Geometry-Dependent Elastic Flow Dynamics in Micropillar Arrays
Abstract
:1. Introduction
2. Materials and Methods
3. Results
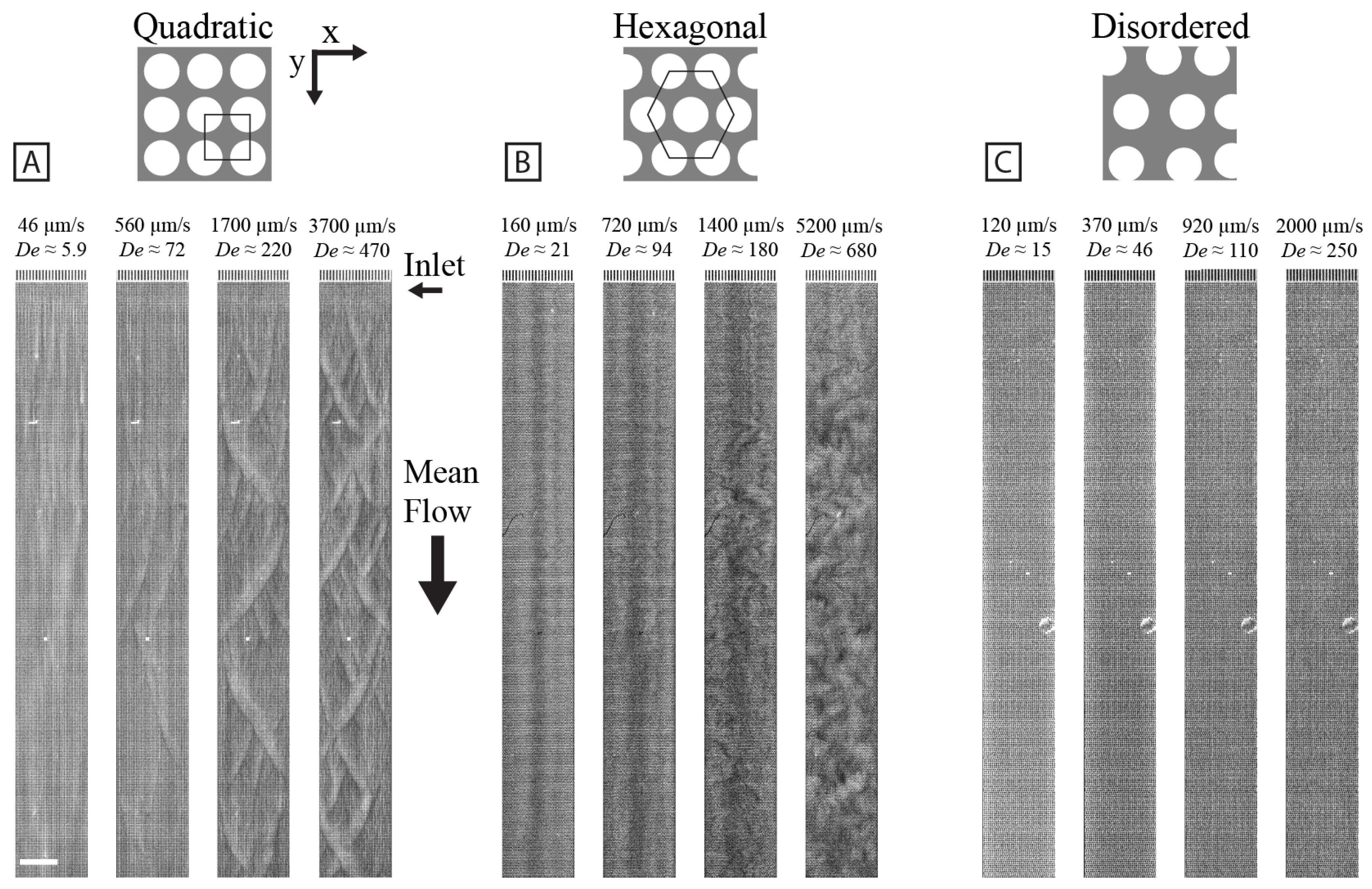
3.1. Array Geometry Dictates the Large-Scale Flow Patterns
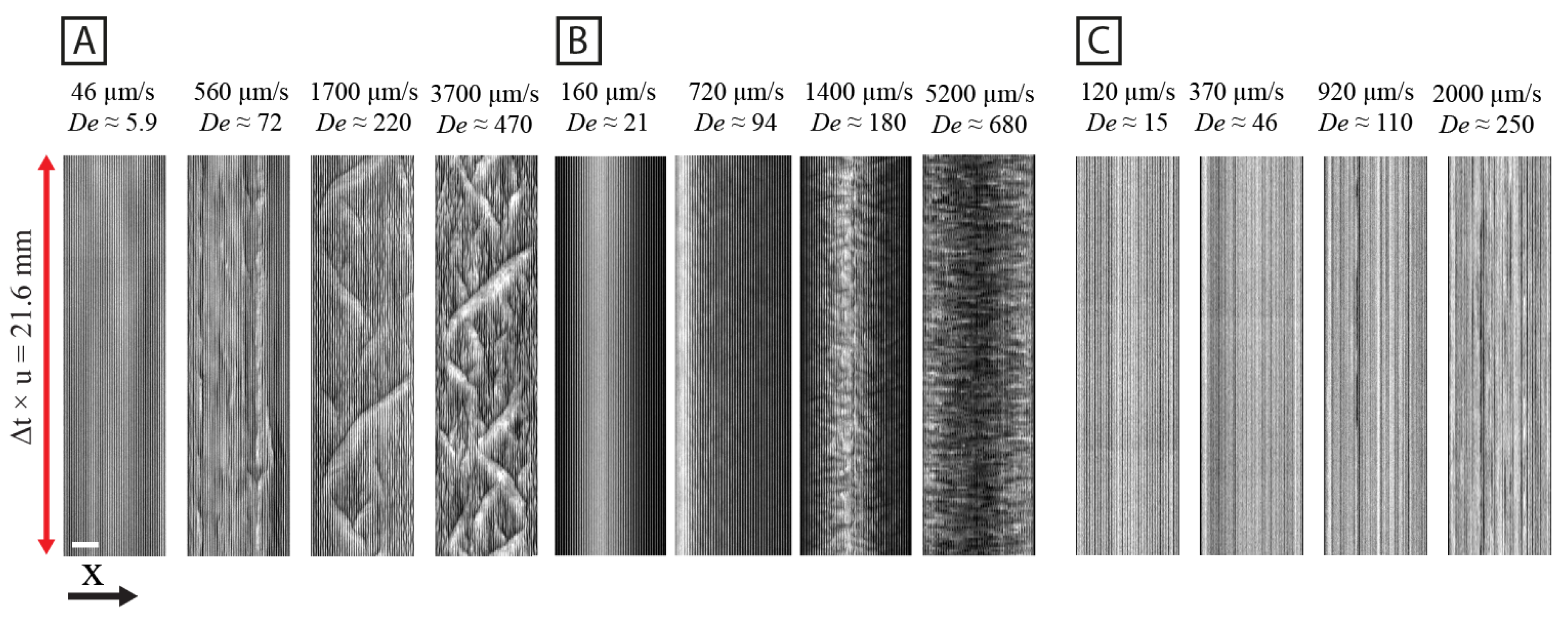
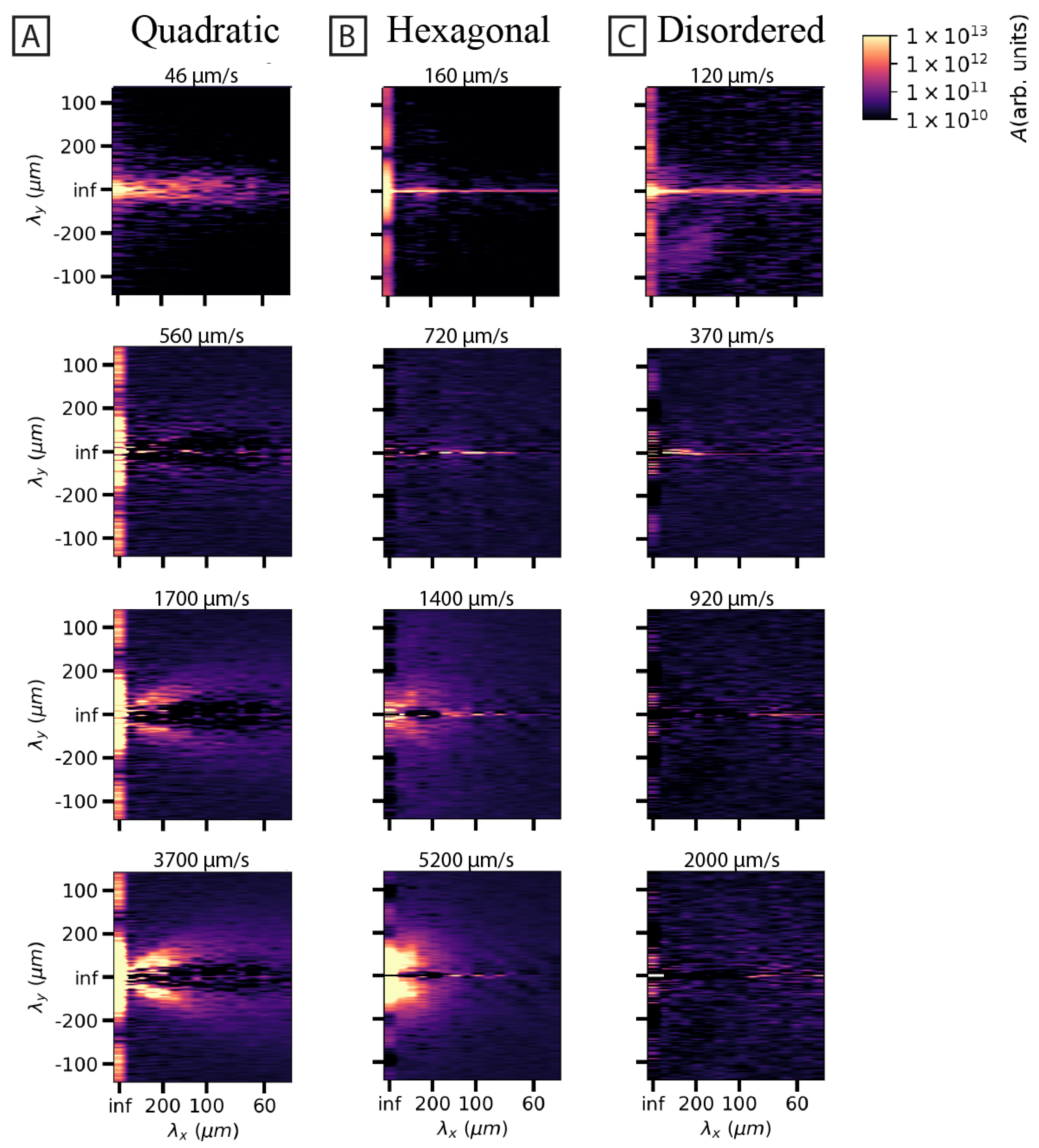
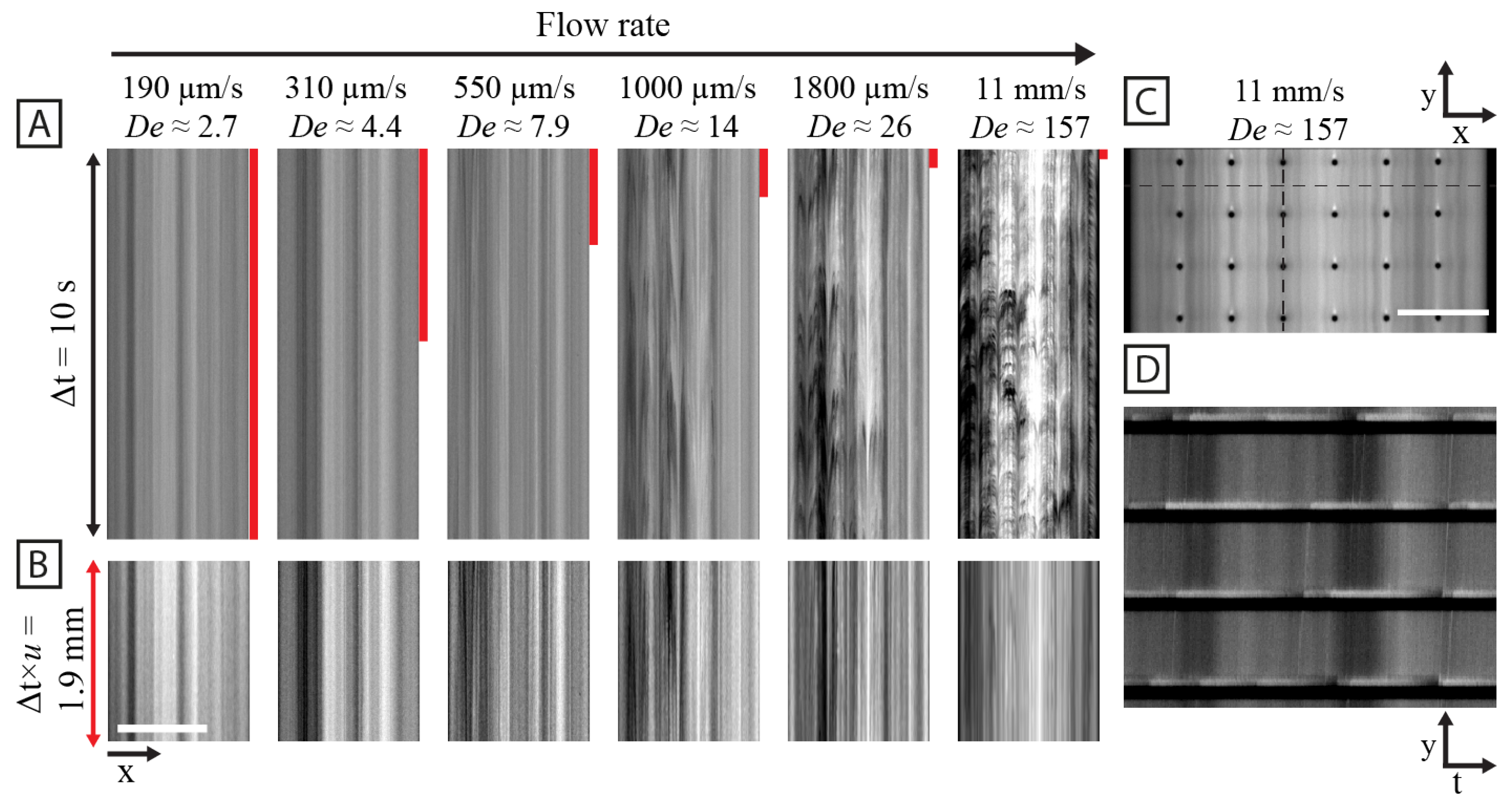
3.2. Spatial and Temporal Frequency Analysis of Concentration Fluctuations
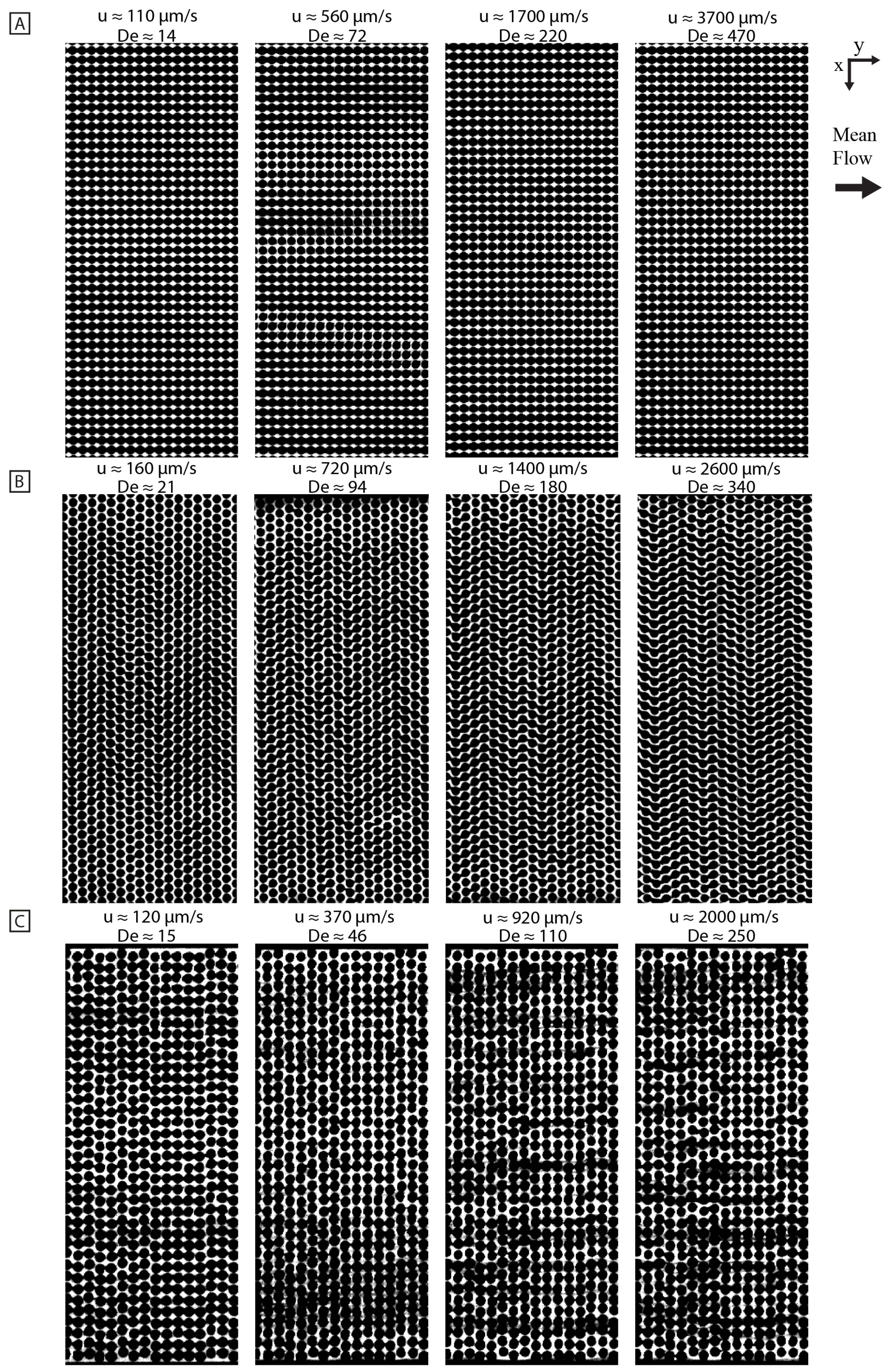
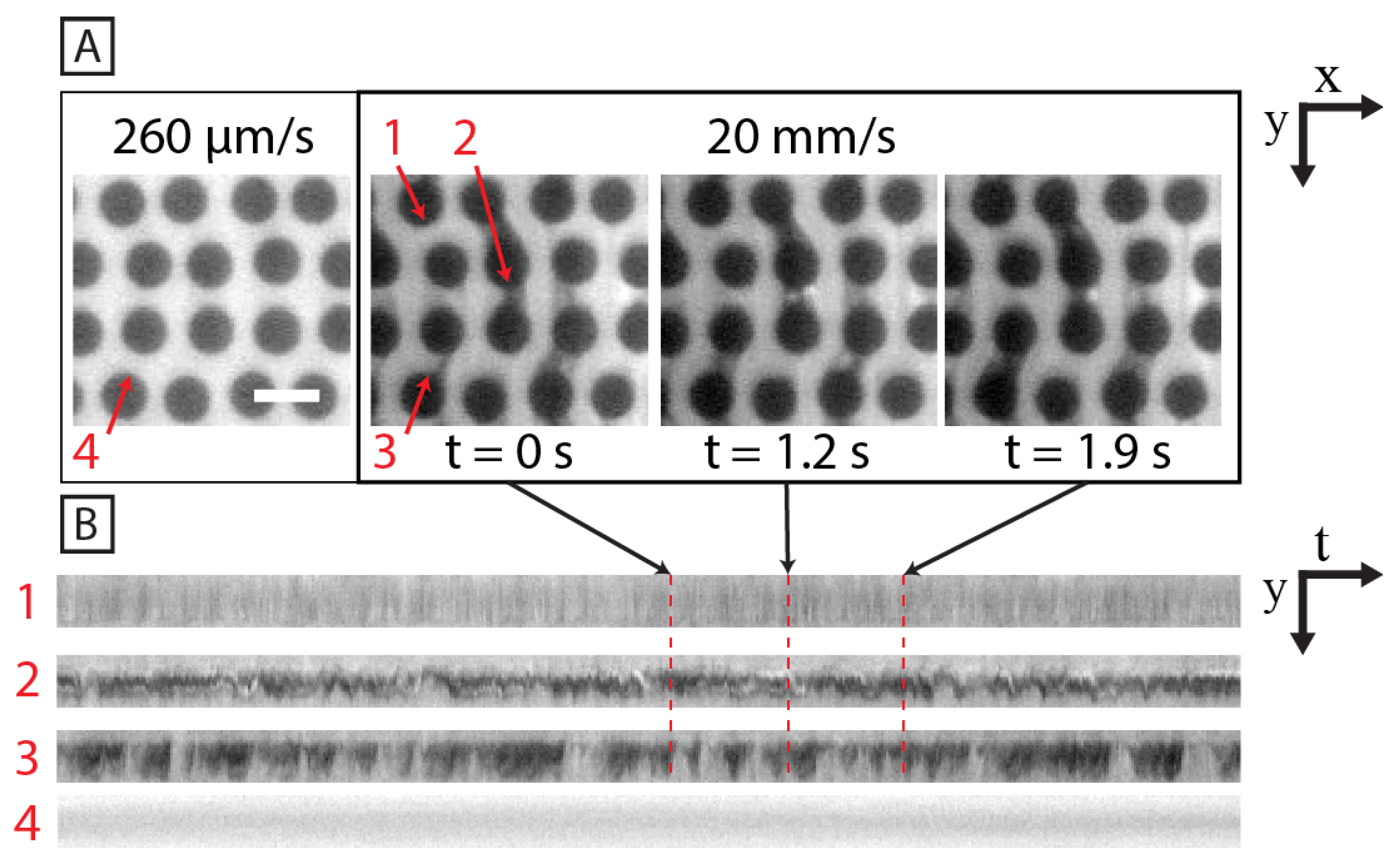
3.3. Persistent Flow Patterns Depend on Geometries
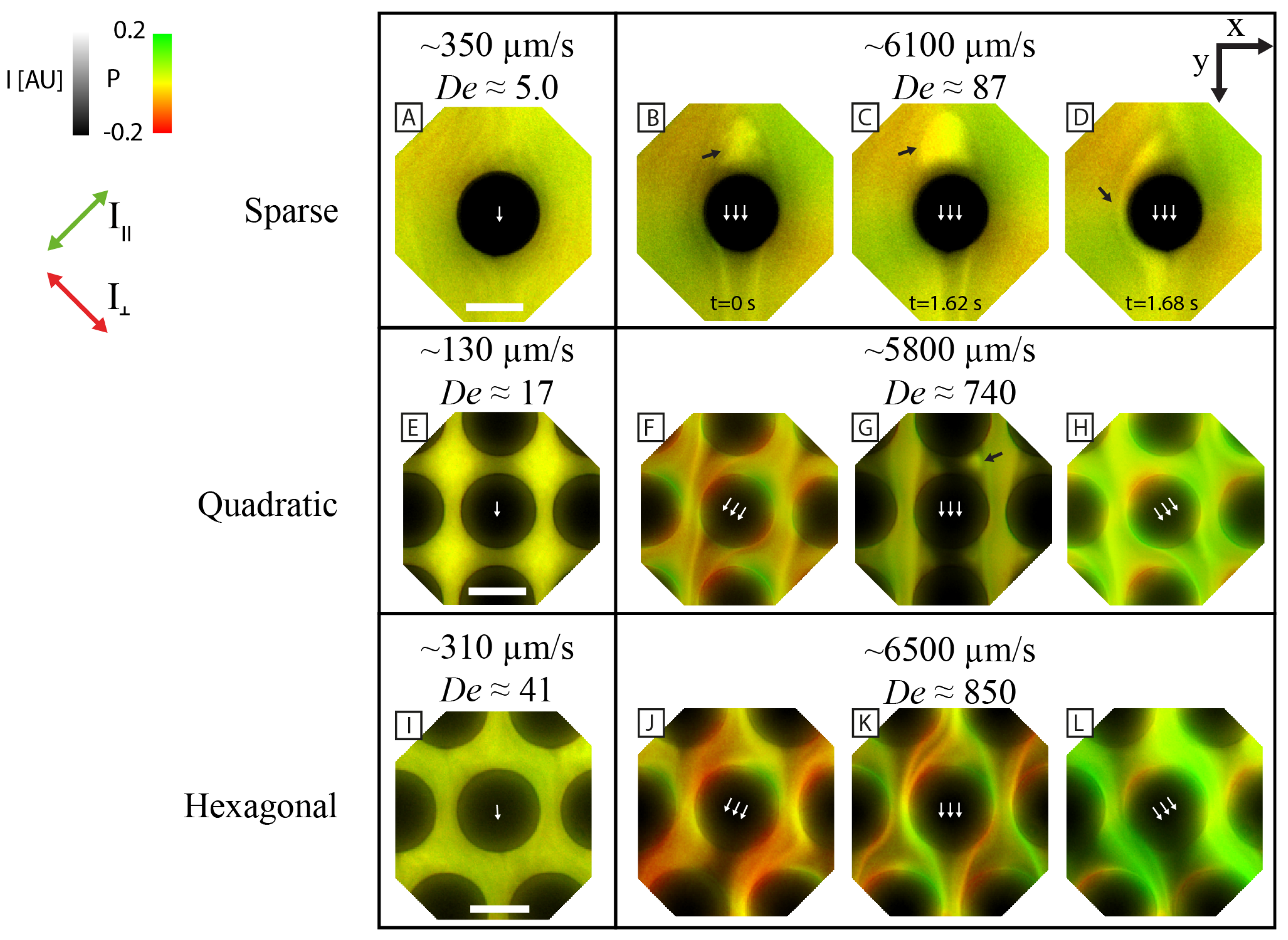
3.4. Microscopic Patterns and Polarization
3.5. The Flow around Sparsely Distributed Pillars Couples Only in the Flow Direction
4. Discussion
4.1. Overall Pattern Formation Is Connected to Flow Behavior around Individual Pillars
4.2. Disordered Arrays Prevent Long-Range Synchronization
4.3. Flow Patterns around the Sparsely Positioned Pillars Are Synchronized in the Flow Direction but Not Laterally
4.4. Cyclic and Random Large-Scale Fluctuations
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Groisman, A.; Steinberg, V. Elastic turbulence in a polymer solution flow. Nature 2000, 405, 53–55. [Google Scholar] [CrossRef]
- Smith, M.M.; Silva, J.A.; Munakata-Marr, J.; McCray, J.E. Compatibility of polymers and chemical oxidants for enhanced groundwater remediation. Environ. Sci. Technol. 2008, 42, 9296–9301. [Google Scholar] [CrossRef]
- Abed, W.M.; Whalley, R.D.; Dennis, D.J.C.; Poole, R.J. Experimental investigation of the impact of elastic turbulence on heat transfer in a serpentine channel. J. Non-Newton. Fluid Mech. 2016, 231, 68–78. [Google Scholar] [CrossRef]
- Sorbie, K.S. Polymer-Improved Oil Recovery; Springer Science & Business Media: Berlin/Heidelberg, Germany, 2013. [Google Scholar]
- Groisman, A.; Steinberg, V. Efficient mixing at low Reynolds numbers using polymer additives. Nature 2001, 410, 905–908. [Google Scholar] [CrossRef] [PubMed]
- Carrel, M.; Morales, V.L.; Beltran, M.A.; Derlon, N.; Kaufmann, R.; Morgenroth, E.; Holzner, M. Biofilms in 3D porous media: Delineating the influence of the pore network geometry, flow and mass transfer on biofilm development. Water Res. 2018, 134, 280–291. [Google Scholar] [CrossRef]
- Randell, S.H.; Boucher, R.C. Effective mucus clearance is essential for respiratory health. Am. J. Respir. Cell Mol. Biol. 2006, 35, 20–28. [Google Scholar] [CrossRef] [PubMed]
- Hopkins, C.C.; Haward, S.J.; Shen, A.Q. Purely elastic fluid–structure interactions in microfluidics: Implications for mucociliary flows. Small 2020, 16, 1903872. [Google Scholar] [CrossRef] [PubMed]
- Browne, C.A.; Datta, S.S. Elastic turbulence generates anomalous flow resistance in porous media. Sci. Adv. 2021, 7, eabj2619. [Google Scholar] [CrossRef] [PubMed]
- Browne, C.A.; Shih, A.; Datta, S.S. Pore-scale flow characterization of polymer solutions in microfluidic porous media. Small 2020, 16, 1903944. [Google Scholar] [CrossRef]
- Ström, O.E.; Beech, J.P.; Tegenfeldt, J.O. Short and long-range cyclic patterns in flows of DNA solutions in microfluidic obstacle arrays. Lab Chip 2023, 23, 1779–1793. [Google Scholar] [CrossRef]
- Chmielewski, C.; Jayaraman, K. Elastic instability in crossflow of polymer solutions through periodic arrays of cylinders. J. Non-Newton. Fluid Mech. 1993, 48, 285–301. [Google Scholar] [CrossRef]
- Shi, X.; Christopher, G.F. Growth of viscoelastic instabilities around linear cylinder arrays. Phys. Fluids 2016, 28, 124102. [Google Scholar] [CrossRef]
- Browne, C.A.; Shih, A.; Datta, S.S. Bistability in the unstable flow of polymer solutions through pore constriction arrays. J. Fluid Mech. 2020, 890, A2. [Google Scholar] [CrossRef]
- Rems, L.; Kawale, D.; Lee, L.J.; Boukany, P.E. Flow of DNA in micro/nanofluidics: From fundamentals to applications. Biomicrofluidics 2016, 10, 043403. [Google Scholar] [CrossRef]
- Kawale, D.; Marques, E.; Zitha, P.L.; Kreutzer, M.T.; Rossen, W.R.; Boukany, P.E. Elastic instabilities during the flow of hydrolyzed polyacrylamide solution in porous media: Effect of pore-shape and salt. Soft Matter 2017, 13, 765–775. [Google Scholar] [CrossRef] [PubMed]
- Haward, S.J.; Hopkins, C.C.; Shen, A.Q. Stagnation points control chaotic fluctuations in viscoelastic porous media flow. Proc. Natl. Acad. Sci. USA 2021, 118, e2111651118. [Google Scholar] [CrossRef] [PubMed]
- Walkama, D.M.; Waisbord, N.; Guasto, J.S. Disorder suppresses chaos in viscoelastic flows. Phys. Rev. Lett. 2020, 124, 164501. [Google Scholar] [CrossRef] [PubMed]
- Machado, A.; Bodiguel, H.; Beaumont, J.; Clisson, G.; Colin, A. Extra dissipation and flow uniformization due to elastic instabilities of shear-thinning polymer solutions in model porous media. Biomicrofluidics 2016, 10, 043507. [Google Scholar] [CrossRef] [PubMed]
- Howe, A.M.; Clarke, A.; Giernalczyk, D. Flow of concentrated viscoelastic polymer solutions in porous media: Effect of MW and concentration on elastic turbulence onset in various geometries. Soft Matter 2015, 11, 6419–6431. [Google Scholar] [CrossRef]
- Eberhard, U.; Seybold, H.; Secchi, E.; Jiménez-Martínez, J.; Rühs, P.A.; Ofner, A.; Andrade, J.S.; Holzner, M. Mapping the local viscosity of non-Newtonian fluids flowing through disordered porous structures. Sci. Rep. 2020, 10, 11733. [Google Scholar] [CrossRef]
- Ström, O.E.; Beech, J.P.; Tegenfeldt, J.O. High-Throughput Separation of Long DNA in Deterministic Lateral Displacement Arrays. Micromachines 2022, 13, 1754. [Google Scholar] [CrossRef]
- Beech, J.P.; Ström, O.E.; Turato, E.; Tegenfeldt, J.O. Using symmetry to control viscoelastic waves in pillar arrays. RSC Adv. 2023, 13, 31497–31506. [Google Scholar] [CrossRef]
- Bakajin, O.; Duke, T.A.; Tegenfeldt, J.; Chou, C.F.; Chan, S.S.; Austin, R.H.; Cox, E.C. Separation of 100-kilobase DNA molecules in 10 s. Anal. Chem. 2001, 73, 6053–6056. [Google Scholar] [CrossRef]
- Huang, L.R.; Tegenfeldt, J.O.; Kraeft, J.J.; Sturm, J.C.; Austin, R.H.; Cox, E.C. A DNA prism for high-speed continuous fractionation of large DNA molecules. Nat. Biotechnol. 2002, 20, 1048–1051. [Google Scholar] [CrossRef]
- Xia, Y.; Whitesides, G.M. Soft lithography. Angew. Chem. Int. Ed. 1998, 37, 550–575. [Google Scholar] [CrossRef]
- Hung, L.H.; Lee, A.P. Optimization of droplet generation by controlling PDMS surface hydrophobicity. In Proceedings of the ASME International Mechanical Engineering Congress and Exposition, Anaheim, CA, USA, 13–19 November 2004; Volume 47039, pp. 47–48. [Google Scholar]
- Perkins, T.T.; Smith, D.E.; Chu, S. Single polymer dynamics in an elongational flow. Science 1997, 276, 2016–2021. [Google Scholar] [CrossRef]
- Smith, D.E.; Babcock, H.P.; Chu, S. Single-polymer dynamics in steady shear flow. Science 1999, 283, 1724–1727. [Google Scholar] [CrossRef] [PubMed]
- Varshney, A.; Steinberg, V. Elastic wake instabilities in a creeping flow between two obstacles. Phys. Rev. Fluids 2017, 2, 051301. [Google Scholar] [CrossRef]
- Kawale, D.; Bouwman, G.; Sachdev, S.; Zitha, P.L.; Kreutzer, M.T.; Rossen, W.R.; Boukany, P.E. Polymer conformation during flow in porous media. Soft Matter 2017, 13, 8745–8755. [Google Scholar] [CrossRef] [PubMed]
- Qin, B.; Salipante, P.F.; Hudson, S.D.; Arratia, P.E. Upstream vortex and elastic wave in the viscoelastic flow around a confined cylinder. J. Fluid Mech. 2019, 864, R2. [Google Scholar] [CrossRef]
- Varshney, A.; Steinberg, V. Elastic Alfven waves in elastic turbulence. Nat. Commun. 2019, 10, 652. [Google Scholar] [CrossRef] [PubMed]
- De, S.; Kuipers, J.A.; Peters, E.A.; Padding, J.T. Viscoelastic flow past mono-and bidisperse random arrays of cylinders: Flow resistance, topology and normal stress distribution. Soft Matter 2017, 13, 9138–9146. [Google Scholar] [CrossRef]
- Kenney, S.; Poper, K.; Chapagain, G.; Christopher, G.F. Large Deborah number flows around confined microfluidic cylinders. Rheol. Acta 2013, 52, 485–497. [Google Scholar] [CrossRef]
- Pan, L.; Morozov, A.; Wagner, C.; Arratia, P. Nonlinear elastic instability in channel flows at low Reynolds numbers. Phys. Rev. Lett. 2013, 110, 174502. [Google Scholar] [CrossRef] [PubMed]
- De, S.; Van Der Schaaf, J.; Deen, N.G.; Kuipers, J.; Peters, E.; Padding, J. Lane change in flows through pillared microchannels. Phys. Fluids 2017, 29, 113102. [Google Scholar] [CrossRef]
- Qin, B.; Arratia, P.E. Characterizing elastic turbulence in channel flows at low Reynolds number. Phys. Rev. Fluids 2017, 2, 083302. [Google Scholar] [CrossRef]
- Ekanem, E.M.; Berg, S.; De, S.; Fadili, A.; Bultreys, T.; Rücker, M.; Southwick, J.; Crawshaw, J.; Luckham, P.F. Signature of elastic turbulence of viscoelastic fluid flow in a single pore throat. Phys. Rev. E 2020, 101, 042605. [Google Scholar] [CrossRef]
- Fouxon, A.; Lebedev, V. Spectra of turbulence in dilute polymer solutions. Phys. Fluids 2003, 15, 2060–2072. [Google Scholar] [CrossRef]
- Boffetta, G.; Ecke, R.E. Two-Dimensional Turbulence. Annu. Rev. Fluid Mech. 2012, 44, 427–451. [Google Scholar] [CrossRef]
- McKinley, G.H.; Armstrong, R.C.; Brown, R.A. The wake instability in viscoelastic flow past confined circular-cylinders. Philos. Trans. R. Soc. Lond. Ser. A—Math. Phys. Eng. Sci. 1993, 344, 265–304. [Google Scholar]
- De, S.; Koesen, S.P.; Maitri, R.V.; Golombok, M.; Padding, J.T.; van Santvoort, J.F.M. Flow of viscoelastic surfactants through porous media. AIChE J. 2018, 64, 773–781. [Google Scholar] [CrossRef]
- Cadot, O.; Kumar, S. Experimental characterization of viscoelastic effects on two- and three-dimensional shear instabilities. J. Fluid Mech. 2000, 416, 151–172. [Google Scholar] [CrossRef]
- Datta, S.S.; Ardekani, A.M.; Arratia, P.E.; Beris, A.N.; Bischofberger, I.; McKinley, G.H.; Eggers, J.G.; López-Aguilar, J.E.; Fielding, S.M.; Frishman, A.; et al. Perspectives on viscoelastic flow instabilities and elastic turbulence. Phys. Rev. Fluids 2022, 7, 080701. [Google Scholar] [CrossRef]
- Bakajin, O.B.; Duke, T.A.J.; Chou, C.F.; Chan, S.S.; Austin, R.H.; Cox, E.C. Electrohydrodynamic stretching of DNA in confined environments. Phys. Rev. Lett. 1998, 80, 2737–2740. [Google Scholar] [CrossRef]
- Lin, J.; Persson, F.; Fritzsche, J.; Tegenfeldt, J.O.; Saleh, O.A. Bandpass Filtering of DNA Elastic Modes Using Confinement and Tension. Biophys. J. 2012, 102, 96–100. [Google Scholar] [CrossRef]
- Reisner, W.; Morton, K.J.; Riehn, R.; Wang, Y.M.; Yu, Z.N.; Rosen, M.; Sturm, J.C.; Chou, S.Y.; Frey, E.; Austin, R.H. Statics and dynamics of single DNA molecules confined in nanochannels. Phys. Rev. Lett. 2005, 94, 196101. [Google Scholar] [CrossRef]
- de Blois, C.; Haward, S.J.; Shen, A.Q. Canopy elastic turbulence: Spontaneous formation of waves in beds of slender microposts. Phys. Rev. Fluids 2023, 8, 023301. [Google Scholar] [CrossRef]
- Livolant, F.; Leforestier, A. Condensed phases of DNA: Structures and phase transitions. Prog. Polym. Sci. 1996, 21, 1115–1164. [Google Scholar] [CrossRef]
- Livolant, F. Ordered phases of DNA in vivo and in vitro. Phys. A—Stat. Mech. Its Appl. 1991, 176, 117–137. [Google Scholar] [CrossRef]
- Miyoshi, D.; Sugimoto, N. Molecular crowding effects on structure and stability of DNA. Biochimie 2008, 90, 1040–1051. [Google Scholar] [CrossRef] [PubMed]
- Collette, D.; Dunlap, D.; Finzi, L. Macromolecular Crowding and DNA: Bridging the Gap between In Vitro and In Vivo. Int. J. Mol. Sci. 2023, 24, 17502. [Google Scholar] [CrossRef] [PubMed]
- Afik, E.; Steinberg, V. On the role of initial velocities in pair dispersion in a microfluidic chaotic flow. Nat. Commun. 2017, 8, 468. [Google Scholar] [CrossRef]
- Garg, H.; Calzavarini, E.; Mompean, G.; Berti, S. Particle-laden two-dimensional elastic turbulence. Eur. Phys. J. E 2018, 41, 115. [Google Scholar] [CrossRef] [PubMed]
- Esmaily, M.; Villafane, L.; Banko, A.J.; Iaccarino, G.; Eaton, J.K.; Mani, A. A benchmark for particle-laden turbulent duct flow: A joint computational and experimental study. Int. J. Multiph. Flow 2020, 132, 103410. [Google Scholar] [CrossRef]


| Array Pattern | Pillar Radius (m) | 1, 2 | Porosity 3 | Lateral Pillar Gap (m) | Lateral Gap-to-Contour-Length Ratio 4 () |
|---|---|---|---|---|---|
| Quadratic | 44, 444 | 0.52 | 4.1 ± 0.4 | 0.25 | |
| Hexagonal | 44, 444 | 0.52 | 3.9 ± 0.2 | 0.25 | |
| Disordered | 37, 381 | 0.54 ± 7.5 | 4.5 ± 1.5 | 0.27 ± 0.09 | |
| Sparse | 6, 70 | 0.99 | 100 | 6.06 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ström, O.E.; Beech, J.P.; Tegenfeldt, J.O. Geometry-Dependent Elastic Flow Dynamics in Micropillar Arrays. Micromachines 2024, 15, 268. https://doi.org/10.3390/mi15020268
Ström OE, Beech JP, Tegenfeldt JO. Geometry-Dependent Elastic Flow Dynamics in Micropillar Arrays. Micromachines. 2024; 15(2):268. https://doi.org/10.3390/mi15020268
Chicago/Turabian StyleStröm, Oskar E., Jason P. Beech, and Jonas O. Tegenfeldt. 2024. "Geometry-Dependent Elastic Flow Dynamics in Micropillar Arrays" Micromachines 15, no. 2: 268. https://doi.org/10.3390/mi15020268
APA StyleStröm, O. E., Beech, J. P., & Tegenfeldt, J. O. (2024). Geometry-Dependent Elastic Flow Dynamics in Micropillar Arrays. Micromachines, 15(2), 268. https://doi.org/10.3390/mi15020268






