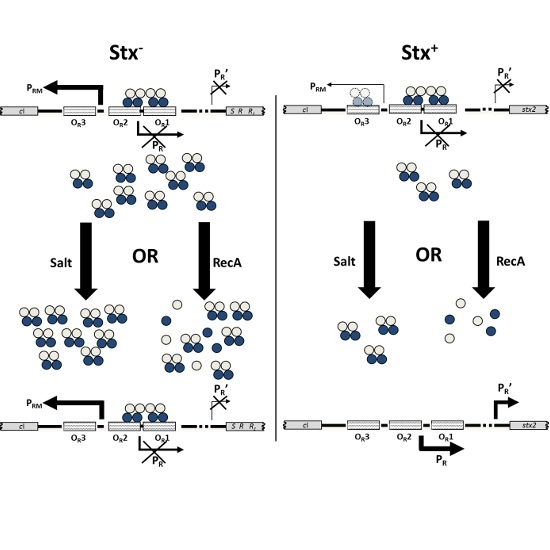
Mechanisms that Determine the Differential Stability of Stx+ and Stx− Lysogens
Abstract
1. Introduction
2. Results
2.1. Spontaneous Induction of Stx+ and Stx− Prophages
2.2. Maturation and Production of Stx+ and Stx− Phages
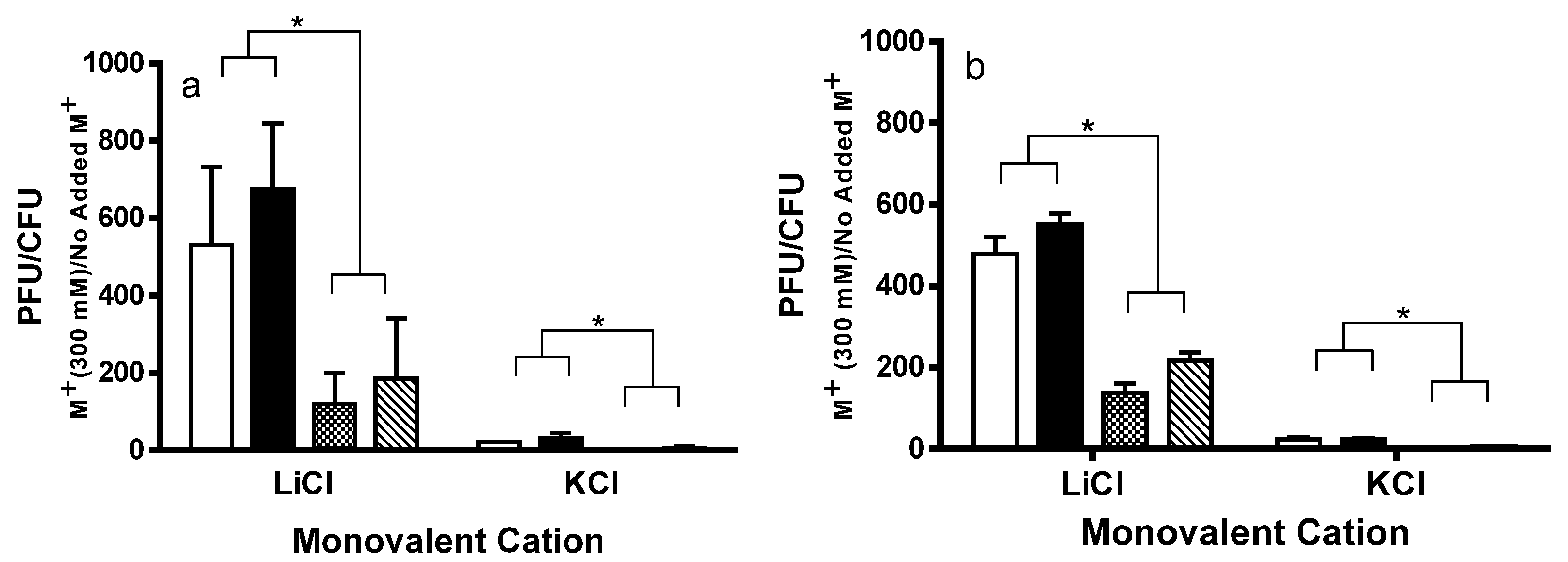
2.3. Effect of DNA Damaging Agents on Stx+ and Stx− Lysogens
2.4. RecA-Independent Induction of Stx+ and Stx− Prophages
2.5. Intracellular cI mRNA Transcript Levels
3. Discussion
4. Materials and Methods
4.1. Strains and Growth Conditions
4.2. Measurement of Spontaneous and Salt Induction
4.3. Measurement of DNA Damaging Agents on Prophage Induction
4.4. Determination of Adsorption Rate
4.5. Determination of Eclipse Period, Latent Period and Burst Size
4.6. Determination of Phage Fitness
4.7. qRT-PCR
| BAA2326 | Sense | CGTTACACTCAATCATGTCGAATC |
| Antisense | CCCTGAAGGTATGCTGATACTC | |
| 933W | Sense | GGGCTAAATTCTTCAACGCTAAC |
| Antisense | ATCTGTCGCAGACAAGATGG | |
| 434 | Sense | AGCAGCCAGTCAACACTTAC |
| Antisense | GGGACTACCCAGCAGTCTAT | |
| λ | Sense | TCATTGCATGGGATCATTGGG |
| Antisense | CCAGGTGATTTCTGCATAGCC | |
| UID | Sense | GTGTGATATCTACCCGCTTCGC |
| Antisense | AGAACGGTTTGTGGTTAATCAGGA |
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Clokie, M.R.J.; Kropinski, A.M. Bacteriophages: Methods and Protocols; Humana Press: New York, NY, USA, 2009. [Google Scholar]
- Jonczyk, E.; Klak, M.; Miedzybrodzki, R.; Gorski, A. The influence of external factors on bacteriophages-review. Folia Microbiol. 2011, 56, 191–200. [Google Scholar] [CrossRef] [PubMed]
- Lainhart, W.; Stolfa, G.; Koudelka, G.B. Shiga toxin as a bacterial defense against a eukaryotic predator, tetrahymena thermophila. J. Bacteriol. 2009, 191, 5116–5122. [Google Scholar] [CrossRef] [PubMed]
- Clokie, M.R.; Millard, A.D.; Letarov, A.V.; Heaphy, S. Phages in nature. Bacteriophage 2011, 1, 31–45. [Google Scholar] [CrossRef] [PubMed]
- Brussow, H.; Canchaya, C.; Hardt, W.-D. Phages and the evolution of bacterial pathogens: From genomic rearrangements to lysogenic conversion. Microbiol. Mol. Biol. Rev. 2004, 68, 560–602. [Google Scholar] [CrossRef] [PubMed]
- Friedman, D.I.; Court, D.L. Bacteriophage lambda: Alive and well and still doing its thing. Curr. Opin. Microbiol. 2001, 4, 201–207. [Google Scholar] [CrossRef]
- Waldor, M.K.; Friedman, D.I. Phage regulatory circuits and virulence gene expression. Curr. Opin. Microbiol. 2005, 8, 459–465. [Google Scholar] [CrossRef] [PubMed]
- Kimmitt, P.T.; Harwood, C.R.; Barer, M.R. Toxin gene expression by Shiga toxin-producing Escherichia coli: The role of antibiotics and the bacterial SOS response. Emerg. Infect. Dis. 2000, 6, 458–465. [Google Scholar] [CrossRef] [PubMed]
- Newland, J.W.; Strockbine, N.A.; Miller, S.F.; O'Brien, A.D.; Holmes, R.K. Cloning of Shiga-like toxin structural genes from a toxin converting phage of Escherichia coli. Science 1985, 230, 179–181. [Google Scholar] [CrossRef] [PubMed]
- O'Brien, A.D.; Newland, J.W.; Miller, S.F.; Holmes, R.K.; Smith, H.W.; Formal, S.B. Shiga-like toxin-converting phages from Escherichia coli strains that cause hemorrhagic colitis or infantile diarrhea. Science 1984, 226, 694–696. [Google Scholar] [CrossRef] [PubMed]
- Neely, M.N.; Friedman, D.I. Functional and genetic analysis of regulatory regions of coliphage H-19b: Location of Shiga-like toxin and lysis genes suggest a role for phage functions in toxin release. Mol. Microbiol. 1998, 28, 1255–1267. [Google Scholar] [CrossRef] [PubMed]
- Neely, M.N.; Friedman, D.I. Arrangement and functional identification of genes in the regulatory region of lambdoid phage H-19b, a carrier of a Shiga-like toxin. Gene 1998, 223, 105–113. [Google Scholar] [CrossRef]
- Wagner, P.L.; Neely, M.N.; Zhang, X.; Acheson, D.W.; Waldor, M.K.; Friedman, D.I. Role for a phage promoter in Shiga toxin 2 expression from a pathogenic Escherichia coli strain. J. Bacteriol. 2001, 183, 2081–2085. [Google Scholar] [CrossRef] [PubMed]
- Wagner, P.L.; Livny, J.; Neely, M.N.; Acheson, D.W.; Friedman, D.I.; Waldor, M.K. Bacteriophage control of Shiga toxin 1 production and release by Escherichia coli. Mol. Microbiol. 2002, 44, 957–970. [Google Scholar] [CrossRef] [PubMed]
- Shimizu, T.; Ohta, Y.; Noda, M. Shiga toxin 2 is specifically released from bacterial cells by two different mechanisms. Infect. Immun. 2009, 77, 2813–2823. [Google Scholar] [CrossRef] [PubMed]
- Chekabab, S.M.; Daigle, F.; Charette, S.J.; Dozois, C.M.; Harel, J. Shiga toxins decrease enterohaemorrhagic Escherichia coli survival within acanthamoeba castellanii. FEMS Microbiol. Lett. 2013, 344, 86–93. [Google Scholar] [CrossRef] [PubMed]
- Stolfa, G.; Koudelka, G.B. Entry and killing of tetrahymena thermophila by bacterially produced Shiga toxin. mBio 2012, 4, e00416-12. [Google Scholar] [CrossRef] [PubMed]
- Arnold, J.W.; Koudelka, G.B. The trojan horse of the microbiological arms race: Phage-encoded toxins as a defence against eukaryotic predators. Environ. Microbiol. 2014, 16, 454–466. [Google Scholar] [CrossRef] [PubMed]
- Crane, J.K.; Byrd, I.W.; Boedeker, E.C. Virulence inhibition by zinc in Shiga-toxigenic Escherichia coli. Infect. Immun. 2011, 79, 1696–1705. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Crane, J.K.; Broome, J.E.; Reddinger, R.M.; Werth, B.B. Zinc protects against Shiga-toxigenic Escherichia coli by acting on host tissues as well as on bacteria. BMC Microbiol. 2014, 14. [Google Scholar] [CrossRef] [PubMed]
- Anderson, J.E. Ph.D. Thesis, Harvard University, Cambridge, MA, USA, 1984.
- Daniels, D.L.; Schroeder, J.L.; Szybalski, W.; Sanger, F.; Coulson, A.R.; Hong, G.F.; Hill, D.F.; Petersen, G.B.; Blattner, F.R. APPENDIX II Complete Annotated Lambda Sequence. In Lambda II; Cold Spring Harbor Laboratory: Cold Spring Harbor, NY, USA, 1983; pp. 519–676. [Google Scholar]
- Rozanov, D.V.; D'Ari, R.; Sineoky, S.P. Reca-independent pathways of lambdoid prophage induction in Escherichia coli. J. Bacteriol. 1998, 180, 6306–6315. [Google Scholar] [PubMed]
- Imamovic, L.; Muniesa, M. Characterizing RecA-independent induction of Shiga toxin2-encoding phages by edta treatment. PLoS ONE 2012, 7, e32393. [Google Scholar] [CrossRef] [PubMed]
- Shkilnyj, P.; Koudelka, G.B. Effect of salt shock on stability of λimm434 lysogens. J. Bacteriol. 2007, 189, 3115–3123. [Google Scholar] [CrossRef] [PubMed]
- Livny, J.; Friedman, D.I. Characterizing spontaneous induction of Stx encoding phages using a selectable reporter system. Mol. Microbiol. 2004, 51, 1691–1704. [Google Scholar] [CrossRef] [PubMed]
- Bullwinkle, T.J.; Koudelka, G.B. The lysis-lysogeny decision of bacteriophage 933W: A 933W repressor-mediated long-distance loop has no role in regulating 933W PRM activity. J. Bacteriol. 2011, 193, 3313–3323. [Google Scholar] [CrossRef] [PubMed]
- Winans, S.C.; Elledge, S.J.; Krueger, J.H.; Walker, G.C. Site-directed insertion and deletion mutagenesis with cloned fragments in Escherichia coli. J. Bacteriol. 1985, 161, 1219–1221. [Google Scholar] [PubMed]
- Shkilnyj, P.; Colon, M.P.; Koudelka, G.B. Bacteriophage 434 Hex protein prevents RecA-mediated repressor autocleavage. Viruses-Basel 2013, 5, 111–126. [Google Scholar] [CrossRef] [PubMed]
- Shao, Y.P.; Wang, I.N. Bacteriophage adsorption rate and optimal lysis time. Genetics 2008, 180, 471–482. [Google Scholar] [CrossRef] [PubMed]
- Wang, I.N. Lysis timing and bacteriophage fitness. Genetics 2006, 172, 17–26. [Google Scholar] [CrossRef] [PubMed]
- Chakraborty, D.; Koudelka, G.B.; University at Buffalo, NY, USA. unpublished work. 2016.
- Xu, J.; Koudelka, G.B. Mutually exclusive utilization of PR and PRM promoters in bacteriophage 434 OR. J. Bacteriol. 2000, 182, 3165–3174. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Guarente, L.; Nye, J.S.; Hochschild, A.; Ptashne, M. Mutant lambda phage repressor with a specific defect in its positive control function. Proc. Natl. Acad. Sci. USA 1982, 79, 2236–2239. [Google Scholar] [CrossRef] [PubMed]
- Muniesa, M.; Serra-Moreno, R.; Jofre, J. Free Shiga toxin bacteriophages isolated from sewage showed diversity although the stx genes appeared conserved. Environ. Microbiol. 2004, 6, 716–725. [Google Scholar] [CrossRef] [PubMed]
- Muniesa, M.; Blanco, J.E.; de Simón, M.; Serra-Moreno, R.; Blanch, A.R.; Jofre, J. Diversity of stx2 converting bacteriophages induced from shiga-toxin-producing Escherichia coli strains isolated from cattle. Microbiology 2004, 150, 2959–2971. [Google Scholar] [CrossRef] [PubMed]
- Beckett, D.; Koblan, K.S.; Ackers, G.K. Quantitative study of protein association at picomolar concentrations: The lambda phage cl repressor. Anal. Biochem. 1991, 196, 69–75. [Google Scholar] [CrossRef]
- Koblan, K.S.; Ackers, G.K. Cooperative protein-DNA interactions: Effects of KCl on lambda cI binding to or. Biochemistry 1991, 30, 7882–7827. [Google Scholar] [CrossRef]
- Koblan, K.S.; Ackers, G.K. Energetics of subunit dimerization in bacteriophage lambda cI repressor: Linkage to protons, temperature, and KCl. Biochemistry 1991, 30, 7817–7821. [Google Scholar] [CrossRef] [PubMed]
- Wharton, R.P.; Brown, E.L.; Ptashne, M. Substituting an à-helix switches the sequence specific DNA interactions of a repressor. Cell 1985, 38, 361–369. [Google Scholar] [CrossRef]
- Bullwinkle, T.J.; Samorodnitsky, D.; Rosati, R.C.; Koudelka, G.B. Determinants of bacteriophage 933W repressor DNA binding specificity. PLoS One 2012, 7, e34563. [Google Scholar] [CrossRef] [PubMed]
- Bushman, F.D.; Ptashne, M. Activation of transcription by the bacteriophage 434 repressor. Proc. Natl. Acad. Sci. USA 1986, 83, 9353–9357. [Google Scholar] [CrossRef] [PubMed]
- Meyer, B.J.; Kleid, D.G.; Ptashne, M. Lambda repressor turns off transcription of its own gene. Proc. Natl. Acad. Sci. USA 1975, 72, 4785–4789. [Google Scholar] [CrossRef] [PubMed]
- Bushman, F.D. The bacteriophage 434 right operator roles of OR 1, OR 2 and OR 3. J. Mol. Biol. 1993, 230, 28–40. [Google Scholar] [CrossRef] [PubMed]
- Johnson, A.D.; Meyer, B.J.; Ptashne, M. Interaction between DNA-bound repressors govern regulation by the lambda repressor. Proc. Natl. Acad. Sci. USA 1979, 76, 5061–5065. [Google Scholar] [CrossRef] [PubMed]
- Koudelka, A.P.; Hufnagel, L.A.; Koudelka, G.B. Purification and characterization of the repressor of the Shiga toxin-encoding bacteriophage 933W: DNA binding, gene regulation, and autocleavage. J. Bacteriol. 2004, 186, 7659–7669. [Google Scholar] [CrossRef] [PubMed]
- Wallace, I.M.; O'Sullivan, O.; Higgins, D.G.; Notredame, C. M-coffee: Combining multiple sequence alignment methods with T-coffee. Nucl. Acids Res. 2006, 34, 1692–1699. [Google Scholar] [CrossRef] [PubMed]
- ExPASy-SIB Bioinformatics Resource Portal. Available online: http://www.ch.embnet.org/software/BOX_form.html (accessed on 23 March 2016).
- Little, J.W.; Shepley, D.P.; Wert, D.W. Robustness of a gene regulatory circuit. EMBO J. 1999, 18, 4299–4307. [Google Scholar] [CrossRef] [PubMed]
- Schlesinger, M. Adsorption of bacteriophages to homologous bacteria. II. Quantitative investigation of adsorption velocity and saturation. Estimation of the particle size of the bacteriophage. Immunitaetsforschung 1932, 114, 149–160. [Google Scholar]
- Bull, J.J. Optimality models of phage life history and parallels in disease evolution. J. Theor. Biol. 2006, 241, 928–938. [Google Scholar] [CrossRef] [PubMed]
- Arber, W.; Enquist, L.; Hohn, B.; Murray, N.E.; Murray, K. Experimental Methods for Use with Lambda. In Lambda II; Cold Spring Harbor Laboratory: Cold Spring Harbor, NY, USA, 1983; pp. 433–466. [Google Scholar]
- Little, J.W. LexA cleavage and other self-processing reactions. J. Bacteriol. 1993, 175, 4943–4950. [Google Scholar] [PubMed]
- Little, J.W.; Mount, D.W.; Yanischperron, C.R. Purified lexA protein is a repressor of the recA and lexA genes. Proc. Natl. Acad. Sci. USA -Biol. Sci. 1981, 78, 4199–4203. [Google Scholar] [CrossRef]
- Tyler, J.S.; Beeri, K.; Reynolds, J.L.; Alteri, C.J.; Skinner, K.G.; Friedman, J.H.; Eaton, K.A.; Friedman, D.I. Prophage induction is enhanced and required for renal disease and lethality in an EHEC mouse model. PLoS Pathogens 2013, 9, e1003236. [Google Scholar] [CrossRef] [PubMed]
- Hendrix, R.W.; Duda, R.L. Bacteriophage lambda papa: Not the mother of all lambda phages. Science 1992, 258, 1145–1148. [Google Scholar] [CrossRef] [PubMed]
- Ellis, E.L.; Delbrück, M. The growth of bacteriophage. J. Gen. Physiol. 1939, 22, 365–384. [Google Scholar] [CrossRef] [PubMed]
- Delbrück, M. The growth of bacteriophage and lysis of the host. J. Gen. Physiol. 1940, 23, 643–660. [Google Scholar] [CrossRef] [PubMed]
- Chang, C.Y.; Nam, K.; Young, R. S gene expression and the timing of lysis by bacteriophage lambda. J. Bacteriol. 1995, 177, 3283–3294. [Google Scholar] [PubMed]





| Phage | Adsorption Rate (mL/min) | Eclipse Period (min) | Latent Period (min) | Burst Size | Fitness (h−1) |
|---|---|---|---|---|---|
| 933W | 1.64 (0.12) × 10−9 | 0–20 | 55 | 159 (12.4) | 2.46 (0.15) |
| BAA2326 | 1.71 (0.59) × 10−9 | 0–20 | 55 | 120 (26.5) | 2.21 (0.09) |
| λ | 1.78 (0.73) × 10−9 | 0–20 | 55 | 155 (31.1) | 2.34 (0.08) |
| 434 | 1.84 (0.12) × 10−9 | 0–20 | 55 | 126 (12.8) | 2.66 (0.17) |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons by Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Colon, M.P.; Chakraborty, D.; Pevzner, Y.; Koudelka, G.B. Mechanisms that Determine the Differential Stability of Stx+ and Stx− Lysogens. Toxins 2016, 8, 96. https://doi.org/10.3390/toxins8040096
Colon MP, Chakraborty D, Pevzner Y, Koudelka GB. Mechanisms that Determine the Differential Stability of Stx+ and Stx− Lysogens. Toxins. 2016; 8(4):96. https://doi.org/10.3390/toxins8040096
Chicago/Turabian StyleColon, Michael P., Dolonchapa Chakraborty, Yonatan Pevzner, and Gerald B. Koudelka. 2016. "Mechanisms that Determine the Differential Stability of Stx+ and Stx− Lysogens" Toxins 8, no. 4: 96. https://doi.org/10.3390/toxins8040096
APA StyleColon, M. P., Chakraborty, D., Pevzner, Y., & Koudelka, G. B. (2016). Mechanisms that Determine the Differential Stability of Stx+ and Stx− Lysogens. Toxins, 8(4), 96. https://doi.org/10.3390/toxins8040096