A Modular, Reconfigurable Microfabricated Assembly Platform for Microfluidic Transport and Multitype Cell Culture and Drug Testing
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials and Reagents
2.2. Fabrication of Devices
2.2.1. SUEX Master Fabrication
2.2.2. PDMS Mold Fabrication
2.2.3. Microelement Fabrication
2.3. Modular Assembly
2.4. Out-of-Plane Microfluidic Connections
2.5. Modular Assembly of Coupled Microwells from Double-Layer Blocks
2.6. Cell Culture
2.6.1. Cell Culture for Studies of Drug Toxicity in Microwell Arrays
2.6.2. Cell Culture for Studies of Cell Migration and Drug Toxicity in Coupled Microwells
2.7. Cell Viability Assays
2.8. Filamentous Actin (F-actin) Staining
2.9. Drug Toxicity Assay
2.9.1. Drug Toxicity Assay for Single-Layer Array Microchips
2.9.2. Drug Toxicity Assay for Double-Layer Connected Microwells
2.10. Statistical Analysis
3. Results and Discussions
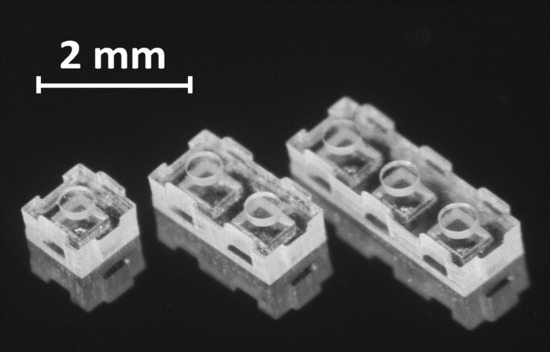
3.1. Fabricated Device Structures
3.2. Mechanical Performance of Fabricated Devices
3.3. Microfluidic Interface Performance
3.4. Cell Behaviors
3.5. Effects of Acetaminophen on Liver Cell Behaviors
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Kim, P.; Kwon, K.W.; Park, M.C.; Lee, S.H.; Kim, S.M.; Suh, K.Y. Soft lithography for microfluidics: A review. BioChip J. 2008, 2, 1–11. [Google Scholar]
- Thorsen, T.; Maerkl, S.J.; Quake, S.R. Microfluidic large-scale integration. Science 2002, 298, 580–584. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cohn, M.B.; Boehringer, K.F.; Noworolski, J.M.; Singh, A.; Keller, C.G.; Goldberg, K.A.; Howe, R.T. Microassembly technologies for MEMS. In Proceedings of the Micromachining and Microfabrication, Santa Clara, CA, USA, 20–24 September 1998; Volume 3513, pp. 2–17. [Google Scholar]
- Hou, X.; Zhang, Y.S.; Santiago, G.T.-D.; Alvarez, M.M.; Ribas, J.; Jonas, S.J.; Weiss, P.S.; Andrews, A.M.; Aizenberg, J.; Khademhosseini, A. Interplay between materials and microfluidics. Nat. Rev. Mater. 2017, 2, 17016. [Google Scholar] [CrossRef]
- Sackmann, E.K.; Fulton, A.L.; Beebe, D.J. The present and future role of microfluidics in biomedical research. Nature 2014, 507, 181. [Google Scholar] [CrossRef]
- Sameith, K.; Antczak, P.; Marston, E.; Turan, N.; Maier, D.; Stankovic, T.; Falciani, F. Functional modules integrating essential cellular functions are predictive of the response of leukaemia cells to DNA damage. Bioinformatics 2008, 24, 2602–2607. [Google Scholar] [CrossRef] [Green Version]
- Liu, J.; Hansen, C.; Quake, S.R.J.A.c. Solving the “world-to-chip” interface problem with a microfluidic matrix. Anal. Chem. 2003, 75, 4718–4723. [Google Scholar] [CrossRef]
- Marcus, J.S.; Anderson, W.F.; Quake, S.R. Microfluidic single-cell mRNA isolation and analysis. Anal. Chem. 2006, 78, 3084–3089. [Google Scholar] [CrossRef] [Green Version]
- Gu, W.; Zhu, X.; Futai, N.; Cho, B.S.; Takayama, S. Computerized microfluidic cell culture using elastomeric channels and Braille displays. Proc. Natl. Acad. Sci.USA 2004, 101, 15861–15866. [Google Scholar] [CrossRef] [Green Version]
- Skelley, A.M.; Scherer, J.R.; Aubrey, A.D.; Grover, W.H.; Ivester, R.H.; Ehrenfreund, P.; Grunthaner, F.J.; Bada, J.L.; Mathies, R.A. Development and evaluation of a microdevice for amino acid biomarker detection and analysis on Mars. Proc. Natl. Acad. Sci.USA 2005, 102, 1041–1046. [Google Scholar] [CrossRef] [Green Version]
- Owens, C.E.; Hart, A.J. High-precision modular microfluidics by micromilling of interlocking injection-molded blocks. Lab Chip 2018, 18, 890–901. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.G.; Park, K.J.; Seok, S.; Shin, S.; Kim, D.H.; Park, J.Y.; Heo, Y.S.; Lee, S.J.; Lee, T.J. 3D printed modules for integrated microfluidic devices. RSC Adv. 2014, 4, 32876–32880. [Google Scholar] [CrossRef]
- Nie, J.; Gao, Q.; Qiu, J.-J.; Sun, M.; Liu, A.; Shao, L.; Fu, J.-Z.; Zhao, P.; He, Y. 3D printed Lego®-like modular microfluidic devices based on capillary driving. Biofabrication 2018, 10, 035001. [Google Scholar] [CrossRef] [PubMed]
- Jivani, R.R.; Lakhtaria, G.J.; Patadiya, D.D.; Patel, L.D.; Jivani, N.P.; Jhala, B.P. Biomedical microelectromechanical systems (BioMEMS): Revolution in drug delivery and analytical techniques. Saudi Pharm. J. 2016, 24, 1–20. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Duffy, D.C.; McDonald, J.C.; Schueller, O.J.; Whitesides, G.M. Rapid prototyping of microfluidic systems in poly (dimethylsiloxane). Anal. Chem. 1998, 70, 4974–4984. [Google Scholar] [CrossRef]
- Morra, M.; Occhiello, E.; Marola, R.; Garbassi, F.; Humphrey, P.; Johnson, D. On the aging of oxygen plasma-treated polydimethylsiloxane surfaces. J. Colloid Interface Sci. 1990, 137, 11–24. [Google Scholar] [CrossRef]
- Quake, S.R.; Scherer, A. From micro-to nanofabrication with soft materials. Science 2000, 290, 1536–1540. [Google Scholar] [CrossRef] [Green Version]
- Martinez-Duarte, R.; Madou, M. SU-8 Photolithography and Its Impact on Microfluidics, 1st ed.; CRC Press: Boca Raton, FL, USA, 2011; pp. 231–268. [Google Scholar]
- Conradie, E.H.; Moore, D.F. SU-8 thick photoresist processing as a functional material for MEMS applications. J. Micromech. Microeng. 2002, 12, 368. [Google Scholar] [CrossRef]
- Eunsung, L.; Woonbae, K.; Insang, S.; Changyoul, M.; Moon Koo, K.; Hyeon Cheol, K.; Kukjin, C. A morphology-independent wafer level rivet packaging with Lego-like assembly. In Proceedings of the 13th International Conference on Solid-State Sensors, Actuators and Microsystems, Digest of Technical Papers. TRANSDUCERS ’05, Seoul, Korea, 5–9 June 2005. [Google Scholar]
- Hsieh, Y.F.; Yang, A.S.; Chen, J.W.; Liao, S.K.; Su, T.W.; Yeh, S.H.; Chen, P.J.; Chen, P.H. A Lego®-like swappable fluidic module for bio-chem applications. Sens. Actuators B Chem. 2014, 204, 489–496. [Google Scholar] [CrossRef]
- Chen, Y.W.; Wang, H.; Hupert, M.; Witek, M.; Dharmasiri, U.; Pingle, M.R.; Barany, F.; Soper, S.A. Modular microfluidic system fabricated in thermoplastics for the strain-specific detection of bacterial pathogens. Lab Chip 2012, 12, 3348–3355. [Google Scholar] [CrossRef]
- Li, W.; Greener, J.; Voicu, D.; Kumacheva, E. Multiple modular microfluidic (M3) reactors for the synthesis of polymer particles. Lab Chip 2009, 9, 2715–2721. [Google Scholar] [CrossRef] [PubMed]
- Huber, J.; Fleck, N.; Ashby, M. The selection of mechanical actuators based on performance indices. Proc. R. Soc. Lond. Ser. A Math. Phys. Eng. Sci. 1997, 453, 2185–2205. [Google Scholar] [CrossRef]
- Uddin, A.; Milaninia, K.; Chen, C.-H.; Theogarajan, L. Wafer scale integration of CMOS chips for biomedical applications via self-aligned masking. IEEE Trans. Compon. Packag. Manuf. Technol. 2011, 1, 1996–2004. [Google Scholar] [CrossRef]
- Slocum, A.H.; Weber, A.C. Precision passive mechanical alignment of wafers. J. Microelectromech. Syst. 2003, 12, 826–834. [Google Scholar] [CrossRef]
- Fernandez, J.G.; Khademhosseini, A. Micro-masonry: Construction of 3D structures by microscale self-assembly. Adv. Mater. 2010, 22, 2538–2541. [Google Scholar] [CrossRef] [Green Version]
- Rhee, M.; Burns, M.A. Microfluidic assembly blocks. Lab Chip 2008, 8, 1365–1373. [Google Scholar] [CrossRef]
- Lee, M.Y.; Dordick, J.S. High-throughput human metabolism and toxicity analysis. Curr. Opin. Biotechnol. 2006, 17, 619–627. [Google Scholar] [CrossRef]
- Hung, P.J.; Lee, P.J.; Sabounchi, P.; Lin, R.; Lee, L.P. Continuous perfusion microfluidic cell culture array for high-throughput cell-based assays. Biotechnol. Bioeng. 2005, 89, 1–8. [Google Scholar] [CrossRef]
- Dominguez, R.; Holmes, K.C. Actin structure and function. Annu. Rev. Biophys. 2011, 40, 169–186. [Google Scholar] [CrossRef] [Green Version]
- De La Cruz, E.M.; Pollard, T.D. Kinetics and thermodynamics of phalloidin binding to actin filaments from three divergent species. Biochemistry 1996, 35, 14054–14061. [Google Scholar] [CrossRef]
- Saljoughian, M. Acetaminophen Intoxication: A Critical-Care Emergency. US Pharm 2016, 41, 38–41. [Google Scholar]
- Bunchorntavakul, C.; Reddy, K.R. Acetaminophen-related hepatotoxicity. Clin. Liver Dis. 2013, 17, 587–607. [Google Scholar] [CrossRef] [PubMed]
- Yoon, E.; Babar, A.; Choudhary, M.; Kutner, M.; Pyrsopoulos, N. Acetaminophen-Induced Hepatotoxicity: A Comprehensive Update. J. Clin. Transl. Hepatol. 2016, 4, 131–142. [Google Scholar] [PubMed] [Green Version]
- Hinson, J.A.; Roberts, D.W.; James, L.P. Mechanisms of acetaminophen-induced liver necrosis. Handb. Exp. Pharmacol. 2010, 196, 369–405. [Google Scholar]
- Amaral, S.S.; Oliveira, A.G.; Marques, P.E.; Quintao, J.L.; Pires, D.A.; Resende, R.R.; Sousa, B.R.; Melgaco, J.G.; Pinto, M.A.; Russo, R.C.; et al. Altered responsiveness to extracellular ATP enhances acetaminophen hepatotoxicity. Cell Commun. Signal. 2013, 11, 10. [Google Scholar] [CrossRef] [Green Version]
- McGill, M.R.; Yan, H.M.; Ramachandran, A.; Murray, G.J.; Rollins, D.E.; Jaeschke, H. HepaRG cells: A human model to study mechanisms of acetaminophen hepatotoxicity. Hepatology 2011, 53, 974–982. [Google Scholar] [CrossRef] [Green Version]
- Kennon-McGill, S.; McGill, M.R. Extrahepatic toxicity of acetaminophen: Critical evaluation of the evidence and proposed mechanisms. J. Clin. Transl. Res. 2018, 3, 297–310. [Google Scholar]
- Mour, G.; Feinfeld, D.A.; Caraccio, T.; McGuigan, M. Acute renal dysfunction in acetaminophen poisoning. Ren. Fail. 2005, 27, 381–383. [Google Scholar] [CrossRef] [Green Version]
- Carpenter, H.M.; Mudge, G.H. Acetaminophen nephrotoxicity: Studies on renal acetylation and deacetylation. J. Pharm. Exp. Ther. 1981, 218, 161–167. [Google Scholar]
- Yu, Y.L.; Yiang, G.T.; Chou, P.L.; Tseng, H.H.; Wu, T.K.; Hung, Y.T.; Lin, P.S.; Lin, S.Y.; Liu, H.C.; Chang, W.J.; et al. Dual role of acetaminophen in promoting hepatoma cell apoptosis and kidney fibroblast proliferation. Mol. Med. Rep. 2014, 9, 2077–2084. [Google Scholar] [CrossRef] [Green Version]





© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xie, X.; Maharjan, S.; Liu, S.; Zhang, Y.S.; Livermore, C. A Modular, Reconfigurable Microfabricated Assembly Platform for Microfluidic Transport and Multitype Cell Culture and Drug Testing. Micromachines 2020, 11, 2. https://doi.org/10.3390/mi11010002
Xie X, Maharjan S, Liu S, Zhang YS, Livermore C. A Modular, Reconfigurable Microfabricated Assembly Platform for Microfluidic Transport and Multitype Cell Culture and Drug Testing. Micromachines. 2020; 11(1):2. https://doi.org/10.3390/mi11010002
Chicago/Turabian StyleXie, Xin, Sushila Maharjan, Sanwei Liu, Yu Shrike Zhang, and Carol Livermore. 2020. "A Modular, Reconfigurable Microfabricated Assembly Platform for Microfluidic Transport and Multitype Cell Culture and Drug Testing" Micromachines 11, no. 1: 2. https://doi.org/10.3390/mi11010002