Monolithically 3D-Printed Microfluidics with Embedded µTesla Pump
Abstract
1. Introduction
1.1. Benchtop Microfluidic Pumps
1.2. On-Chip Integrated Pumps
1.3. Tesla Turbine Miniature Pumps
1.4. 3D Printed Microfluidics
2. Materials and Methods
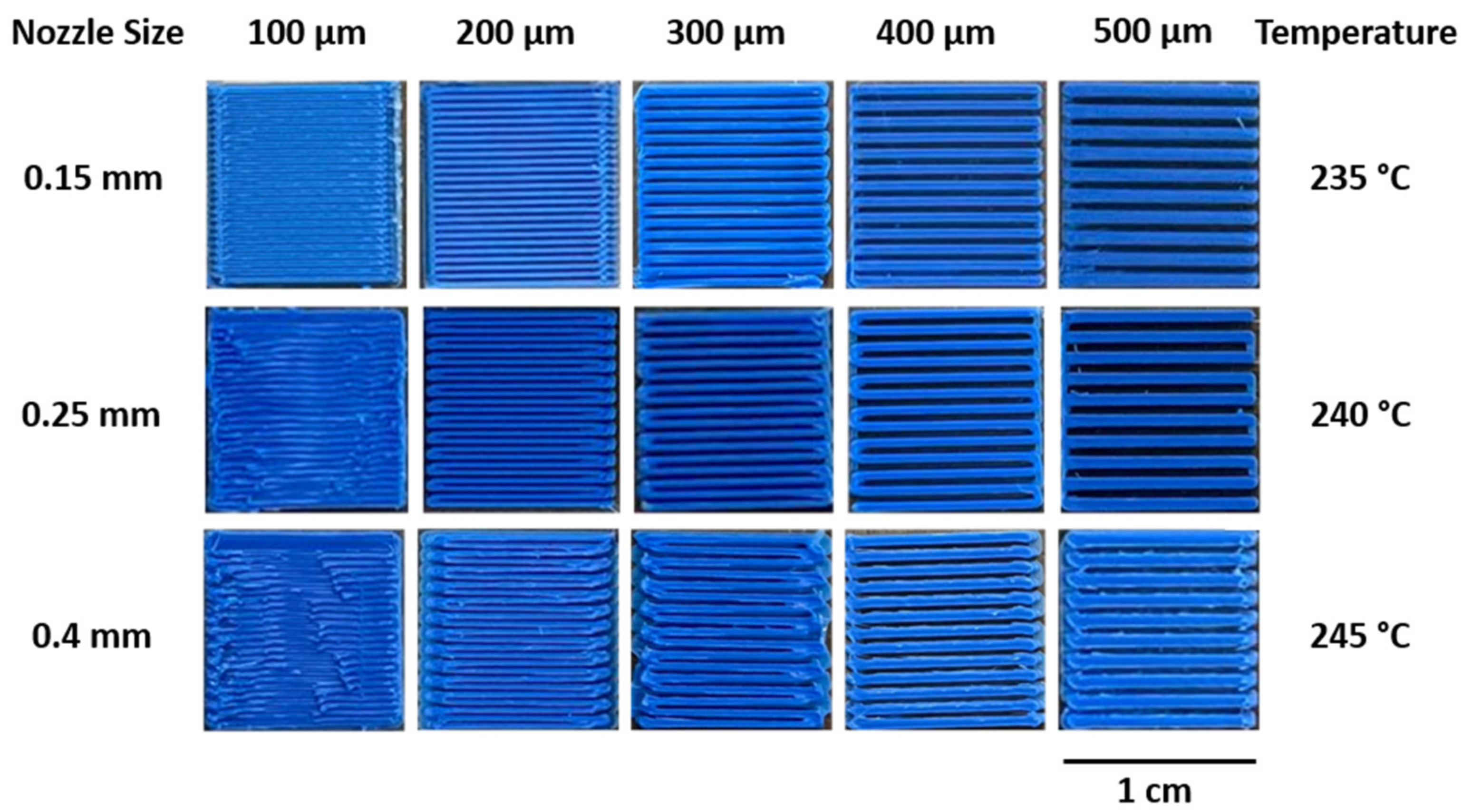
2.1. 3D Printing Optimization
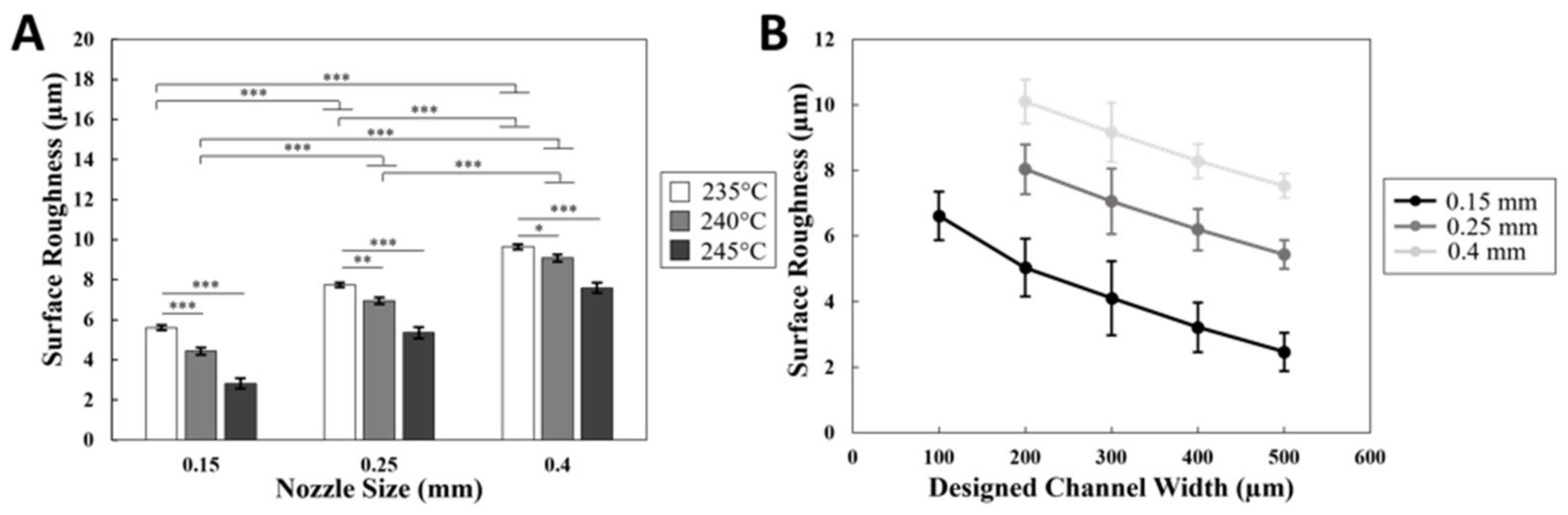
2.2. 3D printing Roughness Measurement
2.3. μTesla Pump Fabrication and Sterilization
2.4. Cell Culture Device Fabrication
2.5. Particle Velocimetry Analysis
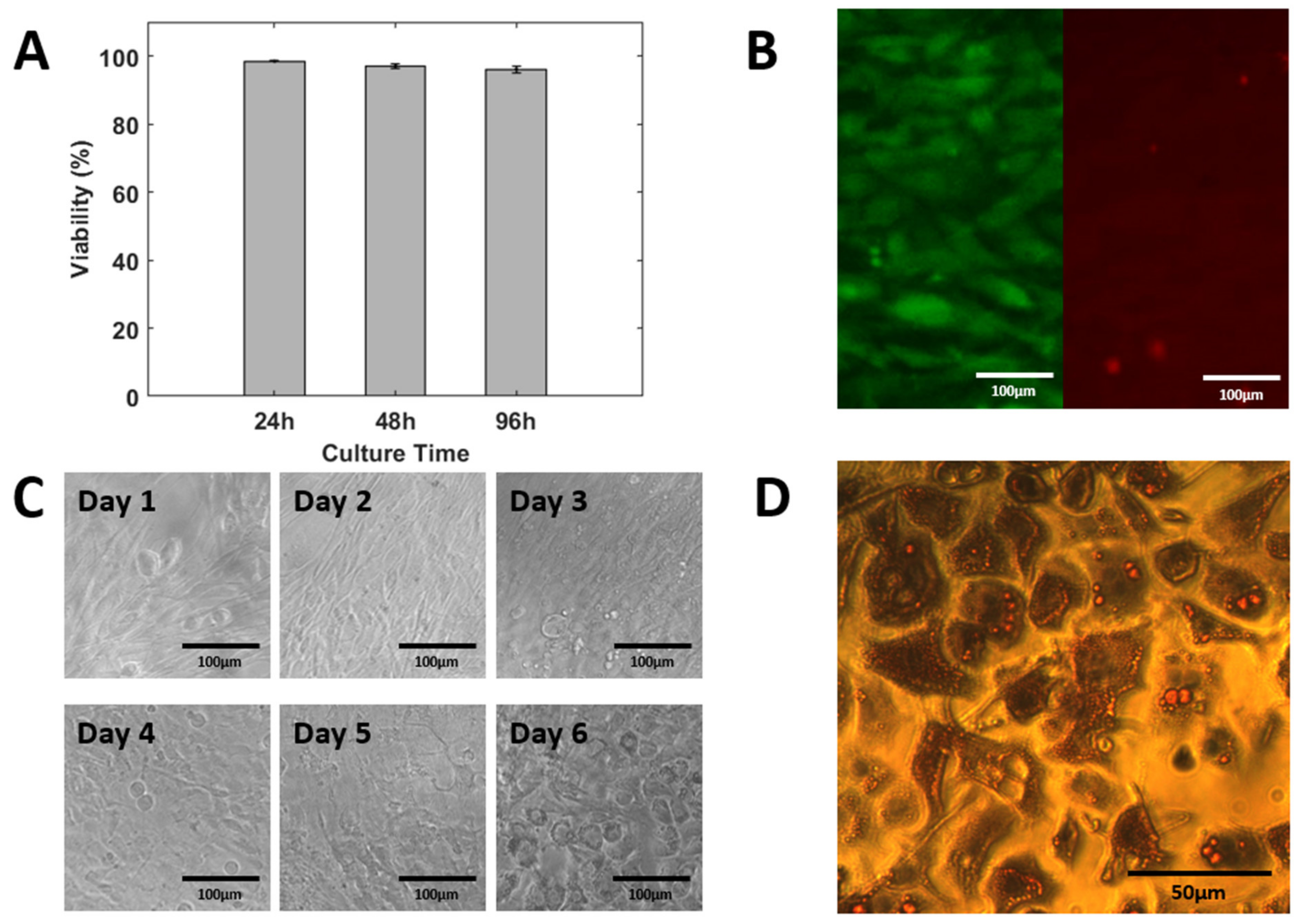
2.6. 3T3 L1 On-Chip Culture and Differentiation
2.7. Adipocytes Staining
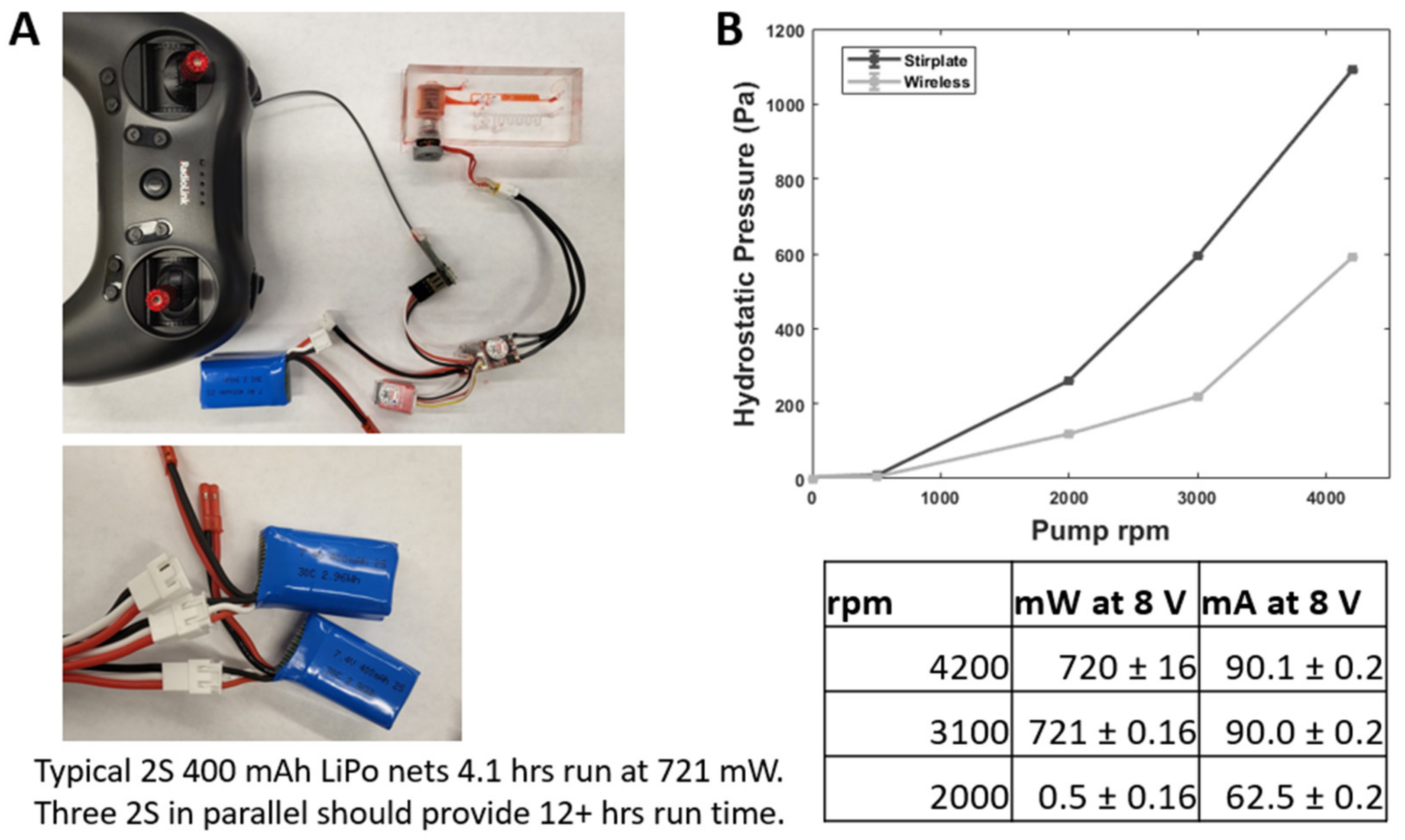
2.8. Wireless µTesla Pump Operations
2.9. Statistical Analysis
3. Results and Discussion
3.1. Optimizing FDM 3D Printing for Integrated Microfluidics
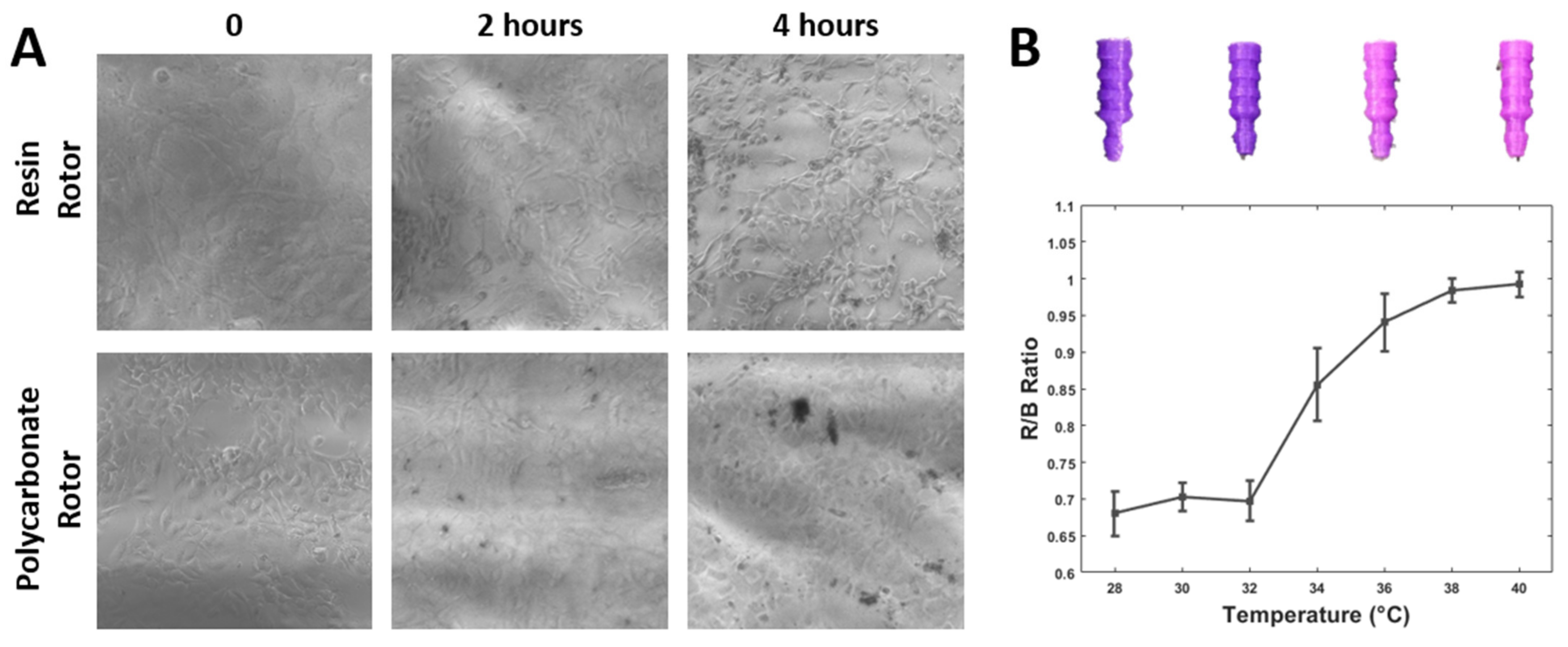
3.2. 3D Printing Material Compatibility for Cell Culture
3.3. Temperature-Sensitive Microfluidic Plugs
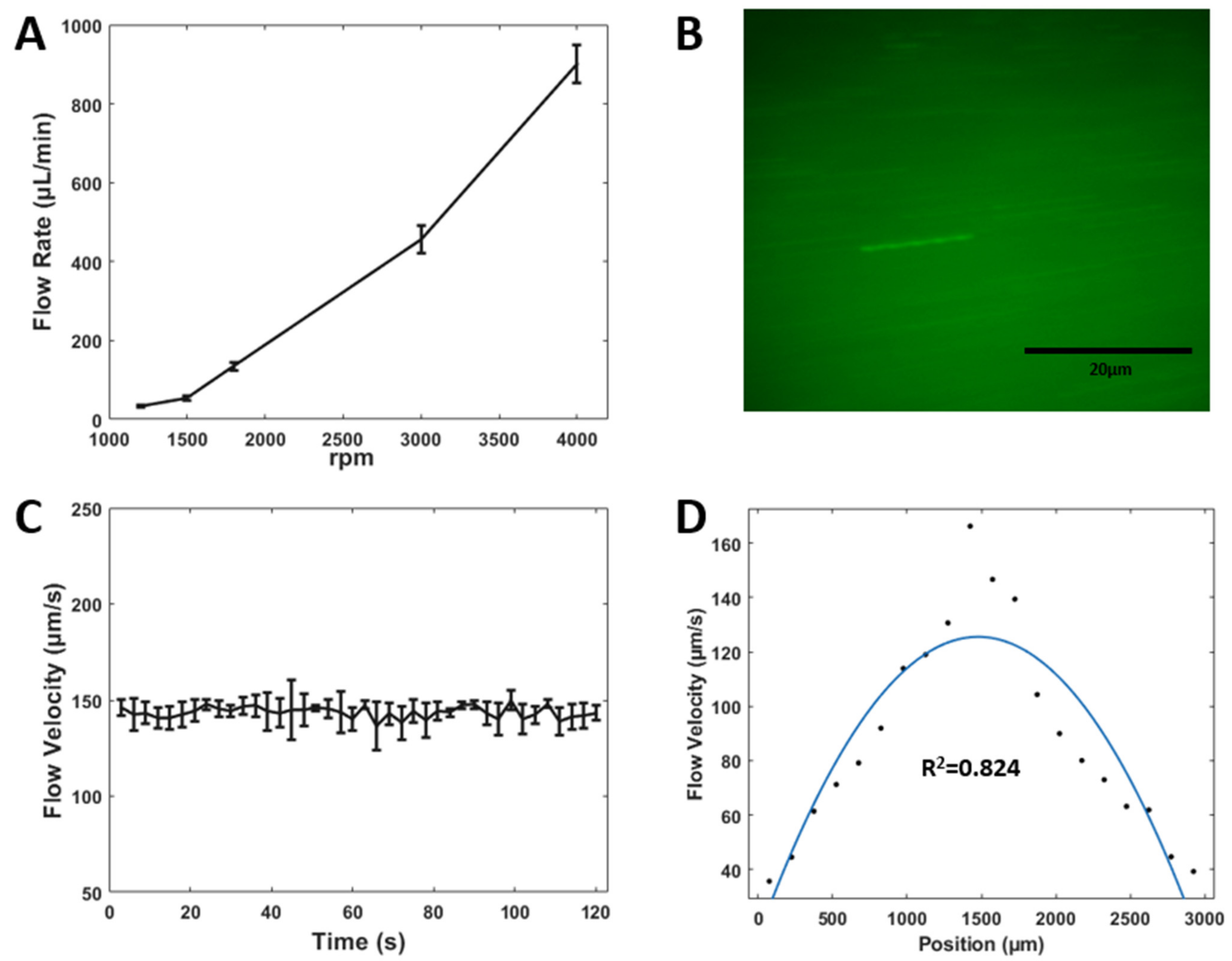
3.4. Flow Rate and Particle Velocimetry Analysis
3.5. On-Chip Cell Culturing and Differentiation
3.6. Wireless Operations
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Byun, C.K.; Abi-Samra, K.; Cho, Y.-K.; Takayama, S. Pumps for microfluidic cell culture. Electrophoresis 2014, 35, 245–257. [Google Scholar] [CrossRef] [PubMed]
- Sung, J.H.; Kam, C.; Shuler, M.L. A microfluidic device for a pharmacokinetic–pharmacodynamic (PK–PD) model on a chip. Lab A Chip 2010, 10, 446–455. [Google Scholar] [CrossRef] [PubMed]
- Beebe, D.J.; Mensing, G.A.; Walker, G.M. Physics and Applications of Microfluidics in Biology. Annu. Rev. Biomed. Eng. 2002, 4, 261–286. [Google Scholar] [CrossRef]
- Hung, P.J.; Lee, P.J.; Sabounchi, P.; Lin, R.; Lee, L.P. Continuous perfusion microfluidic cell culture array for high-throughput cell-based assays. Biotechnol. Bioeng. 2005, 89, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Zeng, W.; Li, S.; Wang, Z. Characterization of syringe-pump-driven versus pressure-driven microfluidic flows. In 2015 International Conference on Fluid Power and Mechatronics (FPM), Harbin, China, 5–7 August 2015; IEEE: Piscataway, NJ, USA, 2015; pp. 711–715. [Google Scholar]
- Schneider, S.; Bubeck, M.; Rogal, J.; Weener, H.J.; Rojas, C.; Weiss, M.; Heymann, M.; van der Meer, A.D.; Loskill, P. Peristaltic on-chip pump for tunable media circulation and whole blood perfusion in PDMS-free organ-on-chip and Organ-Disc systems. Lab A Chip 2021, 21, 3963–3978. [Google Scholar] [CrossRef]
- Skafte-Pedersen, P.; Sabourin, D.; Dufva, M.; Snakenborg, D. Multi-channel peristaltic pump for microfluidic ap-plications featuring monolithic PDMS inlay. Lab A Chip 2009, 9, 3003–3006. [Google Scholar] [CrossRef]
- Ren, Y.; Chow, L.M.-C.; Leung, W.W.-F. Cell culture using centrifugal microfluidic platform with demonstration on Pichia pastoris. Biomed. Microdevices 2013, 15, 321–337. [Google Scholar] [CrossRef]
- Du, X.Y.; Fu, Y.Q.; Luo, J.K.; Flewitt, A.; Milne, W.I. Microfluidic pumps employing surface acoustic waves generated in ZnO thin films. J. Appl. Phys. 2009, 105, 024508. [Google Scholar] [CrossRef]
- Gallas, Q.; Holman, R.; Nishida, T.; Carroll, B.; Sheplak, M.; Cattafesta, L. Lumped element modeling of piezoelec-tric-driven synthetic jet actuators. AIAA J. 2003, 41, 240–247. [Google Scholar] [CrossRef]
- Urbanski, J.P.; Levitan, J.A.; Burch, D.N.; Thorsen, T.; Bazant, M.Z. The effect of step height on the performance of three-dimensional ac electro-osmotic microfluidic pumps. J. Colloid Interface Sci. 2007, 309, 332–341. [Google Scholar] [CrossRef]
- Munyan, J.W.; Fuentes, H.V.; Draper, M.; Kelly, R.T.; Woolley, A.T. Electrically actuated, pressure-driven micro-fluidic pumps. Lab A Chip 2003, 3, 217–220. [Google Scholar] [CrossRef]
- Steigert, J.; Brenner, T.; Grumann, M.; Riegger, L.; Lutz, S.; Zengerle, R.; Ducrée, J. Integrated siphon-based metering and sedimentation of whole blood on a hydrophilic lab-on-a-disk. Biomed. Microdevices 2007, 9, 675–679. [Google Scholar] [CrossRef]
- Hayes, B.; Smith, L.; Kabutz, H.; Hayes, A.C.; Whiting, G.L.; Jayaram, K.; MacCurdy, R. Rapid Fabrication of Low-Cost Thermal Bubble-Driven Micro-Pumps. Micromachines 2022, 13, 1634. [Google Scholar] [CrossRef]
- Park, J.; Park, J.K. Finger-actuated microfluidic display for smart blood typing. Anal. Chem. 2019, 91, 11636–11642. [Google Scholar] [CrossRef]
- Armstrong, J.H. An Investigation of the Performance of a Modified Tesla Turbine. Ph.D. Thesis, Georgia Institute of Technology, Atlanta, GA, USA, 1952. [Google Scholar]
- Rice, W. An analytical and experimental investigation of multiple-disk turbines. J. Eng. Power. 1956, 87, 29–36. [Google Scholar] [CrossRef]
- Couto, H.S.; Duarte, J.B.F.; Bastos-Netto, D. The Tesla turbine revisited. In Proceedings of the Eighth Asia-Pacific International Symposium on Combustion and Energy Utilization, Sochi, Russia, 10–12 October 2006. [Google Scholar]
- Lemma, E.; Deam, R.T.; Toncich, D.; Collins, R. Characterization of a small viscous flow turbine. Exp. Therm. Fluid Sci. 2008, 33, 96–105. [Google Scholar] [CrossRef]
- Engin, T.; Özdemir, M.; Çeşmeci, Ş. Design, testing and two-dimensional flow modeling of a multiple-disk fan. Exp. Therm. Fluid Sci. 2009, 33, 1180–1187. [Google Scholar] [CrossRef]
- Talluri, L.; Fiaschi, D.; Neri, G.; Ciappi, L. Design and optimization of a Tesla turbine for ORC applications. Appl. Energy 2018, 226, 300–319. [Google Scholar] [CrossRef]
- Rusin, K.; Wróblewski, W.; Strozik, M. Comparison of methods for the determination of Tesla turbine performance. J. Theor. Appl. Mech. 2019, 57, 563–575. [Google Scholar] [CrossRef]
- Habhab, M.-B.; Ismail, T.; Lo, J.F. A Laminar Flow-Based Microfluidic Tesla Pump via Lithography Enabled 3D Printing. Sensors 2016, 16, 1970. [Google Scholar] [CrossRef]
- Melchels, F.P.W.; Feijen, J.; Grijpma, D.W. A review on stereolithography and its applications in biomedical engineering. Biomaterials 2010, 31, 6121–6130. [Google Scholar] [CrossRef] [PubMed]
- Gong, H.; Bickham, B.P.; Woolley, A.T.; Nordin, G.P. Custom 3D printer and resin for 18 μm × 20 μm microfluidic flow channels. Lab A Chip 2017, 17, 2899–2909. [Google Scholar] [CrossRef] [PubMed]
- Warr, C.; Hinnen, H.; Avery, S.; Cate, R.; Nordin, G.; Pitt, W. 3D-Printed Microfluidic Droplet Generator with Hydrophilic and Hydrophobic Polymers. Micromachines 2021, 12, 91. [Google Scholar] [CrossRef] [PubMed]
- Monaghan, T.; Harding, M.J.; Harris, R.A.; Friel, R.J.; Christie, S.D.R. Customizable 3D printed microfluidics for integrated analysis and optimization. Lab A Chip 2016, 16, 3362–3373. [Google Scholar] [CrossRef]
- He, Y.; Wu, Y.; Fu, J.-Z.; Gao, Q.; Qiu, J.-J. Developments of 3D Printing Microfluidics and Applications in Chemistry and Biology: A Review. Electroanalysis 2016, 28, 1658–1678. [Google Scholar] [CrossRef]
- Ji, Q.; Zhang, J.M.; Liu, Y.; Li, X.; Lv, P.; Jin, D.; Duan, H. A Modular Microfluidic Device via Multimaterial 3D Printing for Emulsion Generation. Sci. Rep. 2018, 8, 4791. [Google Scholar] [CrossRef]
- Eggleton, B.J.; Palomba, S.; Zhu, F.; Skommer, J.; Friedrich, T.; Kaslin, J.; Wlodkowic, D. 3D printed polymers toxicity profiling: A caution for biodevice applications. In Proceedings of the Micro + Nano Materials, Devices, and Systems, Sydney, Australia, 6–9 December 2015; Volume 9668, pp. 82–88. [Google Scholar] [CrossRef]
- De Almeida Monteiro Melo Ferraz, M.; Nagashima, J.B.; Venzac, B.; Le Gac, S.; Songsasen, N. 3D printed mold leachates in PDMS microfluidic devices. Sci. Rep. 2020, 10, 994. [Google Scholar] [CrossRef]
- Oskui, S.M.; Diamante, G.; Liao, C.; Shi, W.; Gan, J.; Schlenk, D.; Grover, W.H. Assessing and Reducing the Toxicity of 3D-Printed Parts. Environ. Sci. Technol. Lett. 2015, 3, 1–6. [Google Scholar] [CrossRef]
- Musgrove, H.; Catterton, M.; Pompano, R. Applied tutorial for the design and fabrication of biomicrofluidic devices by resin 3D printing. Anal. Chim. Acta 2022, 1209, 339842. [Google Scholar] [CrossRef]
- Lim, K.S.; Levato, R.; Costa, P.F.; Castilho, M.D.; Alcala-Orozco, C.R.; A van Dorenmalen, K.M.; Melchels, F.P.W.; Gawlitta, D.; Hooper, G.J.; Malda, J.; et al. Bio-resin for high resolution lithography-based biofabrication of complex cell-laden constructs. Biofabrication 2018, 10, 034101. [Google Scholar] [CrossRef]
- Nielsen, A.V.; Beauchamp, M.J.; Nordin, G.P.; Woolley, A.T. 3D printed microfluidics. Annu. Rev. Anal. Chem. 2020, 13, 45. [Google Scholar] [CrossRef]
- Swetham, T.; Reddy, K.M.M.; Huggi, A.; Kumar, M.N. A Critical Review on of 3D Printing Materials and Details of Materials used in FDM. Int. J. Sci. Res. Sci. Eng. 2017, 3, 353–361. [Google Scholar]
- Lee, K.G.; Park, K.J.; Seok, S.; Shin, S.; Park, J.Y.; Heo, Y.S.; Lee, T.J. 3D printed modules for integrated micro-fluidic devices. Rsc Adv. 2014, 4, 32876–32880. [Google Scholar] [CrossRef]
- Nelson, M.D.; Ramkumar, N.; Gale, B.K. Flexible, transparent, sub-100 µm microfluidic channels with fused dep-osition modeling 3D-printed thermoplastic polyurethane. J. Micromech. Microeng. 2019, 29, 095010. [Google Scholar] [CrossRef]
- Gottenbos, B.; Grijpma, D.W.; Van Der Mei, H.C.; Feijen, J.; Busscher, H.J. Antimicrobial effects of positively charged surfaces on adhering Gram-positive and Gram-negative bacteria. J. Antimicrob. Chemother. 2001, 48, 7–13. [Google Scholar] [CrossRef]
- Lu, H.; Guo, L.; Kawazoe, N.; Tateishi, T.; Chen, G. Effects of Poly(L-lysine), Poly(acrylic acid) and Poly(ethylene glycol) on the Adhesion, Proliferation and Chondrogenic Differentiation of Human Mesenchymal Stem Cells. J. Biomater. Sci. Polym. Ed. 2009, 20, 577–589. [Google Scholar] [CrossRef]
- Zebisch, K.; Voigt, V.; Wabitsch, M.; Brandsch, M. Protocol for effective differentiation of 3T3-L1 cells to adipocytes. Anal. Biochem. 2012, 425, 88–90. [Google Scholar] [CrossRef]
- Ramirez-Zacarias, J.L.; Castro-Munozledo, F.; Kuri-Harcuch, W. Quantitation of adipose conversion and triglyc-erides by staining intracytoplasmic lipids with Oil Red O. Histochemistry 1992, 97, 493–497. [Google Scholar] [CrossRef]
- Burke, C.; Dalal, A.; Abukhalaf, A.; Noorani, R. Effects of process parameter variation on the surface roughness of polylactic acid (PLA) materials using design of experiments (DOE). IOP Conf. Ser. Mater. Sci. Eng. 2020, 897, 012003. [Google Scholar] [CrossRef]
- Chaidas, D.; Kitsakis, K.; Kechagias, J.; Maropoulos, S. The impact of temperature changing on surface roughness of FFF process. IOP Conf. Ser. Mater. Sci. Eng. 2016, 161, 012033. [Google Scholar] [CrossRef]
- Werner, E.M.; Lam, B.X.; Hui, E.E. Phase-Optimized Peristaltic Pumping by Integrated Microfluidic Logic. Micromachines 2022, 13, 1784. [Google Scholar] [CrossRef] [PubMed]
- Cooney, C.G.; Sipes, D.; Thakore, N.; Holmberg, R.; Belgrader, P. A plastic, disposable microfluidic flow cell for coupled on-chip PCR and microarray detection of infectious agents. Biomed. Microdevices 2011, 14, 45–53. [Google Scholar] [CrossRef] [PubMed]
- Dosh, R.H.; Jordan-Mahy, N.; Sammon, C.; Le Maitre, C.L. Long-term in vitro 3D hydrogel co-culture model of inflammatory bowel disease. Sci. Rep. 2019, 9, 1812. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Duan, K.; Orabi, M.; Warchock, A.; Al-Akraa, Z.; Ajami, Z.; Chun, T.-H.; Lo, J.F. Monolithically 3D-Printed Microfluidics with Embedded µTesla Pump. Micromachines 2023, 14, 237. https://doi.org/10.3390/mi14020237
Duan K, Orabi M, Warchock A, Al-Akraa Z, Ajami Z, Chun T-H, Lo JF. Monolithically 3D-Printed Microfluidics with Embedded µTesla Pump. Micromachines. 2023; 14(2):237. https://doi.org/10.3390/mi14020237
Chicago/Turabian StyleDuan, Kai, Mohamad Orabi, Alexus Warchock, Zaynab Al-Akraa, Zeinab Ajami, Tae-Hwa Chun, and Joe F. Lo. 2023. "Monolithically 3D-Printed Microfluidics with Embedded µTesla Pump" Micromachines 14, no. 2: 237. https://doi.org/10.3390/mi14020237
APA StyleDuan, K., Orabi, M., Warchock, A., Al-Akraa, Z., Ajami, Z., Chun, T.-H., & Lo, J. F. (2023). Monolithically 3D-Printed Microfluidics with Embedded µTesla Pump. Micromachines, 14(2), 237. https://doi.org/10.3390/mi14020237







