ± 0.02 | 1.2 |
| R317P | 356.6 ± 24.2 | 1.3 | 0.38 ± 0.02 | 0.6 | 2858 ± 62 | 1.5 | 0.80 ± 0.01 | 0.9 | 171.3 ± 26.2 | 2.2 | 0.58 ± 0.04 | 0.8 |
| I486V | 293.6 ± 11.6 | 1.1 | 0.67 ± 0.02 | 1.0 | 1876 ± 36 | 1.0 | 1.08 ± 0.01 | 1.2 | 92.6 ± 5.2 | 1.2 | 0.92 ± 0.02 | 1.3 |
| A574T | 247.5 ± 15.7 | 0.9 | 0.57 ± 0.02 | 0.9 | 1783 ± 49 | 0.9 | 0.92 ± 0.01 | 1.1 | 73.4 ± 8.4 | 1.0 | 0.81 ± 0.03 | 1.1 |
| I595T | - | - | - | - | 2026 ± 102 | 1.1 | 0.06 ± 0.01 | 0.1 | 70.3 ± 1.3 | 0.9 | 0.09 ± 0.01 | 0.1 |
| K689Q | - | - | - | - | 5014 ± 220 | 2.7 | 0.41 ± 0.01 | 0.5 | - | - | - | - |
| R838K | 253.5 ± 5.0 | 0.9 | 0.65 ± 0.01 | 1.0 | 1734 ± 24 | 0.9 | 0.99 ± 0.01 | 1.1 | 75.8 ± 1.2 | 1.0 | 0.79 ± 0.05 | 1.1 |
| Thaumatin | Brazzein | |||||||||||
| Variant | EC50 (µM) | R1 | Max ΔF/F0 | R2 | EC50 (mg/L) | R1 | Max ΔF/F0 | R2 | ||||
| WT | 7.6 ± 0.3 | 1.0 | 0.72 ± 0.01 | 1.0 | 1.0 | 1.17 ± 0.04 | 1.0 | |||||
| C9S | 6.6 ± 0.1 | 0.9 | 0.61 ± 0.01 | 0.8 | 95.5 ± 9.7 | 1.2 | 1.14 ± 0.03 | 1.0 | ||||
| P21L | 3.3 ± nd | 0.4 | 0.19 ± 0.04 | 0.3 | 111.8 ± 7.5 | 1.1 | 1.09 ± 0.01 | 0.9 | ||||
| I191V | 7.7 ± 0.9 | 1.0 | 0.34 ± 0.02 | 0.5 | 105.2 ± 35.7 | 1.7 | 1.12 ± 0.02 | 1.0 | ||||
| S212G | 6.1 ± 0.2 | 0.8 | 0.6 ± 0.01 | 0.8 | 161.6 ± 8.6 | 1.4 | 1.18 ± 0.02 | 1.0 | ||||
| I234V | 13.8 ± 0.2 | 1.8 | 1.25 ± 0.01 | 1.7 | 129.6 ± 8.2 | 1.1 | 1.34 ± 0.05 | 1.1 | ||||
| R317G | 10.9 ± 0.3 | 1.4 | 1.16 ± 0.02 | 1.6 | 100.8 ± 13.8 | 1.3 | 1.18 ± 0.03 | 1.0 | ||||
| R317P | 18.9 ± 0.7 | 2.5 | 0.42 ± 0.01 | 0.6 | 124.2 ± 12.4 | 2.5 | 0.91 ± 0.02 | 0.8 | ||||
| I486V | 10.5 ± 0.3 | 1.4 | 1.21 ± 0.02 | 1.7 | 234.2 ± 17.4 | 1.1 | 1.19 ± 0.03 | 1.0 | ||||
| A574T | 14.1 ± 0.3 | 1.9 | 1.37 ± 0.02 | 1.9 | 109.3 ± 9.8 | 1.2 | 1.14 ± 0.01 | 1.0 | ||||
| I595T | 27.8 ± 1.4 | 3.7 | 0.39 ± 0.01 | 0.5 | 116.2 ± 5.1 | 10.8 | 0.13 ± 0.02 | 0.1 | ||||
| K689Q | 33.5 ± nd | 4.4 | 0.87 ± 0.09 | 1.2 | 1035.0 ± 143.2 | 5.7 | 0.64 ± 0.03 | 0.5 | ||||
| R838K | 10.4 ± 0.2 | 1.4 | 1.3 ± 0.02 | 1.8 | 545.7 ± 68.2 | 1.2 | 1.2 ± 0.03 | 1.0 | ||||
| Perillartine (TAS1R2) | Perillartine (TAS1R2/TAS1R3) | |||||||||||
| Variant | EC50 (µM) | R1 | Max ΔF/F0 | R2 | EC50 (µM) | R1 | Max ΔF/F0 | R2 | ||||
| WT | 90.0 ± 17.0 | 1.0 | 1.06 ± 0.06 | 1.0 | 17.5 ± 2.2 | 1.0 | 1.47 ± 0.07 | 1.0 | ||||
| C9S | 189.6 ± 26.6 | 2.1 | 0.44 ± 0.02 | 0.4 | 17.8 ± 1.2 | 1.0 | 1.29 ± 0.03 | 0.9 | ||||
| P21L | 148.6 ± 22.7 | 1.7 | 0.33 ± 0.02 | 0.3 | 26.3 ± 1.1 | 1.5 | 1.35 ± 0.03 | 0.9 | ||||
| I191V | 389.6 ± 47.0 | 4.3 | 0.19 ± 0.01 | 0.2 | 27.9 ± 0.5 | 1.6 | 1.27 ± 0.01 | 0.9 | ||||
| S212G | 177.8 ± 22.7 | 2.0 | 0.47 ± 0.02 | 0.4 | 22.9 ± 0.6 | 1.3 | 1.30 ± 0.01 | 0.9 | ||||
| I234V | 311.3 ± 24.6 | 3.5 | 0.29 ± 0.01 | 0.3 | 20.1 ± 1.1 | 1.1 | 1.52 ± 0.03 | 1.0 | ||||
| R317G | 224.8 ± 31.9 | 2.5 | 0.17 ± 0.01 | 0.2 | 20.7 ± 0.6 | 1.2 | 1.53 ± 0.03 | 1.0 | ||||
| R317P | - | - | - | - | 35.2 ± 2.2 | 2.0 | 1.16 ± 0.03 | 0.8 | ||||
| I486V | 123.9 ± 14.1 | 1.4 | 0.39 ± 0.02 | 0.4 | 17.7 ± 1.2 | 1.0 | 1.49 ± 0.04 | 1.0 | ||||
| ZA574T | 85.3 ± 12.4 | 0.9 | 0.25 ± 0.01 | 0.2 | 19.2 ± 0.9 | 1.1 | 1.39 ± 0.02 | 0.9 | ||||
| I595T | - | - | - | - | 53.2 ± 1.6 | 3.0 | 0.51 ± 0.01 | 0.3 | ||||
| K689Q | - | - | - | - | 132.5 ± 16.9 | 7.6 | 1.36 ± 0.01 | 0.9 | ||||
| R838K | 92.4 ± 19.7 | 1.0 | 0.35 ± 0.03 | 0.3 | 19.1 ± 1.2 | 1.1 | 1.45 ± 0.03 | 1.0 | ||||
| Sucralose | Neotame | Acesulfame K | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Variant | EC50 (µM) | R1 | Max ΔF/F0 | R2 | EC50 (µM) | R1 | Max ΔF/F0 | R2 | EC50 (µM) | R1 | Max ΔF/F0 | R2 |
| WT | 109 ± 4 | 1.0 | 0.65 ± 0.01 | 1.0 | 1.22 ± 0.11 | 1.0 | 0.65 ± 0.02 | 1.0 | 622 ± 45 | 1.0 | 0.28 ± 0.01 | 1.0 |
| A5T | 135 ± 5 | 1.2 | 0.63 ± 0.01 | 1.0 | 1.18 ± 0.05 | 1.0 | 0.65 ± 0.01 | 1.0 | 852 ± 34 | 1.4 | 0.31 ± 0.01 | 1.1 |
| L95P | - | - | - | - | - | - | - | - | - | - | - | - |
| M110T | 282 ± 101 | 2.6 | 0.11 ± 0.02 | 0.2 | 2.70 ± 0.29 | 2.2 | 0.19 ± 0.01 | 0.3 | 1057 ± 648 | 1.7 | 0.20 ± 0.01 | 0.7 |
| R247H | 122 ± 2 | 1.1 | 0.69 ± 0.01 | 1.1 | 1.12 ± 0.07 | 0.9 | 0.71 ± 0.02 | 1.1 | 639 ± 17 | 1.0 | 0.33 ± 0.01 | 1.2 |
| G367C | 182 ± 7 | 1.7 | 0.32 ± 0.01 | 0.5 | 1.95 ± 0.09 | 1.6 | 0.31 ± 0.01 | 0.5 | 443 ± 121 | 0.7 | 0.06 ± 0.01 | 0.2 |
| S446N | 155 ± 11 | 1.4 | 0.68 ± 0.02 | 1.0 | 1.22 ± 0.10 | 1.0 | 0.63 ± 0.02 | 1.0 | 487 ± 78 | 0.8 | 0.19 ± 0.01 | 0.7 |
| F514L | 141 ± 3 | 1.3 | 0.49 ± 0.01 | 0.8 | 1.23 ± 0.06 | 1.0 | 0.45 ± 0.01 | 0.7 | 442 ± 24 | 0.7 | 0.14 ± 0.01 | 0.5 |
| S551N | - | - | - | - | - | - | - | - | - | - | - | - |
| T716M | 161 ± 14 | 1.5 | 0.47 ± 0.02 | 0.7 | 1.30 ± 0.06 | 1.1 | 0.54 ± 0.01 | 0.8 | 739 ± 72 | 1.2 | 0.30 ± 0.01 | 1.1 |
| A735T | 151 ± 4 | 1.4 | 0.70 ± 0.01 | 1.1 | 0.93 ± 0.07 | 0.8 | 0.61 ± 0.02 | 0.9 | 685 ± 56 | 1.1 | 0.47 ± 0.02 | 1.7 |
| F749S | 252 ± 36 | 2.3 | 0.18 ± 0.01 | 0.3 | 1.82 ± 0.12 | 1.5 | 0.22 ± 0.01 | 0.3 | 410 ± 242 | 0.7 | 0.04 ± 0.01 | 0.1 |
| C757R | 113 ± 10 | 1.0 | 0.84 ± 0.03 | 1.3 | 0.88 ± 0.12 | 0.7 | 0.75 ± 0.03 | 1.2 | 468 ± 31 | 0.8 | 0.42 ± 0.01 | 1.5 |
| P780A | 149 ± 12 | 1.4 | 0.76 ± 0.03 | 1.2 | 1.03 ± 0.08 | 0.8 | 0.63 ± 0.02 | 1.0 | 421 ± 31 | 0.7 | 0.30 ± 0.01 | 1.1 |
| R813K | 120 ± 9 | 1.1 | 0.90 ± 0.03 | 1.4 | 0.98 ± 0.07 | 0.8 | 0.79 ± 0.02 | 1.2 | 433 ± 34 | 0.7 | 0.40 ± 0.01 | 1.4 |
| L823F | 134 ± 8 | 1.2 | 0.84 ± 0.02 | 1.3 | 1.10 ± 0.06 | 0.9 | 0.76 ± 0.01 | 1.2 | 376 ± 34 | 0.6 | 0.33 ± 0.01 | 1.2 |
| G832R | 81 ± 4 | 0.7 | 0.91 ± 0.02 | 1.4 | 0.80 ± 0.07 | 0.7 | 0.88 ± 0.02 | 1.4 | 468 ± 64 | 0.8 | 0.55 ± 0.03 | 2.0 |
| Rebaudioside A | Rebaudioside M | Mogroside V | ||||||||||
| Variant | EC50 (µM) | R1 | Max ΔF/F0 | R2 | EC50 (µM) | R1 | Max ΔF/F0 | R2 | EC50 (µM) | R1 | Max ΔF/F0 | R2 |
| WT | 57.8 ± 5.2 | 1.0 | 0.63 ± 0.03 | 1.0 | 40.4 ± 2.9 | 1.0 | 0.69 ± 0.02 | 1.0 | 31.3 ± 0.8 | 1.0 | 0.93 ± 0.01 | 1.0 |
| A5T | 46.9 ± 8.6 | 0.8 | 1.05 ± 0.08 | 1.7 | 30.2 ± 1.5 | 0.7 | 1.06 ± 0.02 | 1.5 | 31.3 ± 1.0 | 1.0 | 0.97 ± 0.01 | 1.0 |
| L95P | - | - | - | - | - | - | - | - | 262.5 ± nd | 8.4 | 0.08 ± nd | 0.1 |
| M110T | 103.9 ± 9.5 | 1.8 | 0.26 ± 0.01 | 0.4 | 103.2 ± 18.0 | 2.6 | 0.38 ± 0.04 | 0.6 | 94.5 ± 8.4 | 3.0 | 0.43 ± 0.02 | 0.5 |
| R247H | 40.7 ± 5.3 | 0.7 | 1.05 ± 0.06 | 1.7 | 29.7 ± 2.6 | 0.7 | 1.15 ± 0.04 | 1.7 | 28.7 ± 1.6 | 0.9 | 1.12 ± 0.02 | 1.2 |
| G367C | 125.1 ± 4.0 | 2.2 | 0.40 ± 0.06 | 0.6 | 47.3 ± 1.9 | 1.2 | 0.35 ± 0.01 | 0.5 | 82.4 ± 5.2 | 2.6 | 0.60 ± 0.02 | 0.6 |
| S446N | 56.1 ± 9.1 | 1.0 | 0.68 ± 0.05 | 1.1 | 36.9 ± 3.6 | 0.9 | 0.64 ± 0.03 | 0.9 | 47.7 ± 1.5 | 1.5 | 0.86 ± 0.01 | 0.9 |
| F514L | 65.2 ± 9.0 | 1.1 | 0.65 ± 0.09 | 1.0 | 32.1 ± 2.5 | 0.8 | 0.64 ± 0.02 | 0.9 | 44.7 ± 2.8 | 1.4 | 0.79 ± 0.02 | 0.8 |
| S551N | - | - | - | - | - | - | - | - | 111.2 ± nd | 3.6 | 0.03 ± nd | 0.0 |
| T716M | 41.3 ± 3.4 | 0.7 | 1.20 ± 0.04 | 1.9 | 30.7 ± 4.0 | 0.8 | 1.10 ± 0.06 | 1.6 | 36.4 ± 1.0 | 1.2 | 1.01 ± 0.01 | 1.1 |
| A735T | 30.6 ± 1.7 | 0.5 | 1.27 ± 0.03 | 2.0 | 23.0 ± 1.9 | 0.6 | 1.24 ± 0.04 | 1.8 | 28.7 ± 2.2 | 0.9 | 1.17 ± 0.03 | 1.3 |
| F749S | 88.9 ± 11.3 | 1.5 | 0.52 ± 0.03 | 0.8 | 71.8 ± 9.6 | 1.8 | 0.54 ± 0.03 | 0.8 | 65.5 ± 5.4 | 2.1 | 0.47 ± 0.02 | 0.5 |
| C757R | 28.4 ± 1.7 | 0.5 | 1.10 ± 0.03 | 1.7 | 24.3 ± 2.2 | 0.6 | 1.13 ± 0.04 | 1.6 | 21.7 ± 1.0 | 0.7 | 1.03 ± 0.02 | 1.1 |
| P780A | 26.3 ± 0.2 | 0.5 | 1.14 ± 0.01 | 1.8 | 23.0 ± 2.6 | 0.6 | 1.21 ± 0.05 | 1.8 | 22.5 ± 2.2 | 0.7 | 1.13 ± 0.04 | 1.2 |
| R813K | 40.6 ± 4.1 | 0.7 | 1.12 ± 0.05 | 1.8 | 27.4 ± 3.4 | 0.7 | 1.18 ± 0.06 | 1.7 | 22.0 ± 1.3 | 0.7 | 1.14 ± 0.02 | 1.2 |
| L823F | 31.8 ± 1.0 | 0.6 | 1.21 ± 0.02 | 1.9 | 33.2 ± 5.4 | 0.8 | 1.35 ± 0.09 | 2.0 | 22.0 ± 1.8 | 0.7 | 1.18 ± 0.04 | 1.3 |
| G832R | 26.7 ± 2.6 | 0.5 | 1.36 ± 0.05 | 2.2 | 22.7 ± 3.2 | 0.6 | 1.38 ± 0.07 | 2.0 | 19.1 ± 1.7 | 0.6 | 1.23 ± 0.04 | 1.3 |
| Saccharin | Cyclamate | NHDC | ||||||||||
| Variant | EC50 (µM) | R1 | Max ΔF/F0 | R2 | EC50 (µM) | R1 | Max ΔF/F0 | R2 | EC50 (µM) | R1 | Max ΔF/F0 | R2 |
| WT | 252.7 ± 6.4 | 1.0 | 0.75 ± 0.01 | 1.0 | 1591 ± 28 | 1.0 | 1.32 ± 0.01 | 1.0 | 50.3 ± 11.0 | 1.0 | 1.13 ± 0.01 | 1.0 |
| A5T | 302.2 ± 25.1 | 1.2 | 0.79 ± 0.04 | 1.1 | 1789 ± 35 | 1.1 | 1.28 ± 0.01 | 1.0 | 76.5 ± 13.1 | 1.5 | 1.24 ± 0.01 | 1.1 |
| L95P | - | - | - | - | - | - | - | - | - | - | - | - |
| M110T | 462.5 ± 41.4 | 1.8 | 0.15 ± 0.06 | 0.2 | 2617 ± 98 | 1.6 | 0.41 ± 0.01 | 0.3 | 202.6 ± 17.1 | 4.0 | 0.48 ± 0.02 | 0.4 |
| R247H | 209.6 ± 3.9 | 0.8 | 0.76 ± 0.01 | 1.0 | 1523 ± 46 | 1.0 | 1.13 ± 0.02 | 0.9 | 65.0 ± 8.9 | 1.3 | 1.14 ± 0.05 | 1.0 |
| G367C | 377.9 ± 28.3 | 1.5 | 0.40 ± 0.01 | 0.5 | 2124 ± 34 | 1.3 | 0.90 ± 0.01 | 0.7 | 81.3 ± 13.1 | 1.6 | 0.75 ± 0.05 | 0.7 |
| S446N | 243.4 ± 12.9 | 1.0 | 0.67 ± 0.02 | 0.9 | 1480 ± 45 | 0.9 | 1.23 ± 0.02 | 0.9 | 49.4 ± 8.6 | 1.0 | 1.07 ± 0.06 | 0.9 |
| F514L | 208.6 ± 5.7 | 0.8 | 0.51 ± 0.01 | 0.7 | 1250 ± 30 | 0.8 | 1.29 ± 0.01 | 1.0 | 37.9 ± 8.4 | 0.8 | 1.21 ± 0.08 | 1.1 |
| S551N | - | - | - | - | - | - | - | - | - | - | - | - |
| T716M | 257.9 ± 31.6 | 1.0 | 0.59 ± 0.04 | 0.8 | 1855 ± 45 | 1.2 | 1.03 ± 0.01 | 0.8 | 55.4 ± 18.4 | 1.1 | 0.84 ± 0.09 | 0.7 |
| A735T | 272.4 ± 24.8 | 1.0 | 0.89 ± 0.04 | 1.2 | 1869 ± 32 | 1.2 | 1.20 ± 0.01 | 0.9 | 42.2 ± 1.4 | 0.8 | 1.00 ± 0.01 | 0.9 |
| F749S | 439.7 ± 39.5 | 1.7 | 0.27 ± 0.01 | 0.4 | 3433 ± 1986 | 2.2 | 1.26 ± 0.04 | 1.0 | 149.8 ± 19.6 | 3.0 | 0.29 ± 0.01 | 0.3 |
| C757R | 170.3 ± 10.4 | 0.7 | 0.75 ± 0.02 | 1.0 | 1150 ± 54 | 0.7 | 1.16 ± 0.03 | 0.9 | 36.5 ± 16.0 | 0.7 | 0.98 ± 0.01 | 0.9 |
| P780A | 247.1 ± 29.0 | 1.0 | 0.89 ± 0.05 | 1.2 | 1166 ± 62 | 0.7 | 1.04 ± 0.03 | 0.8 | 46.4 ± 12.9 | 0.9 | 1.50 ± 0.01 | 1.3 |
| R813K | 156.4 ± 5.9 | 0.6 | 0.83 ± 0.01 | 1.1 | 1331 ± 22 | 0.8 | 1.15 ± 0.01 | 0.9 | 34.2 ± 15.2 | 0.7 | 1.00 ± 0.02 | 0.9 |
| L823F | 162.5 ± 5.2 | 0.6 | 0.83 ± 0.01 | 1.1 | 1369 ± 34 | 0.9 | 1.24 ± 0.01 | 0.9 | 43.1 ± 14.7 | 0.9 | 1.00 ± 0.01 | 0.9 |
| G832R | 186.4 ± 6.6 | 0.7 | 1.02 ± 0.02 | 1.4 | 998 ± 46 | 0.6 | 1.25 ± 0.03 | 0.9 | 28.1 ± 12.3 | 0.6 | 1.03 ± 0.01 | 0.9 |
| Thaumatin | Brazzein | Perillartine | ||||||||||
| Variant | EC50 (µM) | R1 | Max ΔF/F0 | R2 | EC50 (mg/L) | R1 | Max ΔF/F0 | R2 | EC50 (µM) | R1 | Max ΔF/F0 | R2 |
| WT | 10.4 ± 0.6 | 1.0 | 0.57 ± 0.02 | 1.0 | 112.8 ± 11.2 | 1.0 | 0.88 ± 0.03 | 1.0 | 13.8 ± 6.3 | 1.0 | 1.51 ± 0.22 | 1.0 |
| A5T | 9.7 ± 0.8 | 0.9 | 0.73 ± 0.04 | 1.3 | 108.3 ± 11.5 | 1.0 | 1.04 ± 0.04 | 1.2 | 13.2 ± 6.4 | 1.0 | 1.42 ± 0.23 | 0.9 |
| L95P | 27.4 ± 5.8 | 2.6 | 0.89 ± 0.01 | 1.6 | 2178.0 ± 616.0 | 19.3 | 0.10 ± 0.05 | 0.1 | 33.9 ± 2.8 | 2.5 | 0.14 ± 0.02 | 0.1 |
| M110T | 16.3 ± 0.9 | 1.6 | 0.38 ± 0.01 | 0.7 | 221.5 ± 11.3 | 2.0 | 0.68 ± 0.01 | 0.8 | 41.1 ± 19.4 | 3.0 | 1.31 ± 0.21 | 0.9 |
| R247H | 8.2 ± 2.3 | 0.8 | 0.73 ± 0.08 | 1.3 | 70.8 ± 8.1 | 0.6 | 0.95 ± 0.03 | 1.1 | 8.5 ± 3.1 | 0.6 | 1.26 ± 0.15 | 0.8 |
| G367C | 14.8 ± 0.5 | 1.4 | 0.52 ± 0.01 | 0.9 | 181.7 ± 6.9 | 1.6 | 0.72 ± 0.01 | 0.8 | 32.0 ± 17.1 | 2.3 | 1.49 ± 0.24 | 1.0 |
| S446N | 9.8 ± 1.1 | 0.9 | 0.70 ± 0.04 | 1.2 | 120.8 ± 9.3 | 1.1 | 0.98 ± 0.03 | 1.1 | 15.3 ± 8.6 | 1.1 | 1.42 ± 0.26 | 0.9 |
| F514L | 9.8 ± 1 | 0.9 | 0.72 ± 0.04 | 1.3 | 44.4 ± 6.4 | 0.4 | 1.07 ± 0.04 | 1.2 | 13.9 ± 7.3 | 1.0 | 1.40 ± 0.24 | 0.9 |
| S551N | 17.4 ± 2.2 | 1.7 | 0.25 ± 0.01 | 0.4 | 2378.0 ± 1623.0 | 21.1 | 0.21 ± 0.21 | 0.2 | 49.2 ± 13.6 | 3.6 | 0.52 ± 0.04 | 0.3 |
| T716M | 10.0 ± 0.5 | 1.0 | 0.76 ± 0.04 | 1.3 | 112.6 ± 13.6 | 1.0 | 0.94 ± 0.04 | 1.1 | 21.6 ± 11.4 | 1.6 | 1.73 ± 0.33 | 1.1 |
| A735T | 7.2 ± 1.5 | 0.7 | 0.70 ± 0.06 | 1.2 | 82.7 ± 14.9 | 0.7 | 0.90 ± 0.05 | 1.0 | 12.6 ± 5.9 | 0.9 | 1.56 ± 0.23 | 1.0 |
| F749S | 31.2 ± 3.5 | 3.0 | 0.42 ± 0.03 | 0.7 | 212.2 ± 8.6 | 1.9 | 0.65 ± 0.01 | 0.7 | 12.1 ± 6.1 | 0.9 | 1.90 ± 0.30 | 1.3 |
| C757R | 6.7 ± 1.4 | 0.6 | 0.69 ± 0.05 | 1.2 | 74.2 ± 1.9 | 0.7 | 0.86 ± 0.06 | 1.0 | 8.1 ± 2.6 | 0.6 | 1.23 ± 0.13 | 0.8 |
| P780A | 7.8 ± 1.3 | 0.8 | 0.86 ± 0.06 | 1.5 | 80.7 ± 14.8 | 0.7 | 1.06 ± 0.06 | 1.2 | 12.7 ± 5 | 0.9 | 1.44 ± 0.20 | 1.0 |
| R813K | 6.5 ± 0.9 | 0.6 | 0.74 ± 0.04 | 1.3 | 58.0 ± 10.5 | 0.5 | 1.00 ± 0.05 | 1.1 | 10.4 ± 3.6 | 0.8 | 1.28 ± 0.16 | 0.8 |
| L823F | 7.4 ± 1.1 | 0.7 | 0.71 ± 0.04 | 1.2 | 78.9 ± 14.2 | 0.7 | 0.98 ± 0.05 | 1.1 | 10.0 ± 3.1 | 0.7 | 1.39 ± 0.15 | 0.9 |
| G832R | 5.7 ± 0.7 | 0.5 | 0.76 ± 0.03 | 1.3 | 52.8 ± 15.7 | 0.5 | 0.97 ± 0.07 | 1.1 | 7.2 ± 3.0 | 0.5 | 1.18 ± 0.14 | 0.8 |
| Gene | Chromosome 1 Position | Ref | Alt | Variant Name | Tier | Frequency in the Study | Frequency in gnomAD | Rare Variant | Consequence | Impact | Damaging Effects Predicted (%) | Existing Variation |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 18859598 | C | T | P21P | NT | 0.003 | 0.003 | T | Synonymous | L | rs144454001 | ||
| 18854834 | G | A | S212S | NT | 0.003 | <0.001 | T | Synonymous | L | rs371978810 | ||
| 18839767 | G | A | S784S | NT | 0.003 | <0.001 | T | Synonymous | L | rs138899345 | ||
| 18839650 | C | T | T823T | NT | 0.003 | <0.001 | T | Synonymous | L | rs774217162 and COSV64746111 | ||
| 18840172 | G | A | A649A | NT | 0.009 | 0.002 | T | Synonymous | L | rs115672344 | ||
| 18840007 | G | A | P704P | NT | 0.012 | 0.010 | T | Synonymous | L | rs34542537 | ||
| 18840400 | G | A | A573A | NT | 0.018 | 0.046 | T | Synonymous | L | rs11805253 | ||
| 18839749 | G | A | I790I | NT | 0.040 | 0.049 | T | Synonymous | L | rs12075191 | ||
| 18854588 | A | C | T294T | NT | 0.281 | 0.291 | F | Synonymous | L | rs28470550 | ||
| 18857583 | A | G | F77F | NT | 0.293 | 0.320 | F | Synonymous | L | rs68081213 | ||
| 18839800 | G | A | S773S | NT | 0.302 | 0.294 | F | Synonymous | L | rs12033832 | ||
| 18839871 | T | G | K750Q | 2 | 0.003 | - | T | Missense | M | probably damaging (100) | rs933473941 | |
| TAS1R2 | 18841752 | C | T | G523D | 2 | 0.003 | <0.001 | T | Missense | M | probably damaging (100) | rs766687403 |
| 18854355 | G | A | S372F | 2 | 0.003 | - | T | Missense | M | possibly damaging (87.6) | rs535257286 and COSV64747888 | |
| 18854529 | G | T | T314K | 2 | 0.003 | <0.001 | T | Missense | M | possibly damaging (48.4) | rs148292629 | |
| 18840335 | A | G | I595T | 2 | 0.040 | 0.017 | T | Missense | M | benign (7.7) | rs41273167 | |
| 18849382 | T | G | K476Q | 3 | 0.003 | <0.001 | T | Missense | M | benign (10.7) | rs547302644 | |
| 18854723 | C | T | M249I | 3 | 0.003 | 0.003 | T | Missense | M | benign (0.0) | rs148245865 | |
| 18859599 | G | A | P21L | 3 | 0.006 | 0.021 | T | Missense | M | benign (0.3) | rs72953144 | |
| 18849511 | C | T | D433N | 3 | 0.009 | 0.002 | T | Missense | M | benign (3.1) | rs114026861 and COSV64745359 | |
| 18839606 | C | T | R838K | 3 | 0.018 | 0.052 | F | Missense | M | benign (0.0) | rs9988418 | |
| 18840399 | C | T | A574T | 3 | 0.040 | 0.056 | F | Missense | M | benign (0.7) | rs6662276 | |
| 18849352 | T | C | I486V | 3 | 0.197 | 0.190 | F | missense | M | benign (0.0) | rs28374389 | |
| 18854521 | G | C | R317G | 3 | 0.278 | 0.291 | F | missense | M | benign (0.5) | rs34447754 | |
| 18854899 | T | C | I191V | 3 | 0.290 | 0.312 | F | missense | M | benign (0.1) | rs35874116 and CM109811 | |
| 18859635 | G | C | S9C | 3 | 0.799 | 0.787 | F | missense | M | benign (0.0) | rs9701796 | |
| 1333642 | G | A | A579A | NT | 0.003 | 0.004 | T | synonymous | L | . | rs143667857 | |
| 1333882 | C | T | F659F | NT | 0.003 | <0.001 | T | synonymous | L | . | rs142902721 | |
| 1334422 | C | T | G839G | NT | 0.003 | 0.002 | T | synonymous | L | . | rs148758835 | |
| 1332968 | G | A | P441P | NT | 0.003 | 0.024 | T | synonymous | L | . | rs111703380 | |
| 1332116 | G | A | T195T | NT | 0.003 | 0.008 | T | synonymous | L | . | rs146097837 | |
| 1332260 | G | A | V243V | NT | 0.003 | <0.001 | T | synonymous | L | . | rs1258667478 | |
| 1333007 | C | T | Y454Y | NT | 0.003 | 0.001 | T | synonymous | L | . | rs142857537 and COSV59566129 | |
| 1331677 | A | G | K77K | NT | 0.006 | 0.005 | T | synonymous | L | . | rs139515618 | |
| 1333082 | G | A | R479R | NT | 0.006 | 0.007 | T | synonymous | L | . | rs138915131 | |
| 1331905 | C | T | T153T | NT | 0.006 | 0.001 | T | synonymous | L | . | rs147731455 | |
| 1333624 | C | T | L573L | NT | 0.015 | 0.007 | T | synonymous | L | . | rs140035477 and COSV59564228 | |
| 1331360 | T | A | A5A | NT | 0.065 | 0.045 | T | synonymous | L | . | rs141430443 | |
| TAS1R3 | 1332779 | C | T | P416P | NT | 0.068 | 0.122 | F | synonymous | L | . | rs3813210 |
| 1333908 | G | A | W668 | 1 | 0.003 | <0.001 | T | nonsense | H | . | rs147921760 | |
| 1331660 | T | G | W72G | 2 | 0.003 | <0.001 | T | missense | M | possibly damaging (81.1) | rs144594741 | |
| 1331726 | C | T | R94C | 2 | 0.003 | <0.001 | T | missense | M | possibly damaging (86.2) | rs138021134 | |
| 1333116 | G | C | D491H | 2 | 0.003 | <0.001 | T | missense | M | possibly damaging (56.4) | rs1358129448 | |
| 1334232 | TCTC | T | VS776-777V | 2 | 0.003 | <0.001 | T | indel | M | . | rs531899606 | |
| 1333090 | G | A | R482H | 3 | 0.003 | <0.001 | T | missense | M | benign (0.3) | rs143388404 | |
| 1334382 | C | T | P826L | 3 | 0.003 | <0.001 | T | missense | M | benign (0.3) | rs749544965 | |
| 1334108 | G | A | A735T | 3 | 0.006 | 0.063 | F | missense | M | benign (1.1) | rs112507608 | |
| 1332271 | G | A | R247H | 3 | 0.025 | 0.073 | F | missense | M | benign (0.5) | rs111615792 | |
| 1331358 | G | A | A5T | 3 | 0.065 | 0.045 | T | missense | M | benign (2.1) | rs76755863 | |
| 1331358 | GCT | ACA | A5T | 3 | 0.065 | . | T | missense | M | benign (2.1) | . | |
| 1334174 | T | C | C757R | 3 | 0.966 | 0.972 | F | missense | M | benign (0.0) | rs307377 and CM098260 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Belloir, C.; Jeannin, M.; Karolkowski, A.; Briand, L. TAS1R2/TAS1R3 Single-Nucleotide Polymorphisms Affect Sweet Taste Receptor Activation by Sweeteners: The SWEET Project. Nutrients 2025, 17, 949. https://doi.org/10.3390/nu17060949
Belloir C, Jeannin M, Karolkowski A, Briand L. TAS1R2/TAS1R3 Single-Nucleotide Polymorphisms Affect Sweet Taste Receptor Activation by Sweeteners: The SWEET Project. Nutrients. 2025; 17(6):949. https://doi.org/10.3390/nu17060949
Chicago/Turabian StyleBelloir, Christine, Mathilde Jeannin, Adeline Karolkowski, and Loïc Briand. 2025. "TAS1R2/TAS1R3 Single-Nucleotide Polymorphisms Affect Sweet Taste Receptor Activation by Sweeteners: The SWEET Project" Nutrients 17, no. 6: 949. https://doi.org/10.3390/nu17060949
APA StyleBelloir, C., Jeannin, M., Karolkowski, A., & Briand, L. (2025). TAS1R2/TAS1R3 Single-Nucleotide Polymorphisms Affect Sweet Taste Receptor Activation by Sweeteners: The SWEET Project. Nutrients, 17(6), 949. https://doi.org/10.3390/nu17060949






