Cell Migration Assays and Their Application to Wound Healing Assays—A Critical Review
Abstract
:1. Introduction
2. Classification of CMAs
2.1. Creation of CFZs (FR1)
2.1.1. Mechanical Depletion Approach
2.1.2. Other Depletion Approaches
2.2. Measurement of Cell Migration (FR2)
2.3. Creation of Mechanical Conditions (FR3)
2.3.1. Fluid-Induced Pressure
2.3.2. Cell-Generated Force
2.3.3. Environment-Based Force
2.3.4. Characterization of Stress and Strain
2.3.5. Combination of Different Conditions
3. Discussion
3.1. Consistency of CFZs (R1)
- ○
- Technology gap 1: there is no technology available to create many (over 100) CFZs consistently.
3.2. Geometry of CFZs (R2)
- ○
- Knowledge gap 1: there is no knowledge available to explain how and why different shapes of CFZs along with a chemical condition (e.g., calcium concentration) may affect the cell migration behavior in wound healing.
- ○
- Technology gap 2: there is no technology available to make different shapes of CFZs for many CFZs in CMAs.
3.3. Uniformity and Adjustability of Fluid Shear Stresses in Cells (R3 and R4)
- ○
- Technology gap 3: there is no technology available to generate and measure local stress in many cells (around 1 ×/mL in cell density) in nano-scale accuracy.
3.4. Multiple Conditions and Samples (R5)
- ○
- Technology gap 4: the current CMA can provide the 96 conditions only, which may have far more conditions than that which an application needs, e.g., 10, thereby increasing extra costs to the application (due to unused capacity);
- ○
- Technology gap 5: there is no technology available to build a CMA that allows for multiple conditions and, in the meantime, produces multiple samples;
- ○
- Technology gap 6: there is no technology available to build a CMA that allows adjustable multiple conditions and, in the meantime, produces adjustable multiple samples.
4. Limitation of CMAs to Wound Healing Assays
5. Conclusions and Future Research Directions
- (1)
- Further improving consistency of creating many CFZs (Technology gap 1)
- (2)
- Advancing our understanding of the interactions of different geometric shapes of CFZs along with chemical conditions and their effects on cell migration in wound healing (Knowledge gap 1)
- (3)
- Development of devices meet the functional requirement of changing the different shapes of CFZs for many CFZs (Technology gap 2)
- (4)
- Optimization of the structural design of reducing variations (Technology gap 3)
- (5)
- Modularization of existing components in the CMA (Technology gap 4)
- (6)
- Development of CMAs to adjust the number of conditions as well as the number of samples (Technology gap 5,6)
- (7)
- Combination of multiple conditions to create a more biomimetic environment
- (8)
- Extension of CMAs for other biomedical applications
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Bhatia, S.N.; Ingber, D.E. Microfluidic organs-on-chips. Nat. Biotechnol. 2014, 32, 760–772. [Google Scholar] [CrossRef] [PubMed]
- Junaid, A.; Mashaghi, A.; Hankemeier, T.; Vulto, P. An end-user perspective on Organ-on-a-Chip: Assays and usability aspects. Curr. Opin. Biomed. Eng. 2017, 1, 15–22. [Google Scholar] [CrossRef]
- MacQueen, L.; Sun, Y.; Simmons, C.A. Mesenchymal stem cell mechanobiology and emerging experimental platforms. J. R. Soc. Interface 2013, 10, 20130179. [Google Scholar] [CrossRef] [PubMed]
- Rothbauer, M.; Zirath, H.; Ertl, P. Recent advances in microfluidic technologies for cell-to-cell interaction studies. Lab. Chip 2018, 18, 249–270. [Google Scholar] [CrossRef] [PubMed]
- Pijuan, J.; Barceló, C.; Moreno, D.F.; Maiques, O.; Sisó, P.; Marti, R.M.; Macià, A.; Panosa, A. Cell Migration, Invasion, and Adhesion Assays: From Cell Imaging to Data Analysis. Front. Cell Dev. Biol. 2019, 7, 107. [Google Scholar] [CrossRef] [PubMed]
- Hulkower, K.; Herber, R. Cell migration and invasion assays as tools for drug discovery. Pharmaceutics 2011, 3, 107–124. [Google Scholar] [CrossRef] [PubMed]
- Ren, J.Q.; Wang, N.; Guo, P.A.; Fan, Y.P.; Lin, F.; Wu, J.D. Recent advances in microfluidics-based cell migration research. Lab. Chip 2022, 22, 3361–3376. [Google Scholar] [CrossRef] [PubMed]
- Stamm, A.; Reimers, K.; Strauß, S.; Vogt, P.; Scheper, T.; Pepelanova, I. In vitro wound healing assays–state of the art. BioNanoMaterials 2016, 17, 79–87. [Google Scholar] [CrossRef]
- Riahi, R.; Yang, Y.L.; Zhang, D.D.; Wong, P.K. Advances in Wound-Healing Assays for Probing Collective Cell Migration. Jala J. Lab. Autom. 2012, 17, 59–65. [Google Scholar] [CrossRef]
- Liang, C.C.; Park, A.Y.; Guan, J.L. In vitro scratch assay: A convenient and inexpensive method for analysis of cell migration in vitro. Nat. Protoc. 2007, 2, 329–333. [Google Scholar] [CrossRef]
- Nie, F.Q.; Yamada, M.; Kobayashi, J.; Yamato, M.; Kikuchi, A.; Okano, T. On-chip cell migration assay using microfluidic channels. Biomaterials 2007, 28, 4017–4022. [Google Scholar] [CrossRef] [PubMed]
- Gao, A.X.; Tian, Y.L.; Shi, Z.Z.; Yu, L. A Cost-effective Microdevice Bridges Microfluidic and Conventional in vitro Scratch/Wound-healing Assay for Personalized Therapy Validation. Biochip J. 2016, 10, 56–64. [Google Scholar] [CrossRef]
- Poujade, M.; Grasland-Mongrain, E.; Hertzog, A.; Jouanneau, J.; Chavrier, P.; Ladoux, B.; Buguin, A.; Silberzan, P. Collective migration of an epithelial monolayer in response to a model wound. Proc. Natl. Acad. Sci. USA 2007, 104, 15988–15993. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Li, H.J.; Ma, H.P.; Qin, J.H. A simple microfluidic strategy for cell migration assay in an in vitro wound-healing model. Wound Repair. Regen. 2013, 21, 897–903. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Guo, T.F.; Wang, L.R.; Liu, Y.; Chen, G.Y.; Zhou, H.; Zhang, M.Q. Tape-Assisted Photolithographic-Free Microfluidic Chip Cell Patterning for Tumor Metastasis Study. Anal. Chem. 2018, 90, 777–784. [Google Scholar] [CrossRef] [PubMed]
- Gupta, T.; Gupta, R.; Dabaghi, M.; Sahu, R.P.; Hirota, J.A.; Puri, I.K. Label-Free Cell Migration Assay Using Magnetic Exclusion. Adv. Mater. Technol. 2022, 7, 2101033. [Google Scholar] [CrossRef]
- Jonkman, J.E.N.; Cathcart, J.A.; Xu, F.; Bartolini, M.E.; Amon, J.E.; Stevens, K.M.; Colarusso, P. An introduction to the wound healing assay using live-cell microscopy. Cell Adhes. Migr. 2014, 8, 440–451. [Google Scholar] [CrossRef] [PubMed]
- Pahl, G.; Beitz, W.; Feldhusen, J.; Grote, K.-H. Engineering Design: A Systematic Approach, 3rd ed.; Springer: Berlin/Heidelberg, Germany, 2007. [Google Scholar]
- Suh, N.P. The Principles of Design; Oxford University Press: New York, NY, USA, 1990. [Google Scholar]
- Tony, A.; Badea, I.; Yang, C.; Liu, Y.Y.; Wells, G.; Wang, K.M.; Yin, R.X.; Zhang, H.B.; Zhang, W.J. The Additive Manufacturing Approach to Polydimethylsiloxane (PDMS) Microfluidic Devices: Review and Future Directions. Polymers 2023, 15, 1926. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.J.; Wang, J.W. Design theory and methodology for enterprise systems. Enterp. Inf. Syst. 2016, 10, 245–248. [Google Scholar] [CrossRef]
- Zhang, W.J.; Wang, J.W.; Lin, Y.Z. Integrated design and operation management for enterprise systems. Enterp. Inf. Syst. 2019, 13, 424–429. [Google Scholar] [CrossRef]
- Zhao, Y.; Zhu, D.H.; Zhou, F.; Song, J.; Zhang, W.J. An axiomatic design theory for design of apparel products. J. Eng. Fiber Fabr. 2022, 17, 155589250221134350. [Google Scholar] [CrossRef]
- Lin, Y.; Zhang, W.J. Towards a novel interface design framework: Function-behavior-state paradigm. Int. J. Hum. Comput. St. 2004, 61, 259–297. [Google Scholar] [CrossRef]
- Wang, J.W.; Wang, H.F.; Ding, J.L.; Furuta, K.; Kanno, T.; Ip, W.H.; Zhang, W.J. On domain modelling of the service system with its application to enterprise information systems. Enterp. Inf. Syst. 2016, 10, 1–16. [Google Scholar] [CrossRef]
- Dai, Z. Improvement of General Design Theory and Methodology with Its Application to Design of a Retractor for Ventral Hernia Repair Surgery. Ph.D. Thesis, University of Saskatchewan, Saskatoon, SK, Canada, 2019. [Google Scholar]
- Lee, J.; Wang, Y.L.; Ren, F.; Lele, T.P. Stamp Wound Assay for Studying Coupled Cell Migration and Cell Debris Clearance. Langmuir 2010, 26, 16672–16676. [Google Scholar] [CrossRef] [PubMed]
- Yue, P.Y.K.; Leung, E.P.Y.; Mak, N.K.; Wong, R.N.S. A Simplified Method for Quantifying Cell Migration/Wound Healing in 96-Well Plates. J. Biomol. Screen. 2010, 15, 427–433. [Google Scholar] [CrossRef] [PubMed]
- Sticker, D.; Lechner, S.; Jungreuthrnayer, C.; Zanghellini, J.; Erd, P. Microfluidic Migration and Wound Healing Assay Based on Mechanically Induced Injuries of Defined and Highly Reproducible Areas. Anal. Chem. 2017, 89, 2326–2333. [Google Scholar] [CrossRef] [PubMed]
- Monfared, G.S.; Ertl, P.; Rothbauer, M. An on-chip wound healing assay fabricated by xurography for evaluation of dermal fibroblast cell migration and wound closure. Sci. Rep. 2020, 10, 16192. [Google Scholar] [CrossRef] [PubMed]
- Yin, D.; Zhang, H.B.; Yang, C.; Zhang, W.J.; Yang, S.H. A More Biomimetic Cell Migration Assay with High Reliability and Its Applications. Pharmaceuticals 2022, 15, 695. [Google Scholar] [CrossRef] [PubMed]
- Yilmaz, A.; Karavelioglu, Z.; Aydemir, G.; Demircali, A.A.; Varol, R.; Kosar, A.; Uvet, H. Microfluidic wound scratching platform based on an untethered microrobot with magnetic actuation. Sens. Actuators B Chem. 2022, 373, 132643. [Google Scholar] [CrossRef]
- Lin, J.Y.; Lo, K.Y.; Sun, Y.S. A microfluidics-based wound-healing assay for studying the effects of shear stresses, wound widths, and chemicals on the wound-healing process. Sci. Rep. 2019, 9, 20016. [Google Scholar] [CrossRef]
- Keese, C.R.; Wegener, J.; Walker, S.R.; Giaever, L. Electrical wound-healing assay for cells. Proc. Natl. Acad. Sci. USA 2004, 101, 1554–1559. [Google Scholar] [CrossRef] [PubMed]
- Hettler, A.; Werner, S.; Eick, S.; Laufer, S.; Weise, F. A New in vitro model to Study Cellular Responses after Thermomechanical Damage in Monolayer Cultures. PLoS ONE 2013, 8, e82635. [Google Scholar] [CrossRef] [PubMed]
- Zordan, M.D.; Mill, C.P.; Riese, D.J.; Leary, J.F. A High Throughput, Interactive Imaging, Bright-Field Wound Healing Assay. Cytom. Part A 2011, 79, 227–232. [Google Scholar] [CrossRef] [PubMed]
- Monfared, G.S.; Ertl, P.; Rothbauer, M. Microfluidic and Lab-on-a-Chip Systems for Cutaneous Wound Healing Studies. Pharmaceutics 2021, 13, 793. [Google Scholar] [CrossRef] [PubMed]
- Van der Meer, A.D.; Vermeul, K.; Poot, A.A.; Feijen, J.; Vermes, I. A microfluidic wound-healing assay for quantifying endothelial cell migration. Am. J. Physiol. Heart C 2010, 298, H719–H725. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.Y.; Sun, Y.S.; Cheng, K.C.; Lo, K.Y. A Wound-Healing Assay Based on Ultraviolet Light Ablation. SLAS Technol. 2017, 22, 36–43. [Google Scholar] [CrossRef] [PubMed]
- Suarez-Arnedo, A.; Figueroa, F.T.; Clavijo, C.; Arbeláez, P.; Cruz, J.C.; Muñoz-Camargo, C. An image J plugin for the high throughput image analysis of in vitro scratch wound healing assays. PLoS ONE 2020, 15, e0232565. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.J.; Yang, G.S.; Lin, Y.Z.; Ji, C.L.; Gupta, M.M. On Definition of Deep Learning. In Proceedings of the World Automat Congress (WAC), Stevenson, WA, USA, 3–6 June 2018; pp. 232–236. [Google Scholar]
- Javer, A.; Rittscher, J.; Sailem, H.Z. DeepScratch: Single-cell based topological metrics of scratch wound assays. Comput. Struct. Biotechnol. J. 2020, 18, 2501–2509. [Google Scholar] [CrossRef]
- Moen, E.; Bannon, D.; Kudo, T.; Graf, W.; Covert, M.; Van Valen, D. Deep learning for cellular image analysis. Nat. Methods 2019, 16, 1233–1246. [Google Scholar] [CrossRef]
- Oldenburg, J.; Maletzki, L.; Strohbach, A.; Bellé, P.; Siewert, S.; Busch, R.; Felix, S.B.; Schmitz, K.P.; Stiehm, M. Methodology for comprehensive cell-level analysis of wound healing experiments using deep learning in MATLAB. BMC Mol. Cell Biol. 2021, 22, 32. [Google Scholar] [CrossRef]
- Sperber, M.; Hupf, C.; Lemberger, M.-M.; Goricnik, B.; Hinterreiter, N.; Lukic, S.; Oberleitner, M.; Stolwijk, J.A.; Wegener, J. Monitoring the Impact of Nanomaterials on Animal Cells by Impedance Analysis: A Noninvasive, Label-Free, and Multimodal Approach. In Measuring Biological Impacts of Nanomaterials; Wegener, J., Ed.; Springer International Publishing: Cham, Switzerland, 2016; pp. 45–108. [Google Scholar]
- Pena, A.M.; Chen, X.Q.; Pence, I.J.; Bornschlögl, T.; Jeong, S.; Grégoire, S.; Luengo, G.S.; Hallegot, P.; Obeidy, P.; Feizpour, A.; et al. Imaging and quantifying drug delivery in skin—Part 2: Fluorescence and vibrational spectroscopic imaging methods. Adv. Drug Deliver Rev. 2020, 153, 147–168. [Google Scholar] [CrossRef] [PubMed]
- Tian, F.; Cai, L.L.; Chang, J.Q.; Li, S.S.; Liu, C.; Li, T.J.; Sun, J.S. Label-free isolation of rare tumor cells from untreated whole blood by interfacial viscoelastic microfluidics. Lab. Chip 2018, 18, 3436–3445. [Google Scholar] [CrossRef] [PubMed]
- Kurth, F.; Eyer, K.; Franco-Obregón, A.; Dittrich, P.S. A new mechanobiological era: Microfluidic pathways to apply and sense forces at the cellular level. Curr. Opin. Chem. Biol. 2012, 16, 400–408. [Google Scholar] [CrossRef]
- Polacheck, W.J.; Li, R.; Uzel, S.G.M.; Kamm, R.D. Microfluidic platforms for mechanobiology. Lab. Chip 2013, 13, 2252–2267. [Google Scholar] [CrossRef] [PubMed]
- Shemesh, J.; Jalilian, I.; Shi, A.; Yeoh, G.H.; Tate, M.L.K.; Warkiani, M.E. Flow-induced stress on adherent cells in microfluidic devices. Lab. Chip 2015, 15, 4114–4127. [Google Scholar] [CrossRef] [PubMed]
- Kaarj, K.; Yoon, J.Y. Methods of Delivering Mechanical Stimuli to Organ-on-a-Chip. Micromachines 2019, 10, 700. [Google Scholar] [CrossRef] [PubMed]
- Vivas, A.; Van den Berg, A.; Passier, R.; Odijk, M.; Van der Meer, A.D. Fluidic circuit board with modular sensor and valves enables stand-alone, tubeless microfluidic flow control in organs-on-chips. Lab. Chip 2022, 22, 1231–1243. [Google Scholar] [CrossRef]
- Mohith, S.; Karanth, P.N.; Kulkarni, S.M. Recent trends in mechanical micropumps and their applications: A review. Mechatronics 2019, 60, 34–55. [Google Scholar] [CrossRef]
- Saias, L.; Autebert, J.; Malaquin, L.; Viovy, J.L. Design, modeling and characterization of microfluidic architectures for high flow rate, small footprint microfluidic systems. Lab. Chip 2011, 11, 822–832. [Google Scholar] [CrossRef]
- Schimek, K.; Busek, M.; Brincker, S.; Groth, B.; Hoffmann, S.; Lauster, R.; Lindner, G.; Lorenz, A.; Menzel, U.; Sonntag, F.; et al. Integrating biological vasculature into a multi-organ-chip microsystem. Lab. Chip 2013, 13, 3588–3598. [Google Scholar] [CrossRef]
- Li, J.M.; Wei, J.; Liu, Y.C.; Liu, B.; Liu, T.; Jiang, Y.; Ding, L.Q.; Liu, C. A microfluidic design to provide a stable and uniform microenvironment for cell culture inspired by the redundancy characteristic of leaf areoles. Lab. Chip 2017, 17, 3921–3933. [Google Scholar] [CrossRef] [PubMed]
- Bhushan, B.M.; Yoon, J.Y.; Griffith, L.G.; Trumper, D.L. Flux-Biased, Energy-Efficient Electromagnetic Micropumps Utilizing Bistable Magnetic Latching and Energy-Storage Springs. IEEE/ASME Trans. Mechatron. 2021, 26, 2362–2372. [Google Scholar] [CrossRef]
- Liu, B.D.; Sun, J.C.; Li, D.S.; Zhe, J.; Oh, K.W. A high flow rate thermal bubble-driven micropump with induction heating. Microfluid. Nanofluid. 2016, 20, 155. [Google Scholar] [CrossRef]
- Morimoto, Y.; Onoe, H.; Takeuchi, S. Biohybrid robot powered by an antagonistic pair of skeletal muscle tissues. Sci. Robot. 2018, 3, eaat4440. [Google Scholar] [CrossRef]
- Polacheck, W.J.; Chen, C.S. Measuring cell-generated forces: A guide to the available tools. Nat. Methods 2016, 13, 415–423. [Google Scholar] [CrossRef]
- Hackelbusch, S.; Rossow, T.; Steinhilber, D.; Weitz, D.A.; Seiffert, S. Hybrid Microgels with Thermo-Tunable Elasticity for Controllable Cell Confinement. Adv. Healthc. Mater. 2015, 4, 1841–1848. [Google Scholar] [CrossRef] [PubMed]
- Gelmi, A.; Cieslar-Pobuda, A.; de Muinck, E.; Los, M.; Rafat, M.; Jager, E.W.H. Direct Mechanical Stimulation of Stem Cells: A Beating Electromechanically Active Scaffold for Cardiac Tissue Engineering. Adv. Healthc. Mater. 2016, 5, 1471–1480. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.M.; Li, H.; Lam, K.Y. Modeling of a fast-response magnetic-sensitive hydrogel for dynamic control of microfluidic flow. Phys. Chem. Chem. Phys. 2019, 21, 1852–1862. [Google Scholar] [CrossRef] [PubMed]
- Shi, Z.J.; Gao, X.; Ullah, M.W.; Li, S.X.; Wang, Q.; Yang, G. Electroconductive natural polymer-based hydrogels. Biomaterials 2016, 111, 40–54. [Google Scholar] [CrossRef]
- Xin, F.X.; Lu, T.J. Acoustomechanical giant deformation of soft elastomers with interpenetrating networks. Smart Mater. Struct. 2016, 25, 07lt02. [Google Scholar] [CrossRef]
- Gillette, B.M.; Jensen, J.A.; Wang, M.X.; Tchao, J.; Sia, S.K. Dynamic Hydrogels: Switching of 3D Microenvironments Using Two-Component Naturally Derived Extracellular Matrices. Adv. Mater. 2010, 22, 686. [Google Scholar] [CrossRef] [PubMed]
- Guo, W.W.; Lu, C.H.; Qi, X.J.; Orbach, R.; Fadeev, M.; Yang, H.H.; Willner, I. Switchable Bifunctional Stimuli-Triggered Poly-Isopropylacrylamide/DNA Hydrogels. Angew. Chem. Int. Edit 2014, 53, 10134–10138. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.Y.; Korc, M.; Lin, C.C. Biomimetic and enzyme-responsive dynamic hydrogels for studying cell-matrix interactions in pancreatic ductal adenocarcinoma. Biomaterials 2018, 160, 24–36. [Google Scholar] [CrossRef] [PubMed]
- Yin, R.X.; He, J.; Bai, M.R.; Huang, C.; Wang, K.M.; Zhang, H.B.; Yang, S.M.; Zhang, W.J. Engineering synthetic artificial pancreas using chitosan hydrogels integrated with glucose-responsive microspheres for insulin delivery. Mat. Sci. Eng. C-Mater. 2019, 96, 374–382. [Google Scholar] [CrossRef] [PubMed]
- Yoshikawa, H.Y.; Rossetti, F.F.; Kaufmann, S.; Kaindl, T.; Madsen, J.; Engel, U.; Lewis, A.L.; Armes, S.P.; Tanaka, M. Quantitative evaluation of mechanosensing of cells on dynamically tunable hydrogels. J. Am. Chem. Soc. 2011, 133, 1367–1374. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.W.; Sun, N.; Bruce, M.A.; Wu, J.C.; Butte, M.J. Atomic Force Mechanobiology of Pluripotent Stem Cell-Derived Cardiomyocytes. PLoS ONE 2012, 7, e37559. [Google Scholar] [CrossRef] [PubMed]
- Huber, D.; Oskooei, A.; Solvas, X.C.I.; DeMello, A.; Kaigala, G.V. Hydrodynamics in Cell Studies. Chem. Rev. 2018, 118, 2042–2079. [Google Scholar] [CrossRef]
- Papaioannou, T.G.; Stefanadis, C. Vascular wall shear stress: Basic principles and methods. Hell. J. Cardiol. 2005, 46, 9–15. [Google Scholar]
- Schoen, I.; Pruitt, B.L.; Vogel, V. The Yin-Yang of Rigidity Sensing: How Forces and Mechanical Properties Regulate the Cellular Response to Materials. Annu. Rev. Mater. Res. 2013, 43, 589–618. [Google Scholar] [CrossRef]
- Ladoux, B.; Nicolas, A. Physically based principles of cell adhesion mechanosensitivity in tissues. Rep. Prog. Phys. 2012, 75, 116601. [Google Scholar] [CrossRef]
- Saez, A.; Ghibaudo, M.; Buguin, A.; Silberzan, P.; Ladoux, B. Rigidity-driven growth and migration of epithelial cells on microstructured anisotropic substrates. Proc. Natl. Acad. Sci. USA 2007, 104, 8281–8286. [Google Scholar] [CrossRef] [PubMed]
- Leipzig, N.D.; Shoichet, M.S. The effect of substrate stiffness on adult neural stem cell behavior. Biomaterials 2009, 30, 6867–6878. [Google Scholar] [CrossRef] [PubMed]
- Tse, J.R.; Engler, A.J. Stiffness Gradients Mimicking in vivo Tissue Variation Regulate Mesenchymal Stem Cell Fate. PLoS ONE 2011, 6, e15978. [Google Scholar] [CrossRef] [PubMed]
- Chen, A.; Yin, R.X.; Cao, L.; Yuan, C.W.; Ding, H.K.; Zhang, W.J. Soft Robotics: Definition and Research Issues. In Proceedings of the 24th International Conference on Mechatronics and Machine Vision in Practice (M2VIP), Auckland, New Zealand, 21–23 November 2017; pp. 366–370. [Google Scholar] [CrossRef]
- Huang, J.L.; Clement, R.; Sun, Z.H.; Wang, J.Z.; Zhang, W.J. Global stiffness and natural frequency analysis of distributed compliant mechanisms with embedded actuators with a general-purpose finite element system. Int. J. Adv. Manuf. Technol. 2013, 65, 1111–1124. [Google Scholar] [CrossRef]
- Fu, J.P.; Wang, Y.K.; Yang, M.T.; Desai, R.A.; Yu, X.A.; Liu, Z.J.; Chen, C.S. Mechanical regulation of cell function with geometrically modulated elastomeric substrates. Nat. Methods 2010, 7, 733–736. [Google Scholar] [CrossRef]
- Nava, M.M.; Raimondi, M.T.; Pietrabissa, R. Controlling self-renewal and differentiation of stem cells via mechanical cues. J. Biomed. Biotechnol. 2012, 2012, 797410. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.S.; Chen, X.B.; Yang, Q.; Zhang, W.J. Development and characterization of a novel piezoelectric-driven stick-slip actuator with anisotropic-friction surfaces. Int. J. Adv. Manuf. Technol. 2012, 61, 1029–1034. [Google Scholar] [CrossRef]
- Zhang, Z.M.; An, Q.; Zhang, W.J.; Yang, Q.; Tang, Y.J.; Chen, X.B. Modeling of directional friction on a fully lubricated surface with regular anisotropic asperities. Meccanica 2011, 46, 535–545. [Google Scholar] [CrossRef]
- Steward, A.J.; Kelly, D.J. Mechanical regulation of mesenchymal stem cell differentiation. J. Anat. 2015, 227, 717–731. [Google Scholar] [CrossRef]
- Paul, C.D.; Hung, W.-C.; Wirtz, D.; Konstantopoulos, K. Engineered models of confined cell migration. Annu. Rev. Biomed. Eng. 2016, 18, 159–180. [Google Scholar] [CrossRef]
- Yamada, K.M.; Sixt, M. Mechanisms of 3D cell migration. Nat. Rev. Mol. Cell Bio 2019, 20, 738–752. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.B.; Chen, C.S.; Fu, J.P. Forcing Stem Cells to Behave: A Biophysical Perspective of the Cellular Microenvironment. Annu. Rev. Biophys. 2012, 41, 519–542. [Google Scholar] [CrossRef] [PubMed]
- Forero, M.; Yakovenko, O.; Sokurenko, E.V.; Thomas, W.E.; Vogel, V. Uncoiling mechanics of Escherichia coli Type I fimbriae are optimized for catch bonds. PLoS Biol. 2006, 4, e298. [Google Scholar] [CrossRef] [PubMed]
- Sun, Z.Q.; Guo, S.S.; Fässler, R. Integrin-mediated mechanotransduction. J. Cell Biol. 2016, 215, 445–456. [Google Scholar] [CrossRef] [PubMed]
- Huang, G.Y.; Li, F.; Zhao, X.; Ma, Y.F.; Li, Y.H.; Lin, M.; Jin, G.R.; Lu, T.J.; Genin, G.M.; Xu, F. Functional and Biomimetic Materials for Engineering of the Three-Dimensional Cell Microenvironment. Chem. Rev. 2017, 117, 12764–12850. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.Y.; Liu, J.; Lin, S.T.; Zhao, X.H. Hydrogel machines. Mater. Today 2020, 36, 102–124. [Google Scholar] [CrossRef]
- Liu, L.M.; Shadish, J.A.; Arakawa, C.K.; Shi, K.; Davis, J.; DeForest, C.A. Cyclic Stiffness Modulation of Cell-Laden Protein-Polymer Hydrogels in Response to User-Specified Stimuli Including Light. Adv. Biosyst. 2018, 2, 1800240. [Google Scholar] [CrossRef] [PubMed]
- Fu, L.L.; Haage, A.; Kong, N.; Tanentzapf, G.; Li, H.B. Dynamic protein hydrogels with reversibly tunable stiffness regulate human lung fibroblast spreading reversibly. Chem. Commun. 2019, 55, 5235–5238. [Google Scholar] [CrossRef] [PubMed]
- Arkenberg, M.R.; Moore, D.M.; Lin, C.C. Dynamic control of hydrogel crosslinking via sortase-mediated reversible transpeptidation. Acta Biomater. 2019, 83, 83–95. [Google Scholar] [CrossRef]
- Basoli, F.; Giannitelli, S.M.; Gori, M.; Mozetic, P.; Bonfanti, A.; Trombetta, M.; Rainer, A. Biomechanical Characterization at the Cell Scale: Present and Prospects. Front. Physiol. 2018, 9, 1449. [Google Scholar] [CrossRef]
- Roca-Cusachs, P.; Conte, V.; Trepat, X. Quantifying forces in cell biology. Nat. Cell Biol. 2017, 19, 742–751. [Google Scholar] [CrossRef] [PubMed]
- Matellan, C.; Hernández, A.E.D. Where No Hand Has Gone Before: Probing Mechanobiology at the Cellular Level. ACS Biomater. Sci. Eng. 2019, 5, 3703–3719. [Google Scholar] [CrossRef] [PubMed]
- Boldock, L.; Wittkowske, C.; Perrault, C.M. Microfluidic traction force microscopy to study mechanotransduction in angiogenesis. Microcirculation 2017, 24, e12361. [Google Scholar] [CrossRef]
- Style, R.W.; Boltyanskiy, R.; German, G.K.; Hyland, C.; MacMinn, C.W.; Mertz, A.F.; Wilen, L.A.; Xu, Y.; Dufresne, E.R. Traction force microscopy in physics and biology. Soft Matter 2014, 10, 4047–4055. [Google Scholar] [CrossRef] [PubMed]
- Subramaniam, T.; Fauzi, M.B.; Lokanathan, Y.; Law, J.X. The Role of Calcium in Wound Healing. Int. J. Mol. Sci. 2021, 22, 6486. [Google Scholar] [CrossRef] [PubMed]
- Tony, A.; Rasouli, A.; Farahinia, A.; Wells, G.; Zhang, H.; Achenbach, S.; Yang, S.M.; Sun, W.; Zhang, W. Toward a Soft Microfluidic System: Concept and Preliminary Developments. In Proceedings of the 2021 27th International Conference on Mechatronics and Machine Vision in Practice (M2VIP), Shanghai, China, 26–28 November 2021; pp. 755–759. [Google Scholar]
- Rodrigues, M.; Kosaric, N.; Bonham, C.A.; Gurtner, G.C. Wound Healing: A Cellular Perspective. Physiol. Rev. 2019, 99, 665–706. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Chen, J.; Kirsner, R. Pathophysiology of acute wound healing. Clin. Dermatol. 2007, 25, 9–18. [Google Scholar] [CrossRef]
- Velnar, T.; Bailey, T.; Smrkoli, V. The Wound Healing Process: An Overview of the Cellular and Molecular Mechanisms. J. Int. Med. Res. 2009, 37, 1528–1542. [Google Scholar] [CrossRef] [PubMed]
- Gurtner, G.C.; Werner, S.; Barrandon, Y.; Longaker, M.T. Wound repair and regeneration. Nature 2008, 453, 314–321. [Google Scholar] [CrossRef]
- Deal, H.E.; Brown, A.C.; Daniele, M.A. Microphysiological systems for the modeling of wound healing and evaluation of pro-healing therapies. J. Mater. Chem. B 2020, 8, 7062–7075. [Google Scholar] [CrossRef]
- Contardi, M.; Russo, D.; Suarato, G.; Heredia-Guerrero, J.A.; Ceseracciu, L.; Penna, I.; Margaroli, N.; Summa, M.; Spanò, R.; Tassistro, G. Polyvinylpyrrolidone/hyaluronic acid-based bilayer constructs for sequential delivery of cutaneous antiseptic and antibiotic. Chem. Eng. J. 2019, 358, 912–923. [Google Scholar] [CrossRef]
- Sala, F.; Ficorella, C.; Osellame, R.; Käs, J.A.; Vázquez, R.M. Microfluidic Lab-on-a-Chip for Studies of Cell Migration under Spatial Confinement. Biosensors 2022, 12, 604. [Google Scholar] [CrossRef] [PubMed]
- Jin, W.; Lo, K.Y.; Chou, S.E.; Mccue, S.W.; Simpson, M.J. The role of initial geometry in experimental models of wound closing. Chem. Eng. Sci. 2018, 179, 221–226. [Google Scholar] [CrossRef]
- Fan, L.X.; Cai, M.Y.; Lin, Y.; Zhang, W.J. Axiomatic design theory: Further notes and its guideline to applications. Int. J. Mater. Prod. Technol. 2015, 51, 359–374. [Google Scholar] [CrossRef]
- Nair, V.N.; Abraham, B.; MacKay, J.; Box, G.; Kacker, R.N.; Lorenzen, T.J.; Lucas, J.M.; Myers, R.H.; Vining, G.G.; Nelder, J.A. Taguchi’s parameter design: A panel discussion. Technometrics 1992, 34, 127–161. [Google Scholar] [CrossRef]
- Fallahi, H.; Zhang, J.; Phan, H.P.; Nguyen, N.T. Flexible Microfluidics: Fundamentals, Recent Developments, and Applications. Micromachines 2019, 10, 830. [Google Scholar] [CrossRef]
- Kratz, S.R.A.; Eilenberger, C.; Schuller, P.; Bachmann, B.; Spitz, S.; Ertl, P.; Rothbauer, M. Characterization of four functional biocompatible pressure-sensitive adhesives for rapid prototyping of cell-based lab-on-a-chip and organ-on-a-chip systems. Sci. Rep. 2019, 9, 9287. [Google Scholar] [CrossRef] [PubMed]
- Niculescu, A.-G.; Chircov, C.; Bîrcă, A.C.; Grumezescu, A.M. Fabrication and applications of microfluidic devices: A review. Int. J. Mol. Sci. 2021, 22, 2011. [Google Scholar] [CrossRef]
- Bi, Z.M.; Zhang, W.J. Modularity Technology in Manufacture Environment: Taxonomy and Issue. Int. J. Adv. Manuf. Technol. 2001, 18, 381–390. [Google Scholar] [CrossRef]
- Millet, L.J.; Lucheon, J.D.; Standaert, R.F.; Retterer, S.T.; Doktycz, M.J. Modular microfluidics for point-of-care protein purifications. Lab. Chip 2015, 15, 1799–1811. [Google Scholar] [CrossRef]
- Yang, S.M.; Lv, S.S.; Zhang, W.J.; Cui, Y.B. Microfluidic Point-of-Care (POC) Devices in Early Diagnosis: A Review of Opportunities and Challenges. Sensors 2022, 22, 1620. [Google Scholar] [CrossRef]
- Dekker, S.; Buesink, W.; Blom, M.; Alessio, M.; Verplanck, N.; Hihoud, M.; Dehan, C.; César, W.; Le Nel, A.; van den Berg, A.; et al. Standardized and modular microfluidic platform for fast Lab on Chip system development. Sens. Actuators B Chem. 2018, 272, 468–478. [Google Scholar] [CrossRef]
- Sun, Y.S.; Peng, S.W.; Cheng, J.Y. Electrical-stimulated wound-healing chip for studying electric field-assisted wound-healing process. Biomicrofluidics 2012, 6, 034117. [Google Scholar] [CrossRef]
- Lin, J.Y.; Lo, K.Y.; Sun, Y.S. Effects of Substrate-Coating Materials on the Wound-Healing Process. Materials 2019, 12, 2775. [Google Scholar] [CrossRef] [PubMed]
- Jin, W.; Lo, K.Y.; Sun, Y.S.; Ting, Y.H.; Simpson, M.J. Quantifying the role of different surface coatings in experimental models of wound. Chem. Eng. Sci. 2020, 220, 115609. [Google Scholar] [CrossRef]
- Alqurashi, H.O.A., I; Lambert, D.W. The Emerging Potential of Extracellular Vesicles in Cell-Free Tissue Engineering and Regenerative Medicine. Tissue Eng. Part B Rev. 2021, 27, 530–538. [Google Scholar] [CrossRef] [PubMed]
- Yin, L.; Du, G.R.; Zhang, B.; Zhang, H.B.; Yin, R.X.; Zhang, W.J.; Yang, S.M. Efficient Drug Screening and Nephrotoxicity Assessment on Co-culture Microfluidic Kidney Chip. Sci. Rep. 2020, 10, 6568. [Google Scholar] [CrossRef]
- Taylor, M.; Jaunky, T.; Hewitt, K.; Lowe, F.; Fearon, I.; Gaca, M. A comparative assessment of e-cigarette aerosols and cigarette smoke on endothelial cell migration. Toxicol. Lett. 2017, 280, S235–S236. [Google Scholar] [CrossRef]
- Ma, Y.; Pan, J.Z.; Zhao, S.P.; Lou, Q.; Zhu, Y.; Fang, Q. Microdroplet chain array for cell migration assays. Lab. Chip 2016, 16, 4658–4665. [Google Scholar] [CrossRef]
- Ren, J.Q.; Chen, W.F.; Zhong, Z.C.; Wang, N.; Chen, X.; Yang, H.; Li, J.; Tang, P.; Fan, Y.P.; Lin, F.C.; et al. Bronchoalveolar Lavage Fluid from Chronic Obstructive Pulmonary Disease Patients Increases Neutrophil Chemotaxis Measured by a Microfluidic Platform. Micromachines 2023, 14, 1740. [Google Scholar] [CrossRef]
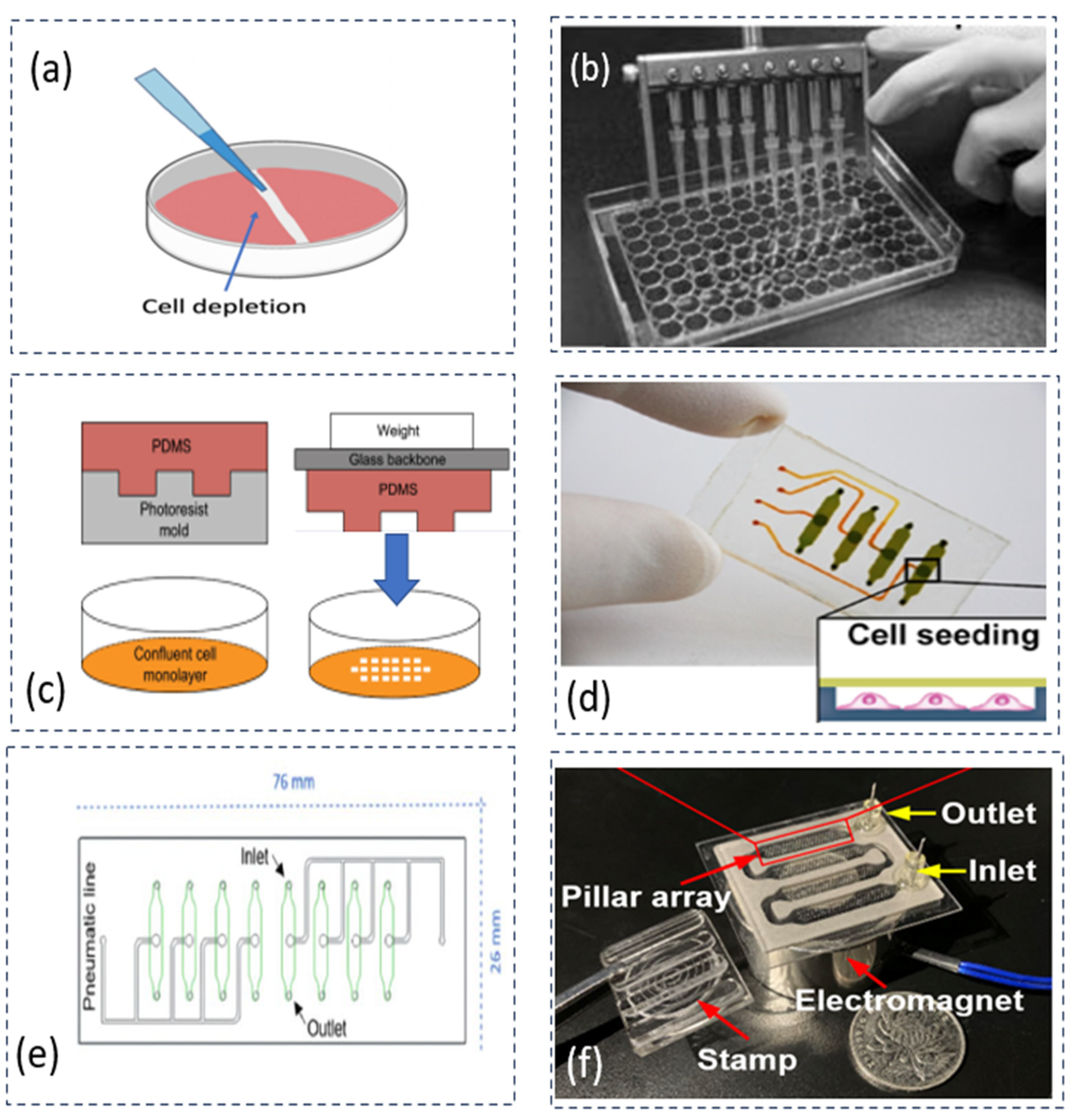


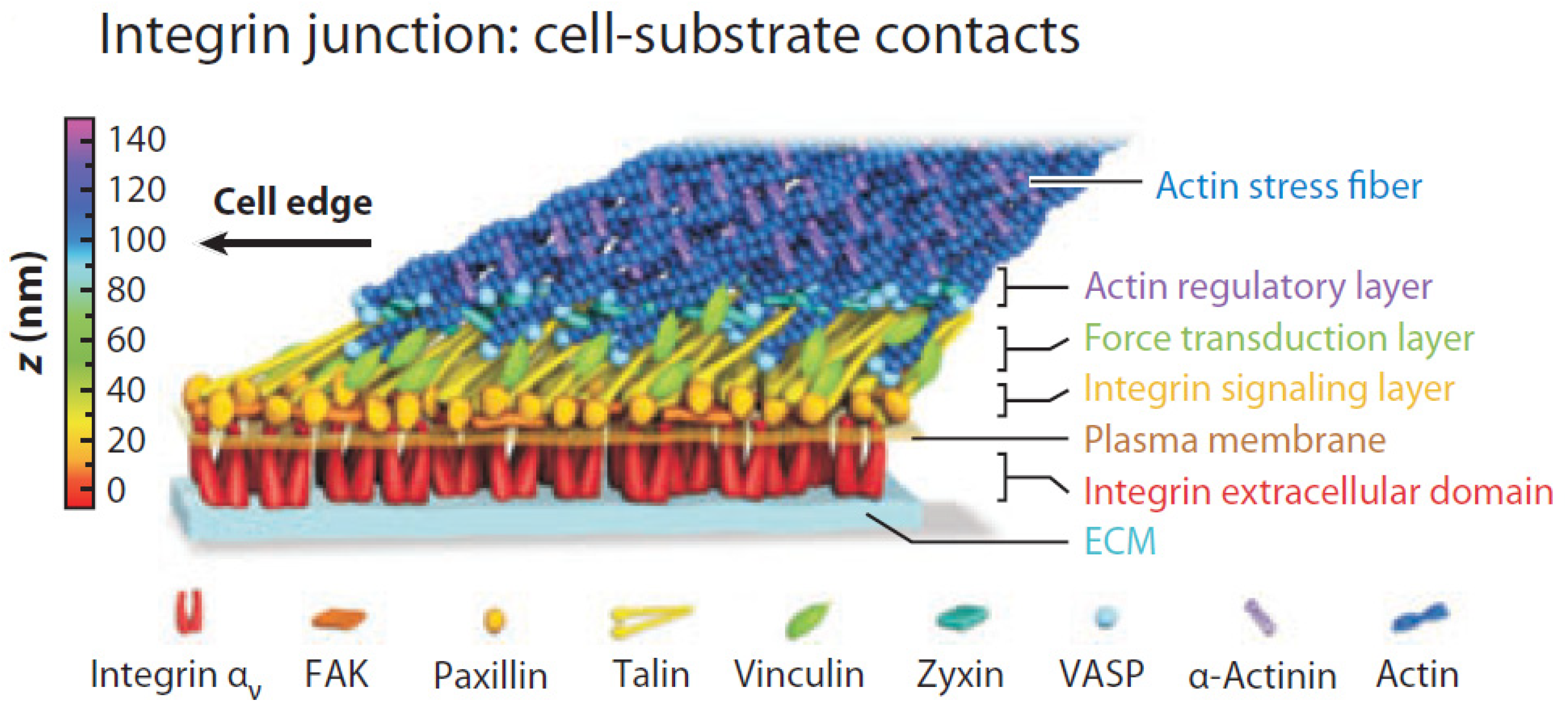
| Principle (Level 0) | Operation Methods | Number of Samples | Size of CFZs | Consistency | Pros | Cons | References |
|---|---|---|---|---|---|---|---|
| Mechanical (DP1-1) | Manual | 1 | 800 μm width | Poor | Simplicity | Irregular size | [10] |
| Manual | 20 | 0.06 mm2 | 6% | Easy fabrication | Cell debris | [27] | |
| Manual | 8 | 600 μm width | Poor | Availability | Lack of shear stress | [28] | |
| Auto | 4 | 0.91 mm2 | 4% | Automation | Bubble formation | [29] | |
| Auto | 40 | 1.5 mm2 | 5% | Controllable force | Bubble formation | [30] | |
| Auto | 400 | 0.126 mm2 | 4% | Large number of samples | Few conditions | [31] | |
| Auto | 1 | 500 μm width | 2% | Different shapes of CFZs | Low efficiency | [32] | |
| Chemical (DP1-2) | n/a | 1 | 3–6 mm width | 5% | Clean CFZs | Affecting cell dynamics | [33] |
| Electrical (DP1-3) | n/a | 1 | 0.05 mm2 | n/a | Easy to control the power | Generating heat | [34] |
| Thermal (DP1-4) | n/a | Few | 5–20 mm2 | Poor | Thermal wounding | Chemical reaction | [35] |
| Stressed cells | |||||||
| Optical (DP1-5) | n/a | 96 | 2 mm2 | Good | Large samples | Costly equipment | [36] |
| Method | Resolution | Pros | Cons | References |
|---|---|---|---|---|
| Label-based (DP2-1) | 0.4–1 μm | Automated imaging Easy to track | Invasive Thin samples (~10 μm) | [45] [46] |
| Label-free (DP2-2) | Poor at single cell (~5 μm) | Samples depth up to 70 μm Non-invasive | Intensive for tracking Low resolution and labor | [47] [48] |
| Principle (Level 1) | Principle (Level 2) | Materials | Stress | Pros | Cons | References |
|---|---|---|---|---|---|---|
| Fluid (DP3a-1) | Electrical | PDMS | <25 dyn/cm2 | Stable power | Non-uniform fluid distribution | [49,50,51,52,53,54,55,56] |
| Electromagnetic | PDMS | 20~60 kpa | Easy to control | Temperature rise at high electric current (~300 mA) | [57] | |
| Pneumatic | PDMS | 102.05 μL/min | Biocompatible | Formation of bubble | [58] | |
| Cells (DP3a-2) | Mechanical | Hydrogels | 2–10 kPa | Biomimetic | Limited force range (~13 mN) | [59] |
| Chemical | Growth factors | 0.1–50 kPa | Chemical conditions | Poor cell dynamics | [60] | |
| Environment (DP3a-3) | Physical | Electrodes magnetics optics acoustics | 2–10 kPa | Sufficient power | External energy Lack of modeling in first principle | [59,61,62,63,64,65] |
| Chemical | PH ions oxygen | 0.1–40 kPa | Easy targeting | Longer reaction time Confounding factors with other chemicals | [66,67,68,69,70] | |
| Probe (DP3b-1) | n/a | Probe | 5 pN to 10 nN | High resolution (0.2 μm) | Few cells Light contamination | [71] |
| Interaction (DP3b-2) | n/a | Fluorescence | 2–120 nN, 0.05–0.6 kPa | Large number of cells (>1000) | Difficult to measure on single cell | [60] |
| Requirements | References | |||||
|---|---|---|---|---|---|---|
| Consistency * (R1) | Geometry (R2) | Shear Stress (R3) | Uniformity (R4) | Number (R5) | ||
| Samples | Conditions | |||||
| Poor | Line | n/a | n/a | 1 | 1 | [10] |
| 6% | Line | n/a | n/a | 8 | 12 | [28] |
| Poor | Square | n/a | n/a | 20 | 1 | [27] |
| n/a | n/a | 0.01 dyn/cm2 | 90% | n/a | 1 | [56] |
| 4% | Circle | 3 μL/min | n/a | 4 | 4 | [29] |
| n/a | Line | 1~18.3 dyn/cm2 | n/a | 3 | 3 | [33] |
| 2~5% | Circle | n/a | n/a | 4 | 8 | [30] |
| 4% | Circle | 1–7 dyn/cm2 | 66% | 400 | 1 | [31] |
| 2% | Multiple | 20 μL/min | n/a | 1 | 1 | [32] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, C.; Yin, D.; Zhang, H.; Badea, I.; Yang, S.-M.; Zhang, W. Cell Migration Assays and Their Application to Wound Healing Assays—A Critical Review. Micromachines 2024, 15, 720. https://doi.org/10.3390/mi15060720
Yang C, Yin D, Zhang H, Badea I, Yang S-M, Zhang W. Cell Migration Assays and Their Application to Wound Healing Assays—A Critical Review. Micromachines. 2024; 15(6):720. https://doi.org/10.3390/mi15060720
Chicago/Turabian StyleYang, Chun, Di Yin, Hongbo Zhang, Ildiko Badea, Shih-Mo Yang, and Wenjun Zhang. 2024. "Cell Migration Assays and Their Application to Wound Healing Assays—A Critical Review" Micromachines 15, no. 6: 720. https://doi.org/10.3390/mi15060720
APA StyleYang, C., Yin, D., Zhang, H., Badea, I., Yang, S. -M., & Zhang, W. (2024). Cell Migration Assays and Their Application to Wound Healing Assays—A Critical Review. Micromachines, 15(6), 720. https://doi.org/10.3390/mi15060720









