A Glycoprotein-Based Surface-Enhanced Raman Spectroscopy–Lateral Flow Assay Method for Abrin and Ricin Detection
Abstract
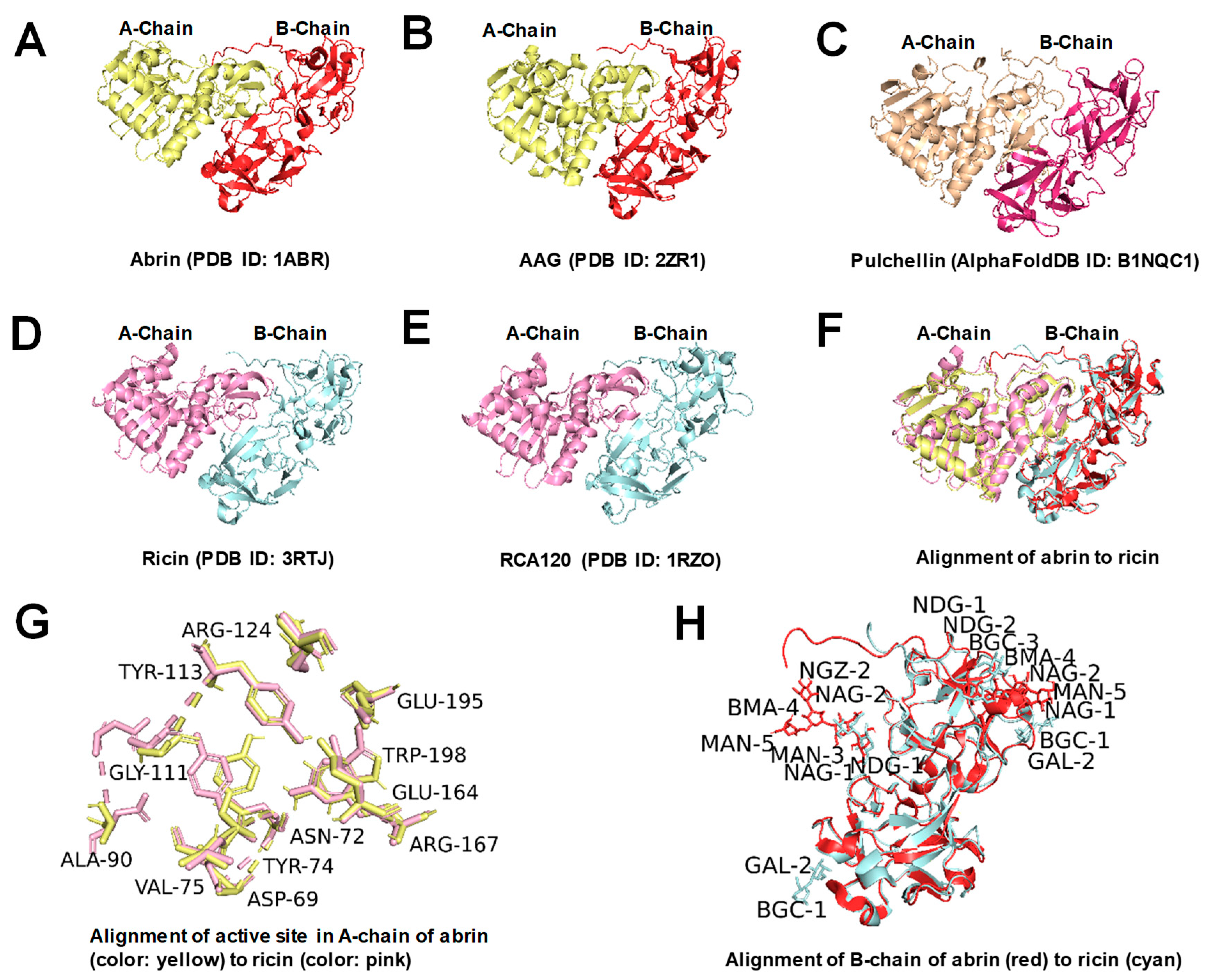
:1. Introduction
2. Results
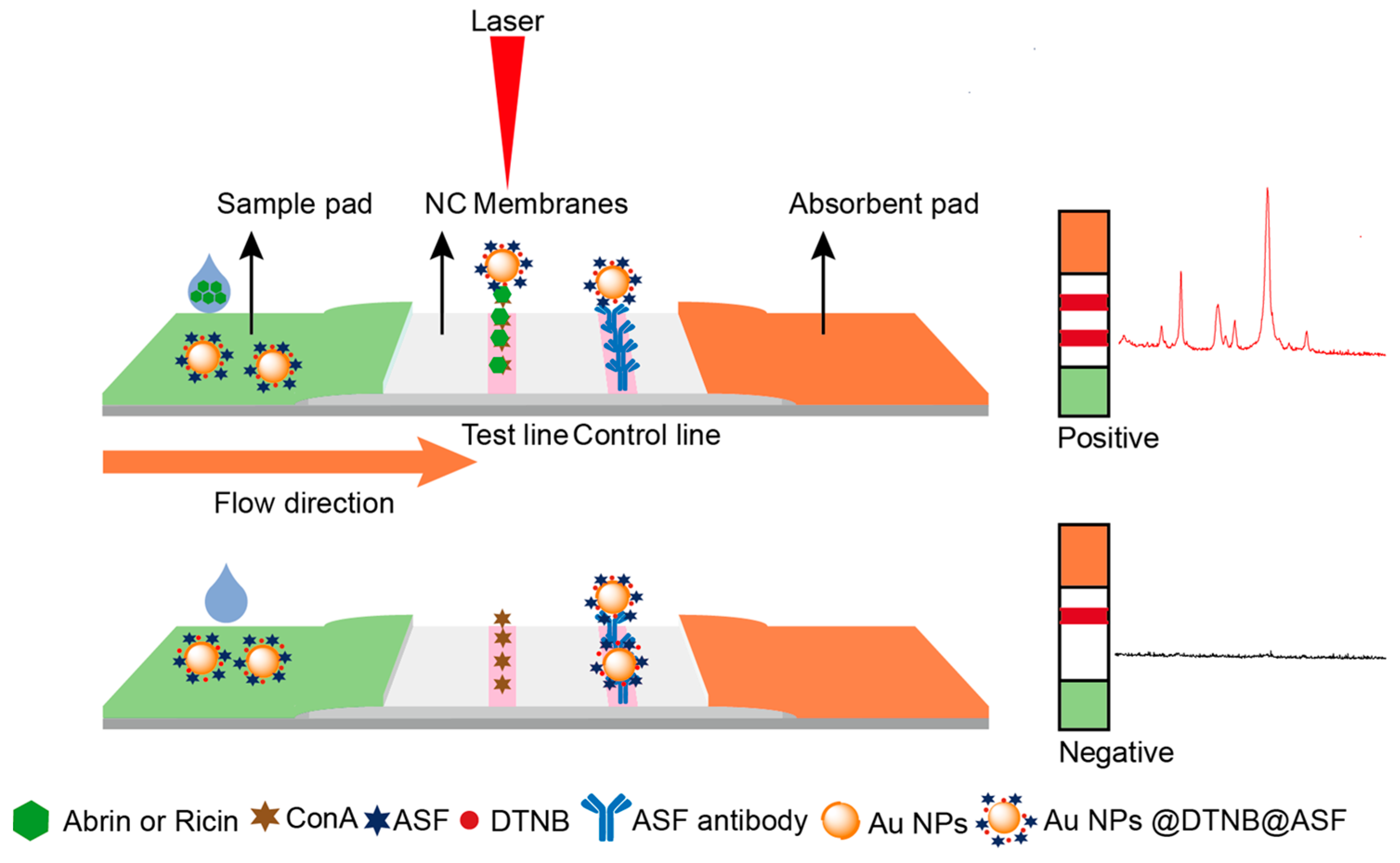
2.1. Schematic Diagram of the SERS-LFA
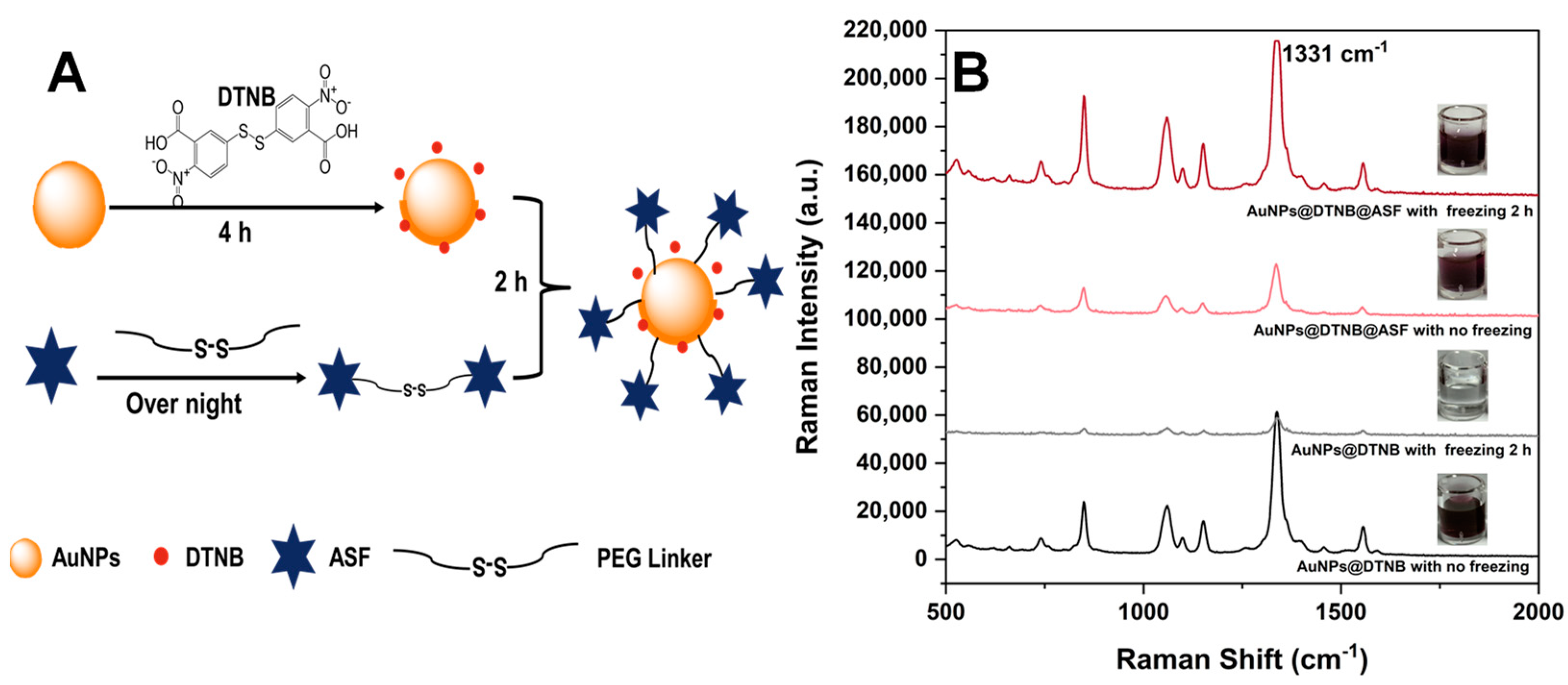
2.2. Preparation and Characterization of SERS NanoGPtags
2.3. Optimization of Experimental Conditions for SERS-LFA Strips Prepared by the Wet Method
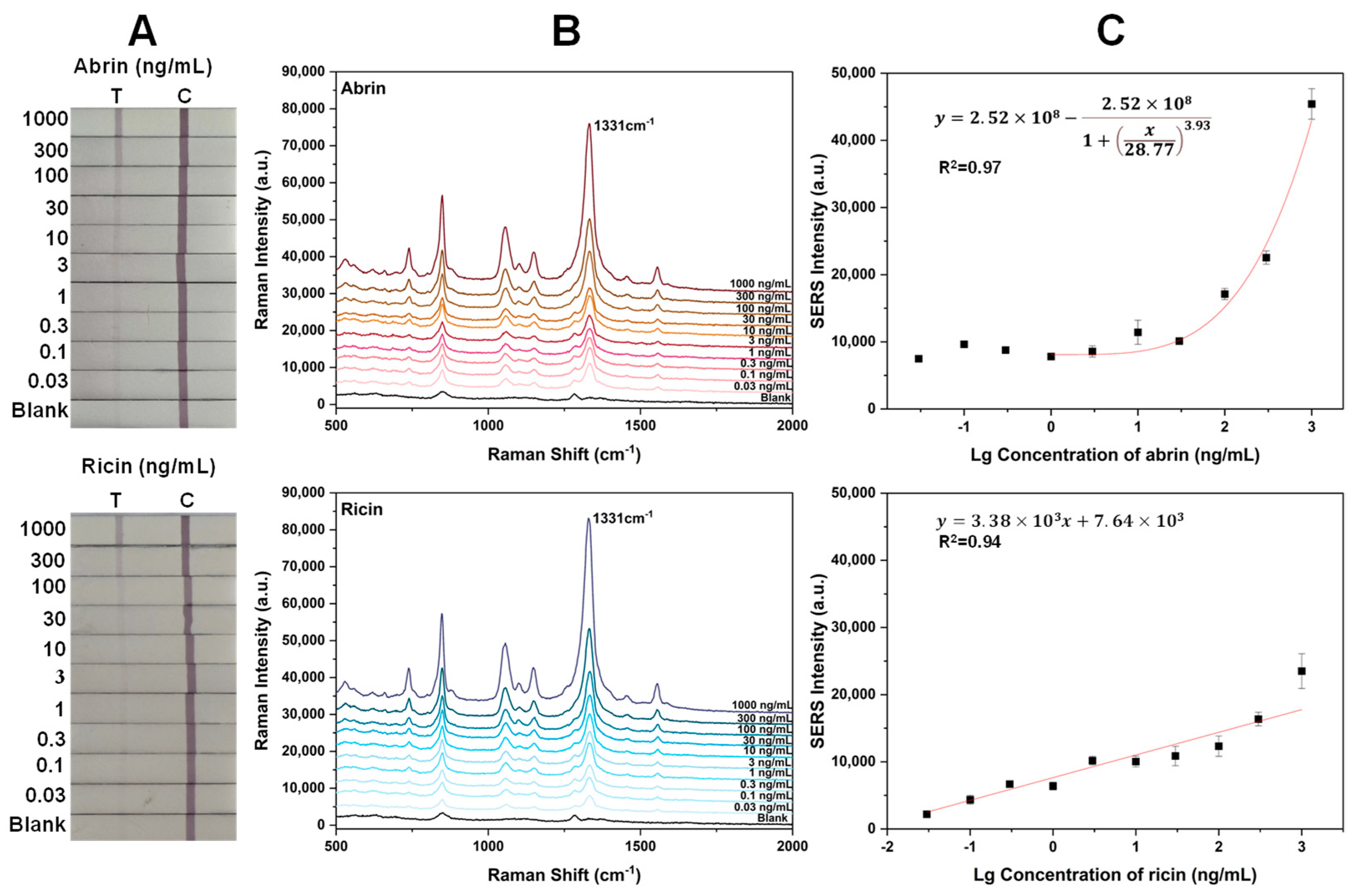
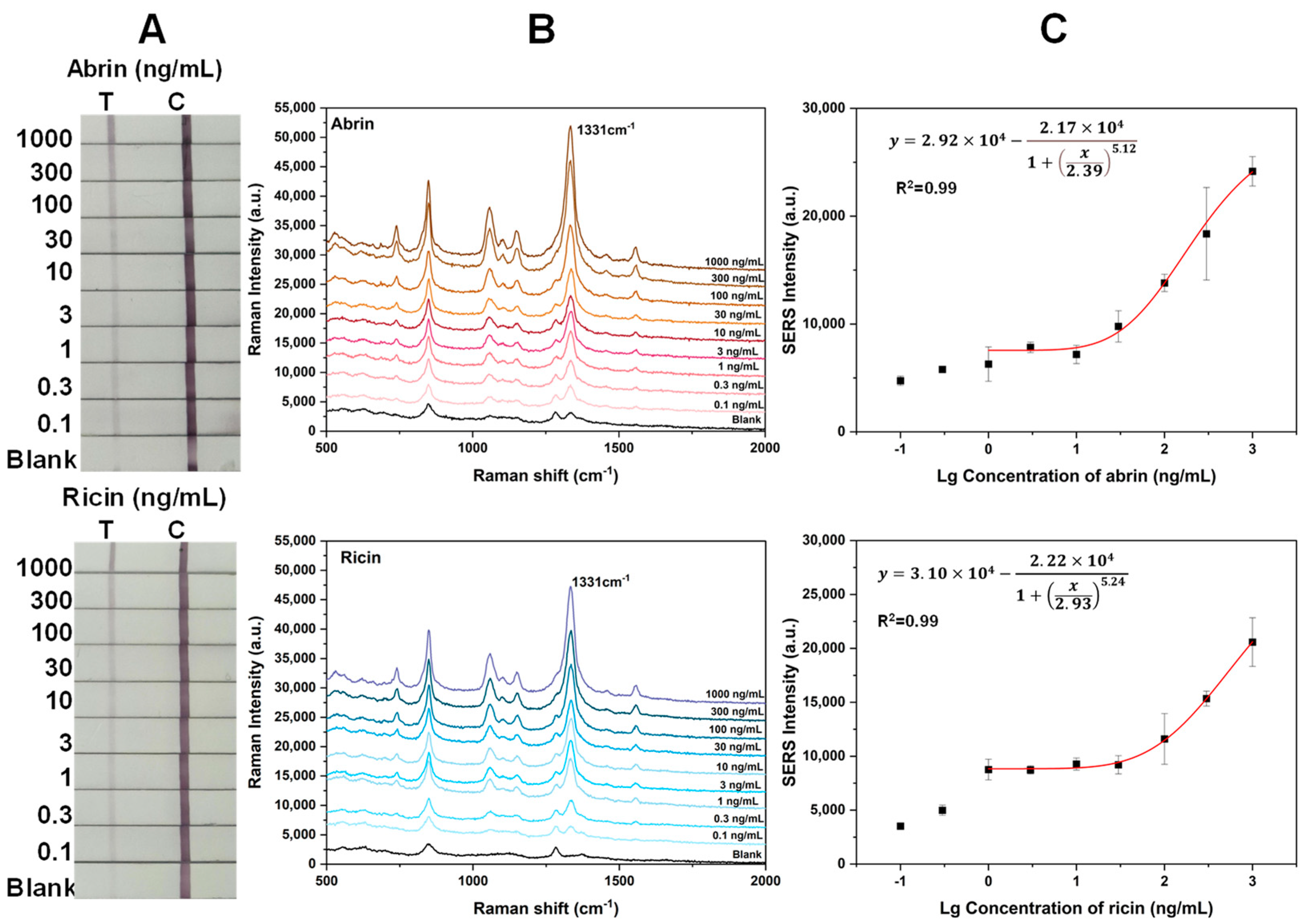
2.4. Sensitivity of the Two Toxins with SERS-LFA Strips Prepared by the Wet Method
2.5. Repeatability of the Two Toxins with SERS-LFA Strips
2.6. Optimization of Experimental Parameters for SERS-LFA Strips Prepared by Dry Method and Method Sensitivity of the Two Toxins
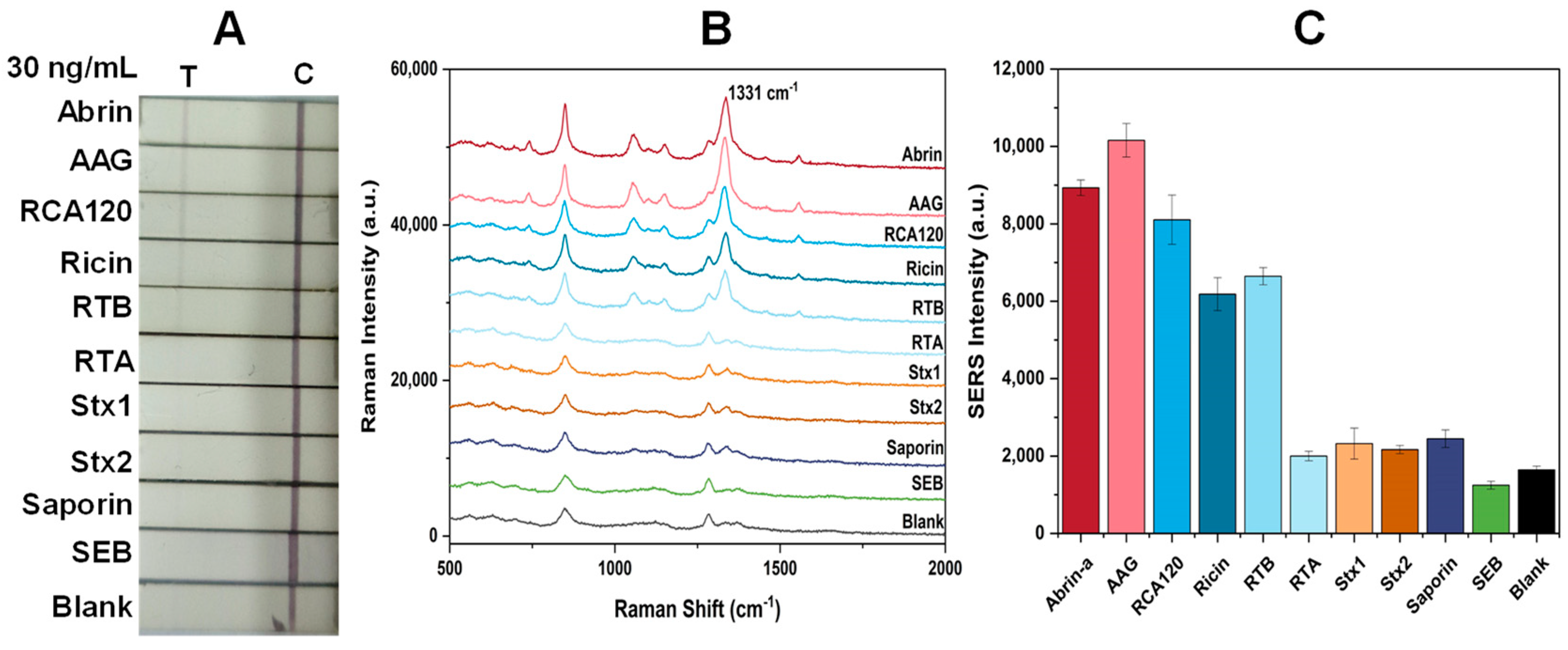
2.7. Specificity of Abrin with Specific SERS-LFA Strips
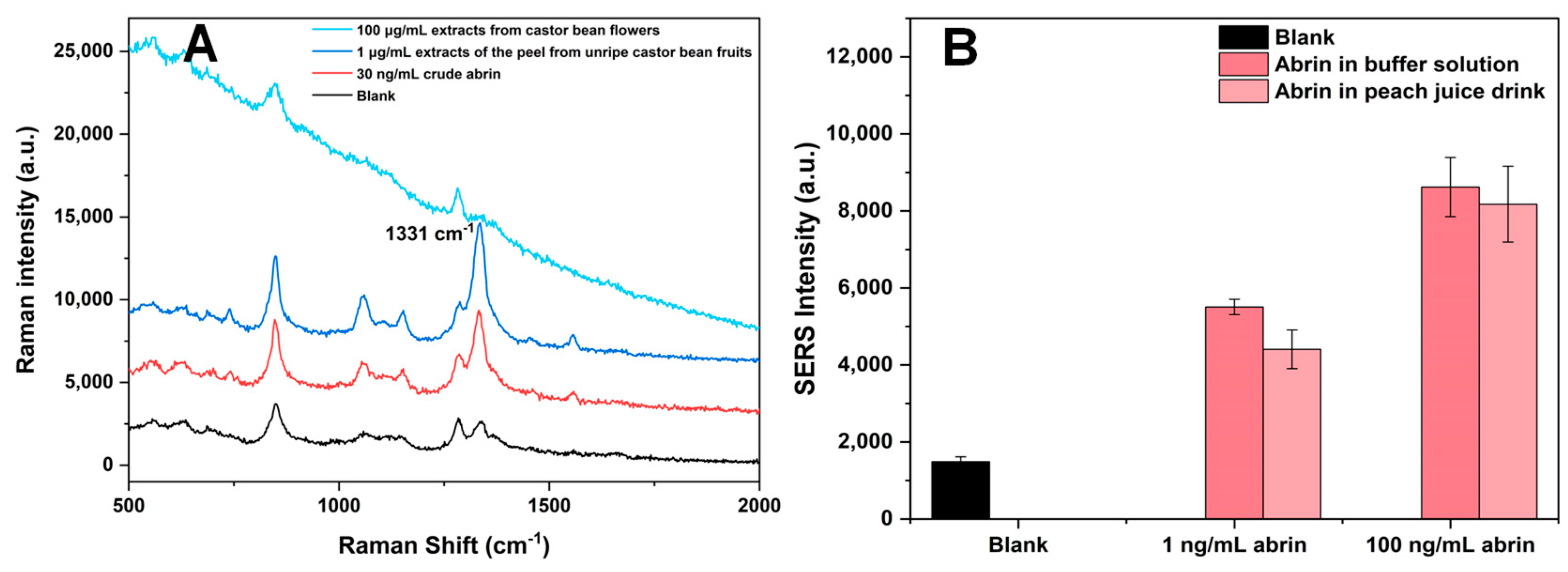
2.8. Examination of White Powder Samples
2.9. Measurements for Actual Botanic Samples and One Spiked Juice Sample
3. Discussion
4. Conclusions
5. Materials and Methods
5.1. Chemicals and Materials
5.2. Synthesis of SERS NanoGPtags
5.3. Preparation of Toxin-Specific SERS–LFA Strips
5.4. Optimization of Experimental Parameters
5.5. SERS-LFA Test Strip Assay
5.6. Evaluation of SERS-LFA Test Strips
5.7. Real Sample Preparation
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Castilho, P.V.; Goto, L.S.; Roberts, L.M.; Araujo, A.P. Isolation and characterization of four type 2 ribosome inactivating pulchellin isoforms from Abrus pulchellus seeds. FEBS J. 2008, 275, 948–959. [Google Scholar] [CrossRef] [PubMed]
- Roy, J.; Som, S.; Sen, A. Isolation, purification, and some properties of a lectin and abrin from Abrus precatorius Linn. Arch. Biochem. Biophys. 1976, 174, 359–361. [Google Scholar] [CrossRef]
- Xiao, L.; Wang, C.; Liu, J.; Liu, L.Y.; Guo, L.; Tang, L. Research progress in analysis and detection techniques, toxicity mechanism, and detoxification countermeasures of Abrin. Mil. Med. Sci. 2024, 48, 294–302. (In Chinese) [Google Scholar] [CrossRef]
- Pitschmann, V.; Hon, Z. Military importance of natural toxins and their analogs. Molecules 2016, 21, 556. [Google Scholar] [CrossRef] [PubMed]
- Gao, S.; Nie, C.; Wang, J.; Wang, J.; Kang, L.; Zhou, Y.; Wang, J.L. Colloidal gold-based immunochromatographic test strip for rapid detection of abrin in food samples. J. Food Prot. 2012, 75, 112–117. [Google Scholar] [CrossRef] [PubMed]
- Ramage, J.G.; Prentice, K.W.; Morse, S.A.; Carter, A.J.; Datta, S.; Drumgoole, R.; Gargis, S.R.; Griffin-Thomas, L.; Hastings, R.; Masri, H.P.; et al. Comprehensive laboratory evaluation of a specific lateral flow assay for the presumptive identification of abrin in suspicious white powders and environmental samples. Biosecur. Bioterror. Biodef. Strategy Pract. Sci. 2014, 12, 49–62. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.; Li, X.B.; Liu, G.W.; Zhang, B.B.; Zhang, Y.; Kong, T.; Tang, J.J.; Li, D.N.; Wang, Z. A colloidal gold probe-based silver enhancement immunochromatographic assay for the rapid detection of abrin-a. Biosens. Bioelectron. 2011, 26, 3710–3713. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Zhao, Y.; Sun, C.; Wang, X.; Wang, X.; Zhang, P.; Qiu, J.; Yang, R.; Zhou, L. Rapid detection of abrin in foods with an up-converting phosphor technology-based lateral flow assay. Sci. Rep. 2016, 6, 34926. [Google Scholar] [CrossRef]
- Asiala, S.M.; Schultz, Z.D. Characterization of hotspots in a highly enhancing SERS substrate. Analyst 2011, 136, 4472–4479. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.C.; Xu, H.B.; Chen, H.Y.; Zhu, S.C.; Huang, W.F.; He, Y.; Hafez, M.E.; Qian, R.C.; Li, D.W. AuNPs-COFs core-shell reversible SERS nanosensor for monitoring intracellular redox dynamics. Anal. Chem. 2022, 94, 14280–14289. [Google Scholar] [CrossRef]
- Khlebtsov, B.; Khlebtsov, N. Surface-enhanced Raman scattering-based lateral-flow immunoassay. Nanomaterials 2020, 10, 2228. [Google Scholar] [CrossRef]
- Hwang, J.; Lee, S.; Choo, J. Application of a SERS-based lateral flow immunoassay strip for the rapid and sensitive detection of staphylococcal enterotoxin B. Nanoscale 2016, 8, 11418–11425. [Google Scholar] [CrossRef]
- Jia, X.; Wang, K.; Li, X.; Liu, Z.; Liu, Y.; Xiao, R.; Wang, S. Highly sensitive detection of three protein toxins via SERS-lateral flow immunoassay based on SiO2@Au nanoparticles. Nanomed.-Nanotechnol. Biol. Med. 2022, 41, 102522. [Google Scholar] [CrossRef] [PubMed]
- Tian, R.; Ren, Y.; Wang, T.; Cao, J.; Li, J.; Deng, A. A SERS-based lateral flow immunochromatographic assay using Raman reporter mediated-gap AuNR@Au nanoparticles as the substrate for the detection of enrofloxacin in food samples. Anal. Chim. Acta 2023, 1257, 341152. [Google Scholar] [CrossRef] [PubMed]
- Yin, L.; You, T.; El-Seedi, H.R.; El-Garawani, I.M.; Guo, Z.; Zou, X.; Cai, J. Rapid and sensitive detection of zearalenone in corn using SERS-based lateral flow immunosensor. Food Chem. 2022, 396, 133707. [Google Scholar] [CrossRef]
- Cao, X.; Song, Q.; Sun, Y.; Mao, Y.; Lu, W.; Li, L. A SERS-LFA biosensor combined with aptamer recognition for simultaneous detection of thrombin and PDGF-BB in prostate cancer plasma. Nanotechnology 2021, 32, 445101. [Google Scholar] [CrossRef] [PubMed]
- Pang, Y.; Li, Q.; Wang, C.; Zhen, S.; Sun, Z.; Xiao, R. CRISPR-cas12a mediated SERS lateral flow assay for amplification-free detection of double-stranded DNA and single-base mutation. Chem. Eng. J. 2022, 429, 132109. [Google Scholar] [CrossRef]
- Tu, Z.; Cheng, S.; Dong, H.; Wang, W.; Yang, X.; Gu, B.; Wang, S.; Wang, C. Universal and ultrasensitive detection of foodborne bacteria on a lateral flow assay strip by using wheat germ agglutinin-modified magnetic SERS nanotags. Rsc Adv. 2022, 12, 27344–27354. [Google Scholar] [CrossRef]
- Liu, B.; Liu, J. Freezing directed construction of bio/nano interfaces: Reagentless conjugation, denser spherical nucleic acids, and better nanoflares. J. Am. Chem. Soc. 2017, 139, 9471–9474. [Google Scholar] [CrossRef]
- Wang, J.; Liu, D.; Wang, Z. Synthesis and cell-surface binding of lectin-gold nanoparticle conjugates. Anal. Methods 2011, 3, 1745–1751. [Google Scholar] [CrossRef]
- Dorner, B.G.; Dorner, M.B.; Worbs, S.; Bossee, A.; Soderstrom, M.; Guo, L. Chapter III. Analysis of ricin/abrin: Analysis strategy. In Recommended Operating Procedures for Analysis in the Verification of Chemical Disarmament, 2023 Edition; Vanninen, P., Ed.; University of Helsinki: Helsinki, Finland, 2023; p. 730. [Google Scholar]
- Yang, J.; Wang, C.; Luo, L.; Li, Z.; Xu, B.; Guo, L.; Xie, J. Highly sensitive MALDI-MS measurement of active ricin: Insight from more potential deoxynucleobase-hybrid oligonucleotide substrates. Analyst 2021, 146, 2955–2964. [Google Scholar] [CrossRef]
- Yang, Z.; Wang, C.; Xiao, L.; Wang, C.; Tang, L.; Guo, L.; Xie, J. SCX-tip-aided LC-MS detection of active ricin via oligonucleotide substrates for depurination kinetics. Analyst 2023, 148, 2782–2792. [Google Scholar] [CrossRef]
- Hodge, D.R.; Prentice, K.W.; Ramage, J.G.; Prezioso, S.; Gauthier, C.; Swanson, T.; Hastings, R.; Basavanna, U.; Datta, S.; Sharma, S.K.; et al. Comprehensive laboratory evaluation of a highly specific lateral flow assay for the presumptive identification of ricin in suspicious white powders and environmental samples. Biosecur. Bioterror. Biodef. Strategy Pract. Sci. 2013, 11, 237–250. [Google Scholar] [CrossRef]
- Hoyt, K.; Barr, J.R.; Kalb, S.R. Detection of ricin activity and structure by using novel galactose-terminated magnetic bead extraction coupled with mass spectrometric detection. Anal. Biochem. 2021, 631, 114364. [Google Scholar] [CrossRef] [PubMed]
- Chen, R.; Wang, H.; Sun, C.; Zhao, Y.; He, Y.; Nisar, M.S.; Wei, W.; Kang, H.; Xie, X.; Du, C.; et al. Au@SiO(2) SERS nanotags based lateral flow immunoassay for simultaneous detection of aflatoxin B(1) and ochratoxin A. Talanta 2023, 258, 124401. [Google Scholar] [CrossRef]
- Bai, X.; Hu, C.; Chen, L.; Wang, J.; Li, Y.; Wan, W.; Jin, Z.; Li, Y.; Xin, W.; Kang, L.; et al. A self-driven microfluidic chip for ricin and abrin detection. Sensors 2022, 22, 3461. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Deng, M.; Ga, S.; Chen, S.; Kang, L.; Wang, J.; Xin, W.; Zhang, T.; You, Z.; An, Y.; et al. Capillary-driven surface-enhanced Raman scattering (SERS)-based microfluidic chip for abrin detection. Nanoscale Res. Lett. 2014, 9, 138. [Google Scholar] [CrossRef]
- Zengin, A.; Tamer, U.; Caykara, T. Fabrication of a SERS based aptasensor for detection of ricin B toxin. J. Mater. Chem. B 2015, 3, 306–315. [Google Scholar] [CrossRef] [PubMed]
- Campos, A.R.; Gao, Z.; Blaber, M.G.; Huang, R.; Schatz, G.C.; Van Duyne, R.P.; Haynes, C.L. Surface-enhanced Raman spectroscopy detection of ricin b chain in human blood. J. Phys. Chem. C 2016, 120, 20961–20969. [Google Scholar] [CrossRef]
- He, L.; Deen, B.; Rodda, T.; Ronningen, I.; Blasius, T.; Haynes, C.; Diez-Gonzalez, F.; Labuza, T.P. Rapid detection of ricin in milk using immunomagnetic separation combined with surface-enhanced Raman spectroscopy. J. Food Sci. 2011, 76, N49–N53. [Google Scholar] [CrossRef]
- Wang, J.; Xu, H.; Luo, L.; Li, N.; Qiao, C.; Wu, J.; Guo, L.; Xie, J. Rapid detection of whole active ricin using a surface-enhanced Raman scattering-based sandwich immunoassay. J. Raman Spectrosc. 2023, 54, 137–149. [Google Scholar] [CrossRef]
- Tang, J.J.; Sun, J.F.; Lui, R.; Zhang, Z.M.; Liu, J.F.; Xie, J.W. New surface-enhanced Raman sensing chip designed for on-site detection of active ricin in complex matrices based on specific depurination. ACS Appl. Mater. Interfaces 2016, 8, 2449–2455. [Google Scholar] [CrossRef] [PubMed]
- Zheng, J.; Zhao, C.; Tian, G.; He, L. Rapid screening for ricin toxin on letter papers using surface enhanced Raman spectroscopy. Talanta 2017, 162, 552–557. [Google Scholar] [CrossRef]
- He, L.; Rodda, T.; Haynes, C.L.; Deschaines, T.; Strother, T.; Diez-Gonzalez, F.; Labuza, T.P. Detection of a foreign protein in milk using surface-enhanced Raman spectroscopy coupled with antibody-modified silver dendrites. Anal. Chem. 2011, 83, 1510–1513. [Google Scholar] [CrossRef] [PubMed]
- Szlag, V.M.; Styles, M.J.; Madison, L.R.; Campos, A.R.; Wagh, B.; Sprouse, D.; Schatz, G.C.; Reineke, T.M.; Haynes, C.L. SERS detection of ricin b-chain via n-acetyl-galactosamine glycopolymers. ACS Sens. 2016, 1, 842–846. [Google Scholar] [CrossRef]
- Karve, S.S.; Weiss, A.A. Glycolipid binding preferences of Shiga toxin variants. PLoS ONE 2014, 9, e101173. [Google Scholar] [CrossRef]
- Millard, C.B.; Leclaire, R.D.; Romano, J.A.; Lukey, B.J.; Salem, H. Ricin and related toxins: Review and perspective. In Chemical Warfare Agents, 2nd ed.; CRC Press: New York, NY, USA, 2008; pp. 423–467. [Google Scholar] [CrossRef]
- Frens, G. Controlled nucleation for the regulation of the particle size in monodisperse gold suspensions. Nat. Phys. Sci. 1973, 241, 20–22. [Google Scholar] [CrossRef]
- Ma, W.; Xu, S.; Nie, H.; Hu, B.; Bai, Y.; Liu, H. Bifunctional cleavable probes for in situ multiplexed glycan detection and imaging using mass spectrometry. Chem. Sci. 2019, 10, 2320–2325. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.Y.; Lee, T.C.; Hu, S.T.; Tung, T.C. Isolation of four isotoxic proteins and one agglutinin from jequiriti bean (Abrus precatorius). Toxicon 1981, 19, 41–51. [Google Scholar] [CrossRef]



| White Powder | Negative | Positive |
|---|---|---|
| 0.5%PBST (Control) | − | + |
| s1: Baby Milk | − | + |
| s2: Baby Talcum Powder | − | + |
| s3: Albumin Powder | − | + (5 μg/mL) |
| s4: Milk Powder with Lactase | − | + |
| s5: Baby Rice Paste | − | + |
| s6: Stevioside | − | + |
| s7: Polyethylene Glycol (PEG) | − | + |
| s8: Starch | − | + |
| Materials Used | Biocomponents | Target Protein | LOD | Reference |
|---|---|---|---|---|
| microfluidic chip | antibody | abrin, ricin | 0.01 ng/mL (buffer) | [27] |
| microfluidic chip with a gold film | antibody | abrin | 0.1 ng/mL (buffer) | [28] |
| AgNPs | aptamer | ricin B chain | 10.2 fg/mL (buffer) | [29] |
| AgFON | aptamer | ricin B chain | 4 μg/mL (blood) | [30] |
| silver dendrite nano substrates | antibody | ricin | 4 μg/mL (milk) | [31] |
| AuNPs | antibody | ricin | 1 ng/mL (buffer) 4 ng/mL (plasma) | [32] |
| AuNPs | oligodeoxynucleotides poly(21dA) | ricin | 8.9 ng/mL (buffer) | [33] |
| silver dendrites | / | ricin B chain | 4.4 μg/mL (water) | [34] |
| silver dendrites | aptamer | ricin B chain | 10 ng/mL (buffer) 50 ng/mL (juice) 100 ng/mL (milk) | [35] |
| gold film over nanospheres | N-acetyl-galactosamine glycopolymer | ricin B chain | 20 ng/mL (water) | [36] |
| SiO2@Au NPs (SERS-LFIA) | antibody | ricin | 0.1 ng/mL (buffer) | [13] |
| AuNPs | glycoprotein | abrin, ricin | 0.1 ng/mL (abrin in buffer) 0.3 ng/mL (ricin in buffer) | Our work |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xiao, L.; Luo, L.; Liu, J.; Liu, L.; Han, H.; Xiao, R.; Guo, L.; Xie, J.; Tang, L. A Glycoprotein-Based Surface-Enhanced Raman Spectroscopy–Lateral Flow Assay Method for Abrin and Ricin Detection. Toxins 2024, 16, 312. https://doi.org/10.3390/toxins16070312
Xiao L, Luo L, Liu J, Liu L, Han H, Xiao R, Guo L, Xie J, Tang L. A Glycoprotein-Based Surface-Enhanced Raman Spectroscopy–Lateral Flow Assay Method for Abrin and Ricin Detection. Toxins. 2024; 16(7):312. https://doi.org/10.3390/toxins16070312
Chicago/Turabian StyleXiao, Lan, Li Luo, Jia Liu, Luyao Liu, Han Han, Rui Xiao, Lei Guo, Jianwei Xie, and Li Tang. 2024. "A Glycoprotein-Based Surface-Enhanced Raman Spectroscopy–Lateral Flow Assay Method for Abrin and Ricin Detection" Toxins 16, no. 7: 312. https://doi.org/10.3390/toxins16070312
APA StyleXiao, L., Luo, L., Liu, J., Liu, L., Han, H., Xiao, R., Guo, L., Xie, J., & Tang, L. (2024). A Glycoprotein-Based Surface-Enhanced Raman Spectroscopy–Lateral Flow Assay Method for Abrin and Ricin Detection. Toxins, 16(7), 312. https://doi.org/10.3390/toxins16070312





