MazEF Homologs in Symbiobacterium thermophilum Exhibit Cross-Neutralization with Non-Cognate MazEFs
Abstract
:1. Introduction
2. Results
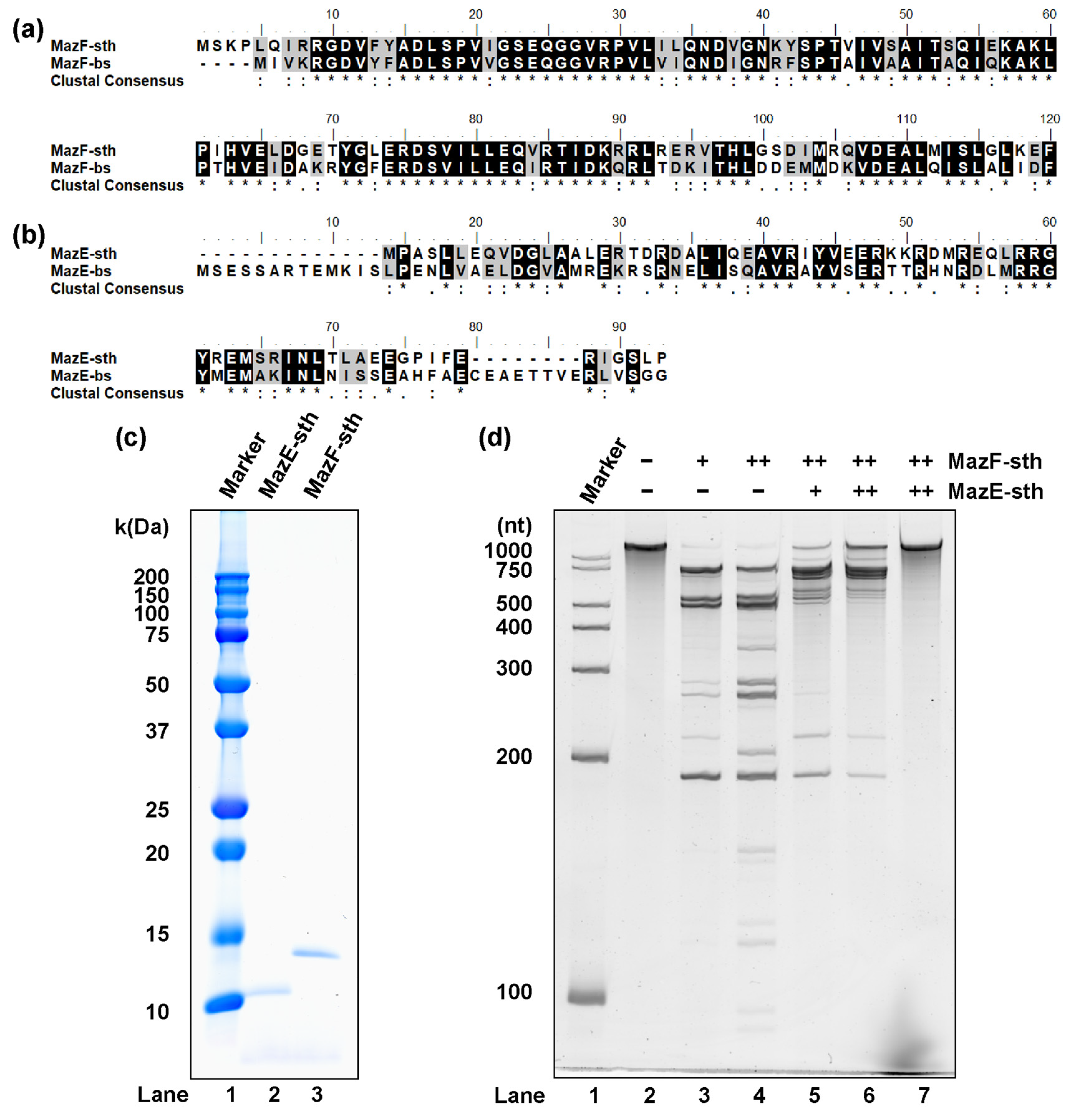
2.1. MazEF-sth Conserved in S. thermophilum Functions as a Ribonuclease and Inhibitor
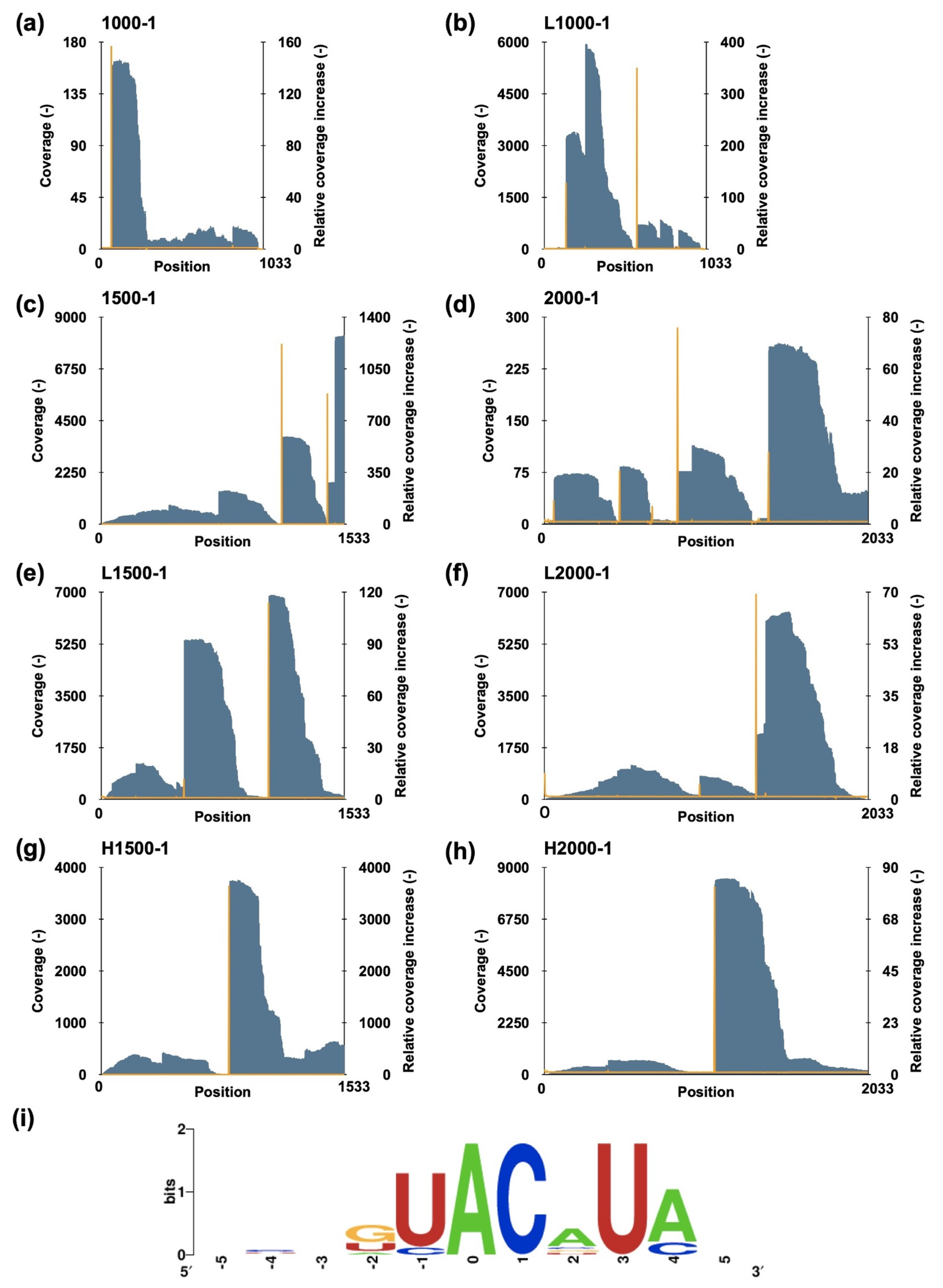
2.2. MazF-sth Preferentially Cleaves the UACAUA Motif
2.3. Estimated Target Coding Sequences of MazF-sth
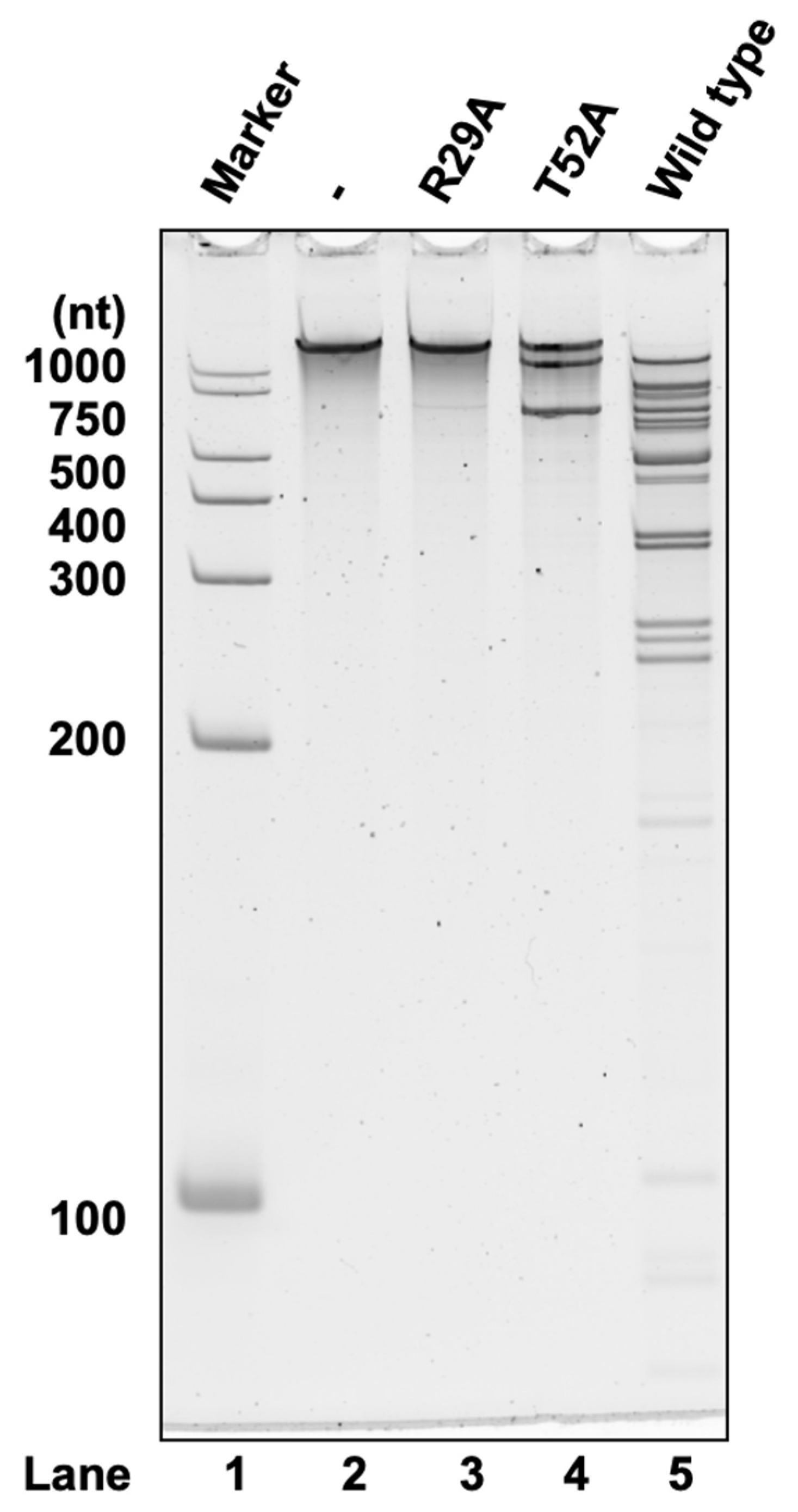
2.4. The Conserved Sites of Arg29 and Thr52 in MazF-sth Are Essential for Cleavage Activity
2.5. MazF-sth Enzymatic Activity Was Suppressed by Non-Cognate MazE-bs
3. Discussion
4. Conclusions
5. Materials and Methods
5.1. Plasmids, RNAs, and Oligonucleotides
5.2. Protein Expression and Purification
5.3. Cleavage Assay
5.4. Massive Parallel Sequencing
5.5. FRET Assay
5.6. Site-Directed Mutagenesis and Cell-Free Expression
5.7. Accession Numbers
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Fraikin, N.; Goormaghtigh, F.; Van Melderen, L. Type II toxin-antitoxin systems: Evolution and revolutions. J. Bacteriol. 2020, 202, e00763-19. [Google Scholar] [CrossRef]
- Jurėnas, D.; Fraikin, N.; Goormaghtigh, F.; Van Melderen, L. Biology and evolution of bacterial toxin–antitoxin systems. Nat. Rev. Microbiol. 2022, 20, 335–350. [Google Scholar] [CrossRef]
- Kamada, K.; Hanaoka, F.; Burley, S.K. Crystal structure of the MazE/MazF complex: Molecular bases of antidote-toxin recognition. Mol. Cell 2003, 11, 875–884. [Google Scholar] [CrossRef]
- Marianovsky, I.; Aizenman, E.; Engelberg-Kulka, H.; Glaser, G. The regulation of the Escherichia coli mazEF promoter involves an unusual alternating palindrome. J. Biol. Chem. 2001, 276, 5975–5984. [Google Scholar] [CrossRef] [PubMed]
- Nikolic, N.; Bergmiller, T.; Vandervelde, A.; Albanese, T.G.; Gelens, L.; Moll, I. Autoregulation of mazEF expression underlies growth heterogeneity in bacterial populations. Nucleic Acids Res. 2018, 46, 2918–2931. [Google Scholar] [CrossRef] [PubMed]
- Aizenman, E.; Engelberg-Kulka, H.; Glaser, G. An Escherichia coli chromosomal “Addiction module” regulated by guanosine [corrected] 3′,5′-bispyrophosphate: A model for programmed bacterial cell death. Proc. Natl. Acad. Sci. USA 1996, 93, 6059–6063. [Google Scholar] [CrossRef] [PubMed]
- Tripathi, A.; Dewan, P.C.; Siddique, S.A.; Varadarajan, R. MazF-induced growth inhibition and persister generation in Escherichia coli. J. Biol. Chem. 2014, 289, 4191–4205. [Google Scholar] [CrossRef] [PubMed]
- Sat, B.; Hazan, R.; Fisher, T.; Khaner, H.; Glaser, G.; Engelberg-Kulka, H. Programmed cell death in Escherichia coli: Some antibiotics can trigger mazEF lethality. J. Bacteriol. 2001, 183, 2041–2045. [Google Scholar] [CrossRef] [PubMed]
- Sat, B.; Reches, M.; Engelberg-Kulka, H. The Escherichia coli mazEF suicide module mediates thymineless death. J. Bacteriol. 2003, 185, 1803–1807. [Google Scholar] [CrossRef] [PubMed]
- Hazan, R.; Sat, B.; Engelberg-Kulka, H. Escherichia coli mazEF-mediated cell death is triggered by various stressful conditions. J. Bacteriol. 2004, 186, 3663–3669. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Zhang, J.; Hoeflich, K.P.; Ikura, M.; Qing, G.; Inouye, M. MazF cleaves cellular mRNAs specifically at ACA to block protein synthesis in Escherichia coli. Mol. Cell 2003, 12, 913–923. [Google Scholar] [CrossRef]
- Culviner, P.H.; Laub, M.T. Global analysis of the E. coli toxin MazF reveals widespread cleavage of mRNA and the inhibition of rRNA maturation and ribosome biogenesis. Mol. Cell 2018, 70, 868–880.e10. [Google Scholar] [CrossRef]
- Miyamoto, T.; Yokota, A.; Ota, Y.; Tsuruga, M.; Aoi, R.; Tsuneda, S.; Noda, N. Nitrosomonas europaea MazF specifically recognises the UGG motif and promotes selective RNA degradation. Front. Microbiol. 2018, 9, 2386. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi, Y.; Nariya, H.; Park, J.H.; Inouye, M. Inhibition of specific gene expressions by protein-mediated mRNA interference. Nat. Commun. 2012, 3, 607. [Google Scholar] [CrossRef] [PubMed]
- Simanshu, D.K.; Yamaguchi, Y.; Park, J.H.; Inouye, M.; Patel, D.J. Structural basis of mRNA recognition and cleavage by toxin MazF and its regulation by antitoxin MazE in Bacillus subtilis. Mol. Cell 2013, 52, 447–458. [Google Scholar] [CrossRef] [PubMed]
- Tamiya-Ishitsuka, H.; Tsuruga, M.; Noda, N.; Yokota, A. Conserved Amino Acid Moieties of Candidatus Desulforudis audaxviator MazF Determine Ribonuclease Activity and Specificity. Front. Microbiol. 2021, 12, 748619. [Google Scholar] [CrossRef] [PubMed]
- Park, J.H.; Yamaguchi, Y.; Inouye, M. Bacillus subtilis MazF-bs (EndoA) is a UACAU-specific mRNA interferase. FEBS Lett. 2011, 585, 2526–2532. [Google Scholar] [CrossRef] [PubMed]
- Zhu, L.; Inoue, K.; Yoshizumi, S.; Kobayashi, H.; Zhang, Y.; Ouyang, M.; Kato, F.; Sugai, M.; Inouye, M. Staphylococcus aureus MazF specifically cleaves a pentad sequence, UACAU, which is unusually abundant in the mRNA for pathogenic adhesive factor SraP. J. Bacteriol. 2009, 191, 3248–3255. [Google Scholar] [CrossRef] [PubMed]
- Rothenbacher, F.P.; Suzuki, M.; Hurley, J.M.; Montville, T.J.; Kirn, T.J.; Ouyang, M.; Woychik, N.A. Clostridium difficile MazF toxin exhibits selective, not global, mRNA cleavage. J. Bacteriol. 2012, 194, 3464–3474. [Google Scholar] [CrossRef] [PubMed]
- Schuster, C.F.; Park, J.H.; Prax, M.; Herbig, A.; Nieselt, K.; Rosenstein, R.; Inouye, M.; Bertram, R. Characterization of a mazEF toxin-antitoxin homologue from Staphylococcus equorum. J. Bacteriol. 2013, 195, 115–125. [Google Scholar] [CrossRef]
- Suzuki, S.; Horinouchi, S.; Beppu, T. Growth of a tryptophanase-producing thermophile, Symbiobacterium thermophilum gen. nov., sp. nov., is dependent on co-culture with a Bacillus sp. Microbiology 1988, 134, 2353–2362. [Google Scholar] [CrossRef]
- Sugihara, T.; Watsuji, T.O.; Kubota, S.; Yamada, K.; Oka, K.; Watanabe, K.; Meguro, M.; Sawada, E.; Yoshihara, K.; Ueda, K.; et al. Distribution of Symbiobacterium thermophilum and related bacteria in the marine environment. Biosci. Biotechnol. Biochem. 2008, 72, 204–211. [Google Scholar] [CrossRef] [PubMed]
- Watsuji, T.O.; Kato, T.; Ueda, K.; Beppu, T. CO2 supply induces the growth of Symbiobacterium thermophilum, a syntrophic bacterium. Biosci. Biotechnol. Biochem. 2006, 70, 753–756. [Google Scholar] [CrossRef] [PubMed]
- Ueda, K.; Yamashita, A.; Ishikawa, J.; Shimada, M.; Watsuji, T.O.; Morimura, K.; Ikeda, H.; Hattori, M.; Beppu, T. Genome sequence of Symbiobacterium thermophilum, an uncultivable bacterium that depends on microbial commensalism. Nucleic Acids Res. 2004, 32, 4937–4944. [Google Scholar] [CrossRef] [PubMed]
- Xie, Y.; Wei, Y.; Shen, Y.; Li, X.; Zhou, H.; Tai, C.; Deng, Z.; Ou, H.Y. TADB 2.0: An updated database of bacterial type II toxin-antitoxin loci. Nucleic Acids Res. 2018, 46, D749–D753. [Google Scholar] [CrossRef] [PubMed]
- Miyamoto, T.; Kato, Y.; Sekiguchi, Y.; Tsuneda, S.; Noda, N. Characterization of MazF-mediated sequence-specific RNA cleavage in Pseudomonas putida using massive parallel sequencing. PLoS ONE 2016, 11, e0149494. [Google Scholar] [CrossRef] [PubMed]
- Akiyama, H.; Ueda, Y.; Nobumasa, H.; Ooshima, H.; Ishizawa, Y.; Kitahiro, K.; Miyagawa, I.; Watanabe, K.; Nakamura, T.; Tanaka, R.; et al. A set of external reference controls/probes that enable quality assurance between different microarray platforms. Anal. Biochem. 2015, 472, 75–83. [Google Scholar] [CrossRef]
- Crooks, G.E.; Hon, G.; Chandonia, J.M.; Brenner, S.E. WebLogo: A sequence logo generator. Genome Res. 2004, 14, 1188–1190. [Google Scholar] [CrossRef]
- Lei, Y.; Oshima, T.; Ogasawara, N.; Ishikawa, S. Functional analysis of the protein Veg, which stimulates biofilm formation in Bacillus subtilis. J. Bacteriol. 2013, 195, 1697–1705. [Google Scholar] [CrossRef]
- Amikam, D.; Galperin, M.Y. PilZ domain is part of the bacterial c-di-GMP binding protein. Bioinformatics 2006, 22, 3–6. [Google Scholar] [CrossRef]
- Kato, F.; Yabuno, Y.; Yamaguchi, Y.; Sugai, M.; Inouye, M. Deletion of mazF increases Staphylococcus aureus biofilm formation in an ica-dependent manner. Pathog. Dis. 2017, 75, ftx026. [Google Scholar] [CrossRef] [PubMed]
- Ma, D.; Mandell, J.B.; Donegan, N.P.; Cheung, A.L.; Ma, W.; Rothenberger, S.; Shanks, R.M.Q.; Richardson, A.R.; Urish, K.L. The toxin-antitoxin MazEF drives Staphylococcus aureus biofilm formation, antibiotic tolerance, and chronic infection. mBio 2019, 10, e01658-19. [Google Scholar] [CrossRef] [PubMed]
- Ramisetty, B.C.M.; Santhosh, R.S. Horizontal gene transfer of chromosomal Type II toxin–antitoxin systems of Escherichia coli. FEMS Microbiol. Lett. 2016, 363, fnv238. [Google Scholar] [CrossRef] [PubMed]
- Fiedoruk, K.; Daniluk, T.; Swiecicka, I.; Sciepuk, M.; Leszczynska, K. Type II toxin–antitoxin systems are unevenly distributed among Escherichia coli phylogroups. Microbiology 2015, 161, 158–167. [Google Scholar] [CrossRef] [PubMed]
- Chopra, N.; Saumitra, A.; Pathak, A.; Bhatnagar, R.; Bhatnagar, S. Linkage, mobility, and selfishness in the MazF family of bacterial toxins: A snapshot of bacterial evolution. Genome Biol. Evol. 2013, 5, 2268–2284. [Google Scholar] [CrossRef] [PubMed]
- Fillol-Salom, A.; Rostøl, J.T.; Ojiogu, A.D.; Chen, J.; Douce, G.; Humphrey, S.; Penadés, J.R. Bacteriophages benefit from mobilizing pathogenicity islands encoding immune systems against competitors. Cell 2022, 185, 3248–3262.e20. [Google Scholar] [CrossRef]
- Oshima, K.; Ueda, K.; Beppu, T.; Nishida, H. Unique evolution of Symbiobacterium thermophilum suggested from gene content and orthologous protein sequence comparisons. Int. J. Evol. Biol. 2011, 2011, 376831. [Google Scholar] [CrossRef]
- Ueda, K.; Beppu, T. Lessons from studies of Symbiobacterium thermophilum, a unique syntrophic bacterium. Biosci. Biotechnol. Biochem. 2007, 71, 1115–1121. [Google Scholar] [CrossRef]
- Ludwig, W.; Schleifer, K.H.; Whitman, W.B. Revised road map to the phylum firmicutes. In Bergey’s Manual® of Systematic Bacteriology, 2nd ed.; Whitman, W.B., Ed.; Springer: New York, NY, USA, 2009; Volume 3, pp. 1–13. [Google Scholar]
- Riedel, T.; Bunk, B.; Thürmer, A.; Spröer, C.; Brzuszkiewicz, E.; Abt, B.; Gronow, S.; Liesegang, H.; Daniel, R.; Overmann, J. Genome resequencing of the virulent and multidrug-resistant reference strain Clostridium difficile 630. Genome Announc. 2015, 3, e00276-15. [Google Scholar] [CrossRef]
- Zhu, L.; Sharp, J.D.; Kobayashi, H.; Woychik, N.A.; Inouye, M. Noncognate Mycobacterium tuberculosis toxin-antitoxins can physically and functionally interact. J. Biol. Chem. 2010, 285, 39732–39738. [Google Scholar] [CrossRef]
- Nariya, H.; Inouye, M. MazF, an mRNA interferase, mediates programmed cell death during multicellular Myxococcus development. Cell 2008, 132, 55–66. [Google Scholar] [CrossRef]
- Miyamoto, T.; Yokota, A.; Tsuneda, S.; Noda, N. AAU-specific RNA cleavage mediated by MazF toxin endoribonuclease conserved in Nitrosomonas europaea. Toxins 2016, 8, 174. [Google Scholar] [CrossRef]
- Aoi, R.; Miyamoto, T.; Yokota, A.; Ota, Y.; Fujitani, H.; Tsuneda, S.; Noda, N. MazF endoribonucleolytic toxin conserved in Nitrospira specifically cleaves the AACU, AACG, and AAUU motifs. Toxins 2020, 12, 287. [Google Scholar] [CrossRef]
- Miyamoto, T.; Ota, Y.; Yokota, A.; Suyama, T.; Tsuneda, S.; Noda, N. Characterization of a Deinococcus radiodurans MazF: A UACA-specific RNA endoribonuclease. Microbiologyopen 2017, 6, e00501. [Google Scholar] [CrossRef]
- Lee, M.W.; Rogers, E.E.; Stenger, D.C. Xylella fastidiosa plasmid-encoded PemK toxin is an endoribonuclease. Phytopathology 2012, 102, 32–40. [Google Scholar] [CrossRef] [PubMed]
- Ishida, Y.; Inouye, K.; Ming, O.; Inouye, M. A CUGGU/UUGGU-specific MazF homologue from Methanohalobium evestigatum. Biochem. Biophys. Res. Commun. 2019, 518, 533–540. [Google Scholar] [CrossRef] [PubMed]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly Accurate Protein Structure Prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef] [PubMed]


| Annotation | Locus Tag | Length (bp) | Product |
|---|---|---|---|
| Biofilm-related genes | RS16150 | 279 | Veg family protein |
| RS07500 | 669 | PilZ domain-containing protein | |
| Cell wall-related genes | RS17880 | 663 | M23 family metallopeptidase |
| RS05630 | 1098 | polysaccharide pyruvyl transferase CsaB | |
| RS02655 | 1101 | D-alanine--D-alanineligase | |
| RS16455 | 1275 | UDP-N-acetylglucosamine1-carboxyvinyl transferase | |
| RS13520 | 1506 | UDP-N-acetylmuramoyl-L-alanyl-D-glutamate—2,6-diaminopimelate ligase | |
| Membrane proteins | RS08320 | 768 | ABC transporter permease |
| RS09880 | 1032 | ABC transporter substrate-binding protein | |
| RS07025 | 1095 | YeeE/YedE family protein | |
| RS12640 | 1797 | ATP-dependent zinc metalloprotease FtsH | |
| Metabolism | RS06785 | 636 | guanylate kinase |
| RS00715 | 813 | NAD(P)binding domain-containing protein | |
| RS14505 | 1461 | IMP dehydrogenase | |
| Transcription and translation | RS16105 | 570 | aminoacyl- tRNA hydrolase |
| RS17470 | 624 | helix-turn-helix transcriptional regulator | |
| RS09835 | 702 | RNA polymerases sporulation sigma factor SigK | |
| RS16160 | 858 | 16SrRNA(adenine(1518)-N(6)/adenine(1519)-N(6))-dimethyl transferase RsmA | |
| RS05755 | 1227 | tyrosine-tRNA ligase | |
| RS12140 | 2562 | single-stranded-DNA-specific exonuclease RecJ | |
| Stress response | RS07085 | 507 | Hsp20/alpha crysalin family protein |
| DNA recombinant | RS09165 | 885 | site-specific tyrosine recombinase XerD |
| Polysaccharides degradation | RS02780 | 1293 | glycoside hydrolase family 18 protein |
| Unknown function | RS18735 | 210 | hypothetical protein |
| RS11335 | 246 | hypothetical protein | |
| RS09245 | 447 | hypothetical protein | |
| RS18275 | 558 | hypothetical protein | |
| RS14425 | 666 | hypothetical protein | |
| RS18470 | 672 | hypothetical protein | |
| RS06565 | 921 | hypothetical protein | |
| RS01700 | 921 | hypothetical protein |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jiang, Y.-N.; Tamiya-Ishitsuka, H.; Aoi, R.; Okabe, T.; Yokota, A.; Noda, N. MazEF Homologs in Symbiobacterium thermophilum Exhibit Cross-Neutralization with Non-Cognate MazEFs. Toxins 2024, 16, 81. https://doi.org/10.3390/toxins16020081
Jiang Y-N, Tamiya-Ishitsuka H, Aoi R, Okabe T, Yokota A, Noda N. MazEF Homologs in Symbiobacterium thermophilum Exhibit Cross-Neutralization with Non-Cognate MazEFs. Toxins. 2024; 16(2):81. https://doi.org/10.3390/toxins16020081
Chicago/Turabian StyleJiang, Yu-Nong, Hiroko Tamiya-Ishitsuka, Rie Aoi, Takuma Okabe, Akiko Yokota, and Naohiro Noda. 2024. "MazEF Homologs in Symbiobacterium thermophilum Exhibit Cross-Neutralization with Non-Cognate MazEFs" Toxins 16, no. 2: 81. https://doi.org/10.3390/toxins16020081
APA StyleJiang, Y.-N., Tamiya-Ishitsuka, H., Aoi, R., Okabe, T., Yokota, A., & Noda, N. (2024). MazEF Homologs in Symbiobacterium thermophilum Exhibit Cross-Neutralization with Non-Cognate MazEFs. Toxins, 16(2), 81. https://doi.org/10.3390/toxins16020081





