Neuronal Activity Reporters as Drug Screening Platforms
Abstract
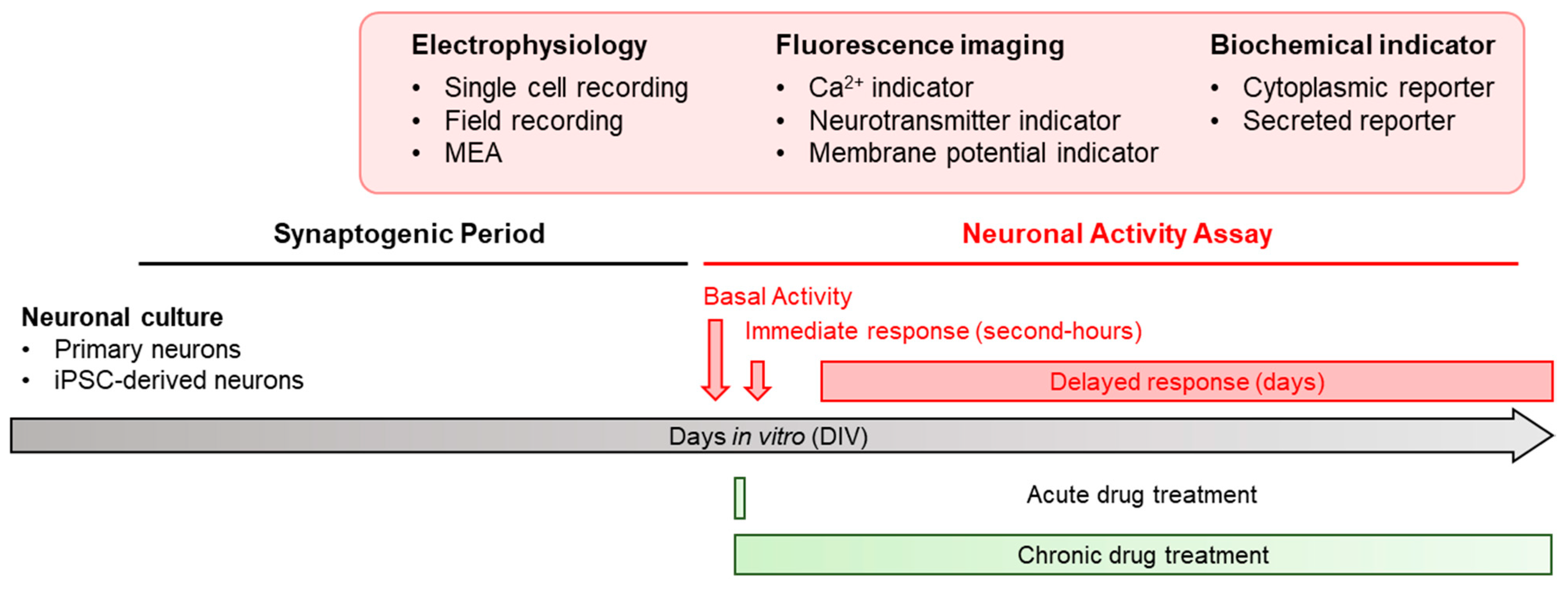
1. Introduction
2. Electrophysiological Recordings
3. Calcium and Neurotransmitter Indicators
4. Membrane Voltage Indicators:
5. Immediate Early Genes
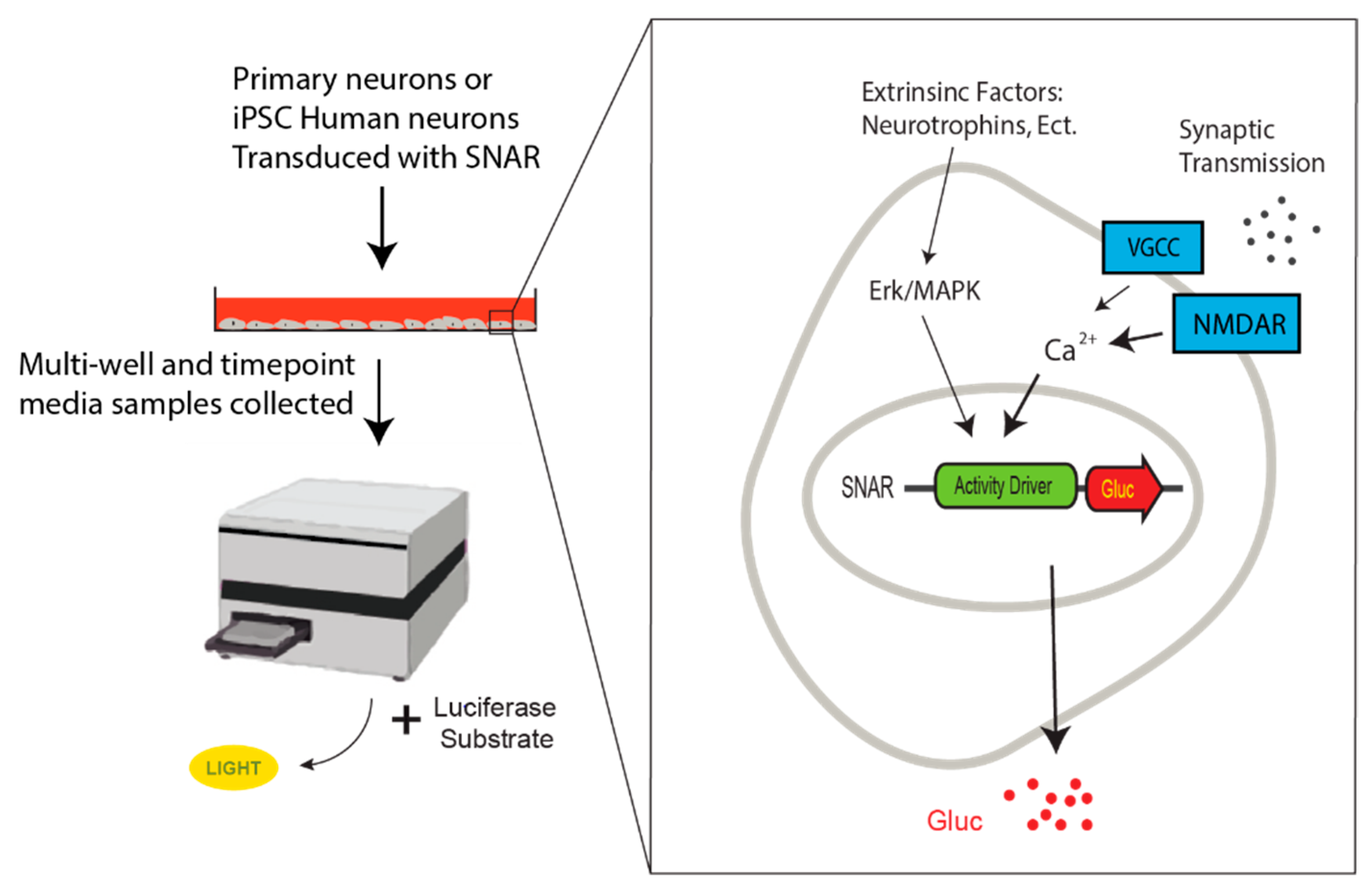
6. SNAR
7. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Eminatohara, K.; Eakiyoshi, M.; Eokuno, H. Role of Immediate-Early Genes in Synaptic Plasticity and Neuronal Ensembles Underlying the Memory Trace. Front. Mol. Neurosci. 2016, 8, 78. [Google Scholar] [CrossRef]
- DeNardo, L.; Luo, L. Genetic strategies to access activated neurons. Curr. Opin. Neurobiol. 2017, 45, 121–129. [Google Scholar] [CrossRef] [PubMed]
- Parenti, I.; Rabaneda, L.G.; Schoen, H.; Novarino, G. Neurodevelopmental Disorders: From Genetics to Functional Pathways. Trends Neurosci. 2020, 43, 608–621. [Google Scholar] [CrossRef]
- Carroll, W.M. The global burden of neurological disorders. Lancet Neurol. 2019, 18, 418–419. [Google Scholar] [CrossRef]
- Fink, J.J.; Levine, E.S. Uncovering True Cellular Phenotypes: Using Induced Pluripotent Stem Cell-Derived Neurons to Study Early Insults in Neurodevelopmental Disorders. Front. Neurol. 2018, 9, 237. [Google Scholar] [CrossRef]
- Nierode, G.; Kwon, P.S.; Dordick, J.S.; Kwon, S.-J. Cell-Based Assay Design for High-Content Screening of Drug Candidates. J. Microbiol. Biotechnol. 2016, 26, 213–225. [Google Scholar] [CrossRef]
- Wang, L.; Yu, C.; Wang, J. Development of reporter gene assays to determine the bioactivity of biopharmaceuticals. Biotechnol. Adv. 2019, 39, 107466. [Google Scholar] [CrossRef] [PubMed]
- Santos, A.C.; Chiola, S.; Yang, G.; Shcheglovitov, A.; Park, S. Secreted Reporter Assay Enables Quantitative and Longitudinal Monitoring of Neuronal Activity. Eneuro 2021, 8, 0518-20. [Google Scholar] [CrossRef]
- Liu, C.; Li, T.; Chen, J. Role of High-Throughput Electrophysiology in Drug Discovery. Curr. Protoc. Pharmacol. 2019, 87, e69. [Google Scholar] [CrossRef] [PubMed]
- Noguchi, A.; Ikegaya, Y.; Matsumoto, N. In Vivo Whole-Cell Patch-Clamp Methods: Recent Technical Progress and Future Perspectives. Sensors 2021, 21, 1448. [Google Scholar] [CrossRef]
- Obergrussberger, A.; Brüggemann, A.; Goetze, T.A.; Rapedius, M.; Haarmann, C.; Rinke, I.; Becker, N.; Oka, T.; Ohtsuki, A.; Stengel, T.; et al. Automated Patch Clamp Meets High-Throughput Screening: 384 Cells Recorded in Parallel on a Planar Patch Clamp Module. J. Lab. Autom. 2016, 21, 779–793. [Google Scholar] [CrossRef]
- Rosholm, K.R.; Badone, B.; Karatsiompani, S.; Nagy, D.; Seibertz, F.; Voigt, N.; Bell, D.C. Adventures and Advances in Time Travel with Induced Pluripotent Stem Cells and Automated Patch Clamp. Front. Mol. Neurosci. 2022, 15, 898717. [Google Scholar] [CrossRef]
- Harris, K.D.; Quiroga, R.Q.; Freeman, J.; Smith, S.L. Improving data quality in neuronal population recordings. Nat. Neurosci. 2016, 19, 1165–1174. [Google Scholar] [CrossRef]
- Barker-Haliski, M.L.; Johnson, K.; Billingsley, P.; Huff, J.; Handy, L.J.; Khaleel, R.; Lu, Z.; Mau, M.J.; Pruess, T.H.; Rueda, C.; et al. Validation of a Preclinical Drug Screening Platform for Pharmacoresistant Epilepsy. Neurochem. Res. 2017, 42, 1904–1918. [Google Scholar] [CrossRef]
- Shabestari, P.S.; Buccino, A.P.; Kumar, S.S.; Pedrocchi, A.; Hierlemann, A. A modulated template-matching approach to improve spike sorting of bursting neurons. IEEE Biomed. Circuits Syst. Conf. 2021, 2021, 9644995. [Google Scholar] [CrossRef]
- Müller, J.; Ballini, M.; Livi, P.; Chen, Y.; Radivojevic, M.; Shadmani, A.; Viswam, V.; Jones, I.L.; Fiscella, M.; Diggelmann, R.; et al. High-resolution CMOS MEA platform to study neurons at subcellular, cellular, and network levels. Lab Chip 2015, 15, 2767–2780. [Google Scholar] [CrossRef]
- Lonardoni, D.; Amin, H.; Zordan, S.; Boi, F.; Lecomte, A.; Angotzi, G.N.; Berdondini, L. Active High-Density Electrode Arrays: Technology and Applications in Neuronal Cell Cultures. Adv. Neurobiol. 2019, 22, 253–273. [Google Scholar] [CrossRef]
- Liu, Y.; Li, X.; Chen, J.; Yuan, C. Micro/Nano Electrode Array Sensors: Advances in Fabrication and Emerging Applications in Bioanalysis. Front. Chem. 2020, 8, 573865. [Google Scholar] [CrossRef]
- Kim, J.; Shin, H.; Kweon, S.-J.; Lee, S.; Ha, S.; Je, M. A Scalable Readout IC Based on Wideband Noise Cancelling for Full-Rate Scanning of High-Density Microelectrode Arrays. Annu. Int. Conf. IEEE Eng. Med. Biol. Soc. 2021, 2021, 7344–7347. [Google Scholar] [CrossRef]
- Vassallo, A.; Chiappalone, M.; Lopes, R.D.C.; Scelfo, B.; Novellino, A.; Defranchi, E.; Palosaari, T.; Weisschu, T.; Ramirez, T.; Martinoia, S.; et al. A multi-laboratory evaluation of microelectrode array-based measurements of neural network activity for acute neurotoxicity testing. NeuroToxicology 2016, 60, 280–292. [Google Scholar] [CrossRef]
- Maccione, A.; Garofalo, M.; Nieus, T.; Tedesco, M.; Berdondini, L.; Martinoia, S. Multiscale functional connectivity estimation on low-density neuronal cultures recorded by high-density CMOS Micro Electrode Arrays. J. Neurosci. Methods 2012, 207, 161–171. [Google Scholar] [CrossRef]
- Emmenegger, V.; Obien, M.E.J.; Franke, F.; Hierlemann, A. Technologies to Study Action Potential Propagation With a Focus on HD-MEAs. Front. Cell. Neurosci. 2019, 13, 159. [Google Scholar] [CrossRef]
- Buccino, A.P.; Yuan, X.; Emmenegger, V.; Xue, X.; Gänswein, T.; Hierlemann, A. An automated method for precise axon reconstruction from recordings of high-density micro-electrode arrays. J. Neural Eng. 2022, 19, 026026. [Google Scholar] [CrossRef]
- Steinmetz, N.A.; Aydin, C.; Lebedeva, A.; Okun, M.; Pachitariu, M.; Bauza, M.; Beau, M.; Bhagat, J.; Böhm, C.; Broux, M.; et al. Neuropixels 2.0: A miniaturized high-density probe for stable, long-term brain recordings. Science 2021, 372, eabf4588. [Google Scholar] [CrossRef]
- Ahfeldt, T.; Litterman, N.K.; Rubin, L.L. Studying human disease using human neurons. Brain Res. 2016, 1656, 40–48. [Google Scholar] [CrossRef]
- Ratner, M.H.; Farb, D.H. Probing the Neural Circuitry Targets of Neurotoxicants In Vivo through High Density Silicon Probe Brain Implants. Front. Toxicol. 2022, 4, 836427. [Google Scholar] [CrossRef]
- Broussard, G.; Liang, R.; Etian, L. Monitoring activity in neural circuits with genetically encoded indicators. Front. Mol. Neurosci. 2014, 7, 97. [Google Scholar] [CrossRef]
- Podor, B.; Hu, Y.-L.; Ohkura, M.; Nakai, J.; Croll, R.; Fine, A. Comparison of genetically encoded calcium indicators for monitoring action potentials in mammalian brain by two-photon excitation fluorescence microscopy. Neurophotonics 2015, 2, 021014. [Google Scholar] [CrossRef]
- Akerboom, J.; Chen, T.-W.; Wardill, T.; Tian, L.; Marvin, J.; Mutlu, S.; Calderón, N.C.; Esposti, F.; Borghuis, B.G.; Sun, X.R.; et al. Optimization of a GCaMP Calcium Indicator for Neural Activity Imaging. J. Neurosci. 2012, 32, 13819–13840. [Google Scholar] [CrossRef]
- Ohkura, M.; Sasaki, T.; Sadakari, J.; Gengyo-Ando, K.; Kagawa-Nagamura, Y.; Kobayashi, C.; Ikegaya, Y.; Nakai, J. Genetically Encoded Green Fluorescent Ca2+ Indicators with Improved Detectability for Neuronal Ca2+ Signals. PLoS ONE 2012, 7, e51286. [Google Scholar] [CrossRef]
- Chen, T.-W.; Wardill, T.J.; Sun, Y.; Pulver, S.R.; Renninger, S.L.; Baohan, A.; Schreiter, E.R.; Kerr, R.A.; Orger, M.B.; Jayaraman, V.; et al. Ultrasensitive fluorescent proteins for imaging neuronal activity. Nature 2013, 499, 295–300. [Google Scholar] [CrossRef]
- Xiao, D.; Vanni, M.P.; Mitelut, C.C.; Chan, A.W.; LeDue, J.M.; Xie, Y.; Chen, A.C.; Swindale, N.V.; Murphy, T.H. Mapping cortical mesoscopic networks of single spiking cortical or sub-cortical neurons. eLife 2017, 6, e19976. [Google Scholar] [CrossRef]
- Leopold, A.; Shcherbakova, D.M.; Verkhusha, V.V. Fluorescent Biosensors for Neurotransmission and Neuromodulation: Engineering and Applications. Front. Cell. Neurosci. 2019, 13, 474. [Google Scholar] [CrossRef]
- Wu, N.; Nishioka, W.K.; Derecki, N.C.; Maher, M.P. High-throughput-compatible assays using a genetically-encoded calcium indicator. Sci. Rep. 2019, 9, 12692. [Google Scholar] [CrossRef]
- Tian, L.; Hires, S.A.; Looger, L.L. Imaging Neuronal Activity with Genetically Encoded Calcium Indicators. Cold Spring Harb. Protoc. 2012, 2012, 647–656. [Google Scholar] [CrossRef]
- St-Pierre, F.; Chavarha, M.; Lin, M.Z. Designs and sensing mechanisms of genetically encoded fluorescent voltage indicators. Curr. Opin. Chem. Biol. 2015, 27, 31–38. [Google Scholar] [CrossRef]
- Lin, M.Z.; Schnitzer, M.J. Genetically encoded indicators of neuronal activity. Nat. Neurosci. 2016, 19, 1142–1153. [Google Scholar] [CrossRef]
- Boivin, B.; Roet, K.C.D.; Huang, X.; Karhohs, K.W.; Rohban, M.H.; Sandoe, J.; Wiskow, O.; Maeda, R.; Grantham, A.; Dornon, M.K.; et al. A multiparametric activity profiling platform for neuron disease phenotyping and drug screening. Mol. Biol. Cell 2022, 33, ar54. [Google Scholar] [CrossRef] [PubMed]
- Verschuuren, M.; Verstraelen, P.; Barriga, G.G.-D.; Cilissen, I.; Coninx, E.; Verslegers, M.; Larsen, P.H.; Nuydens, R.; De Vos, W.H. High-throughput microscopy exposes a pharmacological window in which dual leucine zipper kinase inhibition preserves neuronal network connectivity. Acta Neuropathol. Commun. 2019, 7, 93. [Google Scholar] [CrossRef]
- Van Dyck, M.; Mishra, R.K.; Pestana, F.; Verstraelen, P.; Lavreysen, H.; Pita-Almenar, J.D.; Kashikar, N.D.; De Vos, W.H. High-throughput Analysis of Synaptic Activity in Electrically Stimulated Neuronal Cultures. Neuroinformatics 2021, 19, 737–750. [Google Scholar] [CrossRef]
- Muto, A.; Ohkura, M.; Abe, G.; Nakai, J.; Kawakami, K. Real-Time Visualization of Neuronal Activity during Perception. Curr. Biol. 2013, 23, 307–311. [Google Scholar] [CrossRef]
- Walker, A.S.; Burrone, J.; Meyer, M.P. Functional imaging in the zebrafish retinotectal system using RGECO. Front. Neural Circuits 2013, 7, 34. [Google Scholar] [CrossRef]
- Zhang, T.; Peterson, R.T. Chapter 51—Zebrafish as a Platform for Drug Screening. In The Zebrafish in Biomedical Research; Cartner, S.C., Eisen, J.S., Farmer, S.C., Guillemin, K.J., Kent, M.L., Sanders, G.E., Eds.; Academic Press: Cambridge, MA, USA, 2020; pp. 659–675. [Google Scholar]
- Strange, K. Drug Discovery in Fish, Flies, and Worms. ILAR J. 2016, 57, 133–143. [Google Scholar] [CrossRef]
- Potekhina, E.S.; Bass, D.Y.; Kelmanson, I.V.; Fetisova, E.S.; Ivanenko, A.V.; Belousov, V.V.; Bilan, D.S. Drug Screening with Genetically Encoded Fluorescent Sensors: Today and Tomorrow. Int. J. Mol. Sci. 2020, 22, 148. [Google Scholar] [CrossRef]
- Kim, G.-H.J.; Mo, H.; Liu, H.; Okorie, M.; Chen, S.; Zheng, J.; Li, H.; Arkin, M.; Huang, B.; Guo, S. In Vivo Dopamine Neuron Imaging-Based Small Molecule Screen Identifies Novel Neuroprotective Compounds and Targets. Front. Pharmacol. 2022, 13, 837756. [Google Scholar] [CrossRef]
- Lin, X.; Duan, X.; Jacobs, C.; Ullmann, J.; Chan, C.-Y.; Chen, S.; Cheng, S.-H.; Zhao, W.-N.; Poduri, A.; Wang, X.; et al. High-throughput brain activity mapping and machine learning as a foundation for systems neuropharmacology. Nat. Commun. 2018, 9, 5142. [Google Scholar] [CrossRef]
- Kanyo, R.; Wang, C.K.; Locskai, L.F.; Li, J.; Allison, W.T.; Kurata, H.T. Functional and behavioral signatures of Kv7 activator drug subtypes. Epilepsia 2020, 61, 1678–1690. [Google Scholar] [CrossRef]
- Streit, A.K.; Fan, Y.N.; Masullo, L.; Baines, R.A. Calcium Imaging of Neuronal Activity in Drosophila Can Identify Anticonvulsive Compounds. PLoS ONE 2016, 11, e0148461. [Google Scholar] [CrossRef]
- Xue, Y. Computational optics for high-throughput imaging of neural activity. Neurophotonics 2022, 9, 041408. [Google Scholar] [CrossRef]
- Steinmetz, N.A.; Buetfering, C.; Lecoq, J.; Lee, C.R.; Peters, A.J.; Jacobs, E.; Coen, P.; Ollerenshaw, D.R.; Valley, M.T.; de Vries, S.; et al. Aberrant Cortical Activity in Multiple GCaMP6-Expressing Transgenic Mouse Lines. eNeuro 2017, 4. [Google Scholar] [CrossRef]
- Larsch, J.; Ventimiglia, D.; Bargmann, C.I.; Albrecht, D.R. High-throughput imaging of neuronal activity in Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA 2013, 110, E4266–E4273. [Google Scholar] [CrossRef]
- Chen, Z.; Truong, T.M.; Ai, H.-W. Illuminating Brain Activities with Fluorescent Protein-Based Biosensors. Chemosensors 2017, 5, 32. [Google Scholar] [CrossRef] [PubMed]
- Nakajima, R.; Jung, A.; Yoon, B.-J.; Baker, B.J. Optogenetic Monitoring of Synaptic Activity with Genetically Encoded Voltage Indicators. Front. Synaptic Neurosci. 2016, 8, 22. [Google Scholar] [CrossRef] [PubMed]
- Guzowski, J.F.; McNaughton, B.L.; Barnes, C.A.; Worley, P.F. Environment-specific expression of the immediate-early gene Arc in hippocampal neuronal ensembles. Nat. Neurosci. 1999, 2, 1120–1124. [Google Scholar] [CrossRef]
- Tyssowski, K.; DeStefino, N.R.; Cho, J.-H.; Dunn, C.J.; Poston, R.G.; Carty, C.E.; Jones, R.D.; Chang, S.M.; Romeo, P.; Wurzelmann, M.K.; et al. Different Neuronal Activity Patterns Induce Different Gene Expression Programs. Neuron 2018, 98, 530–546.e11. [Google Scholar] [CrossRef] [PubMed]
- Daberkow, D.P.; Riedy, M.D.; Kesner, R.P.; Keefe, K.A. Arc mRNA induction in striatal efferent neurons associated with response learning. Eur. J. Neurosci. 2007, 26, 228–241. [Google Scholar] [CrossRef]
- Ivanova, T.; Matthews, A.; Gross, C.; Mappus, R.; Gollnick, C.; Swanson, A.; Bassell, G.; Liu, R. Arc/Arg3.1 mRNA expression reveals a subcellular trace of prior sound exposure in adult primary auditory cortex. Neuroscience 2011, 181, 117–126. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Sørensen, A.T.; Cooper, Y.A.; Baratta, M.V.; Weng, F.-J.; Zhang, Y.; Ramamoorthi, K.; Fropf, R.; LaVerriere, E.; Xue, J.; Young, A.; et al. A robust activity marking system for exploring active neuronal ensembles. eLife 2016, 5, e13918. [Google Scholar] [CrossRef]
- Kawashima, T.; Kitamura, K.; Suzuki, K.; Nonaka, M.; Kamijo, S.; Takemoto-Kimura, S.; Kano, M.; Okuno, H.; Ohki, K.; Bito, H. Functional labeling of neurons and their projections using the synthetic activity–dependent promoter E-SARE. Nat. Methods 2013, 10, 889–895. [Google Scholar] [CrossRef]
- Shao, N.; Bock, R. A codon-optimized luciferase from Gaussia princeps facilitates the in vivo monitoring of gene expression in the model alga Chlamydomonas reinhardtii. Curr. Genet. 2008, 53, 381–388. [Google Scholar] [CrossRef]
- Badr, C.E.; Niers, J.M.; Tjon-Kon-Fat, L.-A.; Noske, D.P.; Wurdinger, T.; Tannous, B.A. Real-time monitoring of nuclear factor kappaB activity in cultured cells and in animal models. Mol. Imaging 2009, 8, 278–290. [Google Scholar] [CrossRef]
- Na, Y.; Park, S.; Lee, C.; Kim, D.-K.; Park, J.M.; Sockanathan, S.; Huganir, R.L.; Worley, P.F. Real-Time Imaging Reveals Properties of Glutamate-Induced Arc/Arg 3.1 Translation in Neuronal Dendrites. Neuron 2016, 91, 561–573. [Google Scholar] [CrossRef] [PubMed]
- Inoue, M.; Yagishita-Kyo, N.; Nonaka, M.; Kawashima, T.; Okuno, H.; Bito, H. Synaptic Activity Responsive Element (SARE). Commun. Integr. Biol. 2010, 3, 443–446. [Google Scholar] [CrossRef] [PubMed]
- Sheng, H.Z.; Fields, R.D.; Nelson, P.G. Specific regulation of immediate early genes by patterned neuronal activity. J. Neurosci. Res. 1993, 35, 459–467. [Google Scholar] [CrossRef] [PubMed]
- Neumann-Haefelin, T.; Wießner, C.; Vogel, P.; Back, T.; Hossmann, K.-A. Differential Expression of the Immediate Early Genes c-Fos, c-Jun, Jun B, and NGFI-B in the Rat Brain following Transient Forebrain Ischemia. J. Cereb. Blood Flow Metab. 1994, 14, 206–216. [Google Scholar] [CrossRef]
| Patch-Clamp Recordings | Microelectrode Arrays (MEA) | Genetically Encoded Calcium Indicators | Genetically Encoded Neurotransmitter Indicators | Genetically Encoded Voltage Indicators | SNAR | |
|---|---|---|---|---|---|---|
| Scalability | Low | MEA: medium-high High density-MEA: Low | Medium-high: automation of imaging: 96 wells possible | Medium-high: automation of imaging: 96 wells possible | Low: need to optimize each tool for each new screen | Very high, the whole assay can be automated |
| Dynamic Range, Signal to noise ratio | Very high | Very high | High, continue increasing | Glutamate and dopamine: High Others: low-medium | Low | Very high: luciferases are linearly quantitative |
| Biological disruption | Very high: cells usually die afterward | Minimal, more from high density | Low, some cytotoxic effects | Low | Low | Very low: the reporter is secreted |
| Longitudinal | Low: see above | Extremely high if culture survives plating on electrodes | Medium, some care needs to be taken comparing between days | Medium, some care needs to be taken comparing between days | Medium | Extremely high: can be followed for hours, days, or weeks |
| Simplicity | Skill, time, and equipment intensive | Complicated to manufacture | Variable: automated imaging, optimization of indicator | Variable: automated imaging, optimization of indicator | Complicated: tools are being optimized | Easy to use, simple lab equipment, easy controls, and optimization |
| Temporal resolution | <1 ms | <1 ms | 200–800 ms | 10–800 ms | <1 ms–200 ms | 30 min |
| Spatial scale | Whole-cell to axon | Network to synapse | Brain to synapse | Brain to synapse | Brain to synapse | Network |
| Computational requirements | Low-medium with established software | High: spike sorting and noise deconvolution: new technology | Medium-high depending on screen | Medium-high depending on screen | Medium-high depending on screen | Very low |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sterin, I.; Santos, A.C.; Park, S. Neuronal Activity Reporters as Drug Screening Platforms. Micromachines 2022, 13, 1500. https://doi.org/10.3390/mi13091500
Sterin I, Santos AC, Park S. Neuronal Activity Reporters as Drug Screening Platforms. Micromachines. 2022; 13(9):1500. https://doi.org/10.3390/mi13091500
Chicago/Turabian StyleSterin, Igal, Ana C. Santos, and Sungjin Park. 2022. "Neuronal Activity Reporters as Drug Screening Platforms" Micromachines 13, no. 9: 1500. https://doi.org/10.3390/mi13091500
APA StyleSterin, I., Santos, A. C., & Park, S. (2022). Neuronal Activity Reporters as Drug Screening Platforms. Micromachines, 13(9), 1500. https://doi.org/10.3390/mi13091500






