Molecular Screening for Cyanobacteria and Their Cyanotoxin Potential in Diverse Habitats
Abstract
1. Introduction
2. Results and Discussion
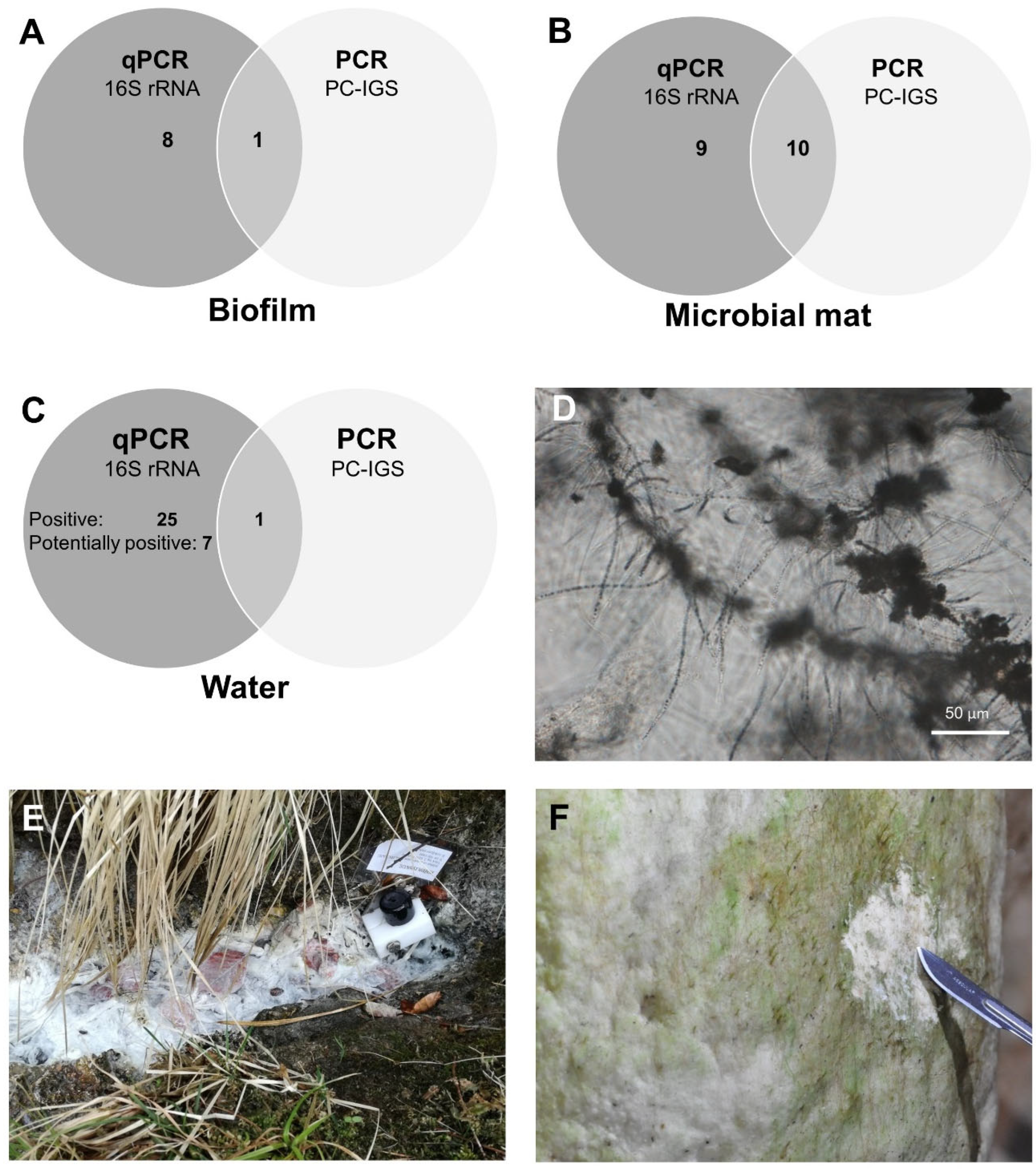
2.1. Molecular Detection of Cyanobacteria: Findings and Constraints
2.2. Cylindrospermopsins: Potential Presence in Sulphidic Springs
3. Conclusions
4. Material and Methods
4.1. Study Sites, Sampling and DNA Extraction
4.2. Molecular Analyses
4.2.1. PCR Screening for PC-IGS
4.2.2. qPCR Reactions for Cyanobacteria and Toxin-Related Genes
4.2.3. Sequencing of PC-IGS and cyrJ
4.3. Microscopy of Sulphidic Biofilm
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Roncero-Ramos, B.; Muñoz-Martín, M.Á.; Chamizo, S.; Fernández-Valbuena, L.; Mendoza, D.; Perona, E.; Cantón, Y.; Mateo, P. Polyphasic evaluation of key cyanobacteria in biocrusts from the most arid region in Europe. PeerJ 2019, 7, e6169. [Google Scholar] [CrossRef] [PubMed]
- Oarga, A. Life in extreme environments. Rev. Biol. Ciências Terra 2009, 9, 1–10. [Google Scholar]
- Shu, W.S.; Huang, L.N. Microbial diversity in extreme environments. Nat. Rev. Microbiol. 2022, 20, 219–235. [Google Scholar] [CrossRef]
- Carré, L.; Zaccai, G.; Delfosse, X.; Girard, E.; Franzetti, B. Relevance of earth-bound extremophiles in the search for extraterrestrial life. Astrobiology 2022, 22, 322–367. [Google Scholar]
- Anwar, U.B.; Zwar, I.P.; de Souza, A.O. Biomolecules produced by extremophiles microorganisms and recent discoveries. In New and Future Developments in Microbial Biotechnology and Bioengineering; Gupta, V.K., Ed.; Elsevier: Amsterdam, The Netherlands, 2020; pp. 247–270. [Google Scholar]
- Xu, H.F.; Dai, G.Z.; Qiu, B.S. Weak red light plays an important role in awakening the photosynthetic machinery following desiccation in the subaerial cyanobacterium Nostoc flagelliforme. Environ. Microbiol. 2019, 21, 2261–2272. [Google Scholar] [CrossRef]
- Bustos-Díaz, E.D.; Barona-Gómez, F.; Cibrián-Jaramillo, A. Cyanobacteria in nitrogen-fixing symbioses. In Cyanobacteria; Mishra, A.K., Tiwari, D.N., Rai, A.N., Eds.; Academic Press: Cambridge, MA, USA, 2019; pp. 29–42. [Google Scholar]
- Nayaka, S.; Toppo, K.; Verma, S. Adaptation in algae to environmental stress and ecological conditions. In Plant Adaptation Strategies in Changing Environment; Shukla, V., Kumar, S., Kumar, N., Eds.; Springer: Singapore, 2017; pp. 103–115. [Google Scholar]
- Billi, D. Challenging the Survival Thresholds of a Desert Cyanobacterium under Laboratory Simulated and Space Conditions. In Extremophiles as Astrobiological Models; Seckbach, J., Stan-Lotter, H., Eds.; Wiley: Hoboken, NJ, USA, 2020; pp. 183–195. [Google Scholar]
- Xu, H.F.; Raanan, H.; Dai, G.Z.; Oren, N.; Berkowicz, S.; Murik, O.; Kaplan, A.; Qiu, B.S. Reading and surviving the harsh conditions in desert biological soil crust: The cyanobacterial viewpoint. FEMS Microbiol. Rev. 2021, 45, fuab036. [Google Scholar] [CrossRef] [PubMed]
- Nelson, C.; Giraldo-Silva, A.; Garcia-Pichel, F. A symbiotic nutrient exchange within the cyanosphere microbiome of the biocrust cyanobacterium, Microcoleus vaginatus. ISME J. 2020, 15, 282–292. [Google Scholar] [CrossRef]
- Dabravolski, S.A.; Isayenkov, S.V. Metabolites facilitating adaptation of desert cyanobacteria to extremely arid environments. Plants 2022, 11, 3225. [Google Scholar] [CrossRef]
- Steinberg, C.E.W.; Schäfer, H.; Beisker, W. Do acid-tolerant cyanobacteria exist? Acta Hydrochim. Et Hydrobiol. 1998, 26, 13–19. [Google Scholar] [CrossRef]
- Cirés, S.; Casero, M.C.; Quesada, A. Toxicity at the Edge of Life: A Review on Cyanobacterial Toxins from Extreme Environments. Mar. Drugs 2017, 15, 233. [Google Scholar] [CrossRef]
- Gärtner, G.; Stoyneva-Gärtner, M.; Uzunov, B. Algal Toxic Compounds and Their Aeroterrestrial, Airborne and other Extremophilic Producers with Attention to Soil and Plant Contamination: A Review. Toxins 2021, 13, 322. [Google Scholar] [CrossRef]
- Cantonati, M.; Komárek, J.; Montejano, G. Cyanobacteria in ambient springs. Biodivers. Conserv. 2015, 24, 865–888. [Google Scholar] [CrossRef]
- Li, Q.; Hu, C.; Yang, H. Responses of cyanobacterial crusts and microbial communities to extreme environments of the stratosphere. Microorganisms 2022, 10, 1252. [Google Scholar] [CrossRef] [PubMed]
- Piano, E.; Bona, F.; Falasco, E.; La Morgia, V.; Badino, G.; Isaia, M. Environmental drivers of phototrophic biofilms in an Alpine show cave (SW-Italian Alps). Sci. Total Environ. 2015, 536, 1007–1018. [Google Scholar] [CrossRef] [PubMed]
- Ai, J.; Guo, J.; Li, Y.; Zhong, X.; Lv, Y.; Li, J.; Yang, A. The diversity of microbes and prediction of their functions in karst caves under the influence of human tourism activities—A case study of Zhijin Cave in Southwest China. Environ. Sci. Pollut. Res. 2022, 29, 25858–25868. [Google Scholar] [CrossRef] [PubMed]
- Buratti, F.M.; Manganelli, M.; Vichi, S.; Stefanelli, M.; Scardala, S.; Testai, E.; Funari, E. Cyanotoxins: Producing organisms, occurrence, toxicity, mechanism of action and human health toxicological risk evaluation. Arch. Toxicol. 2017, 91, 1049–1130. [Google Scholar] [CrossRef]
- Scarlett, K.R.; Kim, S.; Lovin, L.M.; Chatterjee, S.; Scott, J.T.; Brooks, B.W. Global scanning of cylindrospermopsin: Critical review and analysis of aquatic occurrence, bioaccumulation, toxicity and health hazards. Sci. Total Environ. 2020, 738, 139807. [Google Scholar] [CrossRef]
- Chorus, I.; Welker, M. (Eds.) Toxic Cyanobacteria in Water—A Guide to Their Public Health Consequences, Monitoring and Management, 2nd ed.; World Health Organization: Geneva, Switzerland, 2021. [Google Scholar]
- Mehinto, A.C.; Smith, J.; Wenger, E.; Stanton, B.; Linville, R.; Brooks, B.W.; Sutula, M.A.; Howard, M.D. Synthesis of ecotoxicological studies on cyanotoxins in freshwater habitats—Evaluating the basis for developing thresholds protective of aquatic life in the United States. Sci. Total Environ. 2021, 795, 148864. [Google Scholar] [CrossRef]
- Svirčev, Z.; Lalić, D.; Bojadžija Savić, G.; Tokodi, N.; Drobac Backović, D.; Chen, L.; Meriluoto, J.; Codd, G.A. Global geographical and historical overview of cyanotoxin distribution and cyanobacterial poisonings. Arch. Toxicol. 2019, 93, 2429–2481. [Google Scholar] [CrossRef]
- Du, X.; Liu, H.; Yuan, L.; Wang, Y.; Ma, Y.; Wang, R.; Chen, X.; Losiewicz, M.D.; Guo, H.; Zhang, H. The diversity of cyanobacterial toxins on structural characterization, distribution and identification: A systematic review. Toxins 2019, 11, 530. [Google Scholar] [CrossRef]
- Chen, L.; Giesy, J.P.; Adamovsky, O.; Svirčev, Z.; Meriluoto, J.; Codd, G.A.; Mijovic, B.; Shi, T.; Tuo, X.; Li, S.-C.; et al. Challenges of using blooms of Microcystis spp. in animal feeds: A comprehensive review of nutritional, toxicological and microbial health evaluation. Sci. Total Environ. 2021, 764, 142319. [Google Scholar] [CrossRef] [PubMed]
- Yilmaz, S.; Ülger, T.G.; Göktaş, B.; Öztürk, Ş.; Karataş, D.Ö.; Beyzi, E. Cyanotoxin genotoxicity: A review. Toxin Rev. 2021, 41, 699–712. [Google Scholar] [CrossRef]
- Nelson, S.; Young, D.; Toomey, R.; Byl, T. Terrestrial cyanobacteria of Mammoth Cave National Park produce microcystin toxin. In Proceedings of the 28th Tennessee Water Resources Symposium, Montgomery Bell State Park, Burns, TN, USA, 10–12 April 2019. [Google Scholar]
- Mohamed, Z.A. Toxic cyanobacteria and cyanotoxins in public hot springs in Saudi Arabia. Toxicon 2008, 51, 17–27. [Google Scholar] [CrossRef] [PubMed]
- Krienitz, L.; Ballot, A.; Kotut, K.; Wiegand, C.; Pütz, S.; Metcalf, J.S.; Codd, G.A.; Stephan, P. Contribution of hot spring cyanobacteria to the mysterious deaths of Lesser Flamingos at Lake Bogoria, Kenya. FEMS Microbiol. Ecol. 2003, 43, 141–148. [Google Scholar] [CrossRef] [PubMed]
- Strunecký, O.; Kopejtka, K.; Goecke, F.; Tomasch, J.; Lukavský, J.; Neori, A.; Kahl, S.; Pieper, D.H.; Pilarski, P.; Kaftan, D.; et al. High diversity of thermophilic cyanobacteria in Rupite hot spring identified by microscopy, cultivation, single-cell PCR and amplicon sequencing. Extremophiles 2019, 23, 35–48. [Google Scholar] [CrossRef] [PubMed]
- Kleinteich, J.; Wood, S.A.; Puddick, J.; Schleheck, D.; Küpper, F.C.; Dietrich, D. Potent toxins in Arctic environments—Presence of saxitoxins and an unusual microcystin variant in Arctic freshwater ecosystems. Chem. Biol. Interact. 2013, 206, 423–431. [Google Scholar] [CrossRef] [PubMed]
- Mulec, J.; Oarga-Mulec, A.; Schiller, E.; Perşoiu, A.; Holko, L.; Šebela, S. Assessment of the physical environment of epigean invertebrates in a unique habitat: The case of a karst sulfidic spring, Slovenia. Ecohydrology 2014, 8, 1326–1334. [Google Scholar] [CrossRef]
- Mulec, J.; Oarga-Mulec, A.; Holko, L.; Pašić, L.; Kopitar, A.N.; Eleršek, T.; Mihevc, A. Microbiota entrapped in recently-formed ice: Paradana Ice Cave, Slovenia. Sci. Rep. 2021, 11, 1993. [Google Scholar] [CrossRef] [PubMed]
- Neilan, B.A.; Jacobs, D.; Goodman, A.E. Genetic diversity and phylogeny of toxic cyanobacteria determined by DNA polymorphisms within the phycocyanin locus. Appl. Environ. Microbiol. 1995, 61, 3875–3883. [Google Scholar] [CrossRef]
- Taylor, S.C.; Nadeau, K.; Abbasi, M.; Lachance, C.; Nguyen, M.; Fenrich, J. The Ultimate qPCR Experiment: Producing Publication Quality, Reproducible Data the First Time. Trends Biotechnol. 2019, 37, 761–774. [Google Scholar] [CrossRef]
- Zupančič, M.; Kogovšek, P.; Šter, T.; Remec Rekar, Š.; Cerasino, L.; Baebler, Š.; Krivograd Klemenčič, A.; Eleršek, T. Potentially Toxic Planktic and Benthic Cyanobacteria in Slovenian Freshwater Bodies: Detection by Quantitative PCR. Toxins 2021, 13, 133. [Google Scholar] [CrossRef] [PubMed]
- Hughes-Stamm, S.R.; Ashton, K.J.; van Daal, A. Assessment of DNA degradation and the genotyping success of highly degraded samples. Int. J. Leg. Med. 2011, 125, 341–348. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Xiao, P.; Liu, Y.; Wang, J.; Li, R. Targeted deep sequencing reveals high diversity and variable dominance of bloom-forming cyanobacteria in eutrophic lakes. Harmful Algae 2017, 64, 42–50. [Google Scholar] [CrossRef]
- Kim, S.G.; Rhee, S.K.; Ahn, C.Y.; Ko, S.R.; Choi, G.G.; Bae, J.W.; Park, Y.H.; Oh, H.M. Determination of cyanobacterial diversity during algal blooms in Daechung Reservoir, Korea, on the basis of cpcBA intergenic spacer region analysis. Appl. Environ. Microbiol. 2006, 72, 3252–3258. [Google Scholar] [CrossRef] [PubMed]
- Teneva, I.; Dzhambazov, B.; Mladenov, R.; Schirmer, K. Molecular and phylogenetic characterization of Phormidium species (cyanoprokaryota) using the cpcB-IGS-cpcA locus. J. Phycol. 2005, 41, 188–194. [Google Scholar] [CrossRef]
- Piccin-Santos, V.; Brandão, M.M.; Bittencourt-Oliveira, M.D.C. Phylogenetic study of Geitlerinema and Microcystis (Cyanobacteria) using PC-IGS and 16S-23S ITS as markers: Investigation of horizontal gene transfer. J. Phycol. 2014, 50, 736–743. [Google Scholar] [CrossRef] [PubMed]
- Dadheech, P.K.; Ballot, A.; Casper, P.; Kotut, K.; Novelo, E.; Lemma, B.; Pröschold, T.; Krienitz, L. Phylogenetic relationship and divergence among planktonic strains of Arthrospira (Oscillatoriales, Cyanobacteria) of African, Asian and American origin deduced by 16S–23S ITS and phycocyanin operon sequences. Phycologia 2010, 49, 361–372. [Google Scholar] [CrossRef]
- Robertson, B.R.; Tezuka, N.; Watanabe, M.M. Phylogenetic analyses of Synechococcus strains (cyanobacteria) using sequences of 16S rDNA and part of the phycocyanin operon reveal multiple evolutionary lines and reflect phycobilin content. Int. J. Syst. Evol. Microbiol. 2001, 51, 861–871. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Liu, Y.; Xu, Y.; Xiao, P.; Li, R. The divergence of cpcBA-IGS sequences between Dolichospermum and Aphanizomenon (Cyanobacteria) and the molecular detection of Dolichospermum flos-aquae in Taihu Lake, China. Phycologia 2013, 52, 447–454. [Google Scholar] [CrossRef]
- Salmaso, N.; Vasselon, V.; Rimet, F.; Vautier, M.; Elersek, T.; Boscaini, A.; Donati, C.; Moretto, M.; Pindo, M.; Riccioni, G.; et al. DNA sequence and taxonomic gap analyses to quantify the coverage of aquatic cyanobacteria and eukaryotic microalgae in reference databases: Results of a survey in the Alpine region. Sci. Total Environ. 2022, 834, 155175. [Google Scholar] [CrossRef]
- Mulec, J.; Kosi, G.; Vrhovšek, D. Characterization of cave aerophytic algal communities and effects of irradiance levels on production of pigments. J. Caves Karst Stud. 2008, 70, 3–12. [Google Scholar]
- Mulec, J.; Kosi, G.; Vrhovšek, D. Algae promote growth of stalagmites and stalactites in karst caves (Škocjanske jame, Slovenia). Carbonates Evaporites 2007, 22, 6–9. [Google Scholar] [CrossRef]
- Saiz-Jimenez, C. Microbiological and environmental issues in show caves. World J. Microbiol. Biotechnol. 2012, 28, 2453–2464. [Google Scholar] [CrossRef]
- Urzì, C.; De Leo, F.; Bruno, L.; Albertano, P. Microbial Diversity in Paleolithic Caves: A Study Case on the Phototrophic Biofilms of the Cave of Bats (Zuheros, Spain). Microb. Ecol. 2010, 60, 116–129. [Google Scholar] [CrossRef] [PubMed]
- Popović, S.; Krizmanić, J.; Vidaković, D.; Jakovljević, O.; Trbojević, I.; Predojević, D.; Vidović, M.; Subakov Simić, G. Seasonal Dynamics of Cyanobacteria and Algae in Biofilm from the Entrance of Two Caves. Geomicrobiol. J. 2020, 37, 315–326. [Google Scholar] [CrossRef]
- Guiry, M.D.; Guiry, G.M. AlgaeBase. World-Wide Electronic Publication; National University of Ireland: Galway, Ireland, 2023; Available online: https://www.algaebase.org (accessed on 25 July 2023).
- Jeong, Y.; Hong, S.J.; Cho, S.H.; Yoon, S.; Lee, H.; Choi, H.K.; Kim, D.M.; Lee, C.G.; Cho, S.; Cho, B.K. Multi-Omic Analyses Reveal Habitat Adaptation of Marine Cyanobacterium Synechocystis sp. PCC 7338. Front. Microbiol. 2021, 12, 667450. [Google Scholar] [CrossRef] [PubMed]
- Cantonati, M. Cyanoprokaryotes and algae other than diatoms in springs and streams of the Dolomiti Bellunesi National Park (Northern Italy). Algol. Stud. 2008, 126, 113–136. [Google Scholar] [CrossRef]
- Strauss, H.; Chmiel, H.; Christ, A.; Fugmann, A.; Hanselmann, K.; Kappler, A.; Königer, P.; Lutter, A.; Siedenberg, K.; Teichert, B. Multiple sulphur and oxygen isotopes reveal microbial sulphur cycling in spring waters in the Lower Engadin, Switzerland. Isot. Environ. Health Stud. 2015, 52, 75–93. [Google Scholar] [CrossRef]
- Mulec, J.; Summers Engel, A. Karst spring microbial diversity differs across an oxygen-sulphide ecocline and reveals potential for novel taxa discovery. Acta Carsologica 2019, 48, 129–143. [Google Scholar] [CrossRef]
- Klatt, J.M.; de Beer, D.; Häusler, S.; Polerecky, L. Cyanobacteria in Sulfidic Spring Microbial Mats Can Perform Oxygenic and Anoxygenic Photosynthesis Simultaneously during an Entire Diurnal Period. Front. Microbiol. 2016, 7, 1973. [Google Scholar] [CrossRef]
- Hamilton, T.; Klatt, J.; de Beer, D.; Macalady, J.L. Cyanobacterial photosynthesis under sulfidic conditions: Insights from the isolate Leptolyngbya sp. strain hensonii. ISME J. 2018, 12, 568–584. [Google Scholar] [CrossRef] [PubMed]
- Mihali, T.K.; Kellmann, R.; Muenchhoff, J.; Barrow, K.D.; Neilan, B.A. Characterization of the gene cluster responsible for cylindrospermopsin biosynthesis. Appl. Environ. Microbiol. 2008, 74, 716–722. [Google Scholar] [CrossRef] [PubMed]
- Kokociński, M.; Mankiewicz-Boczek, J.; Jurczak, T.; Spoof, L.; Meriluoto, J.; Rejmonczyk, E.; Hautala, H.; Vehniäinen, M.; Pawełczyk, J.; Soininen, J. Aphanizomenon gracile (Nostocales), a cylindrospermopsin-producing cyanobacterium in Polish lakes. Environ. Sci. Pollut. Res. 2013, 20, 5243–5264. [Google Scholar] [CrossRef] [PubMed]
- Mankiewicz-Boczek, J.; Kokociński, M.; Gagała, I.; Pawełczyk, J.; Jurczak, T.; Dziadek, J. Preliminary molecular identification of cylindrospermopsin-producing cyanobacteria in two Polish lakes (Central Europe). FEMS Microbiol. Lett. 2012, 326, 173–179. [Google Scholar] [CrossRef] [PubMed]
- Linz, D.M.; Sienkiewicz, N.; Struewing, I.; Stelzer, E.A.; Graham, J.L.; Lu, J. Metagenomic mapping of cyanobacteria and potential cyanotoxin producing taxa in large rivers of the United States. Sci. Rep. 2023, 13, 2806. [Google Scholar] [CrossRef]
- Kim, K.H.; Yoon, Y.; Hong, W.Y.; Kim, J.; Cho, Y.C.; Hwang, S.J. Application of metagenome analysis to characterize the molecular diversity and saxitoxin-producing potentials of a cyanobacterial community: A case study in the North Han River, Korea. Appl. Biol. Chem. 2018, 61, 153–161. [Google Scholar] [CrossRef]
- Feng, J.; Zhou, L.; Zhao, X.; Chen, J.; Li, Z.; Liu, Y.; Ou, L.; Xie, Z.; Wang, M.; Yin, X.; et al. Evaluation of environmental factors and microbial community structure in an important drinking-water reservoir across seasons. Front. Microbiol. 2023, 14, 1091818. [Google Scholar] [CrossRef]
- Mulec, J. Lampenflora, Definition of. In Encyclopedia of Caves; White, W.B., Culver, D.C., Pipan, T., Eds.; Academic/Elsevier Press: Cambridge, MA, USA, 2019; pp. 635–641. [Google Scholar]
- Mulec, J. Phototrophs in caves. In Cave Ecology; Moldovan, O.T., Kováč, L., Halse, S., Eds.; Springer: Cham, Switzerland, 2018; pp. 91–106. [Google Scholar]
- Al-Tebrineh, J.; Mihali, T.K.; Pomati, F.; Neilan, B.A. Detection of saxitoxin-producing cyanobacteria and Anabaena circinalis in environmental water blooms by quantitative PCR. Appl. Environ. Microbiol. 2010, 76, 7836–7842. [Google Scholar] [CrossRef]
- Campo, E.; Lezcano, M.-Á.; Agha, R.; Cirés, S.; Quesada, A.; El-Shehawy, R. First TaqMan Assay to identify and quantify the cylindrospermopsin-producing cyanobacterium Aphanizomenon ovalisporum in water. Adv. Microbiol. 2013, 03, 430–437. [Google Scholar] [CrossRef]
- Rantala, A.; Fewer, D.P.; Hisbergues, M.; Rouhiainen, L.; Vaitomaa, J.; Börner, T.; Sivonen, K. Phylogenetic evidence for the early evolution of microcystin synthesis. Proc. Natl. Acad. Sci. USA 2004, 101, 568–573. [Google Scholar] [CrossRef]
- Vaitomaa, J.; Rantala, A.; Halinen, K.; Rouhiainen, L.; Tallberg, P.; Mokelke, L.; Sivonen, K. Quantitative Real-Time PCR for Determination of Microcystin Synthetase E Copy Numbers for Microcystis and Anabaena in Lakes. Appl. Environ. Microbiol. 2003, 69, 7289–7297. [Google Scholar] [CrossRef] [PubMed]
- Rantala, A.; Rajaniemi-Wacklin, P.; Lyra, C.; Lepistö, L.; Rintala, J.; Mankiewicz-Boczek, J.; Sivonen, K. Detection of microcystin-producing cyanobacteria in Finnish lakes with genus-specific microcystin synthetase gene E (mcyE) PCR and associations with environmental factors. Appl. Environ. Microbiol. 2006, 72, 6101–6110. [Google Scholar] [CrossRef] [PubMed]
- Lazar, I., Jr.; Lazar, I., Sr. GelAnalyzer 19.1. Available online: www.gelanalyzer.com (accessed on 1 April 2021).
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef] [PubMed]
- Nucleotide; National Library of Medicine (US), National Center for Biotechnology Information: Bethesda, MD, USA, 2004. Available online: https://www.ncbi.nlm.nih.gov/nucleotide/ (accessed on 25 July 2023).
- John, D.M.; Whitton, B.A.; Brook, A.J. The Freshwater Algal Flora of the British Isles: An Identification Guide to Freshwater and Terrestrial Algae; Cambridge University Press: Cambridge, UK, 2011. [Google Scholar]
- Komárek, J. Cyanoprokaryota. 3. Heterocytous genera. In Süswasserflora von Mitteleuropa/Freshwater Flora of Central Europe; Büdel, B., Gärtner, G., Krienitz, L., Schagerl, M., Eds.; Springer Spektrum: Berlin, Germany, 2013; pp. 1–1130. [Google Scholar]
- Komárek, J.; Anagnostidis, K. Modern approach to the classification system of Cyanophytes 4—Nostocales. Arch. Hydrobiol. Suppl. 1989, 82, 247–345. [Google Scholar]

| Sample | Country | Location | Coordinates | Sampling Date [dd/mm/yyyy] | Sample Type | Description | T [°C] | pH | qPCR (16S rRNA) | PCR PC-IGS | Seq. PC-IGS (Genus) | qPCR cyrJ | Seq. cyrJ | qPCR stxA | qPCR mcyE (Dol.) | qPCR mcyE (Mic.) | qPCR mcyE (Pla.) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| B-01 | Serbia | Bela voda, Zlatibor | 43°42′13″ N, 19°34′43″ E | 29/09/2011 | biofilm | alkaline spring | 15.6 | 12.40 | + | + | (+) | − | NS | − | − | − | − |
| B-02 | Slovenia | Hajnsko | 45°12′22″ N, 15°35′26″ E | 13/04/2016 | biofilm | sulphidic spring | 13.6 | 7.13 | + | − | NS | − | NS | − | − | − | − |
| B-03 | Slovenia | Matijeva jama | 45°41′21″ N, 14°15′53″ E | 31/08/2016 | biofilm | biofilm in a cave | NA | NA | + | − | NS | − | NS | − | − | − | − |
| B-04 | Slovenia | Smrdljivec | 45°39′48″ N, 13°59′57″ E | 24/08/2017 | biofilm | sulphidic spring | 19.7 | 7.28 | + | − | NS | − | NS | − | − | − | − |
| B-05 | Slovenia | Smrdljivec | 45°39′48″ N, 13°59′57″ E | 24/08/2017 | biofilm | sulphidic spring | 19.7 | 7.28 | + | − | NS | − | NS | − | − | − | − |
| B-06 | Slovenia | Smrdljivec | 45°39′48″ N, 13°59′57″ E | 24/08/2017 | biofilm | sulphidic spring | 19.7 | 7.28 | + | − | NS | − | NS | − | − | − | − |
| B-07 | Slovenia | Riharjev studenec | 46°18′50″ N, 14°42′07″ E | 22/03/2016 | biofilm | sulphidic spring | 10.2 | 7.76 | + | − | NS | (+) | + Thio-thrix | − | − | − | − |
| B-08 * | Slovenia | Žveplenica | 46°03′59″ N, 13°49′38″ E | 10/04/2016 | biofilm | sulphidic spring | 10.5 | 7.54 | + | − | NS | + | (+) | − | − | − | − |
| B-09 | Slovenia | Žvepovnik | 46°21′00″ N, 14°50′43″ E | 22/03/2016 | biofilm | sulphidic spring | 10.4 | 7.32 | + | − | NS | − | NS | − | − | − | − |
| M-01 | Slovenia | Ljubljana | 46°02′59″ N, 14°29′58″ E | 08/06/2015 | microbial mat | facade, city centre | NA | NA | + | − | NS | − | NS | − | − | − | − |
| M-02 | Slovenia | Ljubljana | 46°04′57″ N, 14°30′47″ E | 08/06/2015 | microbial mat | facade, city outskirt | NA | NA | + | − | NS | − | NS | − | − | − | − |
| M-03 | Slovenia | Ljubljana | 46°04′57″ N, 14°30′47″ E | 08/06/2015 | microbial mat | facade, city outskirt | NA | NA | + | − | NS | − | NS | − | − | − | − |
| M-04 | Slovenia | Ljubljana | 46°04′14″ N, 14°30′51″ E | 05/06/2015 | microbial mat | roof, city outskirt | NA | NA | + | − | NS | − | NS | − | − | − | − |
| M-05 | Slovenia | Ljubljana | 46°04′14″ N, 14°30′51″ E | 05/06/2015 | microbial mat | roof, city outskirt | NA | NA | + | − | NS | − | NS | − | − | − | − |
| M-06 | Slovenia | Matijeva jama | 45°41′21″ N, 14°15′53″ E | 08/07/2019 | microbial mat | microbial mat at a cave entrance | NA | NA | + | + | − | − | NS | − | − | − | − |
| M-07 | Slovenia | Postojnska jama | 45°46′59″ N, 14°12′14″ E | 24/01/2020 | microbial mat | lampenflora in a cave | NA | NA | + | − | NS | − | NS | − | − | − | − |
| M-08 | Slovenia | Postojnska jama | 45°46′59″ N, 14°12′14″ E | 24/01/2020 | microbial mat | lampenflora in a cave | NA | NA | + | − | NS | − | NS | − | − | − | − |
| M-09 | Slovenia | Postojnska jama | 45°46′59″ N, 14°12′14″ E | 24/01/2020 | microbial mat | lampenflora in a cave | NA | NA | + | − | NS | − | NS | − | − | − | − |
| M-10 | Slovenia | Postojnska jama | 45°46′59″ N, 14°12′14″ E | 24/01/2020 | microbial mat | lampenflora in a cave | NA | NA | + | + | (+) | − | NS | − | − | − | − |
| M-11 | Slovenia | Postojnska jama | 45°46′59″ N, 14°12′14″ E | 24/01/2020 | microbial mat | lampenflora in a cave | NA | NA | + | + | − | − | NS | − | − | − | − |
| M-12 * | Slovenia | Škocjanske jame | 45°39′57″ N, 13°59′24″ E | 08/04/2016 | microbial mat | microbial mat at a cave entrance | NA | NA | + | − | NS | − | NS | − | − | − | − |
| M-13 | Slovenia | Škocjanske jame | 45°39′57″ N, 13°59′24″ E | 23/01/2020 | microbial mat | microbial mat at a cave entrance | NA | NA | + | + | − | − | NS | − | − | − | − |
| M-14 | Slovenia | Škocjanske jame | 45°39′57″ N, 13°59′24″ E | 23/01/2020 | microbial mat | microbial mat at a cave entrance | NA | NA | + | + | (+) | (+) | − | − | − | − | − |
| M-15 | Slovenia | Škocjanske jame | 45°39′57″ N, 13°59′24″ E | 23/01/2020 | microbial mat | microbial mat at a cave entrance | NA | NA | + | + | (+) | (+) | − | − | − | − | − |
| M-16 | Slovenia | Škocjanske jame | 45°39′57″ N, 13°59′24″ E | 23/01/2020 | microbial mat | microbial mat at a cave entrance | NA | NA | + | + | − | − | NS | − | − | − | − |
| M-17 | Slovenia | Škocjanske jame | 45°39′57″ N, 13°59′24″ E | 23/01/2020 | microbial mat | microbial mat at a cave entrance | NA | NA | + | + | + Cyano-thece | − | NS | − | − | − | − |
| M-18 * | Slovenia | Škocjanske jame | 45°39′57″ N, 13°59′24″ E | 08/04/2016 | microbial mat | microbial mat at a cave entrance | NA | NA | + | + | − | − | NS | − | − | − | − |
| M-19 * | Slovenia | Škocjanske jame | 45°39′57″ N, 13°59′24″ E | 08/04/2016 | microbial mat | stromatolitic stalagmite at a cave entrance | NA | NA | + | + | + Calothrix | − | NS | − | − | − | − |
| W-01 | Croatia | Slanci, Slanje | 46°13′48″ N, 16°32′56″ E | 24/03/2019 | water | saline spring # | 17.6 | 7.97 | + | + | + Synecho-cystis | − | NS | − | − | − | − |
| W-02 | Georgia | Analisopeli | 41°49′15″ N, 41°47′54″ E | 13/07/2012 | water | discharge from a borehole well | 36.9 | 7.50 | + | − | NS | − | NS | − | − | − | − |
| W-03 | Georgia | Sighnaghi | 41°36′23″ N, 45°56′02″ E | 30/06/2012 | water | spring | 12.3 | 7.50 | + | − | NS | − | NS | − | − | − | − |
| W-04 | Georgia | Tbilisi, Lisi | 41°43′13″ N, 44°45′30″ E | 02/07/2012 | water | discharge from a borehole well | 63.4 | 7.50 | + | − | NS | − | NS | − | − | − | − |
| W-05 | Georgia | Tbilisi, Ortachala | 41°40′25″ N, 44°50′30″ E | 04/07/2012 | water | discharge from a borehole well | 21.5 | 8.00 | + | − | NS | − | NS | − | − | − | − |
| W-06 | Georgia | Vardzia Cave Monastery | 41°23′30″ N, 43°18′33″ E | 08/07/2012 | water | discharge from a borehole well | 51.0 | 9.30 | (+) | − | NS | − | NS | − | − | − | − |
| W-07 | Georgia | Vardzia Cave Monastery | 41°22′52″ N, 43°17′03″ E | 08/07/2012 | water | seeping water in a cave | 10.5 | 7.00 | (+) | − | NS | − | NS | − | − | − | − |
| W-08 | Italy | Soča/Isonzo River | 45°56′17″ N, 13°36′05″ E | 20/06/2019 | water | river | 16.4 | 8.30 | + | − | NS | − | NS | − | − | − | − |
| W-09 | Italy | Vipava/Vipacco River | 45°53′17″ N, 13°35′27″ E | 20/06/2019 | water | river | 20.9 | 8.30 | + | − | NS | − | NS | − | − | − | − |
| W-10 | Italy | Tržič/Monfalcone | 45°47′32″ N, 13°33′57″ E | 20/06/2019 | water | thermal spring | 37.4 | 7.10 | + | − | NS | − | NS | − | − | − | − |
| W-11 | Italy | Adriatic sea | 45°46′51″ N, 13°32′23″ E | 20/06/2019 | water | sea | 25.8 | 8.20 | + | − | NS | − | NS | − | − | − | − |
| W-12 | Italy | Tržič/Monfalcone | 45°47′31″ N, 13°33′56″ E | 20/06/2019 | water | discharge from a borehole well | 39.2 | 7.00 | (+) | − | NS | − | NS | − | − | − | − |
| W-13 | Serbia | Crveno vrelo, Đavolja Varoš | 42°59′26″ N, 21°23′48″ E | 28/09/2012 | water | iron spring | 16.9 | 5.50 | + | − | NS | − | NS | − | − | − | − |
| W-14 | Serbia | Đavolje vrelo, Đavolja Varoš | 42°59′19″ N, 21°23′35″ E | 28/09/2012 | water | iron spring | 25.5 | 2.50 | + | − | NS | − | NS | − | − | − | − |
| W-15 | Serbia | Mokra Gora, Zlatibor | 43°47′28″ N, 19°32′13″ E | 28/09/2010 | water | alkaline spring | 15.4 | 12.03 | (+) | − | NS | − | NS | − | − | − | − |
| W-16 | Serbia | Vranjska Banja | 42°33′00″ N, 22°00′23″ E | 29/09/2012 | water | thermal spring | 83.0 | 8.50 | (+) | − | NS | − | NS | − | − | − | − |
| W-17 | Slovenia | Klariči | 45°48′49″ N, 13°35′54″ E | 20/06/2019 | water | discharge from a borehole well | 14.3 | 7.50 | + | − | NS | − | NS | − | − | − | − |
| W-18 | Slovenia | Reka River | 45°39′21″ N, 14°03′21″ E | 20/06/2019 | water | river | 20.2 | 8.50 | + | − | NS | − | NS | − | − | − | (+) |
| W-19 | Slovenia | Paradana | 45°59′19″ N, 13°50′41″ E | 09/05/2016 04/06/2016 ** | water | ice from cave | NA | 8.21 ** | + | NA | NS | − | NS | − | − | − | − |
| W-20 | Slovenia | Paradana | 45°59′19″ N, 13°50′41″ E | 09/05/2016 04/06/2016 ** | water | ice from cave | NA | 8.62 ** | + | − | NS | − | NS | − | − | − | − |
| W-21 | Slovenia | Paradana | 45°59′19″ N, 13°50′41″ E | 09/05/2016 04/06/2016 ** | water | ice from cave | NA | 8.46 ** | + | − | NS | − | NS | − | − | − | − |
| W-22 | Slovenia | Pivka River, Postojna | 45°46′55″ N, 14°12′14″ E | 02/07/2019 | water | river before ponor in a cave | 22.6 | 7.59 | + | − | NS | − | NS | − | − | − | − |
| W-23 | Slovenia | Planinska jama | 45°49′15″ N, 14°14′48″ E | 17/07/2017 15/11/2023 ** | water | seeping water in a cave | 11.2 ** | 8.12 ** | + | − | NS | − | NS | − | − | − | − |
| W-24 | Slovenia | Planinska jama | 45°49′15″ N, 14°14′48″ E | 11/10/2017 15/11/2023 ** | water | seeping water in a cave | 11.2 ** | 8.12 ** | + | − | NS | − | NS | − | − | − | − |
| W-25 | Slovenia | Planinska jama | 45°49′15″ N, 14°14′48″ E | 11/10/2017 15/11/2023 ** | water | seeping water in a cave | 10.9 ** | 7.95 ** | + | − | NS | − | NS | − | − | − | − |
| W-26 | Slovenia | Planinska jama | 45°49′15″ N, 14°14′48″ E | 11/10/2017 15/11/2023 ** | water | seeping water in a cave | 10.9 ** | 7.88 ** | + | − | NS | − | NS | − | − | − | − |
| W-27 | Slovenia | Planinska jama | 45°49′15″ N, 14°14′48″ E | 14/11/2017 15/11/2023 ** | water | seeping water in a cave | 11.2 ** | 8.12 ** | + | − | NS | − | NS | − | − | − | − |
| W-28 | Slovenia | Planinska jama | 45°49′15″ N, 14°14′48″ E | 14/11/2017 15/11/2023 ** | water | seeping water in a cave | 10.9 ** | 7.95 ** | + | − | NS | − | NS | − | − | − | − |
| W-29 | Slovenia | Planinska jama | 45°49′15″ N, 14°14′48″ E | 14/11/2017 15/11/2023 ** | water | seeping water in a cave | 10.9 ** | 7.88 ** | + | − | NS | − | NS | − | − | − | − |
| W-30 | Slovenia | Postojnska jama | 45°46′59″ N, 14°12′14″ E | 11/10/2017 | water | seeping water in a cave | 10.6 | 8.04 | + | − | NS | − | NS | − | − | − | − |
| W-31 | Slovenia | Postojnska jama | 45°46′59″ N, 14°12′14″ E | 14/11/2017 | water | seeping water in a cave | 10.6 | 8.04 | + | − | NS | − | NS | − | − | − | − |
| W-32 | Slovenia | Smrdljivec | 45°39′48″ N, 13°59′57″ E | 18/07/2017 | water | sulphidic spring | 21.5 | 7.28 | + | − | NS | − | NS | − | − | − | − |
| W-33 | Slovenia | Škocjanske jame | 45°39′57″ N, 13°59′24″ E | 13/12/2017 | water | seeping water in a cave | 18.7 | 8.16 | + | − | NS | − | NS | − | − | − | − |
| Type of Analysis | Target Organisms | Target Gene | Primer Label | Nucleotide Sequence (5′ → 3′) | Amplicon Length [bp] | Reference |
|---|---|---|---|---|---|---|
| qPCR | Cyanobacteria | 16S rRNA | cyano-real16S-F | AGC CAC ACT GGG ACT GAG ACA | 73 | [67] |
| cyano-real16S-R | TCG CCC ATT GCG GAA A | |||||
| PCR, sequencing | Cyanobacteria | PC-IGS | PCβF | GGC TGC TTG TTT ACG CGA CA | 500–740 | [35] |
| PCαR | CCA GTA CCA CCA GCA ACT AA | |||||
| qPCR | Cylindrospermopsin-producers | cyrJ | cyrJ207-F | CCC CTA CAA CCT GAC AAA GCT T | 77 | [68] |
| cyrJ207-R | CCC GCC TGT CAT AGA TGC A | |||||
| PCR, sequencing | Cylindrospermopsin-producers | cyrJ | cynsulF | ACT TCT CTC CTT TCC CTA TC | 586 | [59] |
| cylnamR | GAG TGA AAA TGC GTA GAA CTT G | |||||
| qPCR | Saxitoxin-producers | sxtA | sxtA-F | GAT GAC GGA GTA TTT GAA GC | 125 | [67] |
| sxtA-R | CTG CAT CTT CTG GAC GGT AA | |||||
| qPCR | Microcystin- producers (genus Dolichospermum) | mcyE | mcyE-F2 | GAA ATT TGT GTA GAA GGT GC | 247 | [69] |
| AnamcyE-12R | CAA TCT CGG TAT AGC GGC | [70] | ||||
| qPCR | Microcystin- producers (genus Microcystis) | mcyE | mcyE-F2 | GAA ATT TGT GTA GAA GGT GC | 247 | [69] |
| MicmcyE-R8 | CAA TGG GAG CAT AAC GAG | [70] | ||||
| qPCR | Microcystin- producers (genus Planktothrix) | mcyE | mcyE-F2 | GAA ATT TGT GTA GAA GGT GC | 249 | [69] |
| PlamcyE-R3 | CTC AAT CTG AGG ATA ACG AT | [71] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jablonska, M.; Eleršek, T.; Kogovšek, P.; Skok, S.; Oarga-Mulec, A.; Mulec, J. Molecular Screening for Cyanobacteria and Their Cyanotoxin Potential in Diverse Habitats. Toxins 2024, 16, 333. https://doi.org/10.3390/toxins16080333
Jablonska M, Eleršek T, Kogovšek P, Skok S, Oarga-Mulec A, Mulec J. Molecular Screening for Cyanobacteria and Their Cyanotoxin Potential in Diverse Habitats. Toxins. 2024; 16(8):333. https://doi.org/10.3390/toxins16080333
Chicago/Turabian StyleJablonska, Maša, Tina Eleršek, Polona Kogovšek, Sara Skok, Andreea Oarga-Mulec, and Janez Mulec. 2024. "Molecular Screening for Cyanobacteria and Their Cyanotoxin Potential in Diverse Habitats" Toxins 16, no. 8: 333. https://doi.org/10.3390/toxins16080333
APA StyleJablonska, M., Eleršek, T., Kogovšek, P., Skok, S., Oarga-Mulec, A., & Mulec, J. (2024). Molecular Screening for Cyanobacteria and Their Cyanotoxin Potential in Diverse Habitats. Toxins, 16(8), 333. https://doi.org/10.3390/toxins16080333







