3D Bioprinting of an Endothelialized Liver Lobule-like Construct as a Tumor-Scale Drug Screening Platform
Abstract
1. Introduction
2. Materials and Methods
2.1. Bioprinting Platform
2.2. Bioprinting of Lobule-Like Structures
2.3. SEM Characterization of GelMA Hydrogel and Porosity Analysis
2.4. Mechanical Testing
2.5. Cell Culturing
2.6. Fabrication of Liver Lobule-Like Constructs
2.7. Fabrication of Endothelialized Liver Lobule-Like Constructs
2.8. Evaluation of Cell Viability and Proliferation
2.9. Cell Morphology Analysis
2.10. Drug Treatment
2.11. Statistical Analysis
3. Results and Discussion
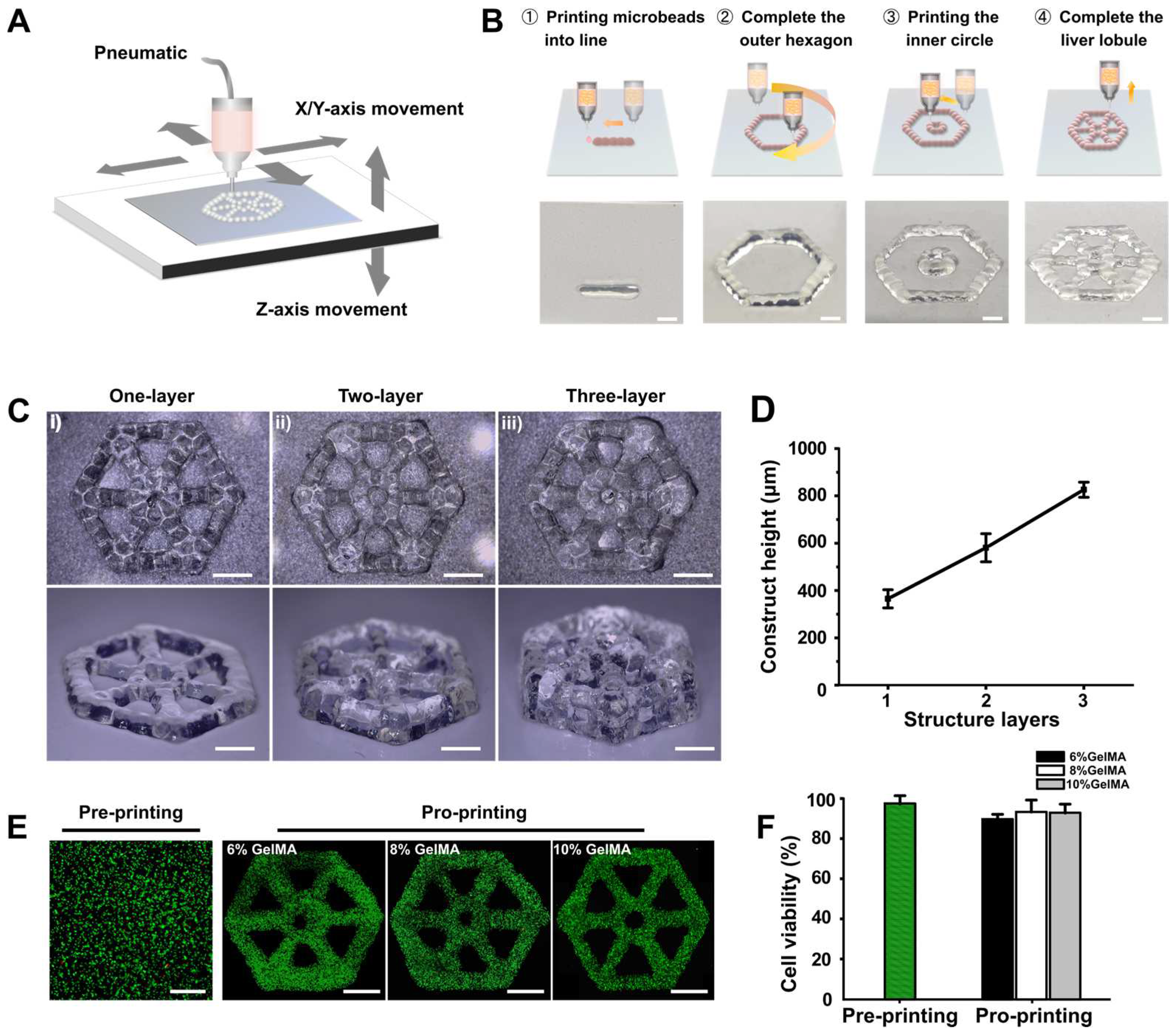
3.1. Patterning Hydrogel Microbeads to Engineer Lobule-Like Structures
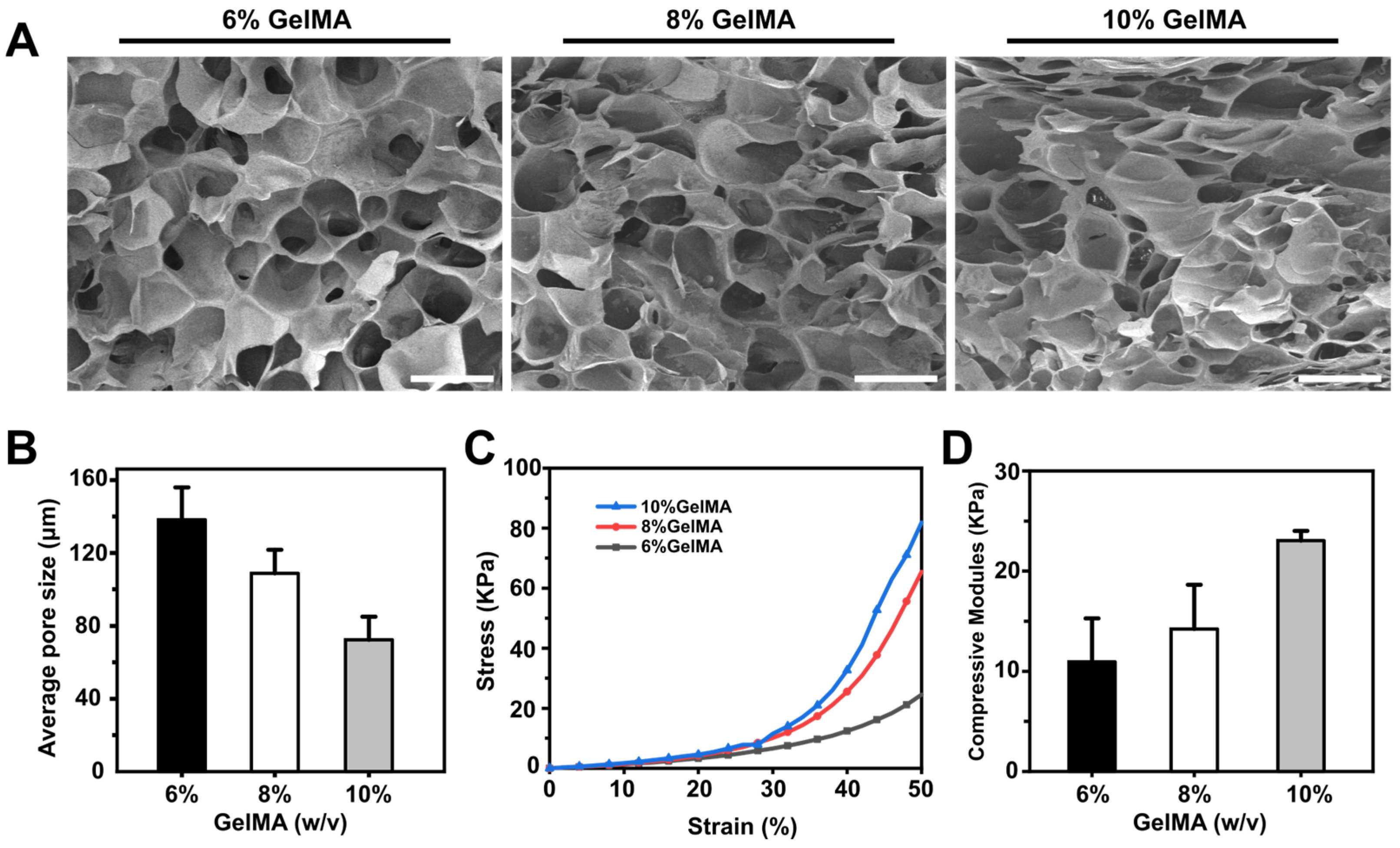
3.2. GelMA Hydrogel Characterization
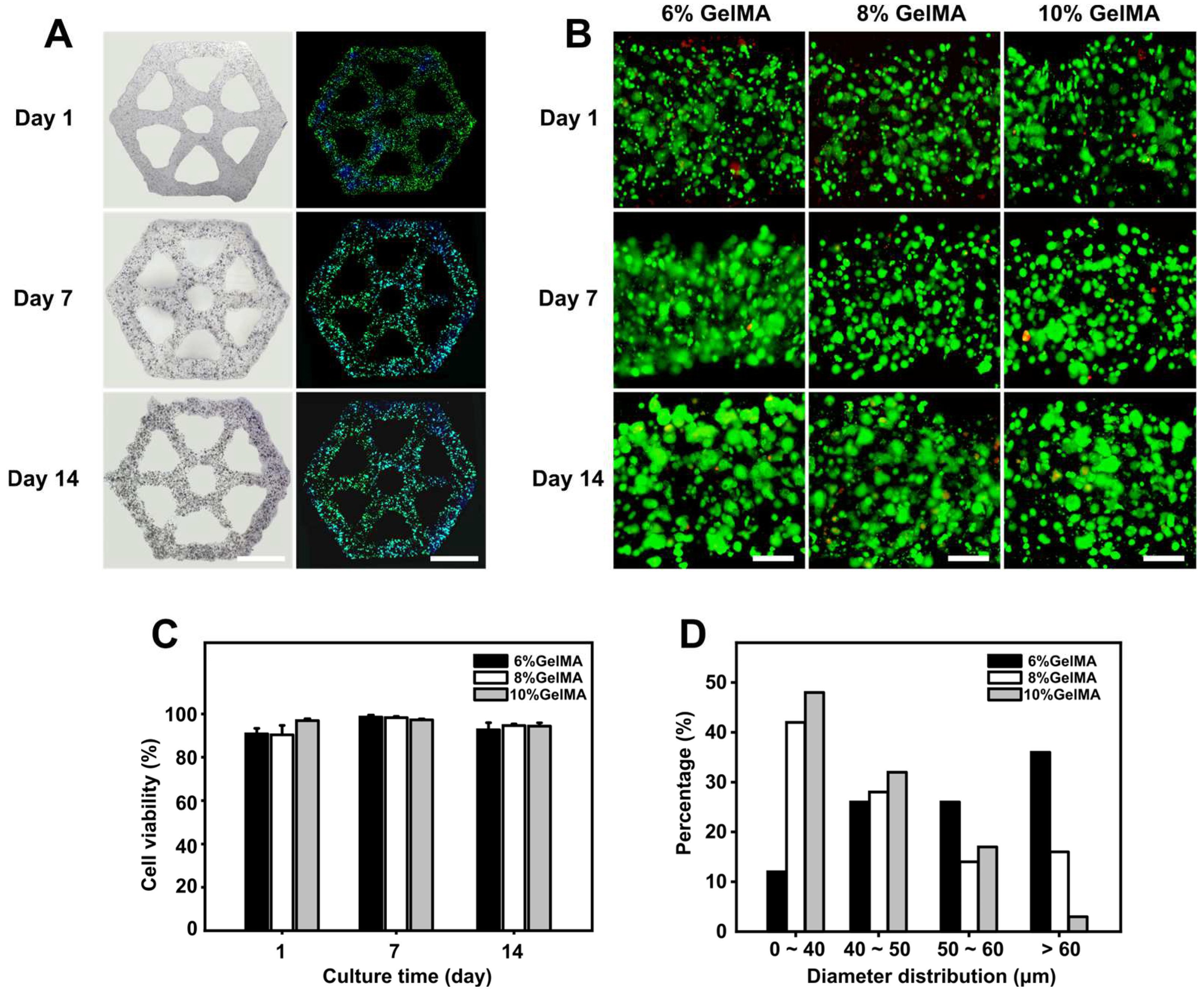
3.3. Liver Lobule-Like Construct Fabrication and Culture
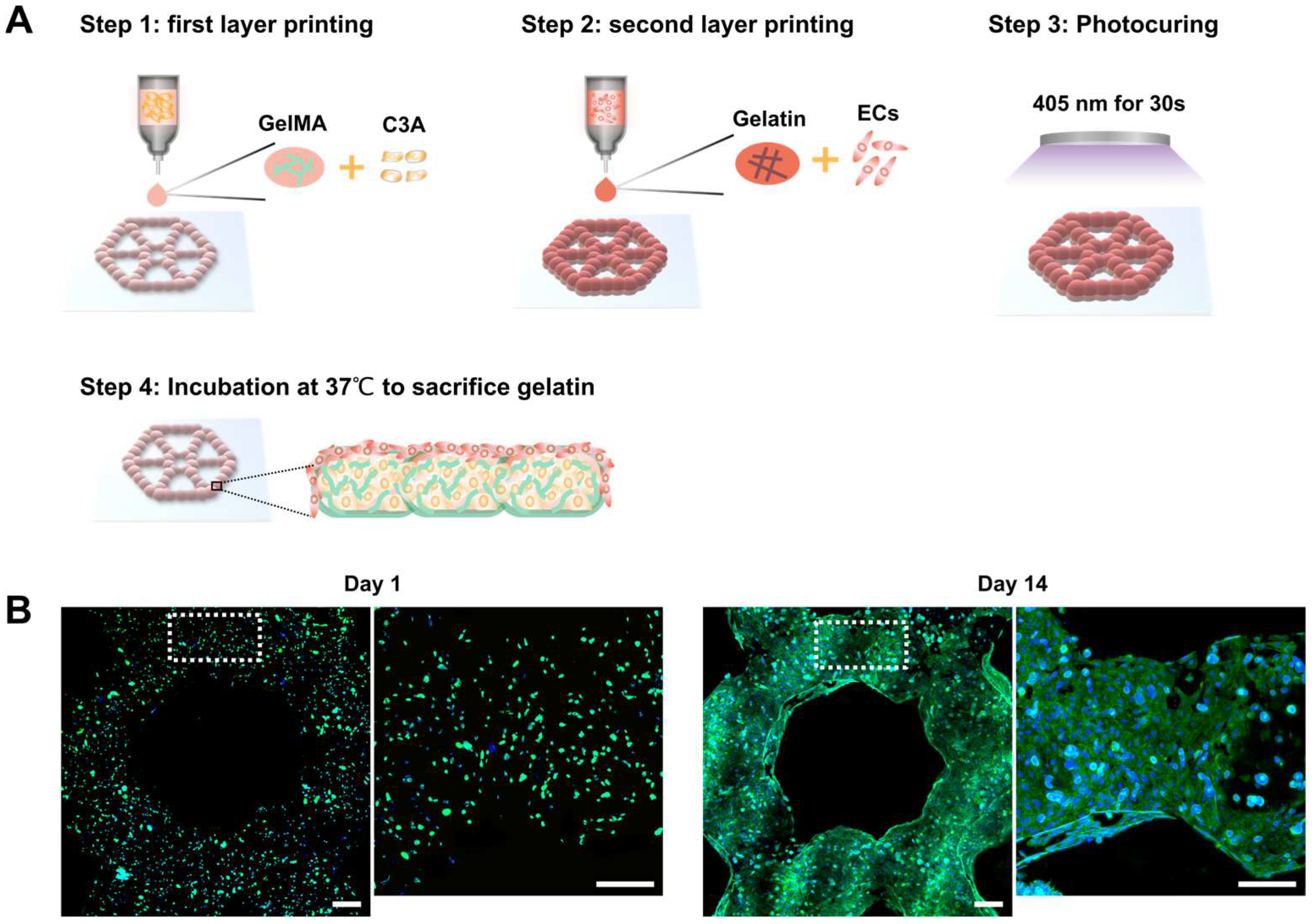
3.4. Development of Endothelialized Liver Lobule-Like Constructs
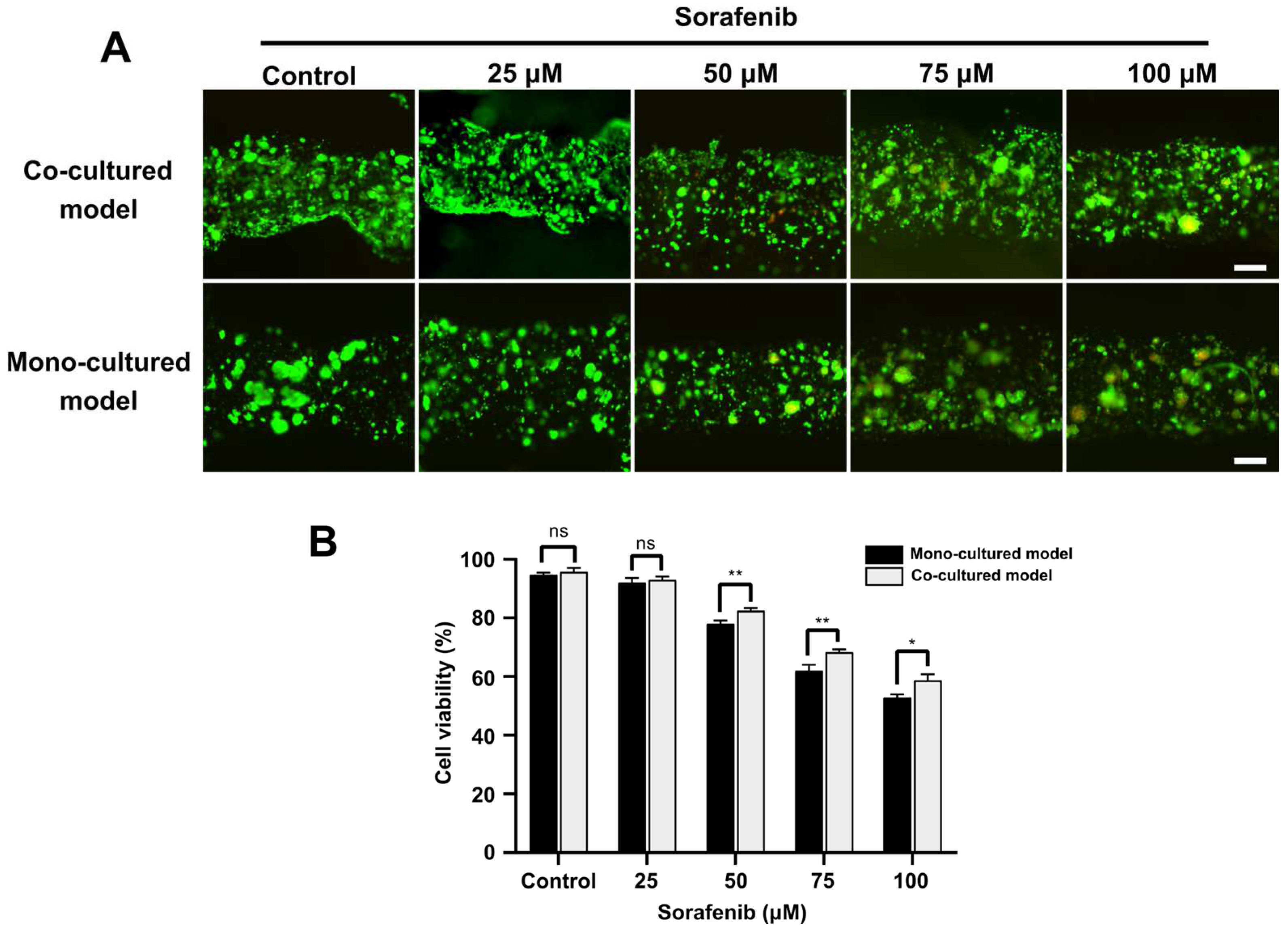
3.5. Drug Screening
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Srivastava, S.; Koay, E.J.; Borowsky, A.D.; De Marzo, A.M.; Ghosh, S.; Wagner, P.D.; Kramer, B.S. Cancer overdiagnosis: A biological challenge and clinical dilemma. Nat. Rev. Cancer. 2019, 19, 349–358. [Google Scholar] [CrossRef] [PubMed]
- Torre, L.A.; Siegel, R.L.; Ward, E.M.; Jemal, A. Global Cancer Incidence and Mortality Rates and Trends—An Update. Cancer Epidemiol. Biomark. Prev. 2016, 25, 16–27. [Google Scholar] [CrossRef] [PubMed]
- Naghavi, M.; Wang, H.; Lozano, R.; Davis, A.; Liang, X.; Zhou, M.; Vollset, S.E.; Ozgoren, A.A.; Abdalla, S.; Abd-Allah, F. Global, regional, and national age-sex specific allcause and cause-specific mortality for 240 causes of death, 1990–2013: A systematic analysis for the Global Burden of Disease Study 2013. Lancet 2015, 385, 117–171. [Google Scholar]
- Li, L.; Wang, H. Heterogeneity of liver cancer and personalized therapy. Cancer Lett. 2016, 379, 191–197. [Google Scholar] [CrossRef]
- Tang, W.; Chen, Z.; Zhang, W.; Cheng, Y.; Zhang, B.; Wu, F.; Wang, Q.; Wang, S.; Rong, D.; Reiter, F.P.; et al. The mechanisms of sorafenib resistance in hepatocellular carcinoma: Theoretical basis and therapeutic aspects. Signal Transduct. Target Ther. 2020, 5, 87. [Google Scholar] [CrossRef]
- Amelian, A.; Wasilewska, K.; Megias, D.; Winnicka, K. Application of standard cell cultures and 3D in vitro tissue models as an effective tool in drug design and development. Pharmacol. Rep. 2017, 69, 861–870. [Google Scholar] [CrossRef]
- Edmondson, R.; Broglie, J.J.; Adcock, A.F.; Yang, L. Three-Dimensional Cell Culture Systems and Their Applications in Drug Discovery and Cell-Based Biosensors. Assay Drug Dev. Technol. 2014, 12, 207–218. [Google Scholar] [CrossRef]
- Brancato, V.; Oliveira, J.M.; Correlo, V.M.; Reis, R.L.; Kundu, S.C. Could 3D models of cancer enhance drug screening? Biomaterials 2020, 232, 119744. [Google Scholar] [CrossRef]
- Heindryckx, F.; Colle, I.; Van Vlierberghe, H. Experimental mouse models for hepatocellular carcinoma research. Int. J. Exp. Pathol. 2009, 90, 367–386. [Google Scholar] [CrossRef]
- Gu, C.Y.; Lee, T.K.W. Preclinical mouse models of hepatocellular carcinoma: An overview and update. Exp. Cell Res. 2022, 412, 113042. [Google Scholar] [CrossRef]
- Paradiso, A.; Volpi, M.; Rinoldi, C.; Celikkin, N.; Contessi Negrini, N.; Bilgen, M.; Dallera, G.; Pierini, F.; Costantini, M.; Święszkowski, W.; et al. In vitro functional models for human liver diseases and drug screening: Beyond animal testing. Biomater. Sci. 2023. advance article. [Google Scholar] [CrossRef]
- Breslin, S.; O’Driscoll, L. Three-dimensional cell culture: The missing link in drug discovery. Drug Discov. Today 2013, 18, 240–249. [Google Scholar] [CrossRef]
- Huang, D.; Gibeley, S.B.; Xu, C.; Xiao, Y.; Celik, O.; Ginsberg, H.N.; Leong, K.W. Engineering Liver Microtissues for Disease Modeling and Regenerative Medicine. Adv. Funct. Mater. 2020, 30, 1909553. [Google Scholar] [CrossRef]
- Lin, R.-Z.; Chang, H.-Y. Recent advances in three-dimensional multicellular spheroid culture for biomedical research. Biotechnol. J. 2008, 3, 1172–1184. [Google Scholar] [CrossRef]
- He, J.; Zhou, C.; Xu, X.; Zhou, Z.; Danoy, M.; Shinohara, M.; Xiao, W.; Zhu, D.; Zhao, X.; Feng, X.; et al. Scalable Formation of Highly Viable and Functional Hepatocellular Carcinoma Spheroids in an Oxygen-Permeable Microwell Device for Anti-Tumor Drug Evaluation. Adv. Healthc. Mater. 2022, 11, 2200863. [Google Scholar] [CrossRef]
- Roopesh, R.P.; Muthusamy, S.; Velayudhan, S.; Sabareeswaran, A.; Anil Kumar, P.R. High-throughput production of liver parenchymal microtissues and enrichment of organ-specific functions in gelatin methacrylamide microenvironment. Biotechnol. Bioeng. 2022, 119, 1018–1032. [Google Scholar] [CrossRef]
- Wu, G.; Wu, J.; Wang, B.; Zhu, X.; Shi, X.; Ding, Y. Importance of tumor size at diagnosis as a prognostic factor for hepatocellular carcinoma survival: A population-based study. Cancer Manag. Res. 2018, 10, 4401–4410. [Google Scholar] [CrossRef]
- Sun, L.; Yang, H.; Wang, Y.; Zhang, X.; Jin, B.; Xie, F.; Jin, Y.; Pang, Y.; Zhao, H.; Lu, X.; et al. Application of a 3D Bioprinted Hepatocellular Carcinoma Cell Model in Antitumor Drug Research. Front. Oncol. 2020, 10, 878. [Google Scholar] [CrossRef]
- Mao, S.; He, J.; Zhao, Y.; Liu, T.; Xie, F.; Yang, H.; Mao, Y.; Pang, Y.; Sun, W. Bioprinting of patient-derived in vitro intrahepatic cholangiocarcinoma tumor model: Establishment, evaluation and anti-cancer drug testing. Biofabrication 2020, 12, 045014. [Google Scholar] [CrossRef]
- Gil, C.J.; Tomov, M.L.; Theus, A.S.; Cetnar, A.; Mahmoudi, M.; Serpooshan, V. In Vivo Tracking of Tissue Engineered Constructs. Micromachines 2019, 10, 474. [Google Scholar] [CrossRef]
- Taymour, R.; Chicaiza-Cabezas, N.A.; Gelinsky, M.; Lode, A. Core–shell bioprinting of vascularized in vitro liver sinusoid models. Biofabrication 2022, 14, 045019. [Google Scholar] [CrossRef] [PubMed]
- Lin, D.S.Y.; Rajasekar, S.; Marway, M.K.; Zhang, B. From Model System to Therapy: Scalable Production of Perfusable Vascularized Liver Spheroids in “Open-Top“ 384-Well Plate. ACS Biomater. Sci. Eng. 2021, 7, 2964–2972. [Google Scholar] [CrossRef] [PubMed]
- Chiew, G.G.Y.; Wei, N.; Sultania, S.; Lim, S.; Luo, K.Q. Bioengineered three-dimensional co-culture of cancer cells and endothelial cells: A model system for dual analysis of tumor growth and angiogenesis. Biotechnol. Bioeng. 2017, 114, 1865–1877. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.; Ohashi, K.; Utoh, R.; Kano, K.; Okano, T. Preserved liver-specific functions of hepatocytes in 3D co-culture with endothelial cell sheets. Biomaterials 2012, 33, 1406–1413. [Google Scholar] [CrossRef]
- Wang, Y.; Kankala, R.K.; Zhang, J.; Hao, L.; Zhu, K.; Wang, S.; Zhang, Y.S.; Chen, A. Modeling Endothelialized Hepatic Tumor Microtissues for Drug Screening. Adv. Sci. 2020, 7, 2002002. [Google Scholar] [CrossRef]
- Teutsch, H.F. The modular microarchitecture of human liver. Hepatology 2005, 42, 317–325. [Google Scholar] [CrossRef]
- Ben-Moshe, S.; Itzkovitz, S. Spatial heterogeneity in the mammalian liver. Nat. Rev. Gastroenterol. Hepatol. 2019, 16, 395–410. [Google Scholar] [CrossRef]
- Wei, X.; Huang, B.; Chen, K.; Fan, Z.; Wang, L.; Xu, M. Dot extrusion bioprinting of spatially controlled heterogenous tumor models. Mater. Des. 2022, 223, 111152. [Google Scholar] [CrossRef]
- Schuppan, D. Structure of the Extracellular Matrix in Normal and Fibrotic Liver: Collagens and Glycoproteins. Semin Liver Dis. 1990, 10, 1–10. [Google Scholar] [CrossRef]
- Bonnans, C.; Chou, J.; Werb, Z. Remodelling the extracellular matrix in development and disease. Nat. Rev. Mol. Cell Biol. 2014, 15, 786–801. [Google Scholar] [CrossRef]
- Kai, F.; Drain, A.P.; Weaver, V.M. The Extracellular Matrix Modulates the Metastatic Journey. Dev. Cell 2019, 49, 332–346. [Google Scholar] [CrossRef]
- Seyedmahmoud, R.; Çelebi-Saltik, B.; Barros, N.; Nasiri, R.; Banton, E.; Shamloo, A.; Ashammakhi, N.; Dokmeci, M.R.; Ahadian, S. Three-Dimensional Bioprinting of Functional Skeletal Muscle Tissue Using Gelatin Methacryloyl-Alginate Bioinks. Micromachines 2019, 10, 679. [Google Scholar] [CrossRef]
- Maloney, E.; Clark, C.; Sivakumar, H.; Yoo, K.; Aleman, J.; Rajan, S.A.P.; Forsythe, S.; Mazzocchi, A.; Laxton, A.W.; Tatter, S.B.; et al. Immersion Bioprinting of Tumor Organoids in Multi-Well Plates for Increasing Chemotherapy Screening Throughput. Micromachines 2020, 11, 208. [Google Scholar] [CrossRef]
- Yue, K.; Trujillo-de Santiago, G.; Alvarez, M.M.; Tamayol, A.; Annabi, N.; Khademhosseini, A. Synthesis, properties, and biomedical applications of gelatin methacryloyl (GelMA) hydrogels. Biomaterials 2015, 73, 254–271. [Google Scholar] [CrossRef]
- Pérez-Cortez, J.E.; Sánchez-Rodríguez, V.H.; Gallegos-Martínez, S.; Chuck-Hernández, C.; Rodriguez, C.A.; Álvarez, M.M.; Trujillo-de Santiago, G.; Vázquez-Lepe, E.; Martínez-López, J.I. Low-Cost Light-Based GelMA 3D Bioprinting via Retrofitting: Manufacturability Test and Cell Culture Assessment. Micromachines 2023, 14, 55. [Google Scholar] [CrossRef]
- Yang, B.; Liu, T.; Gao, G.; Zhang, X.; Wu, B. Fabrication of 3D GelMA Scaffolds Using Agarose Microgel Embedded Printing. Micromachines 2022, 13, 469. [Google Scholar] [CrossRef]
- Klotz, B.J.; Gawlitta, D.; Rosenberg, A.J.W.P.; Malda, J.; Melchels, F.P.W. Gelatin-Methacryloyl Hydrogels: Towards Biofabrication-Based Tissue Repair. Trends Biotechnol. 2016, 34, 394–407. [Google Scholar] [CrossRef]
- Van Den Bulcke, A.I.; Bogdanov, B.; De Rooze, N.; Schacht, E.H.; Cornelissen, M.; Berghmans, H. Structural and rheological properties of methacrylamide modified gelatin hydrogels. Biomacromolecules 2000, 1, 31–38. [Google Scholar] [CrossRef]
- Tang, M.; Tiwari, S.K.; Agrawal, K.; Tan, M.; Dang, J.; Tam, T.; Tian, J.; Wan, X.; Schimelman, J.; You, S.; et al. Rapid 3D Bioprinting of Glioblastoma Model Mimicking Native Biophysical Heterogeneity. Small 2021, 17, 2006050. [Google Scholar] [CrossRef]
- Liu, W.; Heinrich, M.A.; Zhou, Y.; Akpek, A.; Hu, N.; Liu, X.; Guan, X.; Zhong, Z.; Jin, X.; Khademhosseini, A.; et al. Extrusion Bioprinting of Shear-Thinning Gelatin Methacryloyl Bioinks. Adv. Healthcare Mater. 2017, 6, 1601451. [Google Scholar] [CrossRef]
- Xu, P.; Guan, J.; Chen, Y.; Xiao, H.; Yang, T.; Sun, H.; Wu, N.; Zhang, C.; Mao, Y. Stiffness of photocrosslinkable gelatin hydrogel influences nucleus pulposus cell propertiesin vitro. J. Cell Mol. Med. 2021, 25, 880–891. [Google Scholar] [CrossRef] [PubMed]
- Zhuang, Z.; Zhang, Y.; Sun, S.; Li, Q.; Chen, K.; An, C.; Wang, L.; van den Beucken, J.J.J.P.; Wang, H. Control of Matrix Stiffness Using Methacrylate–Gelatin Hydrogels for a Macrophage-Mediated Inflammatory Response. ACS Biomater. Sci. Eng. 2020, 6, 3091–3102. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Yu, K.; Nie, J.; Sun, M.; Fu, J.; Wang, H.; He, Y. Modeling the printability of photocuring and strength adjustable hydrogel bioink during projection-based 3D bioprinting. Biofabrication 2021, 13, 035032. [Google Scholar] [CrossRef] [PubMed]
- Ricken, T.; Werner, D.; Holzhütter, H.; König, M.; Dahmen, U.; Dirsch, O. Modeling function-perfusion behavior in liver lobules including tissue, blood, glucose, lactate and glycogen by use of a coupled two-scale PDE-ODE approach. Biomech. Model. Mechanobiol. 2015, 14, 515–536. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fan, Z.; Wei, X.; Chen, K.; Wang, L.; Xu, M. 3D Bioprinting of an Endothelialized Liver Lobule-like Construct as a Tumor-Scale Drug Screening Platform. Micromachines 2023, 14, 878. https://doi.org/10.3390/mi14040878
Fan Z, Wei X, Chen K, Wang L, Xu M. 3D Bioprinting of an Endothelialized Liver Lobule-like Construct as a Tumor-Scale Drug Screening Platform. Micromachines. 2023; 14(4):878. https://doi.org/10.3390/mi14040878
Chicago/Turabian StyleFan, Zicheng, Xiaoyun Wei, Keke Chen, Ling Wang, and Mingen Xu. 2023. "3D Bioprinting of an Endothelialized Liver Lobule-like Construct as a Tumor-Scale Drug Screening Platform" Micromachines 14, no. 4: 878. https://doi.org/10.3390/mi14040878
APA StyleFan, Z., Wei, X., Chen, K., Wang, L., & Xu, M. (2023). 3D Bioprinting of an Endothelialized Liver Lobule-like Construct as a Tumor-Scale Drug Screening Platform. Micromachines, 14(4), 878. https://doi.org/10.3390/mi14040878





