Venomics Reveals a Non-Compartmentalised Venom Gland in the Early Diverged Vermivorous Conus distans
Abstract
:1. Introduction
2. Results
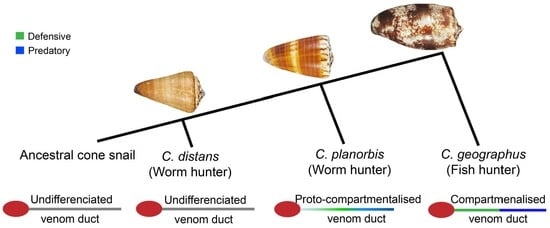
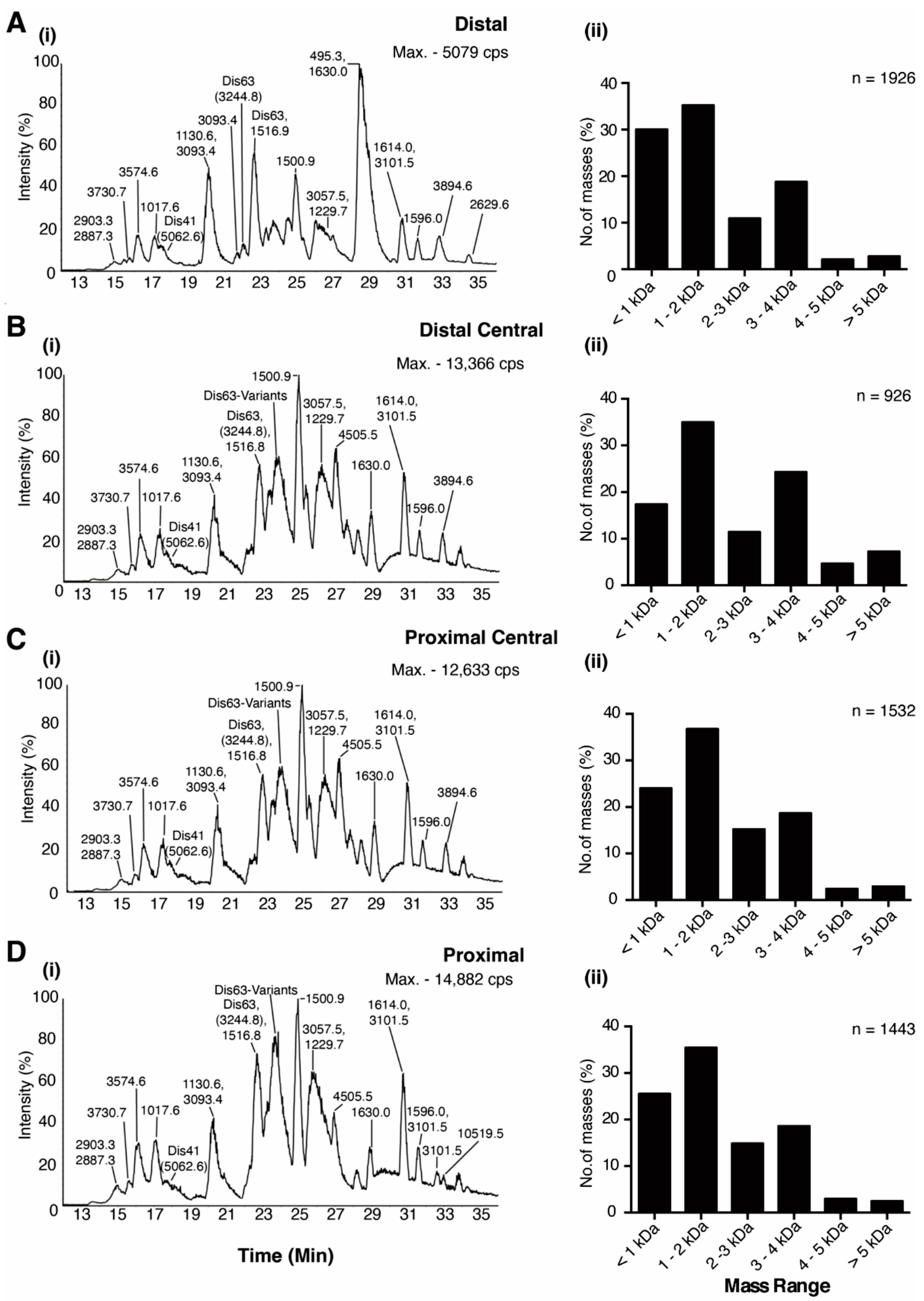
2.1. The Venom Gland of C. distans Is Not Compartmentalised
2.2. C. distans Venom Gland Transcriptome
2.3. Identification of Transcriptomic Sequences in Venom Duct Extracts
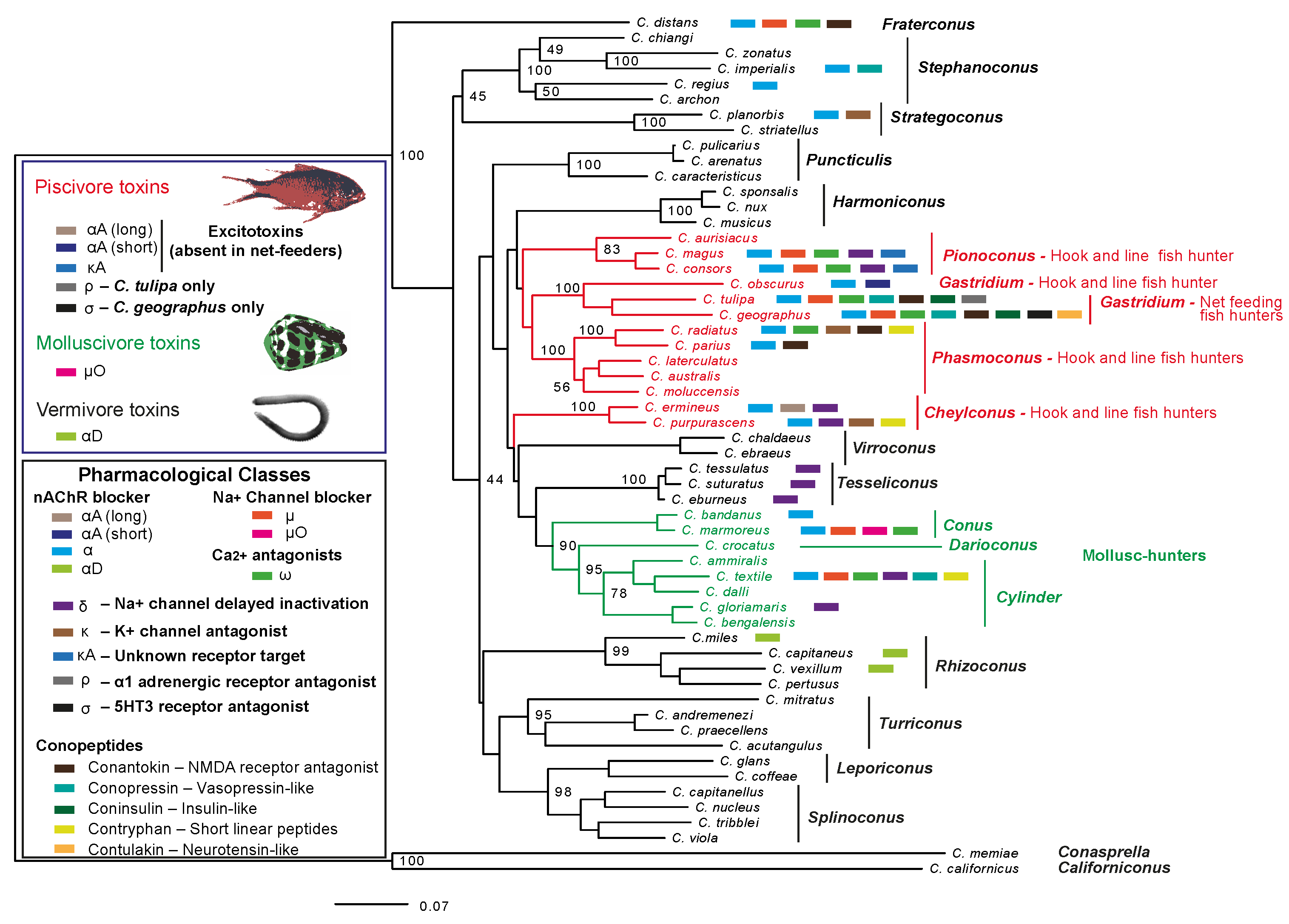
2.4. Comparison of C. distans with C. planorbis
3. Discussion
4. Materials and Methods
4.1. Sample Collection, RNA Extraction and Sequencing
4.2. Transcriptomic Analysis
4.3. Sample Collection for Mass Spectrometry
4.4. Venom Extraction
4.5. Reduction, Alkylation and Enzymatic Digest of Venom Samples
4.6. Venom Gland Mass Spectrometry (LC-ESI-MS and MS/MS)
4.7. MS and MS/MS Data Analysis
4.8. Principal Component Analysis (PCA) of LC-ESI-MS Data
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Casewell, N.R.; Wüster, W.; Vonk, F.J.; Harrison, R.A.; Fry, B.G. Complex cocktails: The evolutionary novelty of venoms. Trends Ecol. Evol. 2013, 28, 219–229. [Google Scholar] [CrossRef]
- Savitzky, A.H. The role of venom delivery strategies in snake evolution. Evolution 1980, 34, 1194–1204. [Google Scholar] [CrossRef]
- Undheim, E.A.; Hamilton, B.R.; Kurniawan, N.D.; Bowlay, G.; Cribb, B.W.; Merritt, D.J.; Fry, B.G.; King, G.F.; Venter, D.J. Production and packaging of a biological arsenal: Evolution of centipede venoms under morphological constraint. Proc. Natl. Acad. Sci. USA 2015, 112, 4026–4031. [Google Scholar] [CrossRef] [Green Version]
- Fry, B.G.; Scheib, H.; van der Weerd, L.; Young, B.; McNaughtan, J.; Ramjan, S.F.R.; Vidal, N.; Poelmann, R.E.; Norman, J.A. Evolution of an arsenal: Structural and functional diversification of the venom system in the advanced snakes (Caenophidia). Mol. Cell. Proteom. 2008, 7, 215–246. [Google Scholar] [CrossRef] [Green Version]
- Li, M.; Fry, B.G.; Kini, R.M. Eggs-only diet: Its implications for the toxin profile changes and ecology of the marbled sea snake (Aipysurus eydouxii). J. Mol. Evol. 2005, 60, 81–89. [Google Scholar] [CrossRef]
- Wright, J.J. Adaptive significance of venom glands in the tadpole madtom Noturus gyrinus (Siluriformes: Ictaluridae). J. Exp. Biol. 2012, 215, 1816–1823. [Google Scholar] [CrossRef] [Green Version]
- Dutertre, S.; Jin, A.-H.; Vetter, I.; Hamilton, B.; Sunagar, K.; Lavergne, V.; Dutertre, V.; Fry, B.G.; Antunes, A.; Venter, D.J. Evolution of separate predation-and defence-evoked venoms in carnivorous cone snails. Nat. Commun. 2014, 5, 3521. [Google Scholar] [CrossRef] [Green Version]
- Walker, A.A.; Mayhew, M.L.; Jin, J.; Herzig, V.; Undheim, E.A.B.; Sombke, A.; Fry, B.G.; Meritt, D.J.; King, G.F. The assassin bug Pristhesancus plagipennis produces two distinct venoms in separate gland lumens. Nat. Commun. 2018, 9, 755. [Google Scholar] [CrossRef] [Green Version]
- Puillandre, N.; Duda, T.F.; Meyer, C.; Olivera, B.M.; Bouchet, P. One, four or 100 genera? A new classification of the cone snails. J. Molluscan Stud. 2015, 81, 1–23. [Google Scholar] [CrossRef] [Green Version]
- Julita, S.I.; Natalie, S.; Olivera, B.M.; Bandyopadhyay, P.K.; Annett, S.; Ferber, M.; Heinrich, T. Using chemistry to reconstruct evolution: On the origins of fish-hunting in venomous cone snails. Proc. Am. Philos. Soc. 2007, 151, 185–200. [Google Scholar]
- Dutertre, S.; Griffin, J.; Lewis, R.J. Phyla Molluska: The venom apparatus of cone snails. In Marine and Freshwater Toxins; Gopalakrishnakone, P., Haddad, J.V., Tubaro, A., Kim, E., Kem, W.R., Eds.; Springer: Dordrecht, The Netherlands, 2016; pp. 327–340. [Google Scholar] [CrossRef]
- Jin, A.H.; Vetter, I.; Himaya, S.W.; Alewood, P.F.; Lewis, R.J.; Dutertre, S. Transcriptome and proteome of Conus planorbis identify the nicotinic receptors as primary target for the defensive venom. Proteomics 2015, 15, 4030–4040. [Google Scholar] [CrossRef] [PubMed]
- Prashanth, J.R.; Dutertre, S.; Jin, A.H.; Lavergne, V.; Hamilton, B.; Cardoso, F.C.; Griffin, J.; Venter, D.J.; Alewood, P.F.; Lewis, R.J. The role of defensive ecological interactions in the evolution of conotoxins. Mol. Ecol. 2016, 25, 598–615. [Google Scholar] [CrossRef]
- Puillandre, N.; Bouchet, P.; Duda, T.; Kauferstein, S.; Kohn, A.; Olivera, B.; Watkins, M.; Meyer, C. Molecular phylogeny and evolution of the cone snails (Gastropoda, Conoidea). Mol. Phylogenet. Evol. 2014, 78, 290–303. [Google Scholar] [CrossRef] [Green Version]
- Saxena, V.; Partoens, P.; De Block, J.; Coen, E.; Vauquelin, G.; De Potter, W. Inhibition of evoked neurotransmitter release from rat hippocampus by a polypeptide toxin isolated from the marine snail Conus distans. Neurochem. Int. 1992, 20, 69–74. [Google Scholar] [CrossRef]
- Partoens, P.; Wang, J.; Coen, E.; Vauquelin, G.; De Potter, W. Two polypeptide toxins with opposite effects on calcium uptake in bovine chromaffin cells: Isolation from the venom of the marine snail Conus distans. Neurochem. Int. 1996, 28, 619–624. [Google Scholar] [CrossRef]
- Chen, P.; Garrett, J.E.; Watkins, M.; Olivera, B.M. Purification and characterization of a novel excitatory peptide from Conus distans venom that defines a novel gene superfamily of conotoxins. Toxicon 2008, 52, 139–145. [Google Scholar] [CrossRef] [Green Version]
- Neves, J.L.B.; Lin, Z.; Imperial, J.S.; Antunes, A.; Vasconcelos, V.; Olivera, B.M.; Schmidt, E.W. Small molecules in the cone snail arsenal. Org. Lett. 2015, 17, 4933–4935. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Biggs, J.S.; Watkins, M.; Puillandre, N.; Ownby, J.-P.; Lopez-Vera, E.; Christensen, S.; Moreno, K.J.; Bernaldez, J.; Licea-Navarro, A.; Corneli, P.S.; et al. Evolution of Conus peptide toxins: Analysis of Conus californicus Reeve, 1844. Mol. Phylogenet. Evol. 2010, 56, 1–12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miles, L.A.; Dy, C.Y.; Nielsen, J.; Barnham, K.J.; Hinds, M.G.; Olivera, B.M.; Bulaj, G.; Norton, R.S. Structure of a novel P-superfamily spasmodic conotoxin reveals an inhibitory cystine knot motif. J. Biol. Chem. 2002, 277, 43033–43040. [Google Scholar] [CrossRef] [Green Version]
- Jin, A.-H.; Dutertre, S.; Kaas, Q.; Lavergne, V.; Kubala, P.; Lewis, R.J.; Alewood, P.F. Transcriptomic messiness in the venom duct of Conus miles contributes to conotoxin diversity. Mol. Cell. Proteom. 2013, 12, 3824–3833. [Google Scholar] [CrossRef] [Green Version]
- Lirazan, M.B.; Hooper, D.; Corpuz, G.P.; Ramilo, C.A.; Bandyopadhyay, P.; Cruz, L.J.; Olivera, B.M. The spasmodic peptide defines a new conotoxin superfamily. Biochemistry 2000, 39, 1583–1588. [Google Scholar] [CrossRef] [PubMed]
- England, L.J.; Imperial, J.; Jacobsen, R.; Craig, A.G.; Gulyas, J.; Akhtar, M.; Rivier, J.; Julius, D.; Olivera, B.M. Inactivation of a serotonin-gated ion channel by a polypeptide toxin from marine snails. Science 1998, 281, 575–578. [Google Scholar] [CrossRef]
- Teichert, R.W.; Jimenez, E.C.; Olivera, B.M. αS-conotoxin RVIIIA: A structurally unique conotoxin that broadly targets nicotinic acetylcholine receptors. Biochemistry 2005, 44, 7897–7902. [Google Scholar] [CrossRef]
- Himaya, S.W.A.; Mari, F.; Lewis, R.J. Accelerated proteomic visualization of individual predatory venoms of Conus purpurascens reveals separately evolved predation-evoked venom cabals. Sci. Rep. 2018, 8, 330. [Google Scholar] [CrossRef] [Green Version]
- Prashanth, J.R.; Lewis, R.J. An efficient transcriptome analysis pipeline to accelerate venom peptide discovery and characterisation. Toxicon 2015, 107, 282–289. [Google Scholar] [CrossRef] [Green Version]
- Mena, E.E.; Gullak, M.F.; Pagnozzi, M.J.; Richter, K.E.; Rivier, J.; Cruz, L.J.; Olivera, B.M. Conantokin-G: A novel peptide antagonist to the N-methyl-D-aspartic acid (NMDA) receptor. Neurosci. Lett. 1990, 118, 241–244. [Google Scholar] [CrossRef]
- Walker, C.S.; Jensen, S.; Ellison, M.; Matta, J.A.; Lee, W.Y.; Imperial, J.S.; Duclos, N.; Brockie, P.J.; Madsen, D.M.; Isaac, J.T. A Novel Conus snail polypeptide causes excitotoxicity by blocking desensitization of AMPA receptors. Curr. Biol. 2009, 19, 900–908. [Google Scholar] [CrossRef] [Green Version]
- Jimenéz, E.C.; Olivera, B.M.; Gray, W.R.; Cruz, L.J. Contryphan is a D-tryptophan-containing Conus peptide. J. Biol. Chem. 1996, 271, 28002–28005. [Google Scholar] [CrossRef] [Green Version]
- Lewis, R.J.; Dutertre, S.; Vetter, I.; Christie, M.D.J. Conus venom peptide pharmacology. Pharmacol. Rev. 2012, 64, 259–298. [Google Scholar] [CrossRef]
- Cruz, L.; Gray, W.; Olivera, B.; Zeikus, R.; Kerr, L.; Yoshikami, D.; Moczydlowski, E. Conus geographus toxins that discriminate between neuronal and muscle sodium channels. J. Biol. Chem. 1985, 260, 9280–9288. [Google Scholar] [CrossRef]
- McIntosh, J.M.; Hasson, A.; Spira, M.E.; Gray, W.R.; Li, W.; Marsh, M.; Hillyard, D.R.; Olivera, B.M. A new family of conotoxins that blocks voltage-gated sodium channels. J. Biol. Chem. 1995, 270, 16796–16802. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, J.; Wu, Q.; Pi, C.; Zhao, Y.; Zhou, M.; Wang, L.; Chen, S.; Xu, A. Isolation and characterization of a T-superfamily conotoxin from Conus litteratus with targeting tetrodotoxin-sensitive sodium channels. Peptides 2007, 28, 2313–2319. [Google Scholar] [CrossRef] [PubMed]
- Zakon, H.H. Convergent evolution on the molecular level. Brain Behav. Evol. 2002, 59, 250–261. [Google Scholar] [CrossRef]
- Terlau, H.; Shon, K.-J.; Grilley, M.; Stocker, M.; Stuehmer, W.; Olivera, B.M. Strategy for rapid immobilization of prey by a fish-hunting marine snail. Nature 1996, 381, 148–151. [Google Scholar] [CrossRef]
- Bayrhuber, M.; Vijayan, V.; Ferber, M.; Graf, R.; Korukottu, J.; Imperial, J.; Garrett, J.E.; Olivera, B.M.; Terlau, H.; Zweckstetter, M.; et al. Conkunitzin-S1is the first member of a new Kunitz-type neurotoxin family: Structural and functional characterization. J. Biol. Chem. 2005, 280, 23766–23770. [Google Scholar] [CrossRef] [Green Version]
- Elliger, C.A.; Richmond, T.A.; Lebaric, Z.N.; Pierce, N.T.; Sweedler, J.V.; Gilly, W.F. Diversity of conotoxin types from Conus californicus reflects a diversity of prey types and a novel evolutionary history. Toxicon 2011, 57, 311–322. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dutertre, S.; Jin, A.-H.; Alewood, P.F.; Lewis, R.J. Intraspecific variations in Conus geographus defence-evoked venom and estimation of the human lethal dose. Toxicon 2014, 91, 135–144. [Google Scholar] [CrossRef]
- Lavergne, V.; Dutertre, S.; Jin, A.-H.; Lewis, R.J.; Taft, R.J.; Alewood, P.F. Systematic interrogation of the Conus marmoreus venom duct transcriptome with ConoSorter reveals 158 novel conotoxins and 13 new gene superfamilies. BMC Genom. 2013, 14, 708. [Google Scholar] [CrossRef] [Green Version]
- Kaas, Q.; Yu, R.; Jin, A.-H.; Dutertre, S.; Craik, D.J. ConoServer: Updated content, knowledge, and discovery tools in the conopeptide database. Nucleic Acids Res. 2012, 40, D325–D330. [Google Scholar] [CrossRef] [PubMed]
- Okonechnikov, K.; Golosova, O.; Fursov, M. Unipro UGENE: A unified bioinformatics toolkit. Bioinformatics 2012, 28, 1166–1167. [Google Scholar] [CrossRef] [Green Version]
- Hale, J.E.; Butler, J.P.; Gelfanova, V.; You, J.-S.; Knierman, M.D. A simplified procedure for the reduction and alkylation of cysteine residues in proteins prior to proteolytic digestion and mass spectral analysis. Anal. Biochem. 2004, 333, 174–181. [Google Scholar] [CrossRef] [PubMed]




| Name (Superfamily) and Predicted Mass (Da) | Predicted Mature Peptide | Framework | Related Conotoxins | Likely Activity |
|---|---|---|---|---|
| Dis2 (A)—3582.4 | RIAEPNTEEEWNECCKDPSCRNNHMDRCAE | CC-C-C | Di1.1, Di1.2 (C. distans) | nAChR antagonists |
| Dis4 (B1)—1888.9 | TITAQEAETARERLSTL | Linear | Conantokin-E1 (C. eburneus) | NMDA receptor antagonist |
| Dis9 (B1)—1234.7 | LVGEVEIIVHK | Linear | Conantokin-V (C. vitulinus) | NMDA receptor antagonist |
| Dis14 (B1)—1947.9 | QTEEEVEESQEKLEEL | Linear | Conantokin-G (C. geographus) | NMDA receptor antagonist |
| Dis17 (Con-ikot-ikot)—12726.0 | GDPTAECCLTLLGCYDTCSHDNTSPDCWGHCKSESTTGCSLDFSLHYCSEFQDCYGPCVTDKDESRCFKACRKEAMIDCLDHGSVECCPGFVNCYQNCRRSREDCITHCLKENC | N/A | Con-ikot-ikot (C. striatus) | AMPA receptor antagonist |
| Dis20 (SFMi2)—2928.9 | DCGRDCVGCDNPANCCCGGQTCVNGNKCE | XXVII | Mi045, Mi046 (C. miles) | N/A |
| Dis22 (M---L-LLTVA)—6305.5 | DQTASGFLRDDESVFPCNSDRCACLPKEGSTTSYQCQSLEASTDDCVNNECITEDEW | IX | Cl9.5 (C. californicus), Turripeptide IX-01 | N/A |
| Dis30 (M---L-LLTVA) (DiXIXA)—4953.8 | GCDPTDGCQTTVCETDTGPCCCKPNFTCQISNSGTKSCSCSGQPSDCPV | XIX | N/A | Hyperexcitability/lethargy (dose-dependent) |
| Dis38 (H)—3984.5 | TDVDCGGVSCTFGCCETVNGEKKCKELDCDVTSDTENS | VI/VII | Teretoxin Tsu6.5 | N/A |
| Dis43 (I1)—3378.3 | CSYSSCKTESCCTGYLCNSVKSCVDPNSGGRF | VI/VII | Im11.14 (C. imperialis) | N/A |
| Dis45 (L)—2026.8 | RCPIACKTCPDENTCIPAP | XIV | Cl14.2 (C. californicus) | N/A |
| Dis46 (M)—3711.5 | TCDPYYCNDGKVCCPEYPTCGDSTGKLICVRVTD | VI/VII | Im6.7 (C. imperialis) | N/A |
| Dis49 (M)—1801.8 | DVKCIGSCDSTVWHRV | N/A | M-conotoxin 6 (C. marmoreus) | N/A |
| Dis53 (NSDis1)—5430.2 | SCEGNDSYCRKPDWISDKPCCDPLVCVCTGPMSGGRRTTCETGKCGPRPSS | XV | N/A | N/A |
| Dis58 (NSDis3)—3668.3 | NACELDSSTGDDCTGTQICCTPSGSMSGECREADEC | VI/VII | Lp7.1 (C. leopardus) | N/A |
| Dis60 (NSG3)—6759.6 | DESQCPECRCHDLKNAICDISEACSDEASCPTSPECNNGNCLCKNFHGGRCVSHSQCNDRHC | XXVIII | G125, G126 (C. geographus) | N/A |
| Dis62 (O1)—3509.3 | CDPPGYPCELRENDCCDACKIVSQNPNVCSDE | VI/VII | MiK42 (C. miles) | ω-conotoxin-like |
| Dis63 (O1)—3244.3 | CLSIGYACGVAISEKCCHVCDNPTGAGTCVYN | VI/VII | Im6.1 (C. imperialis) | ω-conotoxin-like |
| Dis68 (O1)—3205.2 | SCAESGLSCDTRPCCDDKTCVRNGRQSMCS | VI/VII | Mr6.11 (C. marmoreus) | N/A |
| Dis70 (O1)—3078.1 | CTESGLTCWPTNHDCCSGTCNGTMTTGTCT | VI/VII | Mi024 (C. miles) | ω-conotoxin-like |
| Dis71 (O1)—3087.0 | CEDEGSPCQFDSECCSGACTPEGVFDFCE | VI/VII | ω-like peptide (C. capitaneus) | ω-conotoxin-like |
| Dis72 (O1)—2969.1 | SCANYHESCASDPCCEGLECIGAQGGGVCI | VI/VII | ArMKLT2-0322 (C. arenatus) | N/A |
| Dis76 (O1)—2789.9 | CKGTDAPCDDHDECCEHVCDGVCVED | VI/VII | MgJr94P (C. magus) | ω-conotoxin-like |
| Dis80 (O1)—2503.8 | CEDPGEPCGSDHSCCSGSCNHNVCA | VI/VII | ArMKLT2-0322 (C. arenatus) | ω-conotoxin-like |
| Dis81 (O2)—8000.6 | DEDCVTEEGDHVEEGKASLLTTATRVPAMMENCTVRLCTVNNGVTEVLTNHENSVDDSLITWNLWTPCHCFI | XIV | N/A | N/A |
| Dis85 (O2)—4165.4 | QCAPDDFQCDVDEDCCNDLECKCFTSTDCTSGYKCRN | VI/VII | Bt15a (C. betulinus) | N/A |
| Dis91 (O2)— 3759.3 | CKGASALCEEDGECCSGDCKCMHASGCTNDINLRCAA | VI/VII | Di6.5 (C. distans) | N/A |
| Dis97 (O2)— 3768.4 | TCKGRLQSCDHDSECCSPYTCYCGMKQGCNLKCI | XV | Vt15a (C. vitulinus) | N/A |
| Dis98 (O2)—3264.1 | CDTWRDPCTYEHECCWQYHCGFRTCE | VI/VII | Di6.10 (C. distans) | N/A |
| Dis102 (O2—contryphan)—1316.5 | EFDCPWHPWC | N/A | Contryphan-R-like (C. virgo) | Contryphan-like activity |
| Dis104 (O2)—4409.7 | HGGCLNEEGDLVAEDGETIEVECNRCRCEDGDLACTKMACE | IX | Di6.5 (C. distans) | N/A |
| Dis116 (O3)—3535.5 | NVDQECIDACQLEDKNCCGRTDGEPRCAKICL | VI/VII | Di6.6 (C. distans) | N/A |
| Dis119 (O3)—3871.5 | LVEGACTSPSNCPTGQECCPNKLDEPEGSCANDCPFY | VI/VII | Di6.12 (C. distans) | N/A |
| Dis120 (P)—2780.9 | STCPTSCATHMNCWPECTYCTTSGCT | IX | Fla9.1 (C. flavidus), TxIXA (C. textile), GmIXA (C. gloriamaris) | spasmodic |
| Dis121 (S)—3803.1 | SSCSGTCYGSANCDGTCYCREDNCWCTGDSSCACQCA | VIII | Di8.1 (C. distans), GVIIIA, RVIIIA | 5-HT3 receptor and nAChR antagonists |
| Dis124 (T)—8350.9 | TPSEQNLPGELTPADLEGAETTPEESWYSKIKSGVKHASCKLVGYACDDSETEESLLSKIKGGVEHAACKYVNIACED | XIV | Tr5.4 (C. terebra) | N/A |
| Dis134 (T)—6399.6 | APSEPNLQRGLKLGGLKAEPNLQRGLKLGGVKDKLLKVGGNILKGAVQGAVDSLTKEDRKQ | N/A | Vr5.4 (C. varius) | N/A |
| Sequence | |
|---|---|
| Dis1 | MGMRMMFIVFLLVVLATTVVSLRSDRAFNRKNRRIAEPNTEEEWNECCKDPSCRNNHLDRCPE |
| Dis10 | MELYTYLYLLVPLVAFHLIQGTGTRSHGGPLTEGRSADVTALKPEPVLLQKSDARSTDDNGKDKLTRMKRTLKKGGNMARRQTEEEVEESNETLAEAGKR |
| Dis102 | MKKLTILVLVAAVLLSTQVMVQGDGDQPADRNAVPRDDNPGGTSGKLMRVLQGREFDCPWHPWCG |
| Dis103 | MNKLTMLILVATVLLSIQVMVRGDEDCVNEEGDLVAEDGETVKVECNBTrCRCDDGDLACTKMACE |
| Dis11 | MQLYTYLYLLVPLVAFHLIQGTGTRGHGGALTEGRSADVTALKPEPVLLQKSDARSADDNGKDKLTRMRRTLKNKGNMARRQTEEEVEESQEKLEELGKR |
| Dis118 | MSGLGIMVLTLLLLVPMATSQQDGGEKQAMQRDAINAAPGTSITRRETDQECIDTCEQEDKKCCGRTNGEPVCAKICFG |
| Dis121 | MMSKMGAMFVLLLLCPLASNQQEGDIKARRTFWKRDLYGDLAGRSSCSGTCYGSANCDGTCYCREDNCWCTGDSSCACQCA |
| Dis125 | MLCLPVFIILLLLASPAVTTPSEQNLPGELTPADLEGAETTPEESWYSKIKGGVKHASCKLVGYACDDSETEESLLSKIKGGVEHAACKYVNIGCED |
| Dis126 | MLCLPVFIILLLLASPAVTTLSEQNLPGELTPADLEGAETTPEESWYSKIKSGVKHASCKLVGYACDDSETEESLLSKIKGGVEHAACKYVNIACED |
| Dis127 | MLCLPVFIILLLLASPAVTTPSEQNLPGELTPADLEGAETTPEESWYSKIKSGVKHASCKLVGYACDDSETEESLLSKIKGGVEYAACKYVNIACED |
| Dis128 | MLCLPVFIILLLLASPAVTTPSEQNLPGELTPADLEGAETTOEESWYSKIKGGVKHASCKLVGYACDDSETEESLLSKIKGSVEHAACKYVNIGCED |
| Dis132 | MLCLPVFIILLLLASPAVTTPSEQNLPGELTPADLEGAETTPEESWYSKIKGGVKHASCKLVGYACDDSETEESLLSKIKGVSNMLRANTLI |
| Dis133 | MLCLPVFIILLLLAAPAVTAPSEPNLQRGLKLGGLKAEPNLQRGLKLGGVKEGLLKVGASAIKGAVNGALNSITKEDRKK |
| Dis135 | MLCLPVFIILLLLAAPAVTAPSEPNLQRGLKLGGLKAEPNLQRGLKLGGVKDKLLKVGGNIFKGAVQGAVDSLTKEDRKQ |
| Dis16 | MLRLIITAVLASACLALPHRRDAAPADMGALKPFEQQMQPMGMPGSMAGMQGMPGQQAMPGGMLGNQLMPFGPGMGMGAGYRRAADHNQEKRDLPLT |
| Dis18 | MNMWMTPSVLVVVVFTATVVCSTEDERLTRQRRGDPTAECCLTLLGCYDTCSHDNTSPDCWGHCKSESTTGCSLDFSLHYCSEFQDCYGPCVTDKDESRCFKACRKEAMIDCLDHGSVECCPGFVNCYQNCRRSREDCITYCLKENC |
| Dis2 | MGMRMMFIVFLLVVLATTVVSLRSDRAFNRKNRRIAEPNTEEEWNECCKDPSCRNNHMDRCAE |
| Dis20 | MNFYLLLTVTLLLASFTGGDARRIQGMDIYRHFVRRDCGRDCVGCDNOANCCCGGQTCVNGNKCE |
| Dis21 | MNFYLLLTVTLLLASFTGGDARRIQGMDIYRHFVRRDCGKDCVGCDNPANCCCGGQTCVNGNKCE |
| Dis26 | MGFRQLVTVGLLLTFFMSTDASHADQTESGFLRDDETVFPCNSDRCACLPKEGSTTSYQCQSLEASTDDCVNNECITEDEWSGRR |
| Dis27 | MGFRQLVTVGLLLTFFMSTDASHADQTESGFLRDDETVFPCNSDRCACLPKEGSTTSYQCQSLETSTDGCVNNECVTEDEW |
| Dis28 | MRFLLRLTVALFLTWFTETDAAAIGKREVHQVILGEPLTNYATVPPDAFQQKLPEIILGQPLMEYQSTESPEVLS |
| Dis29 | MRFLLRLTVALFLTWFTETDAAAIGKREVHQVILGEPLTNYVPPDAFQQKLPEIILGQPLMEYQSTESPEVLS |
| Dis3 | MQLYTYLYLLVPLVAFHLIQGTGTLGHGGALTEGRSADATAPKPEPVLLQKSDARSADNSKDKLTQMKRTLKKQGHIARTITAEEAERNRERMSTLGKR |
| Dis30 | MSTLGILLLIALLLPLANPAETGDGQAMPRTRNLRSLSFGRTLRRLEKRGCDOTDGCQTTVCETDTGOCCCKONFTCQISNSGTKSCSCSGQOSDCOV |
| Dis43 | MKLSVALLLIVLLLPVVAGEKESGDHVLKKRCSYSSCKTESCCTGYLCNSVKSCVDPNSGGRFGK |
| Dis44 | MKLSVALLLIVLLLPVVAGEKESGDHVLKKRCSYSSCKTESCCTGYLCNSVKDCVDPNSGGRFGK |
| Dis52 | MMTKLGAVTLLSLVIIPQVLLQQHQDGIVDVKSMQRNKGRTAAGSVLSHSLRSTNNEYDAKHERSCEGNNSYCRKPDWVGDKPCCSPLVCVCTGTMSGGRRTTCKRAKCGOHOSSK |
| Dis54 | MMTKLGAVTLLSLVIIPQVLLQQHQDSIADVKSMERNKGRTAAGSVLSHSLRSTNNEYDAKHKRSCEGNDSYCRKPDWISDKOCCDPLVCVCTGOMSGGRRTTCETGKCGPRPSSK |
| Dis55 | MMTKLGAVTLLSLVIIPQVLLQQHQDSIADVKAMERNKGRTAAGSVLSHSLRSTNNEYDTKHKRSCEGNDSYCRKPDWISDKPCCDPLVCVCTGOMSGGRRTTCETGKCGOROSSK |
| Dis56 | MMTKLGAVTLLSLVIIPQVLLQQHQDSIADVKAMERNKGRTAAGSVLSHSLRSTNNEYDAKHKRSCEGNDSYCRKPDWISDKPCCDPLICVCTGPMSGGRRTTCETGKCGPRPSSK |
| Dis57 | MIQALASMAWTSMLCSADQVSTSPSVPTFVMVLMATVLLTGIMETEARTLFQMIARRSSDYPCAGTFADCRGQPDGATCCDTGYCQGNVCHY |
| Dis59 | MQLSVILFVLLLTMPLFNGSVLNAINGRKTFERNDRSTDSSQMFEKRCPTACKSCSOOGTCQPVR |
| Dis60 | MKMYLCLAVVLLLASTIVDSALLDKTETLRNWRRKGRDESQCPECRCHDLKNAICDISEACSDEASCPTSPECNNGNCLCKNFHGGRCVSHSQCNDRHC |
| Dis61 | MKMYLCLAVVLLLASTIVDSALLDKTETLRNWRRKGRDESQCPECRCHELKNAICDISEACNDEASCPTSPGCNNGNCLCKNFHGGRCVSHSECNDRHC |
| Dis64 | MKLTYALIVAVLFLTACQVITTDDSRDKQDLLAMLFSKKRNSRDSKWLTKRCLSIGYACGVAISEKCCHVCDNOTGAGTCVYN |
| Dis65 | MKLTYALIVAVLFLTACQVITTDDSRDKQDLLAMLFNKKRNSRDSKWLTKRCLSIGYACGVAISEKCCHVCDNPTGAGTCVYN |
| Dis66 | MKLTYALIVAVLFLTACQLITTDDSRDKQDLLAMLFNKKRNSRDSKWLAKRCLSIGYACGVAISEKCCHVCDNPTGAGTCVYN |
| Dis67 | MKLTYALIVVVLFLTACQLLTADYSRDKQEYPTMRFRDQMRNAKGPKWIRSCAESGKSCDTKVCCDDMγCIGTPGGSMCNG |
| Dis72 | MKLTCVLVVAVLFLTACQFNTADDSRNKQEYRAARLRVGMQKSNGFRSCANYHESCASDPCCEGLγCIGAQGGGVCI |
| Dis73 | MKLTCVLVVAVLFLTACQFNTADDSRNKQEYRAARLRVGMQKSKGFRSCANYHESCASDPCCEGLECIGAQGGGVCI |
| Dis87 | MKELMILILVATALLSIQVMVRGDGEKPLMGGIKRNAAAGLSALIRGKRCKGTSAICEEDGECCSDDCKCMIASGCSNHINRRCAA |
| Dis88 | MKELMILILVATALLSIQVMVRGDGEKPLMGGIKRNAAAGLSALIRGKRCKGESAICEEDGECCSDDCKCMIASGCSNHINRRCAA |
| Dis89 | MKELMILILVATALLSIQVMVRGDGEKPLMGGIKRNAAAGLSALIRGKRCKGASAICEEDGECCSDDCKCMIASGCSNHINRRCAA |
| Dis90 | MKELMILILVATALLSIQVMVRGDGEKPLMGGVKRNAAAGLSALIRGKRCKGTSAICEEDGECCSDDCKCMIASGCSNHINRRCAA |
| Dis93 | MKELMILILVATTLLSIRVMVRGDGEKPLMGGIKRNAAAGLSALIRGKRCKGASALCEEDGECCSGDCKCMHASGCTNDINLRCAA |
| Dis95 | MKELMILILVATALLSIQVMVRGDGEKPLMGRIKRNAAAGLSALIRGKRCKGTSALCEEDDECCSGDCKCMIASGCTNDINLRCAA |
| Dis96 | MKELMILILVATALLSIQVMVRGDGEKPLMGRIKRNAAAGLSALIRGKRCKGASALCEEDGECCSGDCKCMIASGCTNDINLRCAA |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Prashanth, J.R.; Dutertre, S.; Rai, S.K.; Lewis, R.J. Venomics Reveals a Non-Compartmentalised Venom Gland in the Early Diverged Vermivorous Conus distans. Toxins 2022, 14, 226. https://doi.org/10.3390/toxins14030226
Prashanth JR, Dutertre S, Rai SK, Lewis RJ. Venomics Reveals a Non-Compartmentalised Venom Gland in the Early Diverged Vermivorous Conus distans. Toxins. 2022; 14(3):226. https://doi.org/10.3390/toxins14030226
Chicago/Turabian StylePrashanth, Jutty Rajan, Sebastien Dutertre, Subash Kumar Rai, and Richard J. Lewis. 2022. "Venomics Reveals a Non-Compartmentalised Venom Gland in the Early Diverged Vermivorous Conus distans" Toxins 14, no. 3: 226. https://doi.org/10.3390/toxins14030226
APA StylePrashanth, J. R., Dutertre, S., Rai, S. K., & Lewis, R. J. (2022). Venomics Reveals a Non-Compartmentalised Venom Gland in the Early Diverged Vermivorous Conus distans. Toxins, 14(3), 226. https://doi.org/10.3390/toxins14030226