Droplet Microfluidics for High-Throughput Screening and Directed Evolution of Biomolecules
Abstract
:1. Introduction
1.1. Comparison of Compartmentalization in Droplets and Multi-Well Plates
1.2. Advantages of Microfluidic Platforms in HTS Applications
1.3. Bulk Emulsification versus Droplet Microfluidics
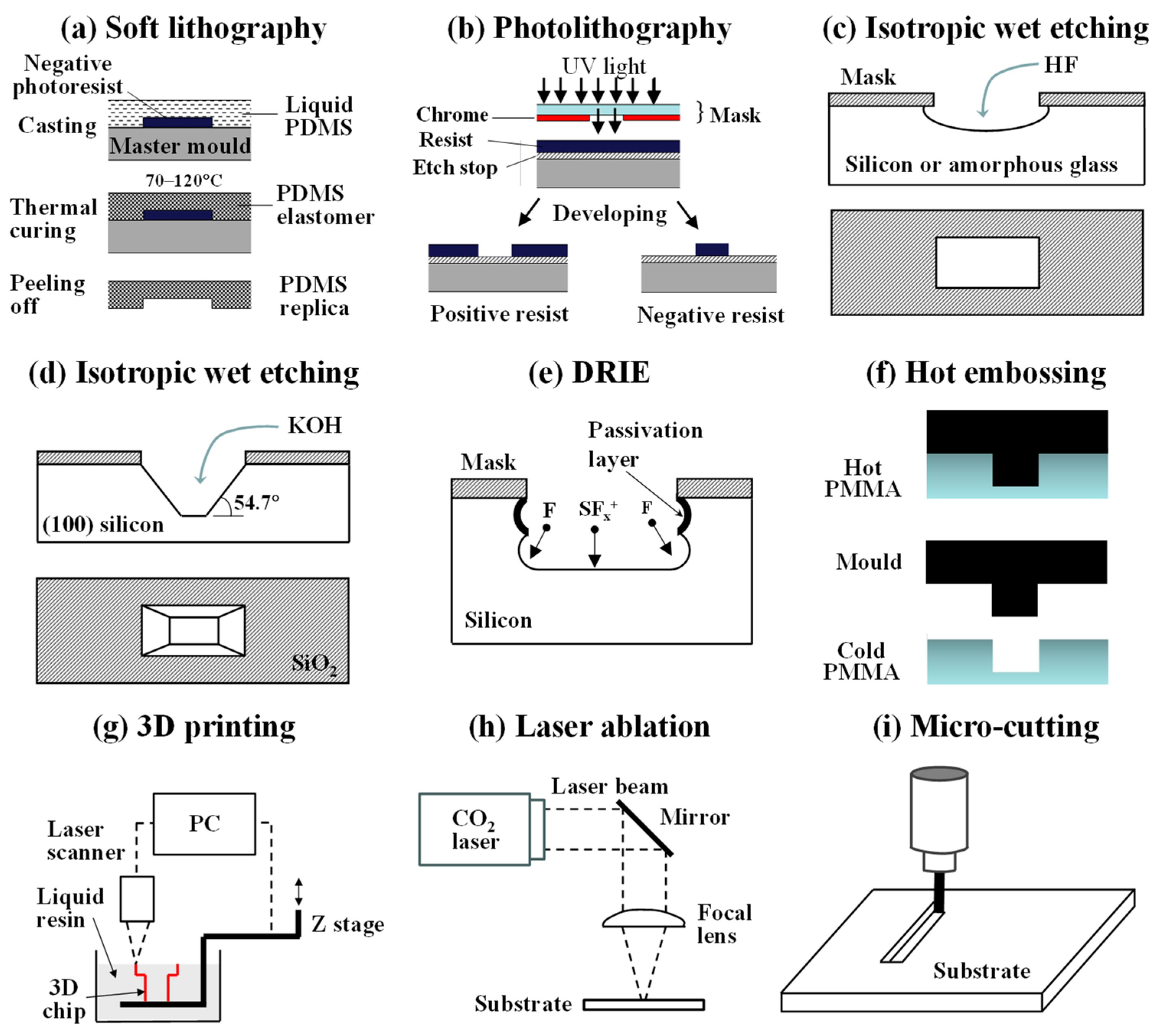
2. Fabrication Methods for Microfluidic Devices
2.1. Soft Lithography
2.2. Photolithography and Etching
2.3. Hot Embossing and Injection Moulding
2.4. Three-Dimensional Printing
2.5. Laser Ablation and Microcutting
2.6. Glass Capillary Pulling
3. Microfluidic Unit Operations for Directed Evolution of Biomolecules
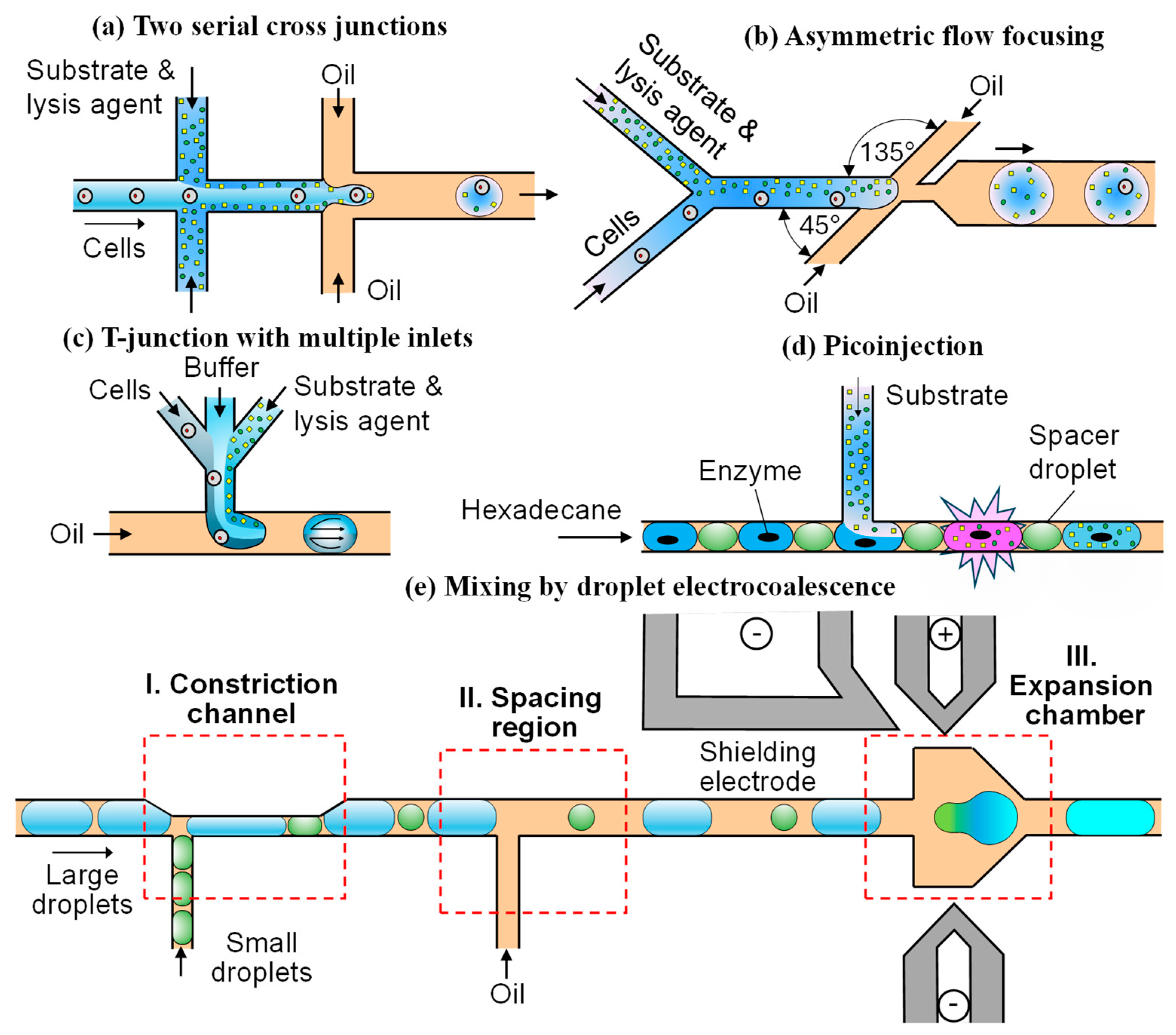
3.1. On-Chip Reagent Mixing
3.1.1. Merging Two Continuous Fluid Streams
3.1.2. Merging a Continuous Fluid Stream and a Droplet
3.1.3. Merging Two Droplets
3.2. Droplet-Based Sample Compartmentalization
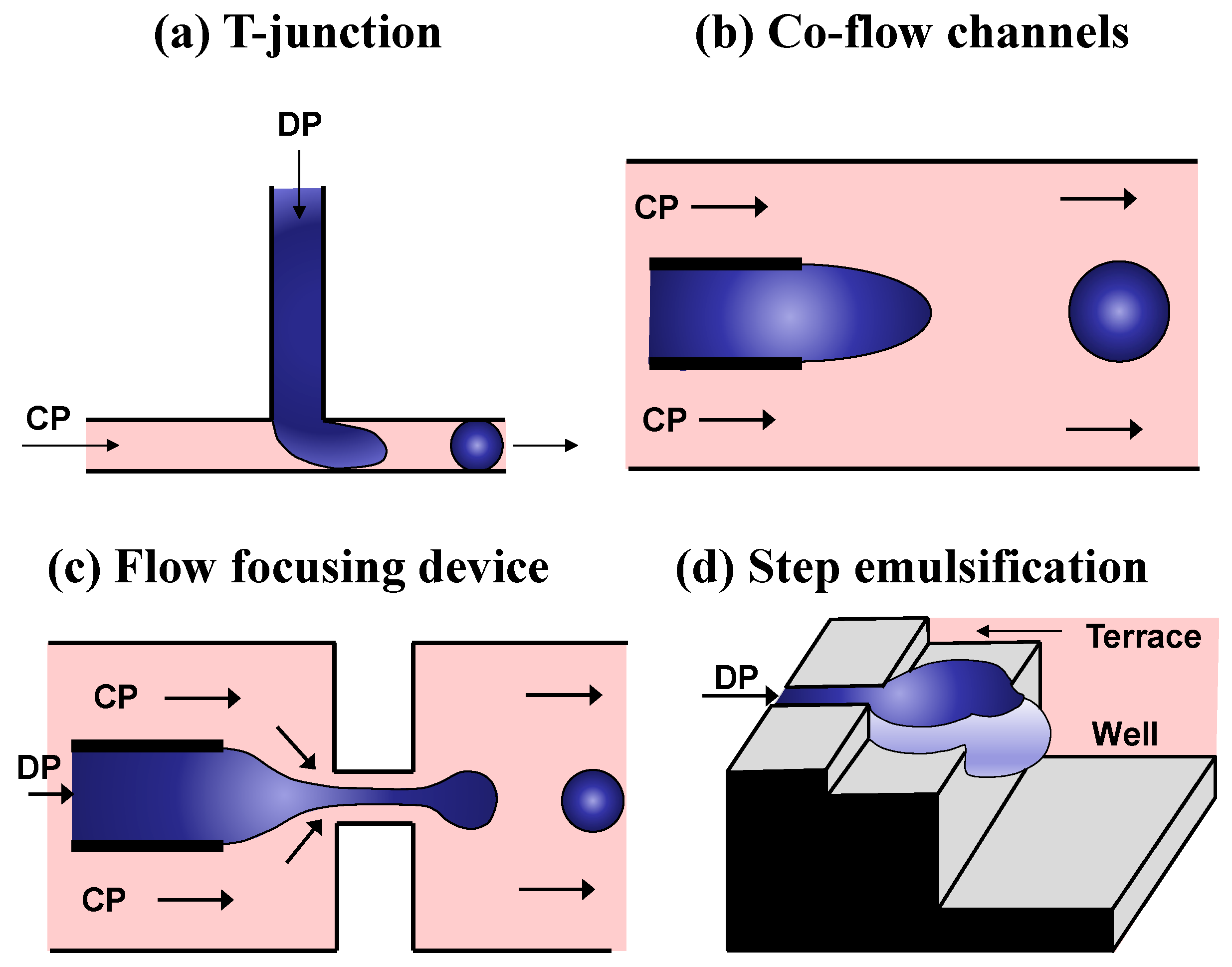
3.3. Droplet Incubation
3.3.1. On-Chip Droplet Incubation in Serpentine Channels
3.3.2. On-Chip Droplet Incubation in Large Chambers
3.3.3. On-Chip Droplet Incubation in Trapping Microwells

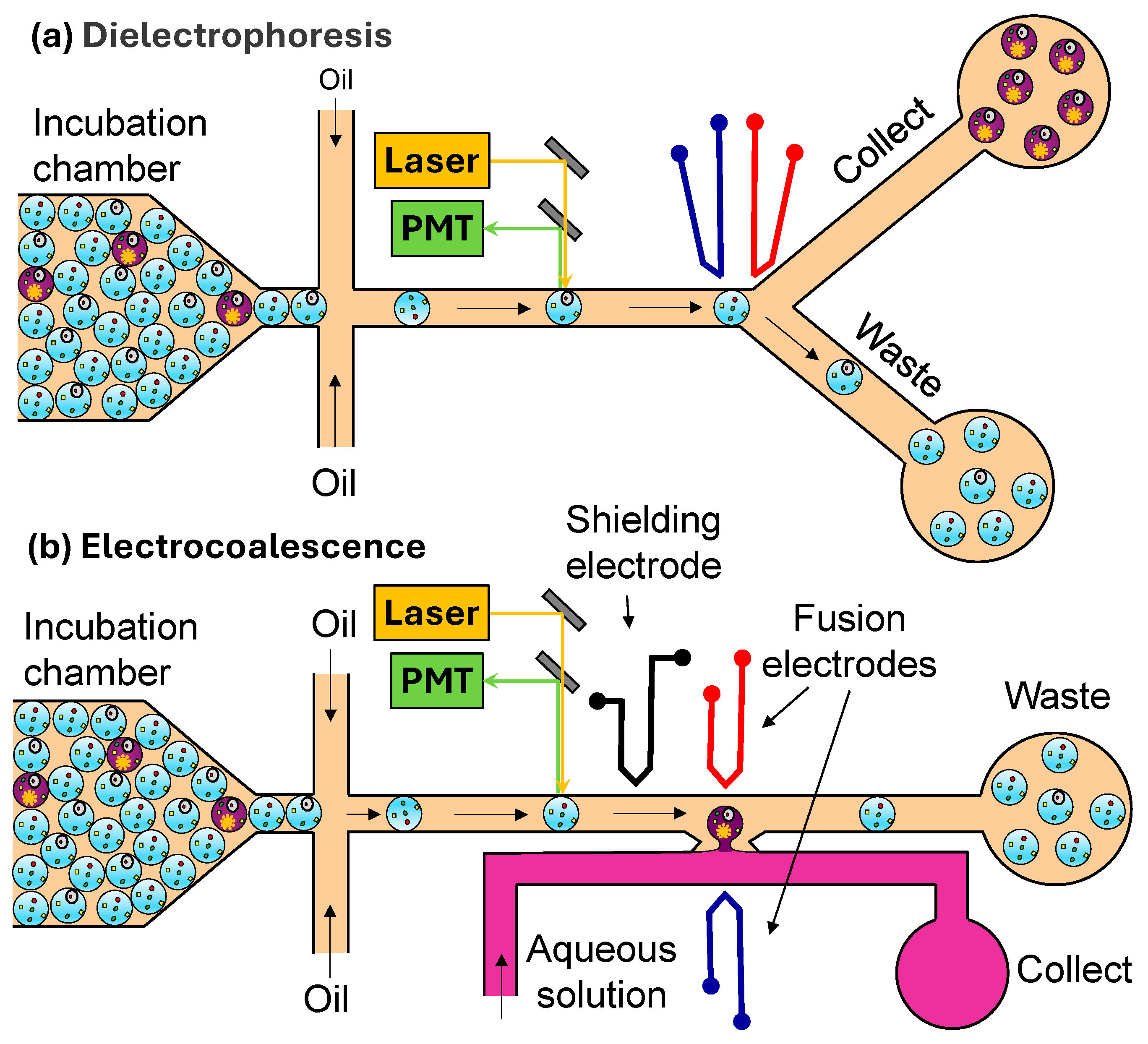
3.4. Droplet Sorting
3.4.1. Signal Detection
3.4.2. Droplet Actuation
4. Directed Evolution of Biomolecules in Water-in-Oil (W/O) Droplets
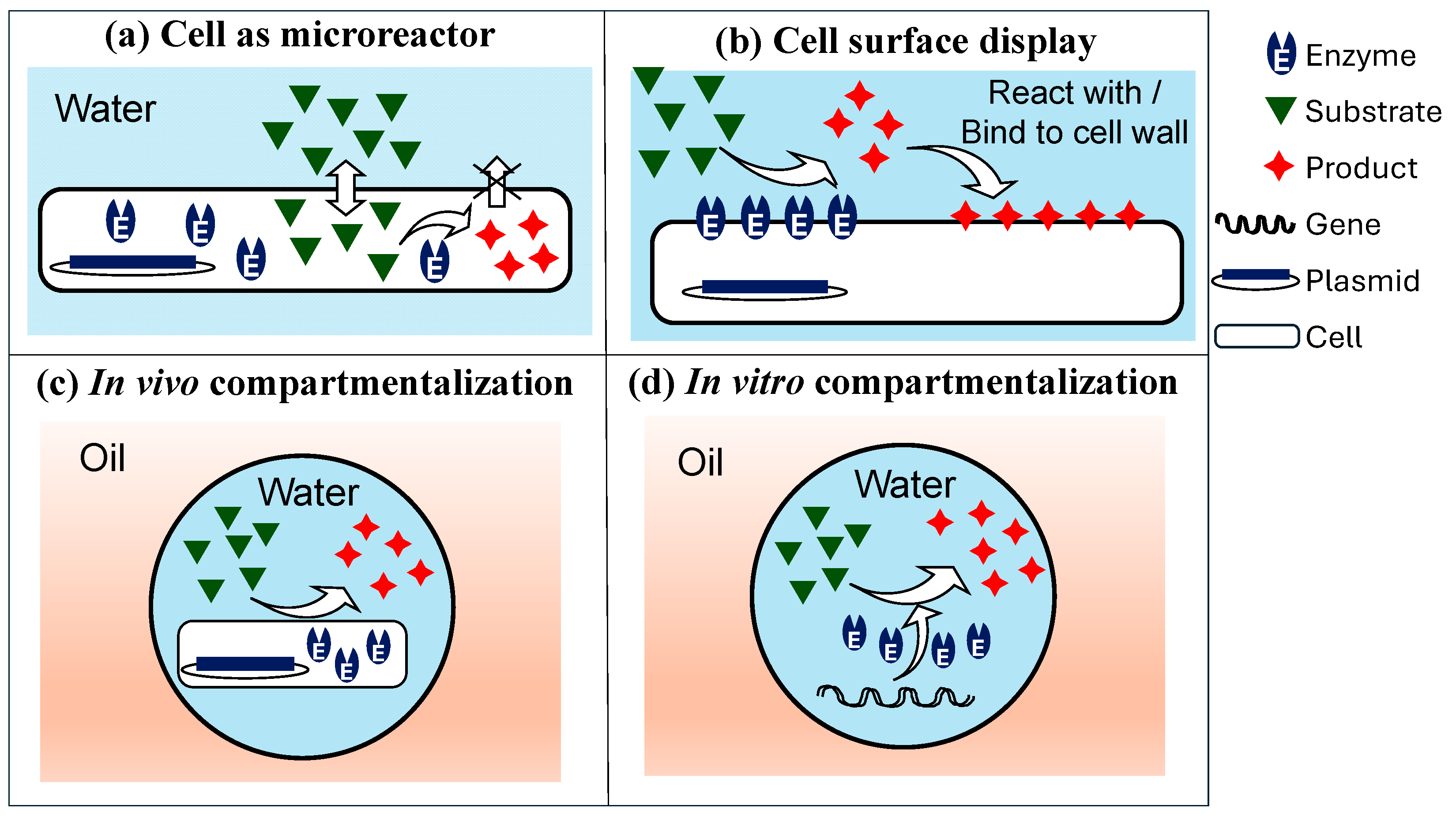
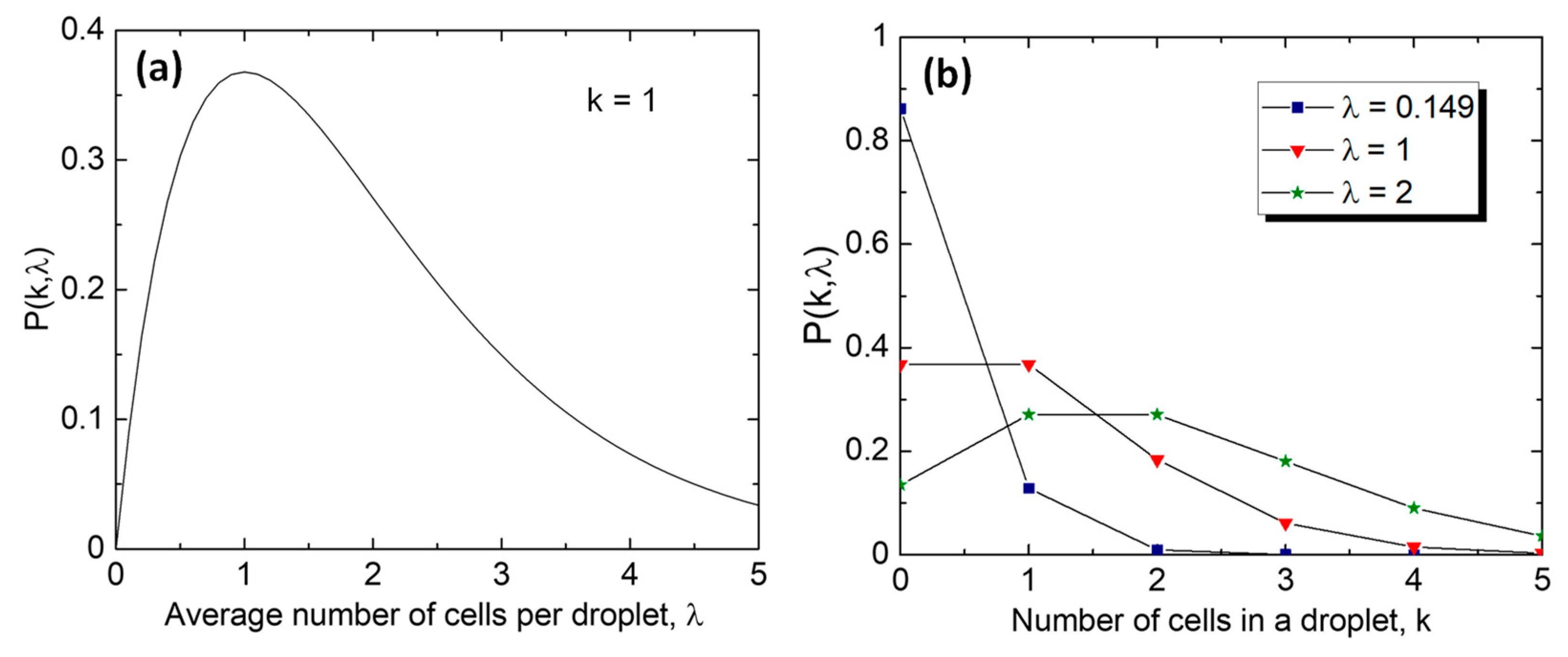
4.1. Cell Compartmentalization in W/O Emulsions
Distribution of Cells in Droplets
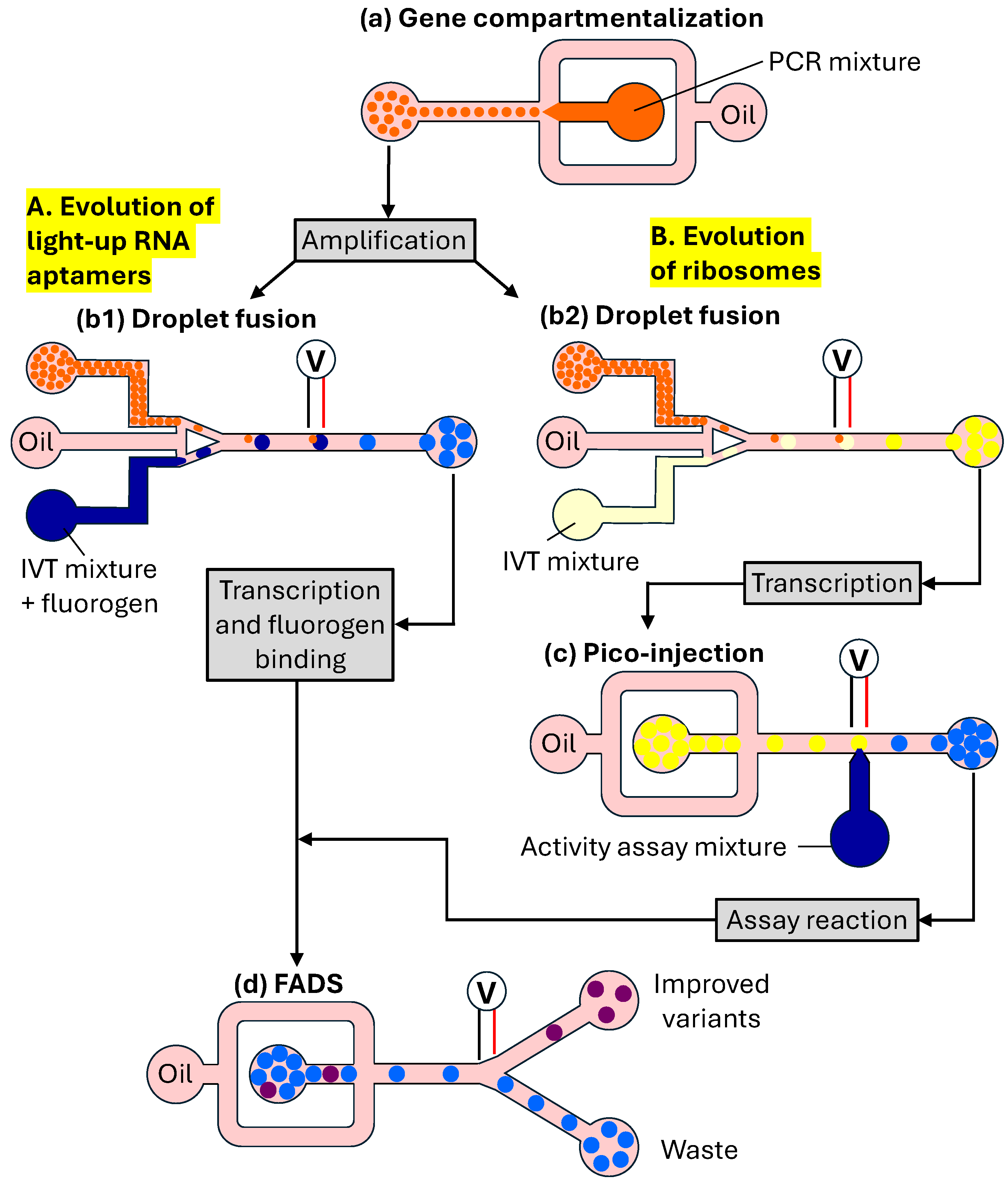
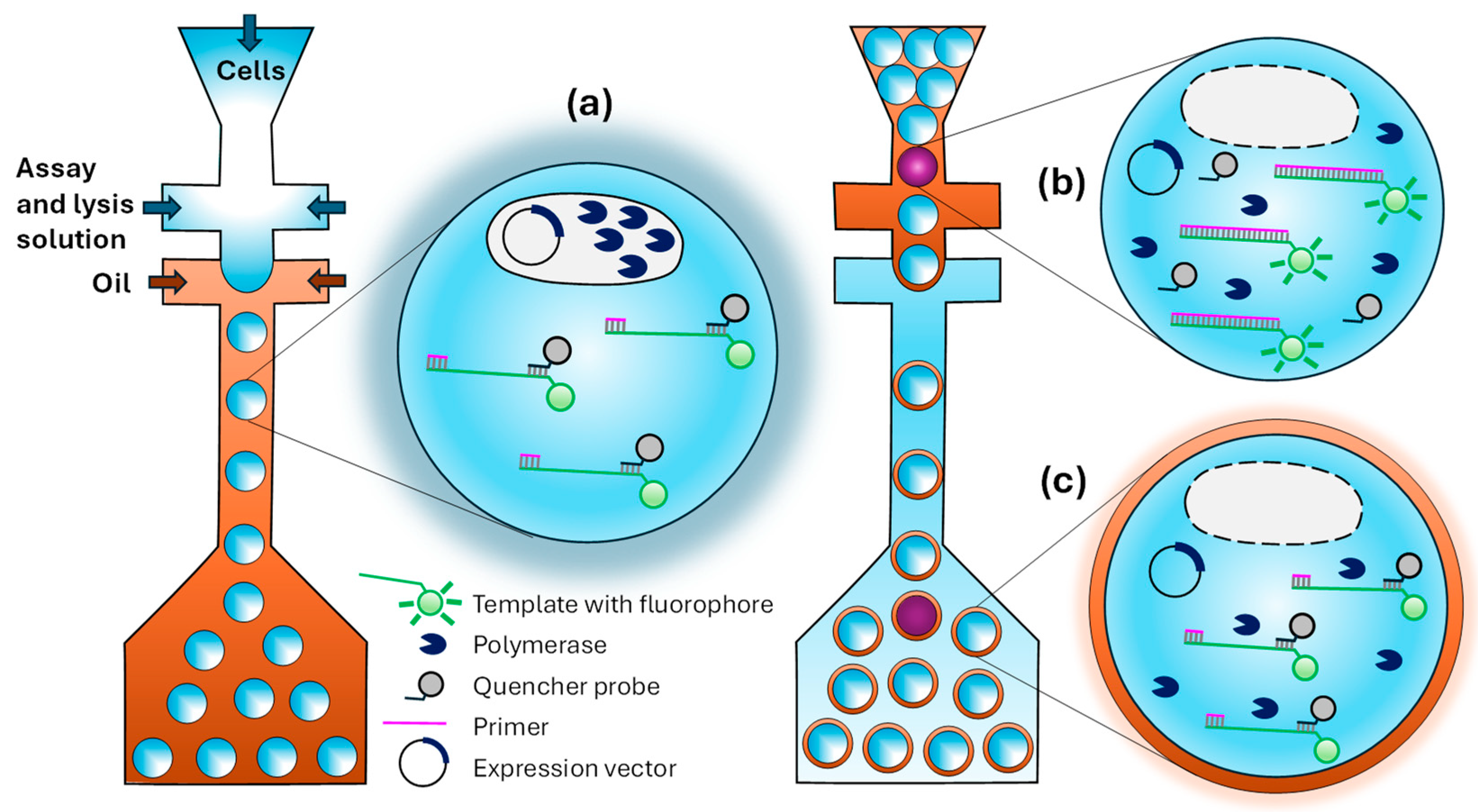
4.2. In Vitro Compartmentalization (IVC) in W/O Emulsions
5. HTS of Enzymes in Double Emulsion Droplets
6. HTS of Biomolecules in Giant Unilamellar Vesicles
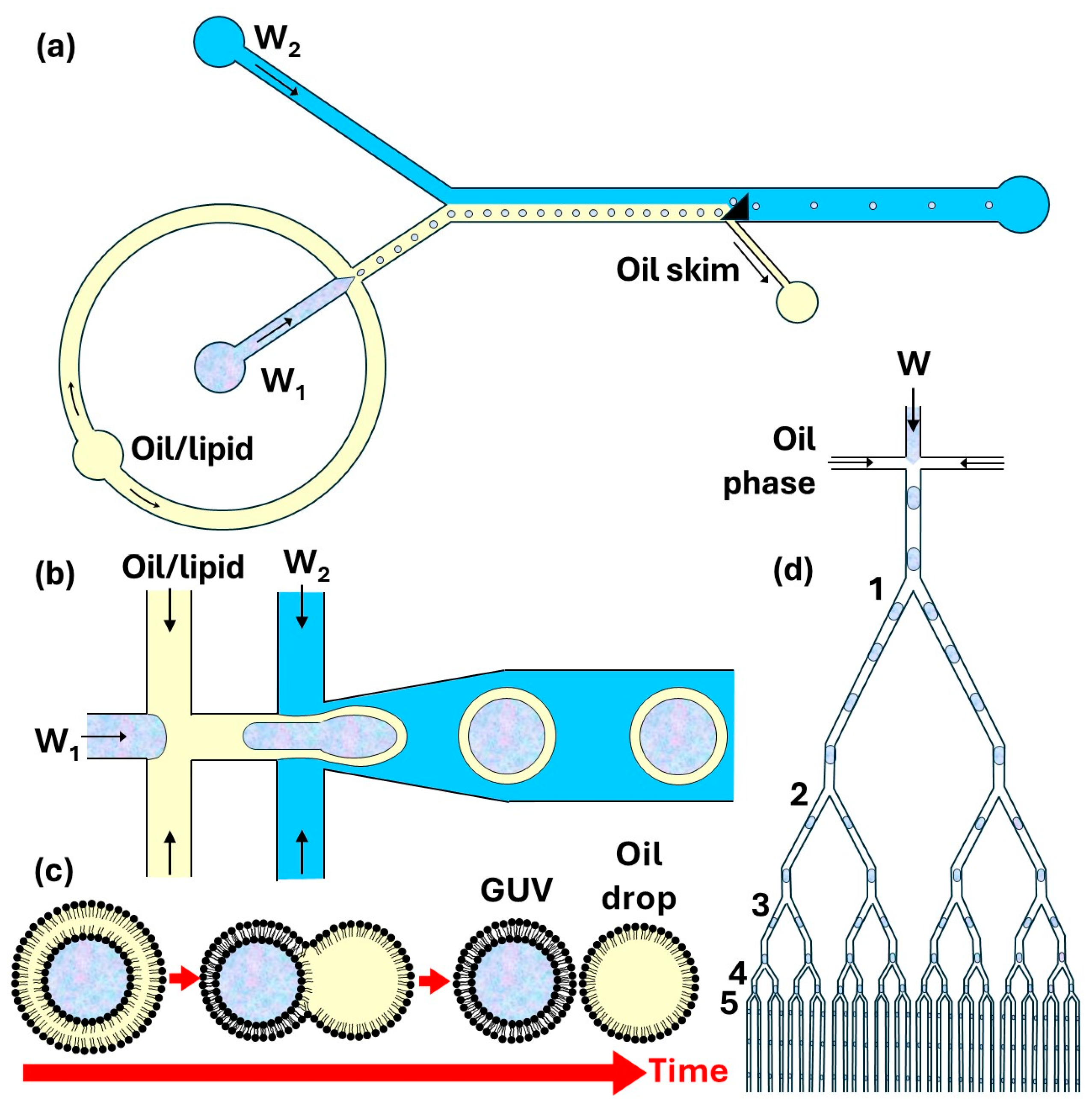
6.1. Microfluidic Production of Giant Unilamellar Vesicles by Phase Transfer Method
6.2. Microfluidic Production of Giant Unilamellar Vesicles by Double Emulsion Templating
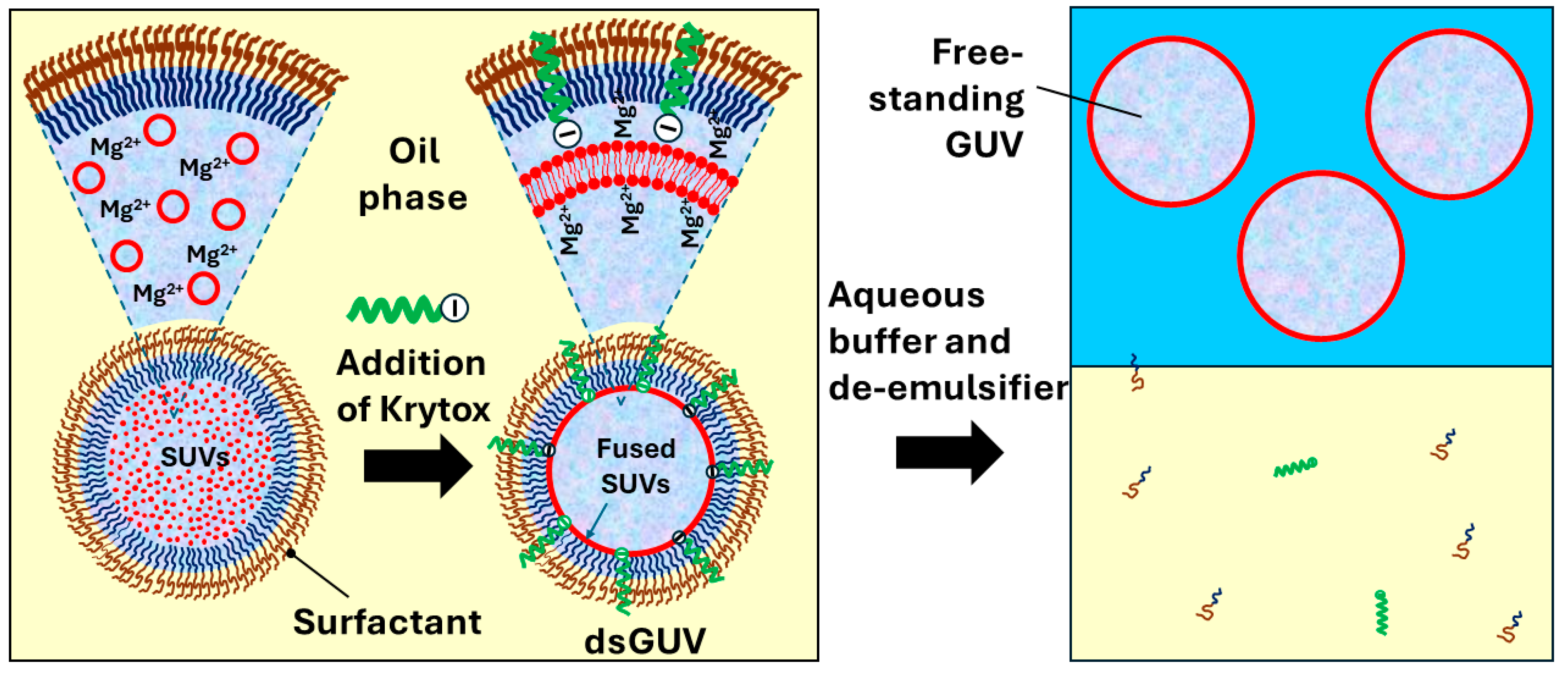
6.3. Microfluidic Production of GUVs by Fusion of Internally Trapped SUVs
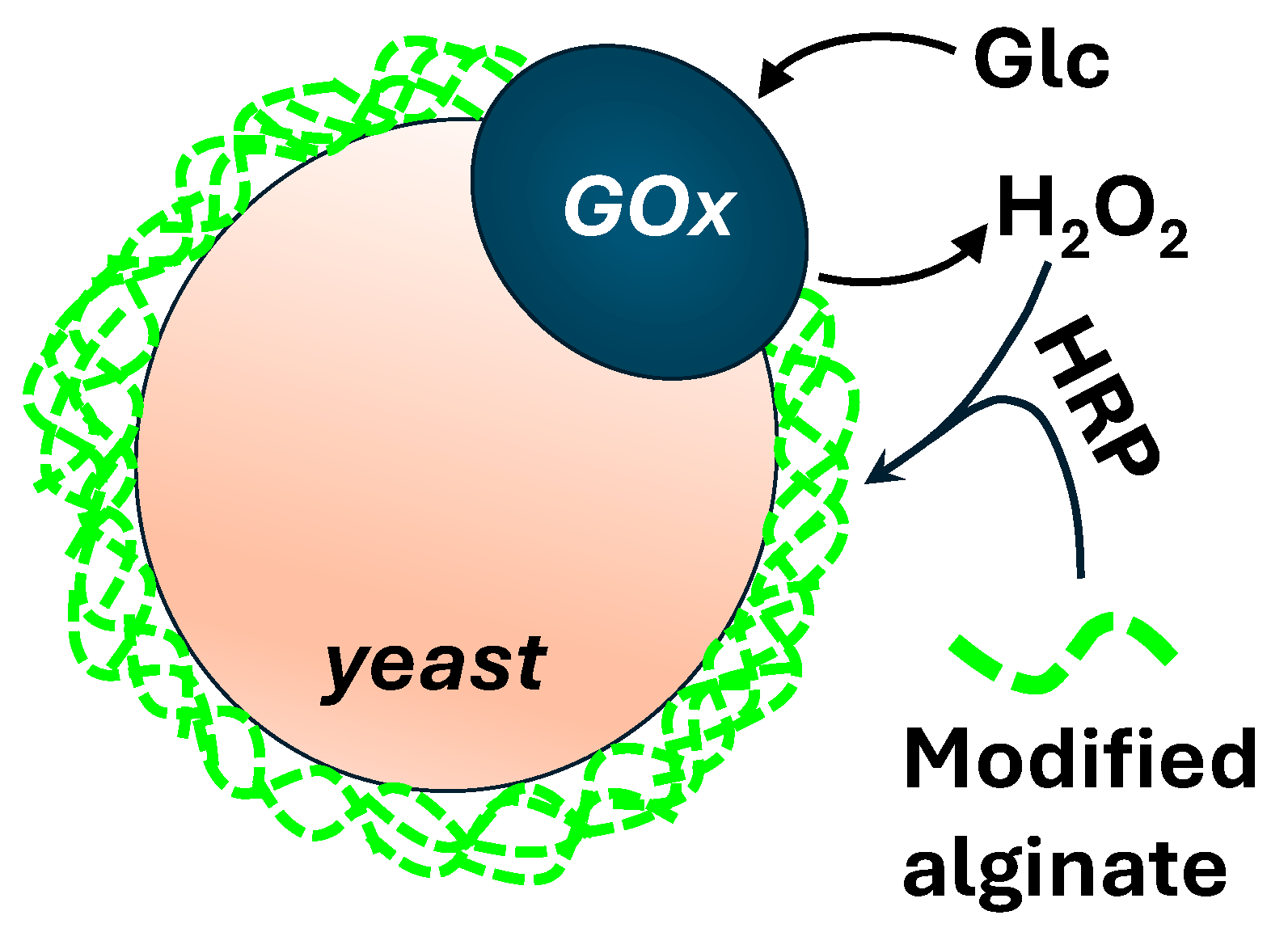
7. Directed Evolution in Microgels
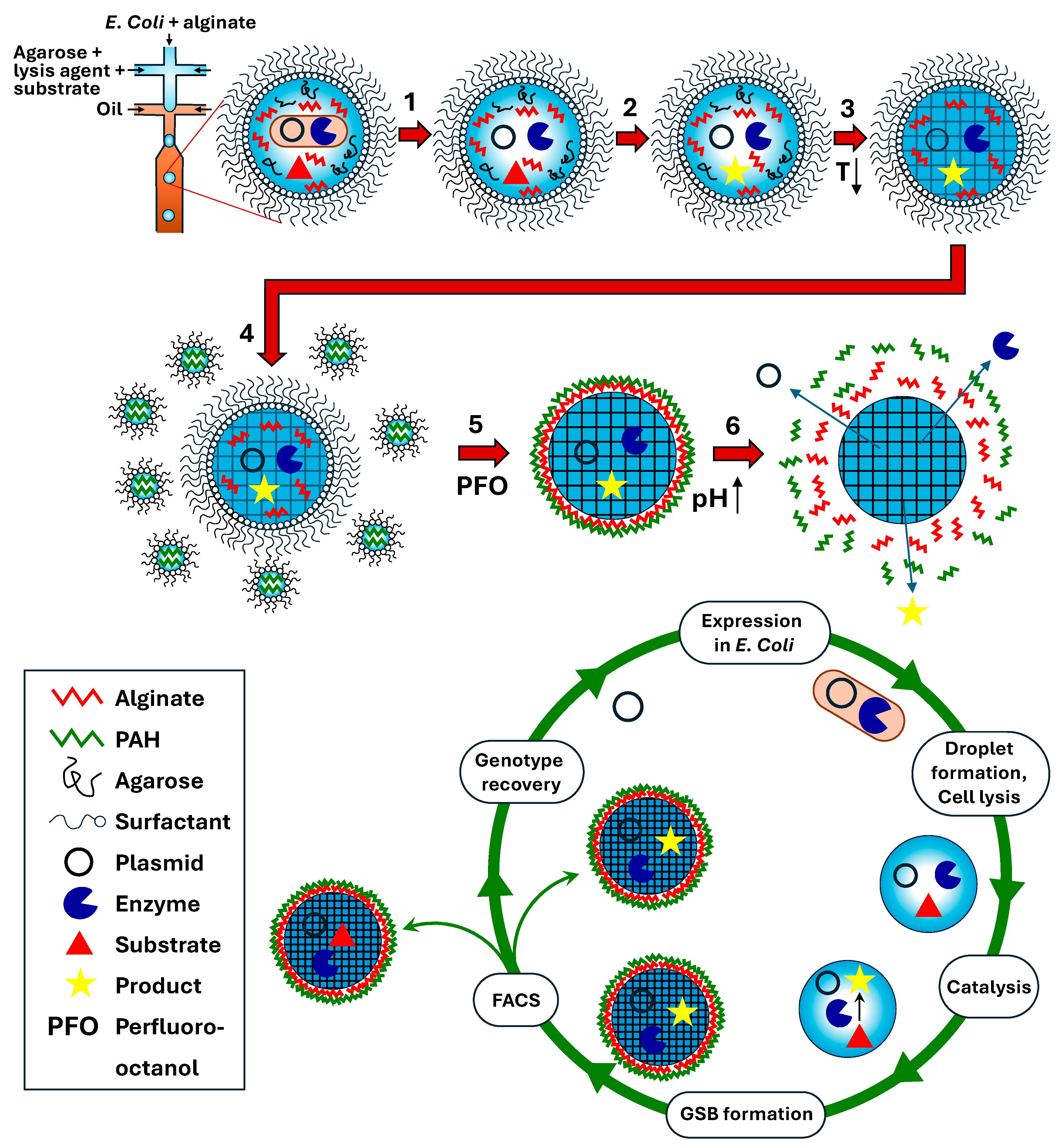
8. HTS in Core–Shell Gel Microcapsules
9. Conclusions
Funding
Conflicts of Interest
References
- Beneyton, T.; Thomas, S.; Griffiths, A.D.; Nicaud, J.-M.; Drevelle, A.; Rossignol, T. Droplet-Based Microfluidic High-Throughput Screening of Heterologous Enzymes Secreted by the Yeast Yarrowia Lipolytica. Microb. Cell Factories 2017, 16, 18. [Google Scholar] [CrossRef]
- Aharoni, A.; Amitai, G.; Bernath, K.; Magdassi, S.; Tawfik, D.S. High-Throughput Screening of Enzyme Libraries: Thiolactonases Evolved by Fluorescence-Activated Sorting of Single Cells in Emulsion Compartment. Chem. Biol. 2005, 12, 1281–1289. [Google Scholar] [CrossRef]
- Chiu, F.W.Y.; Stavrakis, S. High-Throughput Droplet-Based Microfluidics for Directed Evolution of Enzymes. Electrophoresis 2019, 40, 2860–2872. [Google Scholar] [CrossRef]
- Mayr, L.M.; Bojanic, D. Novel Trends in High-Throughput Screening. Curr. Opin. Pharmacol. 2009, 9, 580–588. [Google Scholar] [CrossRef]
- Larry, A.; Sklar, L.A.; Carter, M.B.; Edwards, B.S. Flow Cytometry for Drug Discovery, Receptor Pharmacology and High Throughput Screening. Curr. Opin. Pharmacol. 2007, 7, 527–534. [Google Scholar] [CrossRef]
- Leemhuis, H.; Kelly, R.M.; Dijkhuizen, L. Directed Evolution of Enzymes: Library Screening Strategies. Life 2009, 61, 222–228. [Google Scholar] [CrossRef]
- Autour, A.; Ryckelynck, M. Ultrahigh-Throughput Improvement and Discovery of Enzymes Using Droplet-Based Microfluidic Screening. Micromachines 2017, 8, 128. [Google Scholar] [CrossRef]
- Gantz, M.; Neun, S.; Medcalf, E.J.; van Vliet, L.D.; Hollfelder, F. Ultrahigh-Throughput Enzyme Engineering and Discovery in In Vitro Compartments. Chem. Rev. 2023, 123, 5571–5611. [Google Scholar] [CrossRef]
- Brouzes, E.; Medkova, M.; Savenelli, N.; Samuels, M.L. Droplet Microfluidic Technology for Single-Cell High-Throughput Screening. Proc. Natl. Acad. Sci. USA 2009, 106, 14195–14200. [Google Scholar] [CrossRef] [PubMed]
- Gantz, M.; Aleku, G.A.; Hollfelder, F. Ultrahigh-Throughput Screening in Microfluidic Droplets: A Faster Route to New Enzymes. Trends Biochem. Sci. 2022, 47, 451–452. [Google Scholar] [CrossRef] [PubMed]
- Xia, Y.; Whitesides, G.M. Soft Lithography. Annu. Rev. Mater. Sci. 1998, 28, 153–184. [Google Scholar] [CrossRef]
- Pease, R.F. Maskless Lithography. Microelectron. Eng. 2005, 78–79, 381–392. [Google Scholar] [CrossRef]
- Rein, C.; Toner, M.; Sevenler, D. Rapid Prototyping for High-Pressure Microfluidics. Sci. Rep. 2023, 13, 1232. [Google Scholar] [CrossRef]
- van Meer, B.J.; de Vries, H.; Firth, K.S.A.; van Weerd, J.; Tertoolen, L.G.J.; Karperien, H.B.J.; Jonkheijm, P.; Denning, C.; IJzerman, A.P.; Mummery, C.L. Small Molecule Absorption by PDMS in the Context of Drug Response Bioassays. Biochem. Biophys. Res. Commun. 2017, 482, 323–328. [Google Scholar] [CrossRef]
- Lee, J.N.; Park, C.; Whitesides, G.M. Solvent Compatibility of Poly(Dimethylsiloxane)-Based Microfluidic Devices. Anal. Chem. 2003, 75, 6544–6554. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Hou, S.; Chu, J.; Xiong, Y.; Xiong, P.; Liu, G.; Tian, Y. Observation Interface of PDMS Membrane in a Microfluidic Chip Based on One-Step Molding. Micromachines 2017, 8, 64. [Google Scholar] [CrossRef]
- Yilmaz, A.; Utz, M. Characterisation of Oxygen Permeation into a Microfluidic Device for Cell Culture by In Situ NMR Spectroscopy. Lab Chip 2016, 16, 2079–2085. [Google Scholar] [CrossRef] [PubMed]
- Phan, T.H.T.; Kim, S.J. Super-Hydrophobic Microfluidic Channels Fabricated via Xurography-Based Polydimethylsiloxane (PDMS) Micromolding. Chem. Eng. Sci. 2022, 258, 117768. [Google Scholar] [CrossRef]
- Tropmann, A.; Tanguy, L.; Koltay, P.; Zengerle, R.; Riegger, L. Completely Superhydrophobic PDMS Surfaces for Microfluidics. Langmuir 2012, 28, 8292–8295. [Google Scholar] [CrossRef]
- Harrison, D.J.; Fluri, K.; Seiler, K.; Fan, Z.; Effenhauser, C.S.; Manz, A. Micromachining a Miniaturized Capillary Electrophoresis-Based Chemical Analysis System on a Chip. Science 1993, 261, 895–897. [Google Scholar] [CrossRef]
- Wang, T.; Chen, J.; Zhou, T.; Song, L. Fabricating Microstructures on Glass for Microfluidic Chips by Glass Molding Process. Micromachines 2018, 9, 269. [Google Scholar] [CrossRef] [PubMed]
- Juang, Y.-J.; Chiu, Y.-J. Fabrication of Polymer Microfluidics: An Overview. Polymers 2022, 14, 2028. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, I.; Mukataka, S.; Nakajima, M. Production of Monodisperse Oil-in-Water Emulsions Using a Large Silicon Straight-Through Microchannel Plate. Ind. Eng. Chem. Res. 2005, 44, 5852–5856. [Google Scholar] [CrossRef]
- Lee, U.N.; Su, X.; Guckenberger, D.J.; Dostie, A.M.; Zhang, T.; Berthier, E.; Theberge, A.B. Fundamentals of Rapid Injection Molding for Microfluidic Cell-Based Assays. Lab Chip 2018, 18, 496–504. [Google Scholar] [CrossRef] [PubMed]
- Jeon, J.S.; Chung, S.; Kamm, R.D.; Charest, J.L. Hot Embossing for Fabrication of a Microfluidic 3D Cell Culture Platform. Biomed. Microdevices 2011, 13, 325–333. [Google Scholar] [CrossRef]
- Su, R.; Wang, F.; McAlpine, M.C. 3D Printed Microfluidics: Advances in Strategies, Integration, and Applications. Lab Chip 2023, 23, 1279–1299. [Google Scholar] [CrossRef]
- Salentijn, G.I.J.; Oomen, P.E.; Grajewski, M.; Verpoorte, E. Fused Deposition Modeling 3D Printing for (Bio)analytical Device Fabrication: Procedures, Materials, and Applications. Anal. Chem. 2017, 89, 7053–7061. [Google Scholar] [CrossRef]
- Kim, M.K.; Lee, I.H.; Kim, H.C. Effect of Fabrication Parameters on Surface Roughness of FDM Parts. Int. J. Precis. Eng. Manuf. Green Technol. 2018, 19, 137–142. [Google Scholar] [CrossRef]
- Huang, J.; Qin, Q.; Wang, J. A Review of Stereolithography: Processes and Systems. Processes 2020, 8, 1138. [Google Scholar] [CrossRef]
- Griffin, K.; Pappas, D. 3D Printed Microfluidics for Bioanalysis: A Review of Recent Advancements and Applications. TrAC Trends Anal. Chem. 2023, 158, 116892. [Google Scholar] [CrossRef]
- Mansour, H.; Soliman, E.A.; El-Bab, A.M.F.; Matsushita, Y.; Abdel-Mawgood, A.L. Fabrication and Characterization of Microfluidic Devices Based on Boron-Modified Epoxy Resin Using CO2 Laser Ablation for Bio-Analytical Applications. Sci. Rep. 2023, 13, 12623. [Google Scholar] [CrossRef] [PubMed]
- Waddell, E.A.; Locascio, L.E.; Kramer, G.W. UV Laser Micromachining of Polymers for Microfluidic Applications. SLAS Technol. 2002, 7, 78–82. [Google Scholar] [CrossRef]
- Sarma, U.; Joshi, S.N. Machining of Micro-Channels on Polycarbonate by using Laser-Induced Plasma Assisted Ablation (LIPAA). Opt. Laser Technol. 2020, 128, 106257. [Google Scholar] [CrossRef]
- Wlodarczyk, K.L.; Hand, D.P.; Maroto-Valer, M.M. Maskless, Rapid Manufacturing of Glass Microfluidic Devices Using a Picosecond Pulsed Laser. Sci. Rep. 2019, 9, 20215. [Google Scholar] [CrossRef] [PubMed]
- Wei, Y.; Wang, T.; Wang, Y.; Zeng, S.; Ho, Y.-P.; Ho, H.-P. Rapid Prototyping of Multi-Functional and Biocompatible Parafilm®-Based Microfluidic Devices by Laser Ablation and Thermal Bonding. Micromachines 2023, 14, 656. [Google Scholar] [CrossRef] [PubMed]
- Scott, S.M.; Ali, Z. Micromachines Fabrication Methods for Microfluidic Devices: An Overview. Micromachines 2021, 12, 319. [Google Scholar] [CrossRef] [PubMed]
- Utada, A.S.; Chu, L.Y.; Fernandez-Nieves, A.; Link, D.R.; Holtze, C.; Weitz, D.A. Dripping, Jetting, Drops, and Wetting: The Magic of Microfluidics. MRS Bull. 2007, 32, 702–708. [Google Scholar] [CrossRef]
- Schnettler, J.D.; Klein, O.J.; Kaminski, T.S.; Colin, P.Y.; Hollfelder, F. Ultrahigh-Throughput Directed Evolution of a Metal-Free α/β-Hydrolase with a Cys-His-Asp Triad into an Efficient Phosphotriesterase. J. Am. Chem. Soc. 2023, 145, 1083–1096. [Google Scholar] [CrossRef]
- Ma, F.; Chung, M.T.; Yao, Y.; Nidetz, R.; Lee, L.M.; Liu, A.P.; Feng, Y.; Kurabayashi, K.; Yang, G.Y. Efficient Molecular Evolution to Generate Enantioselective Enzymes Using a Dual-Channel Microfluidic Droplet Screening Platform. Nat. Commun. 2018, 9, 1030. [Google Scholar] [CrossRef]
- Shenoy, V.J.; Edwards, C.E.R.; Helgeson, M.E.; Valentine, M.T. Design and Characterization of a 3D-Printed Staggered Herringbone Mixer. Design and Characterization of a 3D-Printed Staggered Herringbone Mixer. BioTechniques 2021, 70, 285–289. [Google Scholar] [CrossRef]
- Howell, P.B., Jr.; Mott, D.R.; Fertig, S.; Kaplan, C.R.; Golden, J.P.; Oran, E.S.; Ligler, F.S. A Microfluidic Mixer with Grooves Placed on the Top and Bottom of the Channel. Lab Chip 2005, 5, 524–530. [Google Scholar] [CrossRef]
- Stroock, A.D.; Dertinger, S.K.W.; Ajdari, A.; Mezić, I.; Stone, H.A.; Whitesides, G.M. Chaotic Mixer for Microchannels. Science 2002, 295, 647–651. [Google Scholar] [CrossRef] [PubMed]
- Belousov, K.I.; Filatov, N.A.; Kukhtevich, I.V.; Kantsler, V.; Evstrapov, A.A.; Bukatin, A.S. An Asymmetric Flow-Focusing Droplet Generator Promotes Rapid Mixing of Reagents. Sci. Rep. 2021, 11, 8797. [Google Scholar] [CrossRef] [PubMed]
- Samlali, K.; Alves, C.L.; Jezernik, M.; Shih, S.C.C. Droplet Digital Microfluidic System for Screening Filamentous Fungi Based on Enzymatic Activity. Microsyst. Nanoeng. 2022, 8, 123. [Google Scholar] [CrossRef]
- Song, H.; Chen, D.L.; Ismagilov, R.F. Reactions in Droplets in Microfluidic Channels. Angew. Chem. Int. Ed. Engl. 2006, 45, 7336–7356. [Google Scholar] [CrossRef] [PubMed]
- Bringer, M.R.; Gerdts, C.J.; Song, H.; Tice, J.D.; Ismagilov, R.F. Microfluidic Systems for Chemical Kinetics that Rely on Chaotic Mixing in Droplets. Philos. Transact. A Math. Phys. Eng. Sci. 2004, 15, 1087–1104. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Wang, J.; Feng, L.; Lin, T. Fluid Mixing in Droplet-Based Microfluidics with a Serpentine Microchannel. RSC Adv. 2015, 5, 104138. [Google Scholar] [CrossRef]
- Cao, X.; Zhou, B.; Yu, C.; Liu, X. Droplet-Based Mixing Characteristics in Bumpy Serpentine Microchannel. Chem. Eng. Process. 2021, 159, 108246. [Google Scholar] [CrossRef]
- Ahn, K.; Link, D.; Griffiths, A.; Weitz, D. Enzyme-Inhibitor Assay Using Microdroplets. Bull. Am. Phys. Soc. 2005. BAPS.2005.MAR.V37.2. Available online: https://meetings.aps.org/link/BAPS.2005.MAR.V37.2 (accessed on 25 July 2024).
- Abate, A.R.; Hung, T.; Mary, P.; Agresti, J.J.; Weitz, D.A. High-Throughput Injection with Microfluidics Using Picoinjectors. Proc. Natl. Acad. Sci. USA 2010, 107, 19163–19166. [Google Scholar] [CrossRef]
- Ryckelynck, M.; Baudrey, S.; Rick, C.; Marin, A.; Coldren, F.; Westhof, E.; Griffiths, A.D. Using Droplet-Based Microfluidics to Improve the Catalytic Properties of RNA Under Multiple-Turnover Conditions. RNA 2015, 21, 458–469. [Google Scholar] [CrossRef]
- Breukers, J.; de Beeck, H.O.; Rutten, I.; Fernández, M.L.; Eyckermancd, S.; Lammertyn, J. Highly Flexible and Accurate Serial Picoinjection in Droplets by Combined Pressure and Flow Rate Control. Lab Chip 2022, 22, 3475–3488. [Google Scholar] [CrossRef] [PubMed]
- Ng, E.X.; Miller, M.A.; Jing, T.; Lauffenburger, D.A.; Chen, C.H. Low-Volume Multiplexed Proteolytic Activity Assay and Inhibitor Analysis Through a Pico-Injector Array. Lab Chip 2015, 15, 1153–1159. [Google Scholar] [CrossRef] [PubMed]
- Yuan, H.; Pan, Y.; Tian, J.; Chao, Y.; Li, J.; Shum, H.C. Electricity-Free Picoinjection Assisted Droplet Microfluidics. Sens. Actuators B Chem. 2019, 298, 126766. [Google Scholar] [CrossRef]
- Nan, L.; Mao, T.; Shum, H.C. Self-Synchronization of Reinjected Droplets for High-Efficiency Droplet Pairing and Merging. Microsyst. Nanoeng. 2023, 9, 24. [Google Scholar] [CrossRef] [PubMed]
- Frenz, L.; El Harrak, A.; Pauly, M.; Bégin-Colin, S.; Griffiths, A.D.; Baret, J.C. Droplet-Based Microreactors for the Synthesis of Magnetic Iron Oxide Nanoparticles. Angew. Chem. Int. Ed. Engl. 2008, 47, 6817–6820. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Guzman, A.R.; Wippold, J.A.; Li, Y.; Dai, J.; Huang, C.; Han, A. An Ultra High-Efficiency Droplet Microfluidics Platform Using Automatically Synchronized Droplet Pairing and Merging. Lab Chip 2020, 20, 3948–3959. [Google Scholar] [CrossRef] [PubMed]
- Duchamp, M.; Arnaud, M.; Bobisse, S.; Coukos, G.; Harari, A.; Renaud, P. Microfluidic Device for Droplet Pairing by Combining Droplet Railing and Floating Trap Arrays. Micromachines 2021, 12, 1076. [Google Scholar] [CrossRef] [PubMed]
- Thorsen, T.; Roberts, R.W.; Arnold, F.H.; Quake, S.R. Dynamic Pattern Formation in a Vesicle-Generating Microfluidic Device. Phys. Rev. Lett. 2001, 86, 4163. [Google Scholar] [CrossRef] [PubMed]
- Umbanhowar, P.B.; Prasad, V.; Weitz, D.A. Monodisperse Emulsion Generation via Drop Break Off in a Coflowing Stream. Langmuir 2000, 16, 347–351. [Google Scholar] [CrossRef]
- Anna, S.L.; Bontoux, N.; Stone, H.A. Formation of Dispersions Using “Flow Focusing” in Microchannels. Appl. Phys. Lett. 2003, 82, 364–366. [Google Scholar] [CrossRef]
- Li, Z.; Leshansky, A.M.; Pismen, L.M.; Tabeling, P. Step-Emulsification in a Microfluidic Device. Lab Chip 2015, 15, 1023–1031. [Google Scholar] [CrossRef]
- Tice, J.D.; Song, H.; Lyon, A.D.; Ismagilov, R.F. Formation of Droplets and Mixing in Multiphase Microfluidics at Low Values of the Reynolds and the Capillary Numbers. Langmuir 2003, 19, 9127–9133. [Google Scholar] [CrossRef]
- Garstecki, P.; Fuerstman, M.J.; Stone, H.A.; Whitesides, G.M. Formation of Droplets and Bubbles in a Microfluidic T-Junction—Scaling and Mechanism of Break-Up. Lab Chip 2006, 6, 437. [Google Scholar] [CrossRef]
- De Menech, M.; Garstecki, P.; Jousse, F.; Stone, H.A. Transition from Squeezing to Dripping in a Microfluidic T-Shaped Junction. J. Fluid. Mech. 2008, 595, 141–161. [Google Scholar] [CrossRef]
- Utada, A.S.; Fernandez-Nieves, A.; Stone, H.A.; Weitz, D.A. Dripping to Jetting Transitions in Coflowing Liquid Streams. Phys. Rev. Lett. 2007, 99, 094502. [Google Scholar] [CrossRef] [PubMed]
- Utada, A.S.; Lorenceau, E.; Link, D.R.; Kaplan, P.D.; Stone, H.A.; Weitz, D.A. Monodisperse Double Emulsions Generated from a Microcapillary Device. Science 2005, 308, 537–541. [Google Scholar] [CrossRef] [PubMed]
- Yuan, X.; Glidle, A.; Furusho, H.; Yin, H. A 3D Hydrodynamic Flow-Focusing Device for Cell Sorting. Microfluid. Nanofluidics 2021, 25, 23. [Google Scholar] [CrossRef]
- Deydier, T.; Bolognesi, G.; Vladisavljević, G.T. Scaled-up Droplet Generation in Parallelised 3D Flow Focusing Junctions. Colloids Surf. A Physicochem. Eng. 2022, 641, 128439. [Google Scholar] [CrossRef]
- Dewandre, A.; Rivero-Rodriguez, J.; Vitry, Y.; Sobac, B.; Scheid, B. Microfluidic Droplet Generation Based on Non-Embedded Co-Flow-Focusing Using 3D Printed Nozzle. Sci. Rep. 2020, 10, 1001–1002. [Google Scholar] [CrossRef]
- Bandulasena, M.V.; Vladisavljević, G.T.; Benyahia, B. Versatile Reconfigurable Glass Capillary Microfluidic Devices with Lego® Inspired Blocks for Drop Generation and Micromixing. J. Colloid Interface Sci. 2019, 542, 23–32. [Google Scholar] [CrossRef]
- Agresti, J.J.; Antipov, E.; Abate, A.R.; Ahn, K.; Rowat, A.C.; Baret, J.C.; Marquez, M.; Klibanov, A.M.; Griffiths, A.D.; Weitz, D.A. Ultrahigh-Throughput Screening in Drop-Based Microfluidics for Directed Evolution. Proc. Natl. Acad. Sci. USA 2010, 107, 4004–4009. [Google Scholar] [CrossRef]
- Vallejo, D.; Nikoomanzar, A.; Paegel, B.M.; Chaput, J.C. Fluorescence-activated droplet sorting for single-cell directed evolution. ACS Synth. Biol. 2019, 8, 1430–1440. [Google Scholar] [CrossRef] [PubMed]
- Kawakatsu, T.; Kikuchi, Y.; Nakajima, M. Regular-Sized Cell Creation in Microchannel Emulsification by Visual Microprocessing Method. J. Am. Oil Chem. Soc. 1997, 74, 317–321. [Google Scholar] [CrossRef]
- Eggersdorfer, M.L.; Zheng, W.; Nawar, S.; Mercandetti, C.; Ofner, A.; Leibacher, I.; Koehler, S.; Weitz, D.A. Tandem Emulsification for high-Throughput Production of Double Emulsions. Lab Chip 2017, 17, 936–942. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Zhu, C.; Fu, T.; Ma, Y. Effect of nozzle width on droplet formation in wedge-shaped step-emulsification microchannel devices. Colloids Surf. A Physicochem. Eng. 2024, 691, 133879. [Google Scholar] [CrossRef]
- Sugiura, S.; Nakajima, M.; Seki, M. Prediction of Droplet Diameter for Microchannel Emulsification. Langmuir 2002, 18, 3854–3859. [Google Scholar] [CrossRef]
- Vladisavljević, G.T.; Ekanem, E.E.; Zhang, Z.; Khalid, N.; Kobayashi, I.; Nakajima, M. Long-Term Stability of Droplet Production by Microchannel (Step) Emulsification in Microfluidic Silicon Chips with Large Number of Terraced Microchannels. Chem. Eng. J. 2018, 333, 380–391. [Google Scholar] [CrossRef]
- Sugiura, S.; Nakajima, M.; Seki, M. Preparation of Monodispersed Polymeric Microspheres over 50 µm Employing Microchannel Emulsification. Ind. Eng. Chem. Res. 2002, 41, 4043–4047. [Google Scholar] [CrossRef]
- Zhang, Y.; Kobayashi, I.; Wada, Y.; Neves, M.A.; Uemura, K.; Nakajima, M. Asymmetric Straight-through Microchannel Arrays Made of Aluminum for Producing Monodisperse O/W Emulsions. Particul. Sci. Technol. 2020, 38, 747–755. [Google Scholar] [CrossRef]
- Paegel, B.M.; Joyce, G.F. Microfluidic Compartmentalized Directed Evolution. Chem. Biol. 2010, 17, 717–724. [Google Scholar] [CrossRef]
- Vladisavljević, G.T.; Khalid, N.; Neves, M.A.; Kuroiwa, T.; Nakajima, M.; Uemura, K.; Ichikawa, S.; Kobayashi, I. Industrial Lab-on-a-Chip: Design, Applications and Scale-Up for Drug Discovery and Delivery. Adv. Drug Del. Rev. 2013, 65, 1626–1663. [Google Scholar] [CrossRef]
- Stone, Z.B.; Stone, H.A. Imaging and Quantifying Mixing in a Model Droplet Micromixer. Phys. Fluids 2005, 17, 063103. [Google Scholar] [CrossRef]
- Frenz, L.; Blank, K.; Brouzes, E.; Griffiths, A.D. Reliable Microfluidic On-Chip Incubation of Droplets in Delay-Lines. Lab Chip 2009, 9, 1344–1348. [Google Scholar] [CrossRef]
- Zhou, Y.; Yu, Z.; Wu, M.; Lan, Y.; Jia, C.; Zhao, J. Single-Cell Sorting Using Integrated Pneumatic Valve Droplet Microfluidic Chip. Talanta 2023, 253, 124044. [Google Scholar] [CrossRef]
- Kim, H.S.; Hsu, S.C.; Han, S.I.; Thapa, H.R.; Guzman, A.R.; Browne, D.R.; Tatli, M.; Devarenne, T.P.; Stern, D.B.; Han, A. High-Throughput Droplet Microfluidics Screening Platform for selecting Fast-Growing and High Lipid-Producing Microalgae from a Mutant Library. Plant Direct 2017, 1, e00011. [Google Scholar] [CrossRef]
- Shi, W.; Qin, J.; Ye, N.; Lin, B. Droplet-Based Microfluidic System for Individual Caenorhabditis Elegans Assay. Lab Chip 2008, 8, 1432–1435. [Google Scholar] [CrossRef]
- Han, S.H.; Kim, J.; Lee, D. Static Array of Droplets and On-demand Recovery for Biological Assays. Biomicrofluidics 2020, 14, 051302. [Google Scholar] [CrossRef]
- Kartashov, O.O.; Chapek, S.V.; Polyanichenko, D.S.; Belyavsky, G.I.; Alexandrov, A.A.; Butakova, M.A.; Soldatov, A.V. Online Microfluidic Droplets Characterization Using Microscope Data Intelligent Analysis. Big Data Cogn. Comput. 2023, 7, 7. [Google Scholar] [CrossRef]
- Courtney, M.; Chen, X.; Chan, S.; Mohamed, T.; Rao, P.P.N.; Ren, C.L. Droplet Microfluidic System with On-Demand Trapping and Releasing of Droplet for Drug Screening Applications. Anal. Chem. 2017, 89, 910–915. [Google Scholar] [CrossRef]
- Huang, C.; Jiang, Y.; Li, Y.; Zhang, H. Droplet Detection and Sorting System in Microfluidics: A Review. Micromachines 2023, 14, 103. [Google Scholar] [CrossRef]
- Fu, X.; Zhang, Y.; Xu, Q.; Sun, X.; Meng, F. Recent Advances on Sorting Methods of High-Throughput Droplet-Based Microfluidics in Enzyme Directed Evolution. Front. Chem. 2021, 9, 666867. [Google Scholar] [CrossRef] [PubMed]
- Gielen, F.; Hours, R.; Emond, S.; Hollfelder, F. Ultrahigh-Throughput–Directed Enzyme Evolution by Absorbance-Activated Droplet Sorting (AADS). Proc. Natl. Acad. Sci. USA 2016, 113, E7383–E7389. [Google Scholar] [CrossRef] [PubMed]
- Medcalf, E.J.; Gantz, M.; Kaminski, T.S.; Hollfelder, F. Ultra-High-Throughput Absorbance-Activated Droplet Sorting for Enzyme Screening at Kilohertz Frequencies. Anal. Chem. 2023, 95, 4597–4604. [Google Scholar] [CrossRef] [PubMed]
- Kelly, R.T.; Page, J.S.; Marginean, I.; Tang, K.; Smith, R.D. Dilution-Free Analysis from Picoliter Droplets by Nano-Electrospray Ionization Mass Spectrometry. Angew. Chem. Int. Ed. 2009, 48, 6832–6835. [Google Scholar] [CrossRef] [PubMed]
- Haidas, D.; Bachler, S.; Köhler, M.; Blank, L.M.; Zenobi, R.; Dittrich, P.S. Microfluidic Platform for Multimodal Analysis of Enzyme Secretion in Nanoliter Droplet Arrays. Anal. Chem. 2019, 91, 2066–2073. [Google Scholar] [CrossRef] [PubMed]
- Pilát, Z.; Ježek, J.; Kizovský, M.; Klementová, T.; Krátký, S.; Sobota, J.; Samek, O.; Zemánek., P.; Buryška, T.; Damborský, J.; et al. Surface-Enhanced Raman Spectroscopy in Microfluidic Chips for Directed Evolution of Enzymes and Environmental Monitoring. In Proceedings of the 2019 PhotonIcs & Electromagnetics Research Symposium-Spring (PIERS-Spring), Rome, Italy, 17–20 June 2019; pp. 1301–1309. [Google Scholar] [CrossRef]
- Davoodi, H.; Nordin, N.; Bordonali, L.; Korvink, J.G.; MacKinnon, N.; Badilita, V. An NMR-Compatible Microfluidic Platform Enabling: In situ Electrochemistry. Lab Chip 2020, 20, 3202–3212. [Google Scholar] [CrossRef] [PubMed]
- Pan, C.W.; Horvath, D.G.; Braza, S.; Moore, T.; Lynch, A.; Feit, C.; Abbyad, P. Sorting by Interfacial Tension (SIFT): Label-Free Selection of Live Cells Based on Single-Cell Metabolism. Lab Chip 2019, 19, 1344. [Google Scholar] [CrossRef]
- Ahn, K.; Kerbage, C.; Hunt, T.P.; Westervelt, R.M.; Link, D.R.; Weitz, D.A. Dielectrophoretic Manipulation of Drops for High-Speed Microfluidic Sorting Devices. Appl. Phys. Lett. 2006, 88, 024104. [Google Scholar] [CrossRef]
- Frey, C.; Göpfrich, K.; Pashapour, S.; Platzman, I.; Spatz, J.P. Electrocoalescence of Water-in-Oil Droplets with a Continuous Aqueous Phase: Implementation of Controlled Content Release. ACS Omega 2020, 5, 7529–7536. [Google Scholar] [CrossRef]
- Caen, O.; Schütz, S.; Jammalamadaka, M.S.S.; Vrignon, J.; Nizard, P.; Schneider, T.M.; Baret, J.C.; Taly, V. High-Throughput Multiplexed Fluorescence-Activated Droplet Sorting. Microsyst. Nanoeng. 2018, 4, 33. [Google Scholar] [CrossRef]
- Fidalgo, L.M.; Whyte, G.; Bratton, D.; Kaminski, C.F.; Abell, C.; Huck, W.T.S. From Microdroplets to Microfluidics: Selective Emulsion Separation in Microfluidic Devices. Angew. Chem. Int. Ed. Engl. 2008, 47, 2042–2045. [Google Scholar] [CrossRef] [PubMed]
- Payne, E.M.; Holland-Moritz, D.A.; Sun, S.; Kennedy, R.T. High-Throughput Screening by Droplet Microfluidics: Perspective into Key Challenges and Future Prospects. Lab Chip 2020, 20, 2247–2262. [Google Scholar] [CrossRef] [PubMed]
- Courtois, F.; Olguin, L.F.; Whyte, G.; Theberge, A.B.; Huck, W.T.S.; Hollfelder, F.; Abell, C. Controlling the Retention of Small Molecules in Emulsion Microdroplets for Use in Cell-Based Assays. Anal. Chem. 2009, 81, 3008–3016. [Google Scholar] [CrossRef] [PubMed]
- Weng, L.; Spoonamore, J.E. Droplet Microfluidics-Enabled High-Throughput Screening for Protein Engineering. Micromachines 2019, 10, 734. [Google Scholar] [CrossRef]
- Hussain, S.M.S.; Adewunmi, A.A.; Mahboob, A.; Murtaza, M.; Zhou, X.; Kamal, M.S. Fluorinated Surfactants: A Review on Recent Progress on Synthesis and Oilfield Applications. Adv. Colloid Interface Sci. 2022, 303, 102634. [Google Scholar] [CrossRef]
- Gruner, P.; Riechers, B.; Orellana, L.A.C.; Brosseau, Q.; Maes, F.; Beneyton, T.; Pekin, D.; Baret, J.C. Stabilisers for Water-in-Fluorinated-Oil Dispersions: Key Properties for Microfluidic Applications. Curr. Opin. Colloid Interface Sci. 2015, 20, 183–191. [Google Scholar] [CrossRef]
- Holtze, C.; Rowat, A.C.; Agresti, J.J.; Hutchison, J.B.; Angilè, F.E.; Schmitz, C.H.J.; Köster, S.; Duan, H.; Humphry, K.J.; Scanga, R.A.; et al. Biocompatible Surfactants for Water-In-Fluorocarbon Emulsions. Lab Chip 2008, 8, 1632–1639. [Google Scholar] [CrossRef]
- Payne, E.M.; Taraji, M.; Murray, B.E.; Holland-Moritz, D.A.; Moore, J.C.; Haddad, P.R.; Kennedy, R.T. Evaluation of analyte transfer between microfluidic droplets by mass spectrometry. Anal. Chem. 2023, 95, 4662–4670. [Google Scholar] [CrossRef]
- Kintses, B.; Hein, C.; Mohamed, M.F.; Fischlechner, M.; Courtois, F.; Lainé, C.; Hollfelder, F. Picoliter Cell Lysate Assays in Microfluidic Droplet Compartments for Directed Enzyme Evolution. Chem. Biol. 2012, 19, 1001–1009. [Google Scholar] [CrossRef]
- Levin, I.; Aharoni, A. Evolution in Microfluidic Droplet. Chem. Biol. 2012, 19, 929–931. [Google Scholar] [CrossRef]
- Rosenthal, R.G.; Zhang, X.D.; Ilić Đurđić, K.; Collins, J.J.; Weitz, D.A. Controlled Continuous Evolution of Enzymatic Activity Screened at Ultrahigh Throughput Using Drop-Based Microfluidics. Angew. Chem. Int. Ed. 2023, 62, e202303112. [Google Scholar] [CrossRef] [PubMed]
- Kemna, E.W.M.; Schoeman, R.M.; Wolbers, F.; Vermes, I.; Weitz, D.A.; van den Berg, A. High-Yield Cell Ordering and Deterministic Cell-in-Droplet Encapsulation Using Dean Flow in a Curved Microchannel. Lab Chip 2012, 12, 2881–2887. [Google Scholar] [CrossRef] [PubMed]
- Collins, D.J.; Neild, A.; deMello, A.; Liud, A.Q.; Ai, Y. The Poisson Distribution and Beyond: Methods for Microfluidic Droplet Production and Single Cell Encapsulation. Lab Chip 2015, 15, 3439. [Google Scholar] [CrossRef]
- Gao, Y.; Magaud, P.; Baldas, L.; Lafforgue, C.; Abbas, M.; Colin, S. Self-Ordered Particle Trains in Inertial Microchannel Flows. Microfluid. Nanofluidics 2017, 21, 154. [Google Scholar] [CrossRef]
- Dietsche, C.; Mutlu, B.R.; Edd, J.F.; Koumoutsakos, P.; Toner, M. Dynamic Particle Ordering in Oscillatory Inertial Microfluidics. Microfluid. Nanofluidics 2019, 23, 83. [Google Scholar] [CrossRef]
- Hu, X.; Lin, J.; Chen, D.; Ku, X. Stability Condition of Self-Organizing Staggered Particle Trains in Channel Flow. Microfluid. Nanofluidics 2020, 24, 25. [Google Scholar] [CrossRef]
- Liu, J.; Pan, Z. Self-Ordering and Organization of In-Line Particle Chain in a Square Microchannel. Phys. Fluids 2022, 34, 023309. [Google Scholar] [CrossRef]
- Tang, T.; Zhao, H.; Shen, S.; Yang, L.; Lim, C.T. Enhancing single-cell encapsulation in droplet microfluidics with fine-tunable on-chip sample enrichment. Microsyst. Nanoeng. 2024, 10, 3. [Google Scholar] [CrossRef] [PubMed]
- Shahrivar, K.; Del Giudice, F. Beating Poisson Stochastic Particle Encapsulation in Flow-Focusing Microfluidic Devices Using Viscoelastic Liquids. Soft Matter 2022, 18, 5928–5933. [Google Scholar] [CrossRef]
- Del Giudice, F.; D’Avino, G.; Greco, F.; Maffettone, P.L.; Shen, A.Q. Fluid Viscoelasticity Drives Self-Assembly of Particle Trains in a Straight Microfluidic Channel. Phys. Rev. Appl. 2018, 10, 064058. [Google Scholar] [CrossRef]
- D’Avino, G.; Maffettone, P.L. Numerical Simulations on the Dynamics of a Particle Pair in a Viscoelastic Fluid in a Microchannel: Effect of Rheology, Particle Shape, and Confinement. Microfluid. Nanofluidics 2019, 23, 82. [Google Scholar] [CrossRef]
- Jeyasountharan, A.; Shahrivar, K.; D’Avino, G.; Del Giudice, F. Viscoelastic Particle Train Formation in Microfluidic Flows Using a Xanthan Gum Aqueous Solution. Anal. Chem. 2021, 93, 5503–5512. [Google Scholar] [CrossRef] [PubMed]
- Beneyton, T.; Wijaya, I.P.M.; Postros, P.; Najah, M.; Leblond, P.; Couvent, A.; Mayot, E.; Griffiths, A.D.; Drevelle, A. High-Throughput Screening of Filamentous Fungi Using Nanoliter-Range Droplet-Based Microfluidics. Sci. Rep. 2016, 6, 27223. [Google Scholar] [CrossRef] [PubMed]
- Tawfik, D.S.; Griffiths, A.D. Man-Made Cell-like Compartments for Molecular Evolution. Nat. Biotechnol. 1998, 16, 652–656. [Google Scholar] [CrossRef]
- Fallah-Araghi, A.; Baret, J.C.; Ryckelynck, M.; Griffiths, A.D. A Completely In Vitro Ultrahigh-Throughput Droplet-Based Microfluidic Screening System for Protein Engineering and Directed Evolution. Lab Chip 2012, 12, 882–891. [Google Scholar] [CrossRef]
- Cubi, R.; Bouhedda, F.; Collot, M.; Klymchenko, A.S.; Ryckelynck, M. µIVC-Useq: A Microfluidic-Assisted High-Throughput Functional Screening in Tandem with Next-Generation Sequencing and Artificial Neural Network to Rapidly Characterize RNA Molecules. RNA 2021, 27, 841–853. [Google Scholar] [CrossRef]
- Autour, A.; Bouhedda, F.; Cubi, R.; Ryckelynck, M. Optimization of Fluorogenic RNA-Based Biosensors Using Droplet-Based Microfluidic Ultrahigh-Throughput Screening. Methods 2019, 161, 46–53. [Google Scholar] [CrossRef] [PubMed]
- Autour, A.; Westhof, E.; Ryckelynck, M. iSpinach: A Fluorogenic RNA Aptamer Optimized for In Vitro Application. Nucleic Acids Res. 2016, 44, 2491–2500. [Google Scholar] [CrossRef]
- Holstein, J.M.; Gylstorff, C.; Hollfelder, F. Cell-free Directed Evolution of a Protease in Microdroplets at Ultrahigh Throughput. ACS Synth. Biol. 2021, 10, 252–257. [Google Scholar] [CrossRef]
- Mazutis, L.; Baret, J.C.; Treacy, P.; Skhiri, Y.; Araghi, A.F.; Ryckelynck, M.; Taly, V.; Griffiths, A.D. Multi-step microfluidic droplet processing: Kinetic analysis of an in vitro translated enzyme. Lab Chip 2009, 9, 2902–2908. [Google Scholar] [CrossRef]
- Mazutis, L.; Araghi, A.F.; Miller, O.J.; Baret, J.C.; Frenz, L.; Janoshazi, A.; Taly, V.; Miller, B.J.; Hutchison, J.B.; Link, D.; et al. Droplet-Based Microfluidic Systems for High-Throughput Single DNA Molecule Isothermal Amplification and Analysis. Anal. Chem. 2009, 81, 4813–4821. [Google Scholar] [CrossRef] [PubMed]
- Zinchenko, A.; Devenish, S.R.A.; Kintses, B.; Colin, P.Y.; Fischlechner, M.; Hollfelder, F. One in a Million: Flow Cytometric Sorting of Single Cell-Lysate Assays in Monodisperse Picolitre Double Emulsion Droplets for Directed Evolution. Anal. Chem. 2014, 86, 2526–2533. [Google Scholar] [CrossRef] [PubMed]
- Bouzetos, E.; Ganar, K.A.; Mastrobattista, E.; Deshpande, S.; van der Oost, J. (R)evolution-on-a-Chip. Trends Biotechnol. 2022, 40, 60–76. [Google Scholar] [CrossRef] [PubMed]
- Zhuang, S.; Semenec, L.; Nagy, S.S.; Cain, A.K.; Inglis, D.W. High-Precision Screening and Sorting of Double Emulsion Droplets. Cytom. Part A 2024, in press. [CrossRef] [PubMed]
- Terekhov, S.S.; Smirnov, I.V.; Stepanova, A.V.; Bobik, T.V.; Mokrushina, Y.A.; Ponomarenko, N.A.; Belogurov, A.A.; Rubtsova, M.P.; Kartseva, O.V.; Gomzikova, M.O.; et al. Microfluidic Droplet Platform for Ultrahigh-Throughput Single-Cell Screening of Biodiversity. Proc. Natl. Acad. Sci. USA 2017, 114, 2550. [Google Scholar] [CrossRef] [PubMed]
- Larsen, A.C.; Dunn, M.R.; Hatch, A.; Sau, S.P.; Youngbull, C.; Chaput, J.C. A general Strategy for Expanding Polymerase Function by Droplet Microfluidics. Nat. Commun. 2016, 7, 11235. [Google Scholar] [CrossRef] [PubMed]
- Aden, S.; Snoj, T.; Anderluh, G. Chapter Eight-The use of Giant Unilamellar Vesicles to Study Functional Properties of Pore-Forming Toxins. Methods Enzymol. 2021, 649, 219–251. [Google Scholar] [CrossRef] [PubMed]
- Nishimura, K.; Matsuura, T.; Nishimura, K.; Sunami, T.; Suzuki, H.; Yomo, T. Cell-Free Protein Synthesis inside Giant Unilamellar Vesicles Analyzed by Flow Cytometry. Langmuir 2012, 28, 8426–8432. [Google Scholar] [CrossRef] [PubMed]
- Moga, A.; Yandrapalli, N.; Dimova, R.; Robinson, T. Optimization of the Inverted Emulsion Method for High-Yield Production of Biomimetic Giant Unilamellar Vesicles. ChemBioChem 2019, 20, 2674–2682. [Google Scholar] [CrossRef]
- Elani, Y.; Purushothaman, S.; Booth, P.J.; Seddon, J.M.; Brooks, N.J.; Law, R.V.; Ces, O. Measurements of the Effect of Membrane Asymmetry on the Mechanical Properties of lipid Bilayers. Chem. Commun. 2015, 51, 6976–6979. [Google Scholar] [CrossRef]
- Trantidou, T.; Regoutz, A.; Voon, X.N.; Payne, D.J.; Ces, O. A “Cleanroom-Free” and Scalable Manufacturing Technology for the Microfluidic Generation of Lipid-Stabilized Droplets and Cell-Sized Multisomes. Sens. Actuators B 2018, 267, 34–41. [Google Scholar] [CrossRef]
- Matosevic, S.; Paegel, B.M. Stepwise Synthesis of Giant Unilamellar Vesicles on a Microfluidic Assembly Line. J. Am. Chem. Soc. 2011, 133, 2798–2800. [Google Scholar] [CrossRef]
- Lu, L.; Schertzerb, J.W.; Chiarot, P.R. Continuous Microfluidic Fabrication of Synthetic Asymmetric Vesicles. Lab Chip 2015, 15, 3591–3599. [Google Scholar] [CrossRef] [PubMed]
- Maktabi, S.; Schertzer, J.W.; Chiarot, P.R. Dewetting-Induced Formation and Mechanical Properties of Synthetic Bacterial Outer Membrane Models (GUVs) with Controlled Inner-Leaflet Lipid Composition. Soft Matter 2019, 15, 3938–3948. [Google Scholar] [CrossRef] [PubMed]
- Yandrapalli, N.; Petit, J.; Bäumchen, O.; Robinson, T. Surfactant-Free Production of Biomimetic Giant Unilamellar Vesicles using PDMS-Based Microfluidics. Commun. Chem. 2021, 4, 100. [Google Scholar] [CrossRef]
- Staufer, O.; Antona, S.; Zhang, D.; Csatári, J.; Schröter, M.; Janiesch, J.W.; Fabritz, S.; Berger, I.; Platzman, I.; Spatz, J.P. Microfluidic Production and Characterization of Biofunctionalized Giant Unilamellar Vesicles for Targeted Intracellular Cargo Delivery. Biomaterials 2021, 264, 120203. [Google Scholar] [CrossRef] [PubMed]
- Shum, H.C.; Lee, D.; Yoon, I.; Kodger, T.; Weitz, D.A. Double Emulsion Templated Monodisperse Phospholipid Vesicles. Langmuir 2008, 24, 7651–7653. [Google Scholar] [CrossRef]
- Deshpande, S.; Caspi, Y.; Meijering, A.E.C.; Dekker, C. Octanol-Assisted Liposome Assembly on Chip. Nat. Commun. 2016, 7, 10447. [Google Scholar] [CrossRef]
- Petit, J.; Polenz, I.; Baret, J.C.; Herminghaus, S.; Bäumchen, O. Vesicles-on-a-Chip: A Universal Microfluidic Platform for the Assembly of Liposomes and Polymersomes. Eur. Phys. J. E 2016, 39, 59. [Google Scholar] [CrossRef]
- Arriaga, L.R.; Datta, S.S.; Kim, S.H.; Amstad, E.; Kodger, T.E.; Monroy, F.; Weitz, D.A. Ultrathin Shell Double Emulsion Templated Giant Unilamellar Lipid Vesicles with Controlled Microdomain Formation. Small 2014, 10, 950–956. [Google Scholar] [CrossRef]
- Michelon, M.; Huang, Y.; de la Torre, L.G.; Weitz, D.A.; Cunha, R.L. Single-Step Microfluidic Production of W/O/W Double Emulsions as Templates for β-Carotene-Loaded Giant Liposomes Formation. Chem. Eng. J. 2019, 366, 27–32. [Google Scholar] [CrossRef]
- Bao, P.; Paterson, D.A.; Peyman, S.A.; Jones, J.C.; Sandoe, J.A.T.; Gleeson, H.F.; Evans, S.D.; Bushby, R.J. Production of Giant Unilamellar Vesicles and Encapsulation of Lyotropic Nematic Liquid Crystals. Soft Matter 2021, 17, 2234. [Google Scholar] [CrossRef] [PubMed]
- Deng, N.N.; Yelleswarapu, M.; Huck, W.T.S. Monodisperse Uni- and Multicompartment Liposomes. J. Am. Chem. Soc. 2016, 138, 7584–7591. [Google Scholar] [CrossRef] [PubMed]
- Trantidou, T.; Friddin, M.S.; Salehi-Reyhani, A.; Ces, O.; Elani, Y. Droplet Microfluidics for the Construction of Compartmentalised Model Membranes. Lab Chip 2018, 18, 2488–2509. [Google Scholar] [CrossRef]
- Haller, B.; Göpfrich, K.; Schröter, M.; Janiesch, J.W.; Platzman, I.; Spatz, J.P. Charge-Controlled Microfluidic Formation of Lipid-Based Single- and Multicompartment Systems. Lab Chip 2018, 18, 2665. [Google Scholar] [CrossRef]
- Weiss, M.; Frohnmayer, J.P.; Benk, L.T.; Haller, B.; Janiesch, J.W.; Heitkamp, T.; Börsch, M.; Lira, R.B.; Dimova, R.; Lipowsky, R.; et al. Sequential Bottom-Up Assembly of Mechanically Stabilized Synthetic Cells by Microfluidics. Nat. Mater 2018, 17, 89–96. [Google Scholar] [CrossRef] [PubMed]
- Hatch, A.C.; Fisher, J.S.; Tovar, A.R.; Hsieh, A.T.; Lin, R.; Pentoney, S.L.; Yang, D.L.; Lee, A.P. 1-Million Droplet Array with Wide-Field Fluorescence Imaging for Digital PCR. Lab Chip 2011, 11, 3838–3845. [Google Scholar] [CrossRef] [PubMed]
- Deshpande, S.; Spoelstra, W.K.; van Doorn, M.; Kerssemakers, J.; Dekker, C. Mechanical Division of Cell-Sized Liposomes. ACS Nano 2018, 12, 2560–2568. [Google Scholar] [CrossRef]
- Vanella, R.; Ta, D.T.; Nash, M.A. Enzyme-Mediated Hydrogel Encapsulation of Single Cells For High-Throughput Screening and Directed Evolution of Oxidoreductases. Biotechnol. Bioeng. 2019, 116, 1878–1886. [Google Scholar] [CrossRef]
- Fischlechner, M.; Schaerli, Y.; Mohamed, M.F.; Patil, S.; Abell, C.; Hollfelder, F. Evolution of Enzyme catalysts Caged in Biomimetic Gel-Shell Beads. Nat. Chem. 2014, 6, 791–796. [Google Scholar] [CrossRef]
- Vanella, R.; Kovacevic, G.; Doffini, V.; de Santaella, J.F.; Nash, M.A. High-Throughput Screening, Next Generation Sequencing and Machine Learning: Advanced Methods in Enzyme Engineering. Chem. Commun. 2022, 58, 2455–2467. [Google Scholar] [CrossRef]
- Chen, M.; Bolognesi, G.; Vladisavljević, G.T. Crosslinking Strategies for the Microfluidic Production of Microgels. Molecules 2021, 26, 3752. [Google Scholar] [CrossRef]
- Leng, X.; Zhang, W.; Wang, C.; Cui, L.; Yang, C.J. Agarose Droplet Microfluidics for Highly Parallel and Efficient Single Molecule Emulsion PCR. Lab Chip 2010, 10, 2841–2843. [Google Scholar] [CrossRef]
- Luo, D.; Pullela, S.R.; Marquez, M.; Cheng, Z. Cell Encapsules with Tunable Transport and Mechanical Properties. Biomicrofluidics 2007, 1, 034102. [Google Scholar] [CrossRef] [PubMed]
- Chokkalingam, V.; Tel, J.; Wimmers, F.; Liu, X.; Semenov, S.; Thiele, J.; Figdor, C.G.; Huck, W.T.S. Probing Cellular Heterogeneity in Cytokine-Secreting Immune Cells Using Droplet-Based Microfluidics. Lab Chip 2013, 13, 4740–4744. [Google Scholar] [CrossRef]
- Duarte, J.M.; Barbier, I.; Schaerli, Y. Bacterial Microcolonies in Gel Beads for High-Throughput Screening of Libraries in Synthetic Biology. ACS Synth. Biol. 2017, 6, 1988–1995. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.; Gao, X.; Chen, L.; Zhang, M.; Ma, J.; Zhang, X.; Qin, J. High Throughput Generation and Trapping of Individual Agarose Microgel Using Microfluidic Approach. Microfluid. Nanofluidics 2013, 15, 467–474. [Google Scholar] [CrossRef]
- Tanaka, Y.; Nakamura, A.; Suzuki, Y.; Maruta, K.; Shida, Y.; Ogasawara, W. Using Gel Microdroplets to Develop a Simple High-Throughput Screening Platform for Oleaginous Microorganisms. J. Biotech 2022, 358, 46–54. [Google Scholar] [CrossRef]
- Tomal, W.; Ortyl, J. Water-Soluble Photoinitiators in Biomedical Applications. Polymers 2020, 12, 1073. [Google Scholar] [CrossRef]
- Zhang, C.; Wu, X.; Song, F.; Liu, S.; Yu, S.; Zhou, J. Core–Shell Droplet-Based Microfluidic Screening System for Filamentous Fungi. ACS Sens. 2023, 8, 3468–3477. [Google Scholar] [CrossRef]
- Lai, X.; Yang, M.; Wu, H.; Li, D. Modular Microfluidics: Current Status and Future Prospects. Micromachines 2022, 13, 1363. [Google Scholar] [CrossRef] [PubMed]
- Vittayarukskul, K.; Lee, A.P. A Truly Lego®-like Modular Microfluidics Platform. J. Micromech. Microeng. 2017, 27, 035004. [Google Scholar] [CrossRef]
- Fan, Y.Q.; Wang, H.L.; Gao, K.X.; Liu, J.J.; Chai, D.P.; Zhang, Y.J. Applications of Modular Microfluidics Technology. Chin. J. Anal. Chem. 2018, 46, 1863–1871. [Google Scholar] [CrossRef]




Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vladisavljević, G.T. Droplet Microfluidics for High-Throughput Screening and Directed Evolution of Biomolecules. Micromachines 2024, 15, 971. https://doi.org/10.3390/mi15080971
Vladisavljević GT. Droplet Microfluidics for High-Throughput Screening and Directed Evolution of Biomolecules. Micromachines. 2024; 15(8):971. https://doi.org/10.3390/mi15080971
Chicago/Turabian StyleVladisavljević, Goran T. 2024. "Droplet Microfluidics for High-Throughput Screening and Directed Evolution of Biomolecules" Micromachines 15, no. 8: 971. https://doi.org/10.3390/mi15080971
APA StyleVladisavljević, G. T. (2024). Droplet Microfluidics for High-Throughput Screening and Directed Evolution of Biomolecules. Micromachines, 15(8), 971. https://doi.org/10.3390/mi15080971






