A Micro-Optic Stalk (μOS) System to Model the Collective Migration of Retinal Neuroblasts
Abstract
:1. Introduction
2. Materials and Methods
2.1. Drosophila Fly Stocks
2.2. Dissection of Eye–Brain Complexes
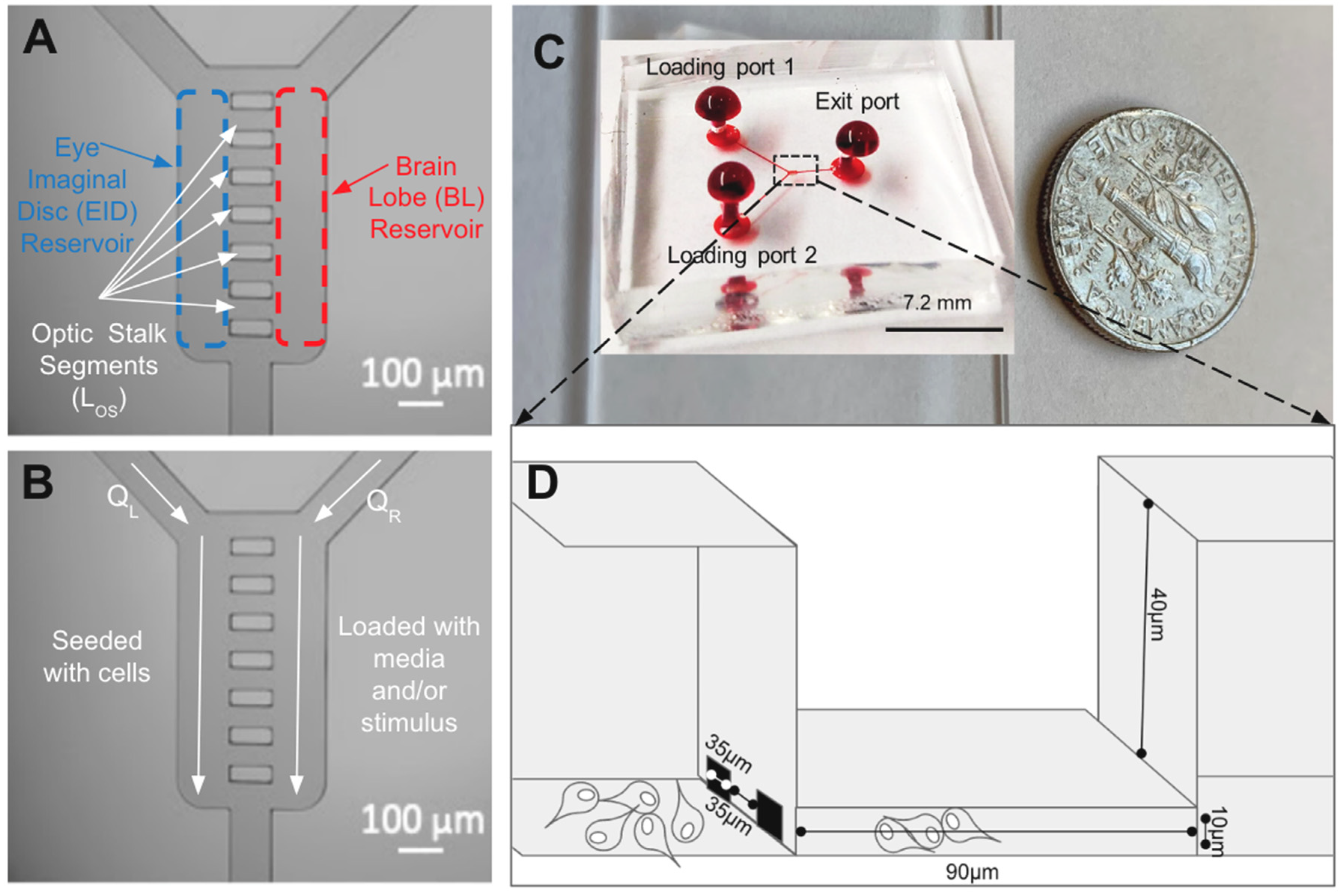
2.3. μOS Design and Operation
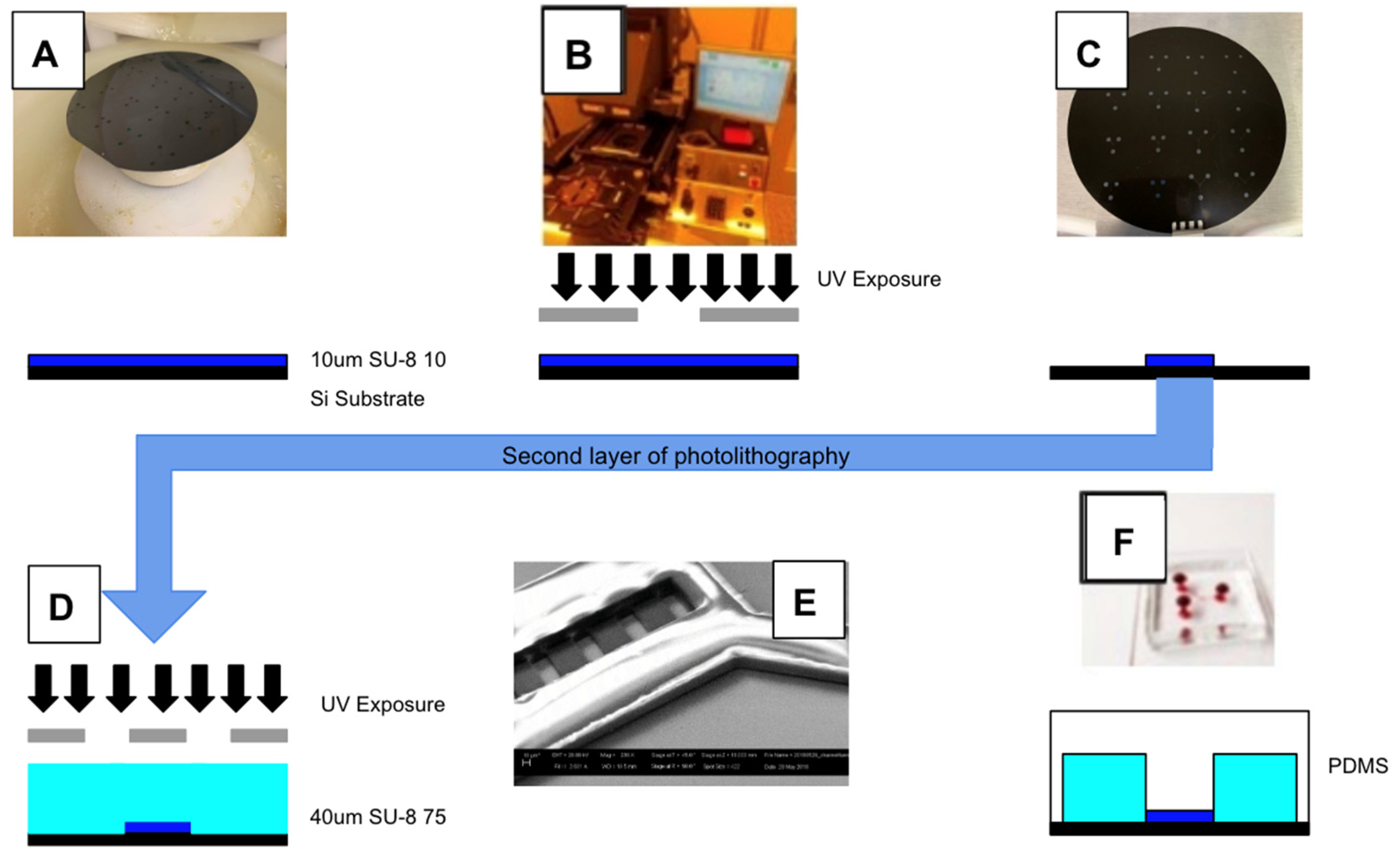
2.4. System Fabrication
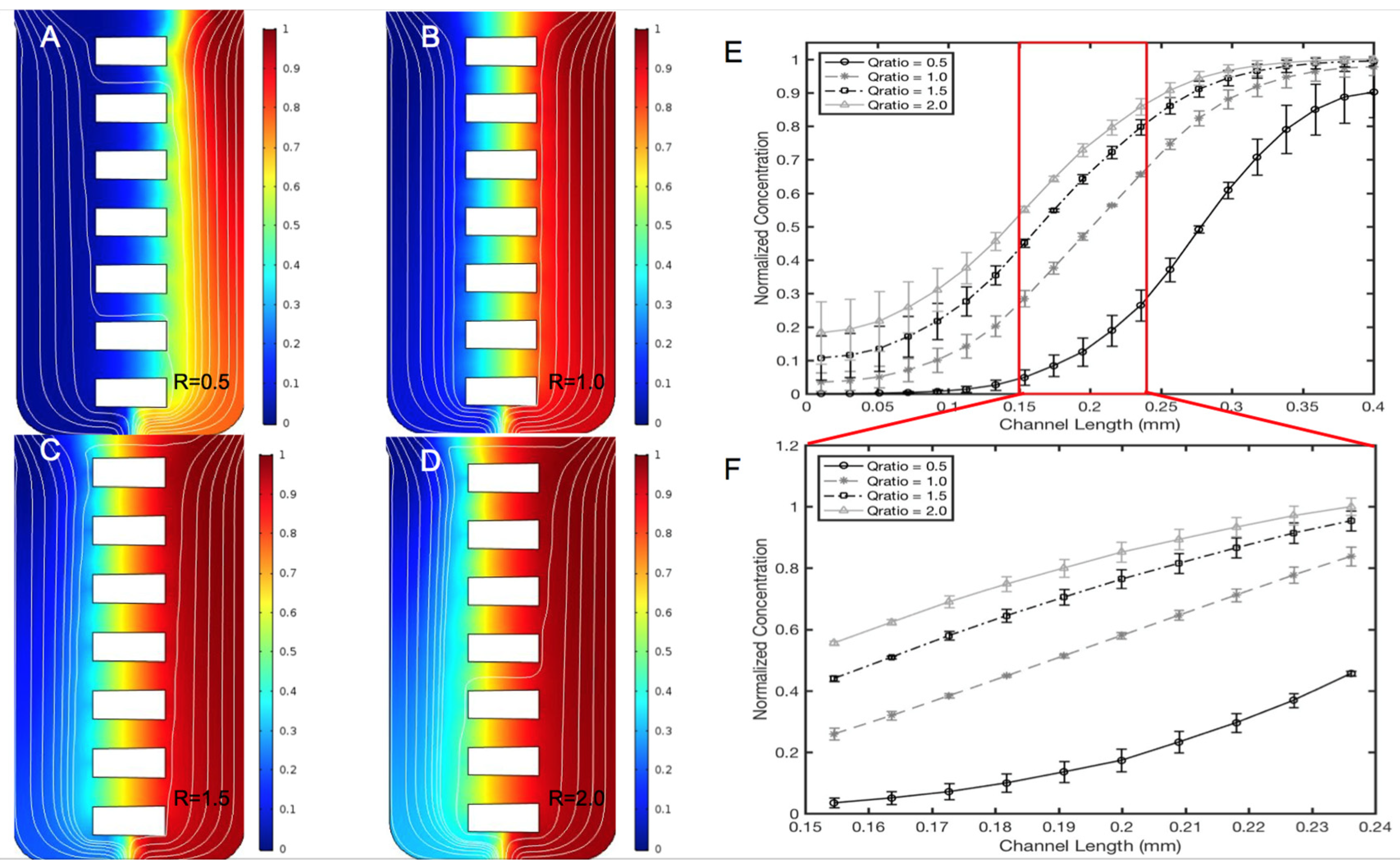
2.5. Computational Transport Model and Experimental Validation
2.6. Microscopy and Imaging
2.7. Device Operation and Measurement of RNB Migration
2.8. Data Analysis and Statistics
3. Results
3.1. Conserved Retinal Development
3.2. Validation of the μOS Chemical Environment
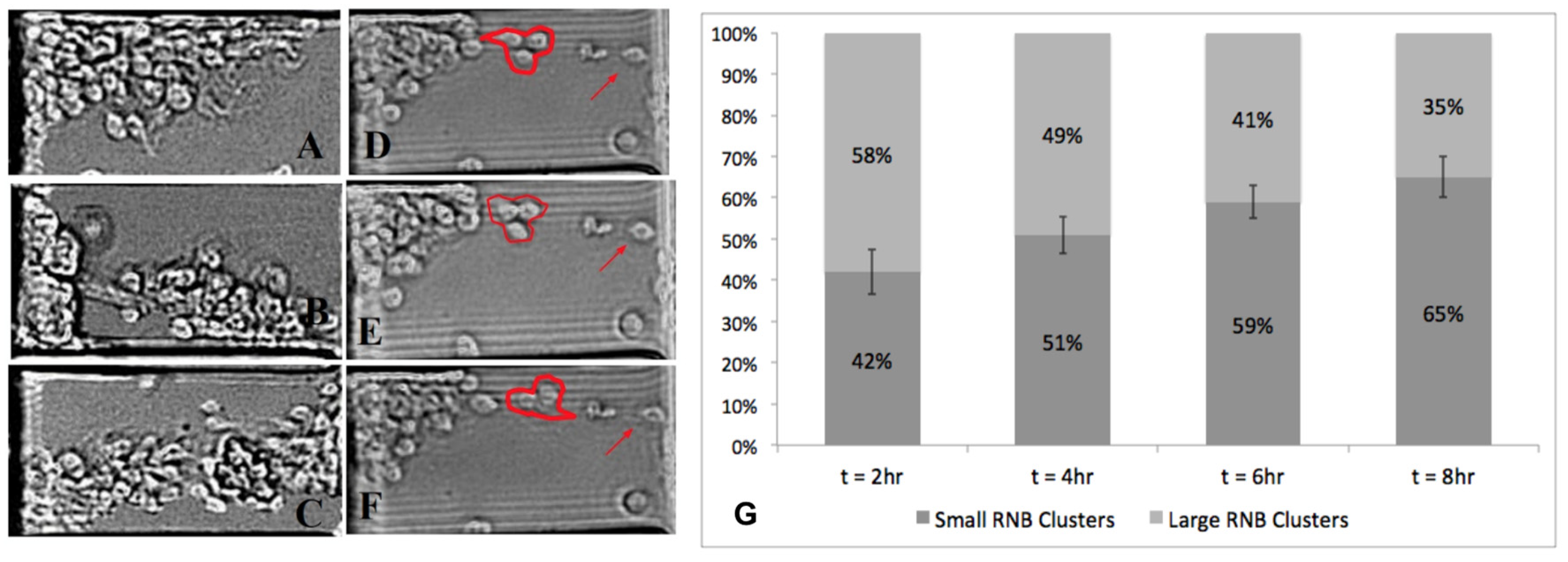
3.3. Collective Migration of RNB Clusters
3.4. Disaggregation of Large RNB Clusters
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Flaxman, S.R.; Bourne, R.R.A.; Resnikoff, S.; Ackland, P.; Braithwaite, T.; Cicinelli, M.V.; Das, A.; Jonas, J.B.; Keeffe, J.; Kempen, J.H.; et al. Vision Loss Expert Group of the Global Burden of Disease, Global causes of blindness and distance vision impairment 1990–2020: A systematic review and meta-analysis. Lancet Glob. Health 2017, 5, e1221–e1234. [Google Scholar] [CrossRef] [Green Version]
- Pascolini, D.; Mariotti, S.P. Global estimates of visual impairment: 2010. Br. J. Ophthalmol. 2012, 96, 614–618. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pena, J.S.; Vazquez, M. Reducing health disparities in adult vision loss via interfaces with emerging technology. Eye (Lond.) 2019, 33, 532–533. [Google Scholar] [CrossRef] [PubMed]
- West, E.L.; Ribeiro, J.; Ali, R.R. Development of Stem Cell Therapies for Retinal Degeneration. Cold Spring Harb. Perspect. Biol. 2019. [Google Scholar] [CrossRef] [PubMed]
- Cuevas, E.; Parmar, P.; Sowden, J.C. Restoring Vision Using Stem Cells and Transplantation. Adv. Exp. Med. Biol. 2019, 1185, 563–567. [Google Scholar]
- Singh, R.K.; Occelli, L.M.; Binette, F.; Petersen-Jones, S.M.; Nasonkin, I.O. Transplantation of Human Embryonic Stem Cell-Derived Retinal Tissue in the Subretinal Space of the Cat Eye. Stem. Cells Dev. 2019, 28, 1151–1166. [Google Scholar] [CrossRef]
- Fischbach, K.F.; Hiesinger, P.R. Optic lobe development. Adv. Exp. Med. Biol. 2008, 628, 115–136. [Google Scholar]
- Courgeon, M.; Desplan, C. Coordination of neural patterning in the Drosophila visual system. Curr. Opin. Neurobiol. 2019, 56, 153–159. [Google Scholar] [CrossRef]
- Kumar, J.P. The fly eye: Through the looking glass. Dev. Dyn. 2018, 247, 111–123. [Google Scholar] [CrossRef] [Green Version]
- Scarpa, E.; Mayor, R. Collective cell migration in development. J. Cell Biol. 2016, 212, 143–155. [Google Scholar] [CrossRef] [Green Version]
- Pascalis, C.D.; Etienne-Manneville, S. Single and collective cell migration: The mechanics of adhesions. Mol. Biol. Cell 2017, 28, 1833–1846. [Google Scholar] [CrossRef] [PubMed]
- Suen, H.C.; Qian, Y.; Liao, J.; Luk, C.S.; Lee, W.T.; Ng, J.K.W.; Chan, T.T.H.; Hou, H.W.; Li, I.; Li, K.; et al. Transplantation of Retinal Ganglion Cells Derived from Male Germline Stem Cell as a Potential Treatment to Glaucoma. Stem. Cells Dev. 2019, 28, 1365–1375. [Google Scholar] [CrossRef] [PubMed]
- Xie, H.; Zhang, W.; Zhang, M.; Akhtar, T.; Li, Y.; Yi, W.; Sun, X.; Zuo, Z.; Wei, M.; Fang, X.; et al. Chromatin accessibility analysis reveals regulatory dynamics of developing human retina and hiPSC-derived retinal organoids. Sci. Adv. 2020, 6, eaay5247. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sridhar, A.; Hoshino, A.; Finkbeiner, C.R.; Chitsazan, A.; Dai, L.; Haugan, A.K.; Eschenbacher, K.M.; Jackson, D.L.; Trapnell, C.; Bermingham-McDonogh, O.; et al. Single-Cell Transcriptomic Comparison of Human Fetal Retina, hPSC-Derived Retinal Organoids, and Long-Term Retinal Cultures. Cell Rep. 2020, 30, 1644–1659. [Google Scholar] [CrossRef]
- Camley, B.A.; Zimmermann, J.; Levine, H.; Rappel, W.J. Emergent Collective Chemotaxis without Single-Cell Gradient Sensing. Phys. Rev. Lett. 2016, 116, 098101. [Google Scholar] [CrossRef]
- Theveneau, E.; Steventon, B.; Scarpa, E.; Garcia, S.; Trepat, X.; Streit, A.; Mayor, R. Chase-and-run between adjacent cell populations promotes directional collective migration. Nat. Cell Biol. 2013, 15, 763–772. [Google Scholar] [CrossRef] [Green Version]
- Fischbach, K.-F.; Hiesinger, P.R. Optic Lobe Development. In Brain Development in Drosophila Melanogaster; Springer: Berlin/Heidelberg, Germany, 2008; pp. 115–136. [Google Scholar]
- Pignoni, F.; Zipursky, S.L. Induction of Drosophila eye development by decapentaplegic. Development 1997, 124, 271–278. [Google Scholar]
- Hwang, H.; Lu, H. Microfluidic tools for developmental studies of small model organisms—Nematodes, fruit flies, and zebrafish. Biotechnol. J. 2013, 8, 192–205. [Google Scholar] [CrossRef] [Green Version]
- Alazzam, A.; Mathew, B.; Khashan, S. Microfluidic Platforms for Bio-Applications. In Advanced Mechatronics and MEMS Devices II; Zhang, D., Wei, B., Eds.; Springer International Publishing: New York, NY, USA, 2017; pp. 253–282. [Google Scholar]
- Yang, F.; Gao, C.; Wang, P.; Zhang, G.J.; Chen, Z. Fish-on-a-chip:Microfluidics for zebrafish research. Lab Chip 2016, 16, 1106–1125. [Google Scholar] [CrossRef]
- Macdonald, N.P.; Zhu, F.; Hall, C.J.; Reboud, J.; Crosier, P.S.; Patton, E.E.; Wlodkowic, D.; Cooper, J.M. Assessment of biocompatibility of 3D printed photopolymers using zebrafish embryo toxicity assays. Lab Chip 2016, 16, 291–297. [Google Scholar] [CrossRef] [Green Version]
- Liu, W.; Li, L.; Wang, X.; Ren, L.; Wang, X.; Wang, J.; Tu, Q.; Huang, X.; Wang, J. An integrated microfluidic system for studying cell—Microenvironmental interactions versatilely and dynamically. Lab Chip 2010, 10, 1717–1724. [Google Scholar] [CrossRef] [PubMed]
- Walsh, C.L.; Babin, B.M.; Kasinskas, R.W.; Foster, J.A.; McGarry, M.J.; Forbes, N.S. A multipurpose microfluidic device designed to mimic microenvironment gradients and develop targeted cancer therapeutics. Lab Chip 2009, 9, 545–554. [Google Scholar] [CrossRef] [PubMed]
- Walker, G.M.; Zeringue, H.C.; Beebe, D.J. Microenvironment design considerations for cellular scale studies. Lab Chip 2004, 4, 91–97. [Google Scholar] [CrossRef] [PubMed]
- Kim, L.; YToh, C.; Voldman, J.; Yu, H. A practical guide to microfluidic perfusion culture of adherent mammalian cells. Lab Chip 2007, 7, 681–694. [Google Scholar] [CrossRef] [PubMed]
- Sackmann, E.K.; Fulton, A.L.; Beebe, D.J. The present and future role of microfluidics in biomedical research. Nature 2014, 507, 181–189. [Google Scholar] [CrossRef] [PubMed]
- Boutin, M.E.; Hampton, C.; Quinn, R.; Ferrer, M.; Song, M.J. 3D Engineering of Ocular Tissues for Disease Modeling and Drug Testing. Adv. Exp. Med. Biol. 2019, 1186, 171–193. [Google Scholar]
- Abdolvand, N.; Tostoes, R.; Raimes, W.; Kumar, V.; Szita, N.; Veraitch, F. Long-Term Retinal Differentiation of Human Induced Pluripotent Stem Cells in a Continuously Perfused Microfluidic Culture Device. Biotechnol. J. 2019, 14, e1800323. [Google Scholar] [CrossRef]
- Dodson, K.H.; Echevarria, F.D.; Li, D.; Sappington, R.M.; Edd, J.F. Retina-on-a-chip: A microfluidic platform for point access signaling studies. Biomed. Microdevices 2015, 17, 114. [Google Scholar] [CrossRef] [Green Version]
- Su, P.J.; Liu, Z.; Zhang, K.; Han, X.; Saito, Y.; Xia, X.; Yokoi, K.; Shen, H.; Qin, L. Retinal synaptic regeneration via microfluidic guiding channels. Sci. Rep. 2015, 5, 13591. [Google Scholar] [CrossRef]
- Rountree, C.M.; Troy, J.B.; Saggere, L. Microfluidics-Based Subretinal Chemical Neuromodulation of Photoreceptor Degenerated Retinas. Investg. Ophthalmol. Vis. Sci. 2018, 59, 418–430. [Google Scholar] [CrossRef]
- Gomis, S.; Labib, M.; Coles, B.L.K.; van der Kooy, D.; Sargent, E.H.; Kelley, S.O. Single-Cell Tumbling Enables High-Resolution Size Profiling of Retinal Stem Cells. Acs Appl. Mater. Interfaces 2018, 10, 34811–34816. [Google Scholar] [CrossRef] [PubMed]
- McUsic, A.C.; Lamba, D.A.; Reh, T.A. Guiding the morphogenesis of dissociated newborn mouse retinal cells and hES cell-derived retinal cells by soft lithography-patterned microchannel PLGA scaffolds. Biomaterials 2012, 33, 1396–1405. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eiraku, M.; Takata, N.; Ishibashi, H.; Kawada, M.; Sakakura, E.; Okuda, S.; Sekiguchi, K.; Adachi, T.; Sasai, Y. Self-organizing optic-cup morphogenesis in three-dimensional culture. Nature 2011, 472, 51–56. [Google Scholar] [CrossRef] [PubMed]
- Pena, C.D.; Zhang, S.; Markey, M.; Venkatesh, T.; Vazquez, M. Collective behaviors of Drosophila-derived retinal progenitors in controlled microenvironments. PLoS ONE 2019, 14, e0226250. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pena, C.D.; Zhang, S.; Majeska, R.; Venkatesh, T.; Vazquez, M. Invertebrate Retinal Progenitors as Regenerative Models in a Microfluidic System. Cells 2019, 8, 1301. [Google Scholar] [CrossRef] [Green Version]
- Abeille, F.; Mittler, F.; Obeid, P.; Huet, M.; Kermarrec, F.; Dolega, M.E.; Navarro, F.; Pouteau, P.; Icard, B.; Gidrol, X.; et al. Continuous microcarrier-based cell culture in a benchtop microfluidic bioreactor. Lab Chip 2014, 14, 3510–3518. [Google Scholar] [CrossRef]
- Zabihihesari, A.; Hilliker, A.J.; Rezai, P. Fly-on-a-Chip: Microfluidics for Drosophila melanogaster Studies. Integr. Biol. (Camb.) 2019, 11, 425–443. [Google Scholar] [CrossRef]
- Beck, C.; Singh, T.; Farooqi, A.; Venkatesh, T.; Vazquez, M. Controlled microfluidics to examine growth-factor induced migration of neural progenitors in the Drosophila visual system. J. Neurosci. Methods 2016, 262, 32–40. [Google Scholar] [CrossRef] [Green Version]
- McCutcheon, S.; Unachukwu, U.; Thakur, A.; Majeska, R.; Redenti, S.; Vazquez, M. In vitro formation of neuroclusters in microfluidic devices and cell migration as a function of stromal-derived growth factor 1 gradients. Cell Adhes. Migr. 2017, 11, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Thakur, A.; Mishra, S.; Pena, J.; Zhou, J.; Redenti, S.; Majeska, R.; Vazquez, M. Collective adhesion and displacement of retinal progenitor cells upon extracellular matrix substrates of transplantable biomaterials. J. Tissue Eng. 2018, 9, 2041731417751286. [Google Scholar] [CrossRef]
- Mishra, A.K.; Campanale, J.P.; Mondo, J.A.; Montell, D.J. Cell interactions in collective cell migration. Development 2019, 146. [Google Scholar] [CrossRef] [PubMed]
- Macabenta, F.; Stathopoulos, A. Sticking to a plan: Adhesion and signaling control spatial organization of cells within migrating collectives. Curr. Opin. Genet. Dev. 2019, 57, 39–46. [Google Scholar] [CrossRef] [PubMed]
- Duffy, J.B. GAL4 system in Drosophila: A fly geneticist’s Swiss army knife. Genesis 2002, 34, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi, M.; Yoshida, H. Drosophila as a Model Organism. Adv. Exp. Med. Biol. 2018, 1076, 1–10. [Google Scholar] [PubMed]
- Moraru, M.M.; Egger, B.; Bao, D.B.; Sprecher, S.G. Analysis of cell identity, morphology, apoptosis and mitotic activity in a primary neural cell culture system in Drosophila. Neural Dev. 2012, 7, 14. [Google Scholar] [CrossRef] [Green Version]
- Luhur, A.; Klueg, K.M.; Zelhof, A.C. Generating and working with Drosophila cell cultures: Current challenges and opportunities. Wiley Interdiscip. Rev. Dev. Biol. 2019, 8, e339. [Google Scholar] [CrossRef]
- McCutcheon, S.; Majeska, R.; Schaffler, M.; Vazquez, M. A multiscale fluidic device for the study of dendrite-mediated cell to cell communication. Biomed. Microdevices 2017, 19, 71. [Google Scholar] [CrossRef]
- Mishra, S.; Thakur, A.; Redenti, S.; Vazquez, M. A model microfluidics-based system for the human and mouse retina. Biomed. Microdevices 2015, 17, 107. [Google Scholar] [CrossRef] [Green Version]
- McCutcheon, S.; Majeska, R.J.; Spray, D.C.; Schaffler, M.B.; Vazquez, M. Apoptotic Osteocytes Induce RANKL Production in Bystanders via Purinergic Signaling and Activation of Pannexin Channels. J. Bone Min. Res. 2020, 2020. [Google Scholar] [CrossRef]
- Lerit, D.A.; Plevock, K.M.; Rusan, N.M. Live imaging of Drosophila larval neuroblasts. J. Vis. Exp. 2014, 89, e51756. [Google Scholar]
- Cafferty, P.; Xie, X.; Browne, K.; Auld, V.J. Live imaging of glial cell migration in the Drosophila eye imaginal disc. J. Vis. Exp. 2009, 29, e1155. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, J.S.; Luo, L. A protocol for dissecting Drosophila melanogaster brains for live imaging or immunostaining. Nat. Protoc. 2006, 1, 2110–2115. [Google Scholar] [CrossRef] [PubMed]
- Coluccio, M.L.; D’Attimo, M.A.; Cristiani, C.M.; Candeloro, P.; Parrotta, E.; Dattola, E.; Guzzi, F.; Cuda, G.; Lamanna, E.; Carbone, E.; et al. A Passive Microfluidic Device for Chemotaxis Studies. Micromachines 2019, 10, 551. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, C.Y.; Chang, C.L.; Wang, Y.N.; Fu, L.M. Microfluidic mixing: A review. Int. J. Mol. Sci. 2011, 12, 3263–3287. [Google Scholar] [CrossRef] [Green Version]
- Mishra, S.; Vazquez, M. A Gal-MmicroS Device to Evaluate Cell Migratory Response to Combined Galvano-Chemotactic Fields. Biosensors 2017, 7, 54. [Google Scholar] [CrossRef] [Green Version]
- Kong, Q.; Able, R.A., Jr.; Dudu, V.; Vazquez, M. A microfluidic device to establish concentration gradients using reagent density differences. J. Biomech. Eng. 2010, 132, 121012. [Google Scholar] [CrossRef] [Green Version]
- Singh, T.; Robles, D.; Vazquez, M. Neuronal substrates alter the migratory responses of non-myelinating Schwann cells to controlled BDNF gradients. J. Tissue Eng. Regen. Med. 2020, 2020. [Google Scholar] [CrossRef]
- Pena, J.S.; Robles, D.; Zhang, S.; Vazquez, M. A Milled Microdevice to Advance Glia-Mediated Therapies in the Adult Nervous System. Micromachines 2019, 10, 513. [Google Scholar] [CrossRef] [Green Version]
- Yu, S.R.; Burkhardt, M.; Nowak, M.; Ries, J.; Petrášek, Z.; Scholpp, S.; Schwille, P.; Brand, M. Fgf8 morphogen gradient forms by a source-sink mechanism with freely diffusing molecules. Nature 2009, 461, 533–536. [Google Scholar] [CrossRef]
- Kong, Q.; Vazquez, M. Flow-induced shear stresses increase the number of cell-cell contacts within extracellular matrix. J. Biomed. Mater. Res. A 2009, 89, 968–979. [Google Scholar] [CrossRef]
- Kong, Q.; Vazquez, M. Internal fluid flow increases cellular interconnects between Medial Collateral Ligament fibroblasts and cellular extensions within three-dimensional collagen matrixes. Cell Commun. Adhes. 2006, 13, 139–149. [Google Scholar] [CrossRef] [PubMed]
- Franzdottir, S.R.; Engelen, D.; Yuva-Aydemir, Y.; Schmidt, I.; Aho, A.; Klambt, C. Switch in FGF signalling initiates glial differentiation in the Drosophila eye. Nature 2009, 460, 758–761. [Google Scholar] [CrossRef] [PubMed]
- Mishra, S.; Pena, J.S.; Redenti, S.; Vazquez, M. A novel electro-chemotactic approach to impact the directional migration of transplantable retinal progenitor cells. Exp. Eye Res. 2019, 185, 107688. [Google Scholar] [CrossRef] [PubMed]
- Vopalensky, P.; Kozmik, Z. Eye evolution: Common use and independent recruitment of genetic components. Philos. Trans. R Soc. Lond. B Biol. Sci. 2009, 364, 2819–2832. [Google Scholar] [CrossRef] [Green Version]
- Mirzoyan, Z.; Sollazzo, M.; Allocca, M.; Valenza, A.M.; Grifoni, D.; Bellosta, P. Drosophila melanogaster: A Model Organism to Study Cancer. Front. Genet. 2019, 10, 51. [Google Scholar] [CrossRef] [Green Version]
- Erclik, T.; Hartenstein, V.; McInnes, R.R.; Lipshitz, H.D. Eye evolution at high resolution: The neuron as a unit of homology. Dev. Biol. 2009, 332, 70–79. [Google Scholar] [CrossRef] [Green Version]
- Doke, S.K.; Dhawale, S.C. Alternatives to animal testing: A review. Saudi Pharm. J. 2015, 23, 223–229. [Google Scholar] [CrossRef] [Green Version]
- Chang, C.-C.; Huang, Z.-X.; Yang, R.-J. Three-dimensional hydrodynamic focusing in two-layer polydimethylsiloxane (PDMS) microchannels. J. Micromechanics Microengineering 2007, 17, 1479. [Google Scholar] [CrossRef]
- Kong, Q.; Majeska, R.J.; Vazquez, M. Migration of connective tissue-derived cells is mediated by ultra-low concentration gradient fields of EGF. Exp. Cell Res. 2011, 317, 1491–1502. [Google Scholar] [CrossRef] [Green Version]
- Rapp, L.; Zimmermann, W. Universal aspects of collective behavior in chemotactic systems. Phys. Rev. E 2019, 100, 032609. [Google Scholar] [CrossRef]
- Ding, H.; Jin, G.H.; Zou, L.Q.; Zhang, X.Q.; Li, H.M.; Tao, X.L.; Zhang, X.H.; Qin, J.B.; Tian, M.L. Stromal derived factor-1α in hippocampus radial glial cells in vitro regulates the migration of neural progenitor cells. Cell Biol. Int. 2015, 39, 750–758. [Google Scholar] [CrossRef] [PubMed]
- Shamloo, A.; Heibatollahi, M.; Mofrad, M.R. Directional migration and differentiation of neural stem cells within three-dimensional microenvironments. Integr. Biol. (Camb.) 2015, 7, 335–344. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Song, A.; Lai, S.; Qiu, L.; Huang, Y.; Chen, Q.; Zhu, B.; Xu, D.; Zheng, J.C. Applications of stripe assay in the study of CXCL12-mediated neural progenitor cell migration and polarization. Biomaterials 2015, 72, 163–171. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Keenan, T.M.; Grinager, J.R.; Procak, A.A.; Svendsen, C.N. In vitro localization of human neural stem cell neurogenesis by engineered FGF-2 gradients. Integr. Biol. (Camb.) 2012, 4, 1522–1531. [Google Scholar] [CrossRef]
- Le Maout, E.; Vecchio, S.L.; Bhat, A.; Riveline, D. Directing cell migration on flat substrates and in confinement with microfabrication and microfluidics. Methods Cell Biol. 2018, 147, 109–132. [Google Scholar]
- Pérez-Rodríguez, S.; Tomás-González, E.; García-Aznar, J.M. 3D Cell Migration Studies for Chemotaxis on Microfluidic-Based Chips: A Comparison between Cardiac and Dermal Fibroblasts. Bioengineering 2018, 5, 45. [Google Scholar] [CrossRef] [Green Version]
- Holan, V.; Hermankova, B.; Krulova, M.; Zajicova, A. Cytokine interplay among the diseased retina, inflammatory cells and mesenchymal stem cells—A clue to stem cell-based therapy. World J. Stem Cells 2019, 11, 957–967. [Google Scholar] [CrossRef]
- Makarevich, P.I.; Efimenko, A.Y.; Tkachuk, V.A. Biochemical Regulation of Regenerative Processes by Growth Factors and Cytokines: Basic Mechanisms and Relevance for Regenerative Medicine. Biochemistry (Mosc.) 2020, 85, 11–26. [Google Scholar] [CrossRef]
- Zaghloul, N.A.; Yan, B.; Moody, S.A. Step-wise specification of retinal stem cells during normal embryogenesis. Biol. Cell 2005, 97, 321–337. [Google Scholar] [CrossRef]
- Muth, C.A.; Steinl, C.; Klein, G.; Lee-Thedieck, C. Regulation of hematopoietic stem cell behavior by the nanostructured presentation of extracellular matrix components. PLoS ONE 2013, 8, e54778. [Google Scholar] [CrossRef] [Green Version]
- Abascal, F.; Zardoya, R. Evolutionary analyses of gap junction protein families. Biochim. Biophys. Acta 2013, 1828, 4–14. [Google Scholar] [CrossRef] [PubMed]


| Key Features of μOS | Dimensions of μOS (μm) | Key Features of Developing Eye | Dimensions of Developing Eye (μm) |
|---|---|---|---|
| Length of Optic Stalk (LOS) | 90 ± 5 | Diameter of Eye Imaginal Disc (DEID) | 500 ± 23 [9,54] |
| Characteristic EID width of Optic Stalk (WOS1) | 37 ± 3 | Diameter of Brain Lobe (DBL) | 800 ± 14 [9,54] |
| Characteristic BL width of Optic Stalk (WOS2) | 35 ± 3 | Length of Optic Stalk (LOS) | 90 ± 2 [9,54] |
| Type | Definition | Percentage of RNBs (%) | Average Total Path Length, LT (μm) | Average Net Displacement, DN (μm) | Directionality, DR |
|---|---|---|---|---|---|
| Single Cells | 1–2 cells | 16% | 2.67 ± 0.58 | 0.67 ± 0.58 | 0 |
| Small Clusters | 3–5 cells | 65% | 17.7 ± 1.83 | 10.1 ± 0.99 | 0.72 ± 0.15 |
| Large Clusters | >5 cells | 21% | 14.2 ± 2.94 | 4.44 ± 2.01 | 0.79 ± 0.22 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, S.; Markey, M.; Pena, C.D.; Venkatesh, T.; Vazquez, M. A Micro-Optic Stalk (μOS) System to Model the Collective Migration of Retinal Neuroblasts. Micromachines 2020, 11, 363. https://doi.org/10.3390/mi11040363
Zhang S, Markey M, Pena CD, Venkatesh T, Vazquez M. A Micro-Optic Stalk (μOS) System to Model the Collective Migration of Retinal Neuroblasts. Micromachines. 2020; 11(4):363. https://doi.org/10.3390/mi11040363
Chicago/Turabian StyleZhang, Stephanie, Miles Markey, Caroline D. Pena, Tadmiri Venkatesh, and Maribel Vazquez. 2020. "A Micro-Optic Stalk (μOS) System to Model the Collective Migration of Retinal Neuroblasts" Micromachines 11, no. 4: 363. https://doi.org/10.3390/mi11040363
APA StyleZhang, S., Markey, M., Pena, C. D., Venkatesh, T., & Vazquez, M. (2020). A Micro-Optic Stalk (μOS) System to Model the Collective Migration of Retinal Neuroblasts. Micromachines, 11(4), 363. https://doi.org/10.3390/mi11040363





