Emodin and Aloe-Emodin Reduce Cell Growth and Disrupt Metabolic Plasticity in Human Melanoma Cells
Highlights
- The natural compounds emodin and aloe-emodin disrupt the metabolic plasticity of melanoma cells by impairing glycolysis, mitochondrial function, and redox homeostasis.
- These bioactive compounds target metabolic vulnerabilities across diverse melanoma pheno-types.
- Emodin and aloe-emodin demonstrate therapeutic potential in overcoming resistance mecha-nisms and advancing melanoma treatment strategies.
- Our findings contribute to the growing recognition of metabolic rewiring as a cornerstone of cancer therapy.
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culture
2.2. Cell Proliferation Analysis
2.3. Oxygen Consumption Rate Analysis
2.4. ROS Level Measurements
2.5. Autophagy
2.6. Metabolites Quantification in the Media Samples
2.7. Metabolite Extraction from Cell Culture
2.8. LC-MS Metabolic Profiling
2.9. Metabolomics Statistical Data Analysis
2.10. 13C Metabolic Flux Analysis
2.11. Lactate Dehydrogenase and Pyruvate Dehydrogenase Quantification
2.12. Western Blot Analysis
2.13. Statistical Data Analysis
3. Results
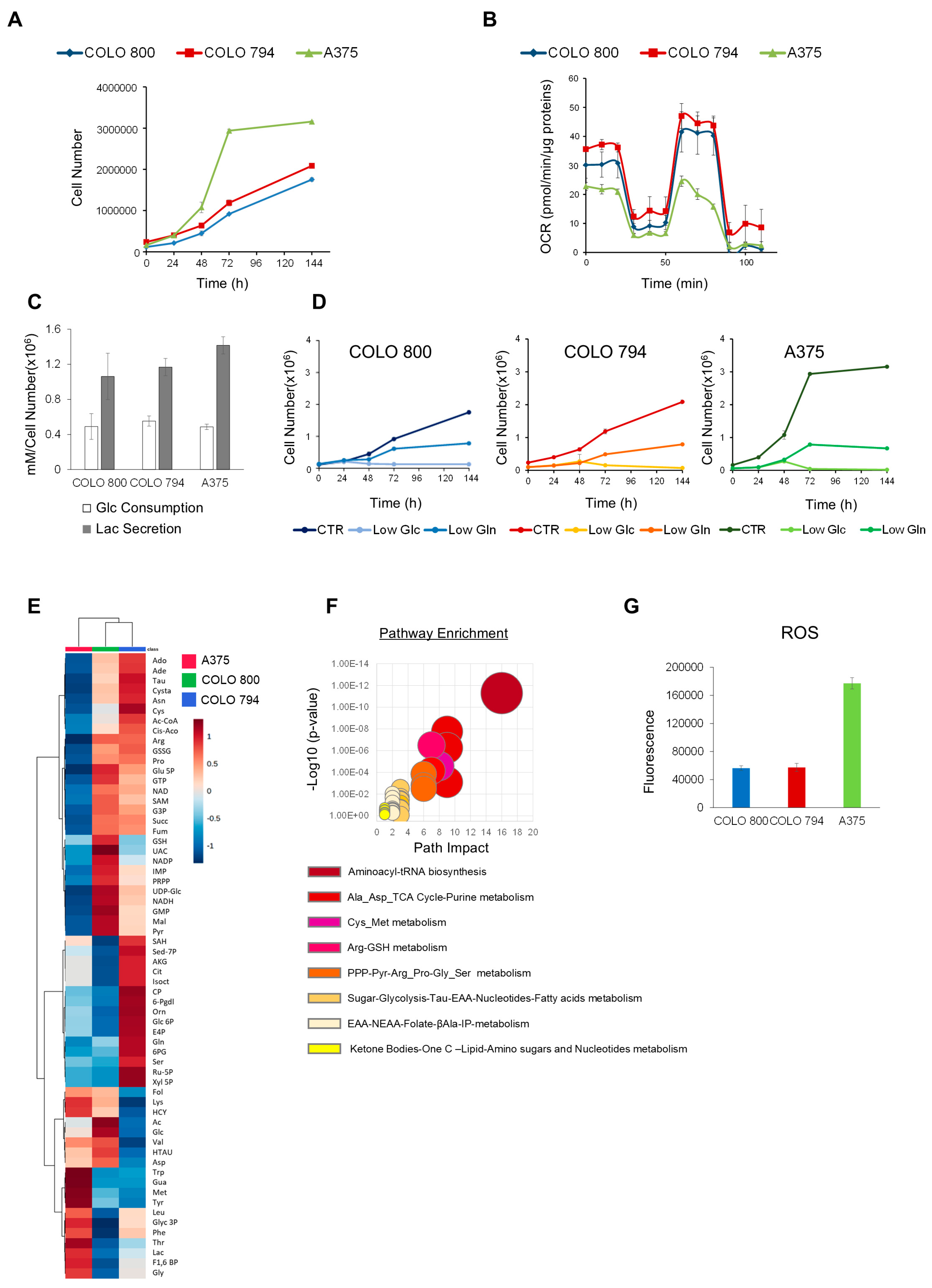
3.1. Melanoma Cancer Cell Lines: A Model of Cancer Metabolic Heterogeneity
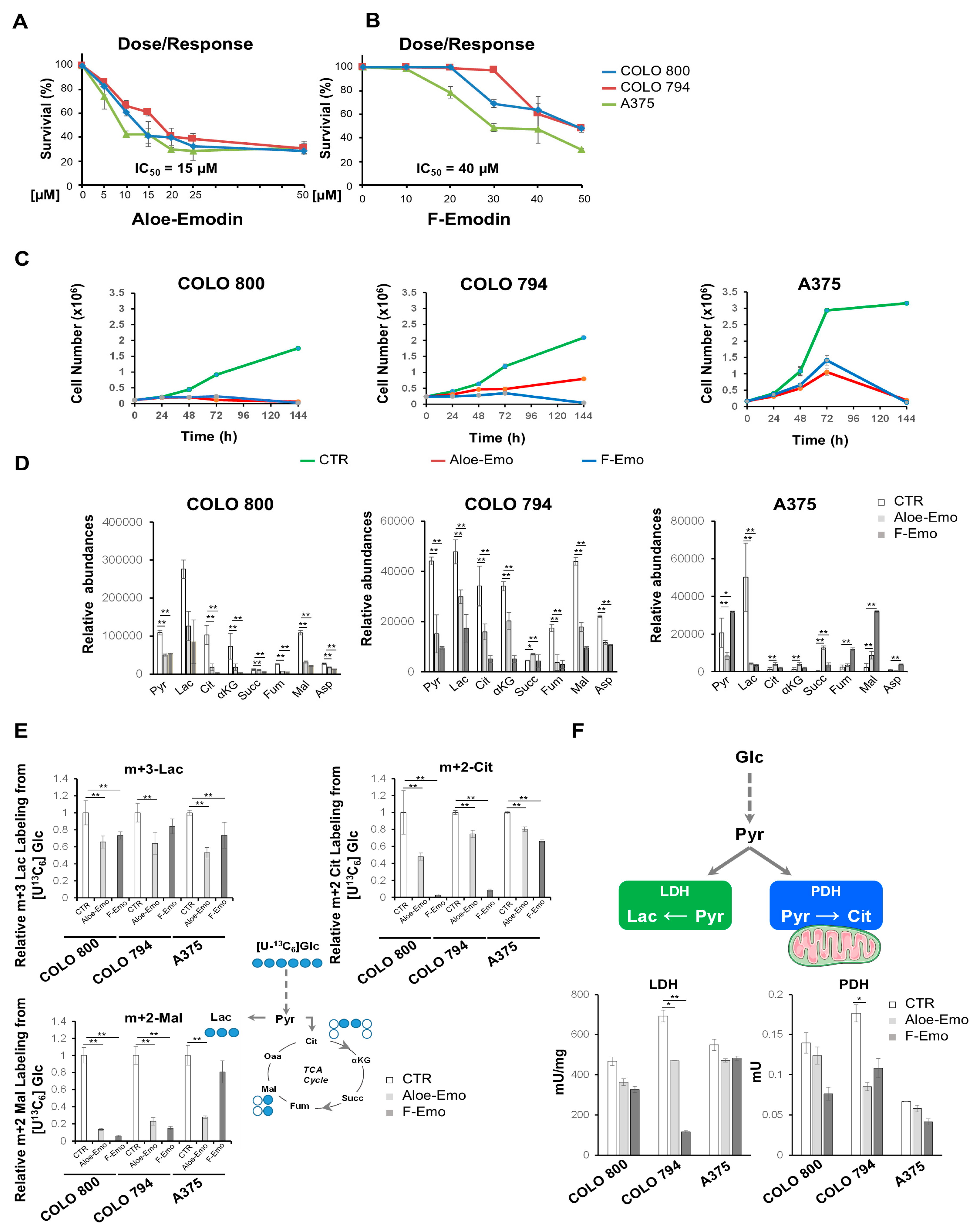
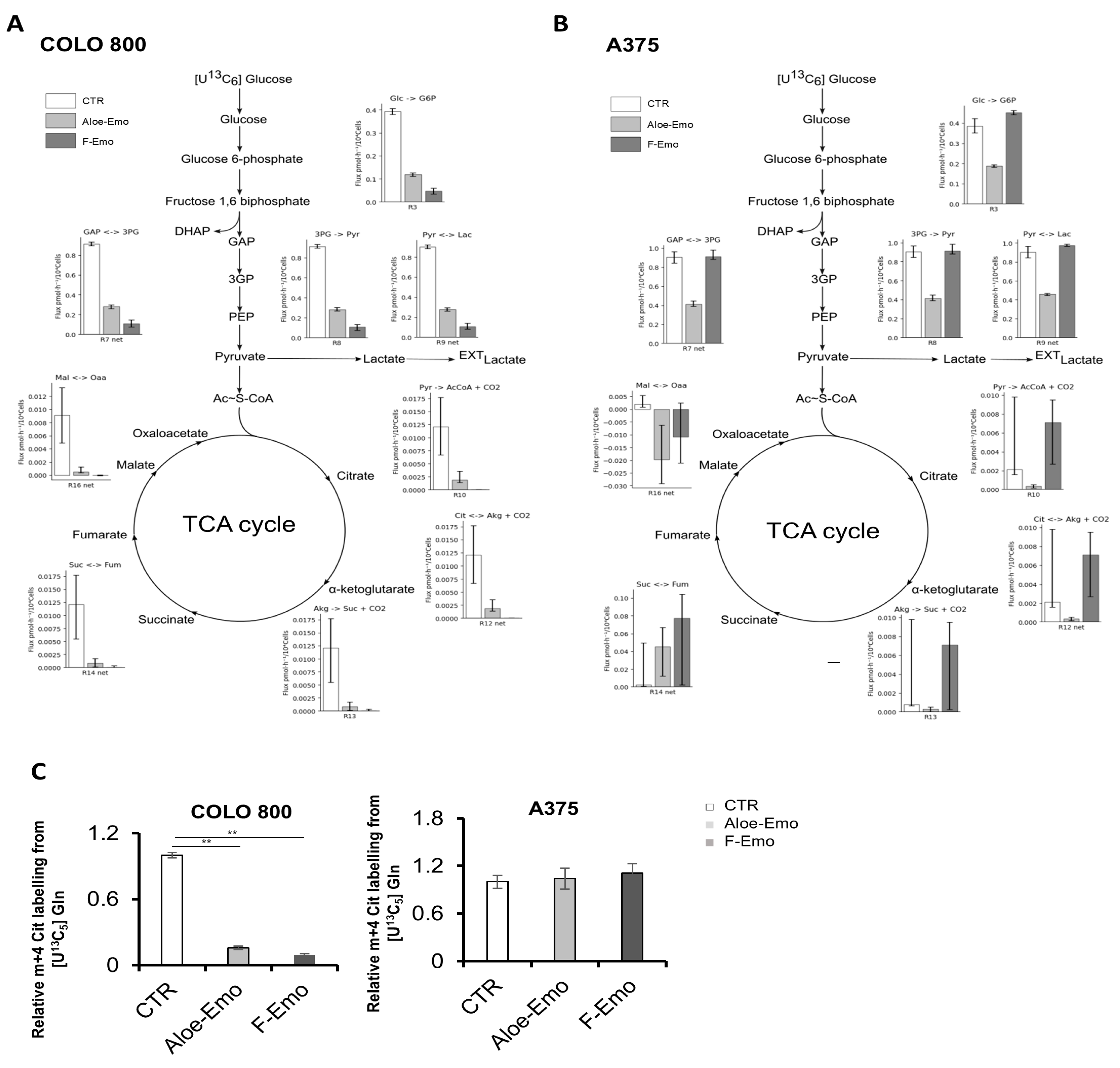
3.2. Emodin and Aloe-Emodin Inhibit Energetic Metabolic Pathways Essential for Melanoma Cell Proliferation
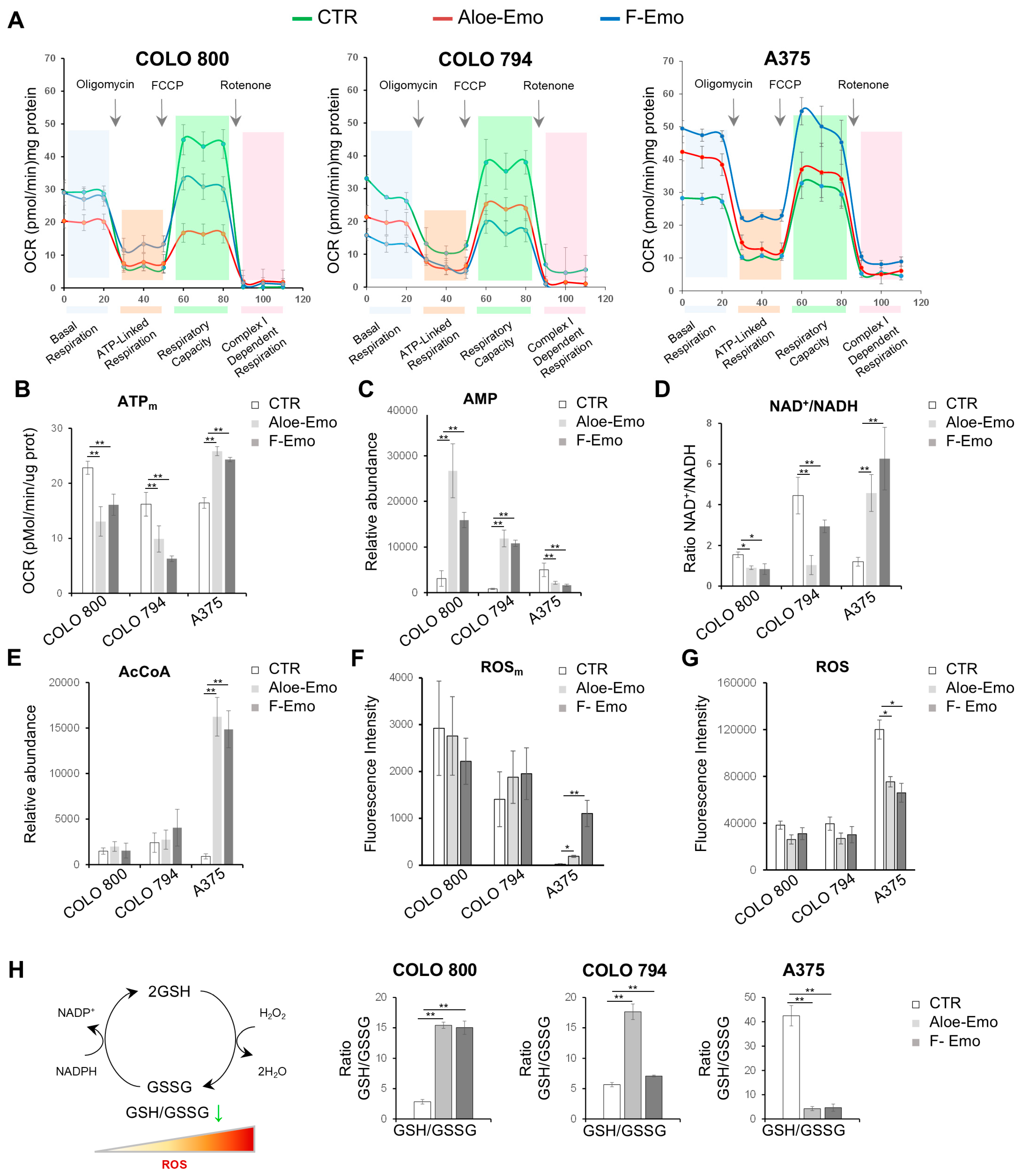
3.3. Emodin and Aloe-Emodin Inhibit Mitochondrial Activity and Alter the Cellular Redox Balance
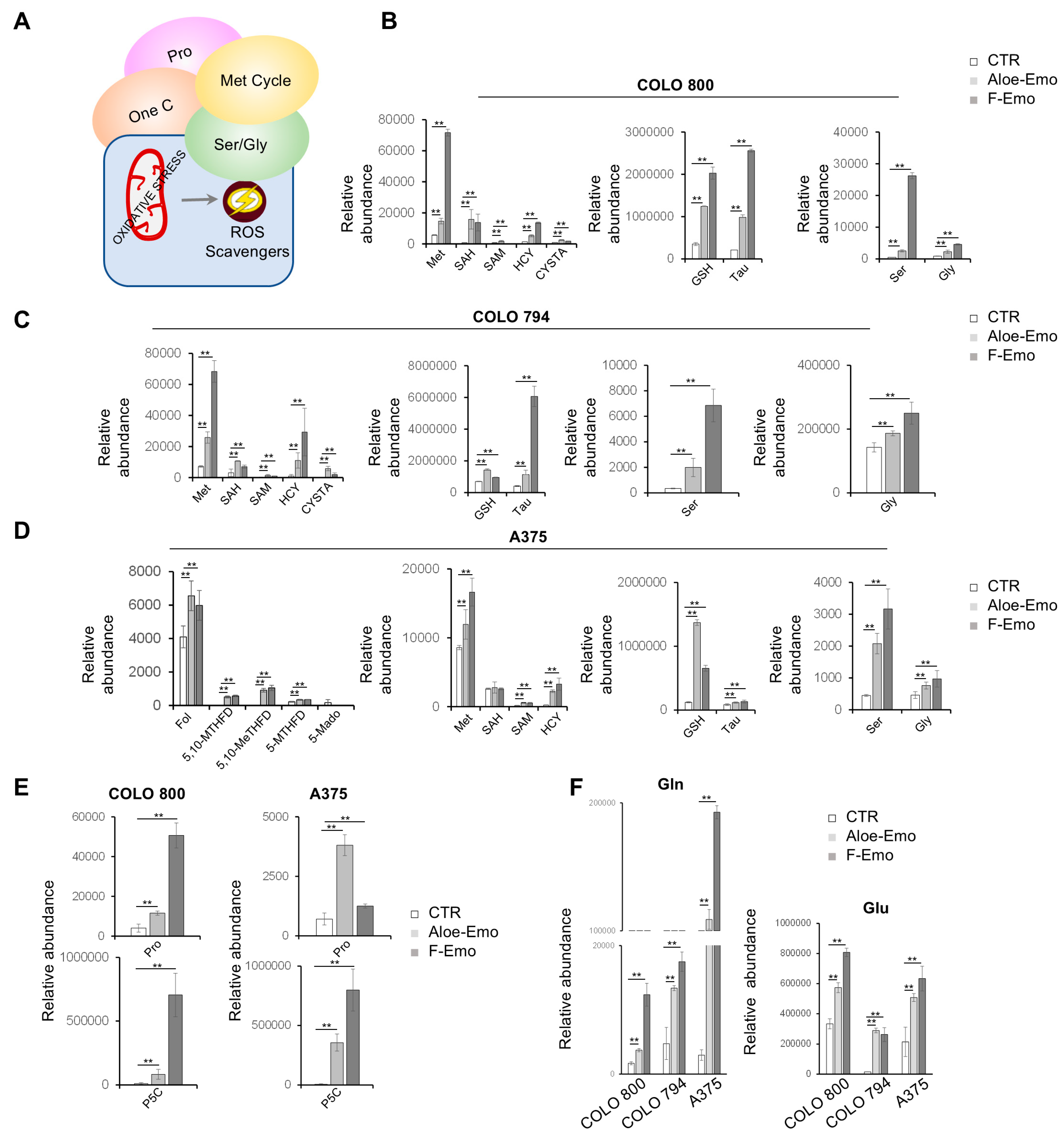
3.4. Metabolic Pathway Activation to Support Redox Homeostasis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| 3PG | 3-phospho glycerate |
| 5,10-MTHFD | 5,10-methyltetrahydrofolate |
| 5,10-MeTHFD | 5,10-methyltetrahydrofuran |
| 6-PG | 6-phosphogluconate |
| 6-PGDL | 6-phosphonoglucono-D-lactone |
| AC | acetic acid |
| AcCoA | acetyl-CoA |
| ADE | adenine |
| ADO | 2-aminoethanethiol dioxygenase |
| AKG | α-ketoglutarate |
| ALA | alanine |
| AMPK | AMP-activated protein kinase |
| ANOVA | Analysis of Variance |
| ARG | arginine |
| ASN | asparagine |
| ASP | aspartate |
| ATCC | American Type Culture Collection |
| ATP | adenosine triphosphate |
| BSA | bovine serum albumin |
| CDP | Cytidine diphosphate |
| CIS-ACO | cis-aconitic acid |
| CIT | citrate |
| CO2 | carbon dioxide |
| CYS | cysteine |
| CYSTA | cystathionine |
| CP | carbamoyl phosphate |
| CTR | control |
| DCFDA | dichloro-dihydro-fluoresceine-diacetate |
| DHAP | dihydroxyacetone phosphate |
| GAP | glyceraldehyde phosphate |
| DMEM | Dulbecco’s modified Eagle’s medium |
| E4P | erythrose 4-phosphate |
| EAA | essential amino acid |
| EMU | elementary metabolite unit |
| F1,6BP | fructose 1,6-bisphosphate |
| FBS | fetal bovine serum |
| FCCP | carbonyl cyanide p-(trifluoromethoxy)phenylhydrazone |
| FOL | folate |
| FUM | fumarate |
| G3P | glyceraldehyde 3-phosphate |
| GLC | glucose |
| GLC.x | extracellular glucose |
| GLC-6P | glucose-6-phosphate |
| GLN | glutamine |
| GLN.x | extracellular glutamine |
| GLU | L-glutamate |
| GLU.x | extracellular L-glutamate |
| GLU 5P | glutamate-5phosphate |
| GLY | glycine |
| GLYC-3P | glycerol 3-phosphate |
| GMP | guanosine monophosphate |
| GSH | glutathione |
| GSSG | glutathione disulfide |
| GUA | Guanine |
| HCY | homocysteine |
| HTAU | hypotaurine |
| IMP | inosine monophosphate |
| ISOCT | isocitrate |
| LAC | lactate |
| LAC.x | extracellular lactate |
| LB | lower bound |
| LC-MS | liquid chromatography–mass spectrometry |
| LDH | lactate dehydrogenase |
| LEU | leucine |
| LYS | lysine |
| MAL | malate |
| MAPK | mitogen-activated protein kinase |
| MET | methionine |
| MFA | metabolic flux analysis |
| NAD | nicotinamide adenine dinucleotide |
| NEAA | non-essential amino acids |
| NF1 | neurofibromin 1 |
| OAA | oxaloacetate |
| OCR | oxygen consumption rate |
| OD | optical density |
| ORA | Over Representation Analysis |
| ORN | ornithine |
| OXPHOS | oxidative phosphorylation |
| P5C | pyrroline-5-carboxylate |
| PDH | pyruvate dehydrogenase |
| PFP | pentafluorophenyl |
| PHE | phenylalanine |
| PPP | pentose phosphate pathway |
| PRO | proline |
| PRPP | phosphoribosyl pyrophosphate |
| PYR | pyruvate |
| RIPA | Radio-Immunoprecipitation Assay |
| ROS | reactive oxygen species |
| RPMI | Roswell Park Memorial Institute |
| RU-5P | ribulose-5-phosphate |
| SAH | S-adenosyl homocysteine |
| SAM | S-adenosyl methionine |
| SED-7P | sedoheptulose 7-phosphate |
| SER | serine |
| SMPDB | Small Molecule Pathway Database |
| STDERR | standard error |
| SUCC | succinate |
| TAU | taurine |
| TBS | Tris-Buffered Saline |
| TCA | tricarboxylic acid |
| THR | threonine |
| TRP | tryptophan |
| TYR | tyrosine |
| UAC | uric acid |
| UB | upper bound |
| UDP-GLC | uridine diphosphate glucose |
| UHPLC-QTOF | ultra-high-performance liquid chromatography–quadrupole time-of-flight |
| VAL | valine |
| XYL-5P | xylulose 5-phosphate |
References
- Zhang, X.; Tai, Z.; Miao, F.; Huang, H.; Zhu, Q.; Bao, L.; Chen, Z. Metabolism Heterogeneity in Melanoma Fuels Deactivation of Immunotherapy: Predict before Protect. Front. Oncol. 2022, 12, 1046102. [Google Scholar] [CrossRef]
- Hossain, S.M.; Eccles, M.R. Phenotype Switching and the Melanoma Microenvironment; Impact on Immunotherapy and Drug Resistance. Int. J. Mol. Sci. 2023, 24, 1601. [Google Scholar] [CrossRef] [PubMed]
- Guo, W.; Wang, H.; Li, C. Signal Pathways of Melanoma and Targeted Therapy. Signal Transduct. Target. Ther. 2021, 6, 424. [Google Scholar] [CrossRef]
- Thielmann, C.M.; Chorti, E.; Matull, J.; Murali, R.; Zaremba, A.; Lodde, G.; Jansen, P.; Richter, L.; Kretz, J.; Möller, I.; et al. NF1-Mutated Melanomas Reveal Distinct Clinical Characteristics Depending on Tumour Origin and Respond Favourably to Immune Checkpoint Inhibitors. Eur. J. Cancer 2021, 159, 113–124. [Google Scholar] [CrossRef] [PubMed]
- Avagliano, A.; Fiume, G.; Pelagalli, A.; Sanità, G.; Ruocco, M.R.; Montagnani, S.; Arcucci, A. Metabolic Plasticity of Melanoma Cells and Their Crosstalk With Tumor Microenvironment. Front. Oncol. 2020, 10, 722. [Google Scholar] [CrossRef]
- Bristot, I.J.; Kehl Dias, C.; Chapola, H.; Parsons, R.B.; Klamt, F. Metabolic Rewiring in Melanoma Drug-Resistant Cells. Crit. Rev. Oncol. Hematol. 2020, 153, 102995. [Google Scholar] [CrossRef]
- Atanasov, A.G.; Zotchev, S.B.; Dirsch, V.M.; Supuran, C.T. Natural Products in Drug Discovery: Advances and Opportunities. Nat. Rev. Drug Discov. 2021, 20, 200–216. [Google Scholar] [CrossRef]
- Seigler, D.S. Benzoquinones, naphthoquinones, and anthraquinones. In Plant Secondary Metabolism; Seigler, D.S., Ed.; Springer: Boston, MA, USA, 1998; pp. 76–93. ISBN 978-1-4615-4913-0. [Google Scholar]
- Huang, Q.; Lu, G.; Shen, H.-M.; Chung, M.C.M.; Ong, C.N. Anti-Cancer Properties of Anthraquinones from Rhubarb. Med. Res. Rev. 2007, 27, 609–630. [Google Scholar] [CrossRef]
- Pecere, T.; Gazzola, M.V.; Mucignat, C.; Parolin, C.; Vecchia, F.D.; Cavaggioni, A.; Basso, G.; Diaspro, A.; Salvato, B.; Carli, M.; et al. Aloe-Emodin Is a New Type of Anticancer Agent with Selective Activity against Neuroectodermal Tumors. Cancer Res. 2000, 60, 2800–2804. [Google Scholar]
- Gunaydin-Akyildiz, A.; Yanikoglu, R.S.; Gulec, M.; Alim-Toraman, G.O.; Kuran, E.D.; Atasoy, S.; Olgun, A.; Topcu, G. Emodin and Aloe-Emodin, Two Potential Molecules in Regulating Cell Migration of Skin Cells through the MAP Kinase Pathway and Affecting Caenorhabditis Elegans Thermotolerance. BMC Mol. Cell Biol. 2023, 24, 23. [Google Scholar] [CrossRef]
- Tran, N.K.S.; Nguyen, N.Q.; Lee, S.; Kim, S.H.; Jeong, D.; Seo, E.; Park, J.J.; Cho, J.; Kang, K.S. Anticancer Effects of Aloe-Emodin from Rheum Undulatum L. through Activation of the P53 Pathway in Human Prostate Cancer Cells. Appl. Biol. Chem. 2024, 67, 102. [Google Scholar] [CrossRef]
- Bonanomi, M.; Salmistraro, N.; Fiscon, G.; Conte, F.; Paci, P.; Bravatà, V.; Forte, G.I.; Volpari, T.; Scorza, M.; Mastroianni, F.; et al. Transcriptomics and Metabolomics Integration Reveals Redox-Dependent Metabolic Rewiring in Breast Cancer Cells. Cancers 2021, 13, 5058. [Google Scholar] [CrossRef] [PubMed]
- Antoniewicz, M.R. A Guide to 13C Metabolic Flux Analysis for the Cancer Biologist. Exp. Mol. Med. 2018, 50, 1–13. [Google Scholar] [CrossRef]
- Antoniewicz, M.R.; Kelleher, J.K.; Stephanopoulos, G. Elementary Metabolite Units (EMU): A Novel Framework for Modeling Isotopic Distributions. Metab. Eng. 2007, 9, 68–86. [Google Scholar] [CrossRef] [PubMed]
- Antoniewicz, M.R.; Kelleher, J.K.; Stephanopoulos, G. Determination of Confidence Intervals of Metabolic Fluxes Estimated from Stable Isotope Measurements. Metab. Eng. 2006, 8, 324–337. [Google Scholar] [CrossRef] [PubMed]
- Young, J.D. INCA: A Computational Platform for Isotopically Non-Stationary Metabolic Flux Analysis. Bioinformatics 2014, 30, 1333–1335. [Google Scholar] [CrossRef]
- Shen, F.; Ge, C.; Yuan, P. Aloe-Emodin Induces Autophagy and Apoptotic Cell Death in Non-Small Cell Lung Cancer Cells via Akt/mTOR and MAPK Signaling. Eur. J. Pharmacol. 2020, 886, 173550. [Google Scholar] [CrossRef] [PubMed]
- Yu, L.; Zhao, Y.; Zhao, Y. Advances in the Pharmacological Effects and Molecular Mechanisms of Emodin in the Treatment of Metabolic Diseases. Front. Pharmacol. 2023, 14, 1240820. [Google Scholar] [CrossRef]
- Trachootham, D.; Alexandre, J.; Huang, P. Targeting Cancer Cells by ROS-Mediated Mechanisms: A Radical Therapeutic Approach? Nat. Rev. Drug Discov. 2009, 8, 579–591. [Google Scholar] [CrossRef]
- Warburg, O. On the Origin of Cancer Cells. Science 1956, 123, 309–314. [Google Scholar] [CrossRef]
- Li, T.; Shi, L.; Liu, W.; Hu, X.; Hui, Y.; Di, M.; Xue, S.; Zheng, Y.; Yao, M.; Li, C.; et al. Aloe-Emodin Induces Mitochondrial Dysfunction and Pyroptosis by Activation of the Caspase-9/3/Gasdermin E Axis in HeLa Cells. Front. Pharmacol. 2022, 13, 854526. [Google Scholar] [CrossRef] [PubMed]
- Lin, L.; Liu, Y.; Fu, S.; Qu, C.; Li, H.; Ni, J. Inhibition of Mitochondrial Complex Function—The Hepatotoxicity Mechanism of Emodin Based on Quantitative Proteomic Analyses. Cells 2019, 8, 263. [Google Scholar] [CrossRef] [PubMed]
- Song, P.; Kim, J.H.; Ghim, J.; Yoon, J.H.; Lee, A.; Kwon, Y.; Hyun, H.; Moon, H.-Y.; Choi, H.-S.; Berggren, P.-O.; et al. Emodin Regulates Glucose Utilization by Activating AMP-Activated Protein Kinase*. J. Biol. Chem. 2013, 288, 5732–5742. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Reyes, I.; Chandel, N.S. Mitochondrial TCA Cycle Metabolites Control Physiology and Disease. Nat. Commun. 2020, 11, 102. [Google Scholar] [CrossRef]
- Paudel, B.B.; Lewis, J.E.; Hardeman, K.N.; Hayford, C.E.; Robbins, C.J.; Stauffer, P.E.; Codreanu, S.G.; Sherrod, S.D.; McLean, J.A.; Kemp, M.L.; et al. An Integrative Gene Expression and Mathematical Flux Balance Analysis Identifies Targetable Redox Vulnerabilities in Melanoma Cells. Cancer Res. 2020, 80, 4565–4577. [Google Scholar] [CrossRef]
- Wang, H.; Fan, M.; Liu, S.; Qu, M.; Hou, X.; Hou, J.; Xu, Y.; Shang, X.; Liu, C.; He, M.; et al. Redox Homeostasis of One-Carbon Metabolism-Dependent Reprogramming Is Critical for RCC Progression under Exogenous Serine/Glycine-Deprived Conditions. BMC Cancer 2024, 24, 1515. [Google Scholar] [CrossRef]
- Ducker, G.S.; Rabinowitz, J.D. One-Carbon Metabolism in Health and Disease. Cell Metab. 2017, 25, 27–42. [Google Scholar] [CrossRef]
- Becker, A.L.; Indra, A.K. Oxidative Stress in Melanoma: Beneficial Antioxidant and Pro-Oxidant Therapeutic Strategies. Cancers 2023, 15, 3038. [Google Scholar] [CrossRef]
- Peppicelli, S.; Kersikla, T.; Menegazzi, G.; Andreucci, E.; Ruzzolini, J.; Nediani, C.; Bianchini, F.; Calorini, L. The Critical Role of Glutamine and Fatty Acids in the Metabolic Reprogramming of Anoikis-Resistant Melanoma Cells. Front. Pharmacol. 2024, 15, 1422281. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Baldassari, F.; Bonanomi, M.; Mallia, S.; Bonas, M.; Brivio, E.; Aramini, T.; Porro, D.; Gaglio, D. Emodin and Aloe-Emodin Reduce Cell Growth and Disrupt Metabolic Plasticity in Human Melanoma Cells. Nutrients 2025, 17, 1113. https://doi.org/10.3390/nu17071113
Baldassari F, Bonanomi M, Mallia S, Bonas M, Brivio E, Aramini T, Porro D, Gaglio D. Emodin and Aloe-Emodin Reduce Cell Growth and Disrupt Metabolic Plasticity in Human Melanoma Cells. Nutrients. 2025; 17(7):1113. https://doi.org/10.3390/nu17071113
Chicago/Turabian StyleBaldassari, Federica, Marcella Bonanomi, Sara Mallia, Matteo Bonas, Elisa Brivio, Tecla Aramini, Danilo Porro, and Daniela Gaglio. 2025. "Emodin and Aloe-Emodin Reduce Cell Growth and Disrupt Metabolic Plasticity in Human Melanoma Cells" Nutrients 17, no. 7: 1113. https://doi.org/10.3390/nu17071113
APA StyleBaldassari, F., Bonanomi, M., Mallia, S., Bonas, M., Brivio, E., Aramini, T., Porro, D., & Gaglio, D. (2025). Emodin and Aloe-Emodin Reduce Cell Growth and Disrupt Metabolic Plasticity in Human Melanoma Cells. Nutrients, 17(7), 1113. https://doi.org/10.3390/nu17071113







