Polymerase Chain Reaction Chips for Biomarker Discovery and Validation in Drug Development
Abstract
1. Introduction
2. Fundamentals of PCR Chips
2.1. What Are PCR Chips?
2.2. Advantages of PCR Chips
2.3. Types of PCR Chips
3. The Role of PCR Chips in Biomarker Discovery
3.1. Identification of Novel Biomarkers
3.2. Quantitative Analysis of Biomarkers
3.3. Single-Cell Analysis
4. The Role of PCR Chips in Biomarker Validation
4.1. Analytical Validation
4.2. Clinical Validation
5. Applications of PCR Chips in Drug Development
5.1. Target Identification and Validation
5.2. Patient Stratification
5.3. Monitoring Drug Response
5.4. Companion Diagnostics
6. Current Challenges and Limitations
6.1. Technological Barriers
6.2. Biological Challenges
6.3. Regulatory and Ethical Considerations in Biomarker-Based Drug Development
7. Emerging Trends and Future Perspectives
7.1. Integration with Artificial Intelligence and Machine Learning
7.2. Multi-Omics Approaches
7.3. Advancements in Chip Design
7.4. Applications Beyond Drug Development
8. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Mayeux, R. Biomarkers: Potential uses and limitations. NeuroRx 2004, 1, 182–188. [Google Scholar] [CrossRef]
- Chikkaveeraiah, B.V.; Bhirde, A.A.; Morgan, N.Y.; Eden, H.S.; Chen, X. Electrochemical Immunosensors for Detection of Cancer Protein Biomarkers. ACS Nano 2012, 6, 6546–6561. [Google Scholar] [CrossRef]
- Haes, A.J.; Chang, L.; Klein, W.L.; Van Duyne, R.P. Detection of a Biomarker for Alzheimer’s Disease from Synthetic and Clinical Samples Using a Nanoscale Optical Biosensor. J. Am. Chem. Soc. 2005, 127, 2264–2271. [Google Scholar] [CrossRef] [PubMed]
- Frank, R.; Hargreaves, R. Clinical biomarkers in drug discovery and development. Nat. Rev. Drug Discov. 2003, 2, 566–580. [Google Scholar] [CrossRef] [PubMed]
- Hanash, S.M.; Baik, C.S.; Kallioniemi, O. Emerging molecular biomarkers—Blood-based strategies to detect and monitor cancer. Nat. Rev. Clin. Oncol. 2011, 8, 142–150. [Google Scholar] [CrossRef]
- Hartl, D.; de Luca, V.; Kostikova, A.; Laramie, J.; Kennedy, S.; Ferrero, E.; Siegel, R.; Fink, M.; Ahmed, S.; Millholland, J. Translational precision medicine: An industry perspective. J. Transl. Med. 2021, 19, 245. [Google Scholar] [CrossRef] [PubMed]
- Seyhan, A.A.; Carini, C. Are innovation and new technologies in precision medicine paving a new era in patients centric care? J. Transl. Med. 2019, 17, 114. [Google Scholar] [CrossRef]
- Derouane, F.; van Marcke, C.; Berlière, M.; Gerday, A.; Fellah, L.; Leconte, I.; Van Bockstal, M.R.; Galant, C.; Corbet, C.; Duhoux, F.P. Predictive biomarkers of response to neoadjuvant chemotherapy in breast cancer: Current and future perspectives for precision medicine. Cancers 2022, 14, 3876. [Google Scholar] [CrossRef]
- Gohil, S.H.; Iorgulescu, J.B.; Braun, D.A.; Keskin, D.B.; Livak, K.J. Applying high-dimensional single-cell technologies to the analysis of cancer immunotherapy. Nat. Rev. Clin. Oncol. 2021, 18, 244–256. [Google Scholar] [CrossRef] [PubMed]
- Goossens, N.; Nakagawa, S.; Sun, X.; Hoshida, Y. Cancer biomarker discovery and validation. Transl. Cancer Res. 2015, 4, 256. [Google Scholar]
- Ilyin, S.E.; Belkowski, S.M.; Plata-Salamán, C.R. Biomarker discovery and validation: Technologies and integrative approaches. Trends Biotechnol. 2004, 22, 411–416. [Google Scholar] [CrossRef] [PubMed]
- McDermott, J.E.; Wang, J.; Mitchell, H.; Webb-Robertson, B.-J.; Hafen, R.; Ramey, J.; Rodland, K.D. Challenges in biomarker discovery: Combining expert insights with statistical analysis of complex omics data. Expert Opin. Med. Diagn. 2013, 7, 37–51. [Google Scholar] [CrossRef]
- Dieterle, F.; Marrer, E. New technologies around biomarkers and their interplay with drug development. Anal. Bioanal. Chem. 2008, 390, 141–154. [Google Scholar] [CrossRef] [PubMed]
- Gupta, S.; Venkatesh, A.; Ray, S.; Srivastava, S. Challenges and prospects for biomarker research: A current perspective from the developing world. Biochim. Biophys. Acta (BBA)-Proteins Proteom. 2014, 1844, 899–908. [Google Scholar] [CrossRef]
- Solier, C.; Langen, H. Antibody-based proteomics and biomarker research—Current status and limitations. Proteomics 2014, 14, 774–783. [Google Scholar] [CrossRef] [PubMed]
- Rifai, N.; Gillette, M.A.; Carr, S.A. Protein biomarker discovery and validation: The long and uncertain path to clinical utility. Nat. Biotechnol. 2006, 24, 971–983. [Google Scholar] [CrossRef]
- Peng, P.; Liu, C.; Li, Z.; Xue, Z.; Mao, P.; Hu, J.; Xu, F.; Yao, C.; You, M. Emerging ELISA derived technologies for in vitro diagnostics. TrAC Trends Anal. Chem. 2022, 152, 116605. [Google Scholar] [CrossRef]
- Jung, B.; Adeli, K. Clinical laboratory reference intervals in pediatrics: The CALIPER initiative. Clin. Biochem. 2009, 42, 1589–1595. [Google Scholar] [CrossRef] [PubMed]
- Crowther, L.M.; Poms, M.; Plecko, B. Multiomics tools for the diagnosis and treatment of rare neurological disease. J. Inherit. Metab. Dis. 2018, 41, 425–434. [Google Scholar] [CrossRef] [PubMed]
- Misra, B.B.; Langefeld, C.; Olivier, M.; Cox, L.A. Integrated omics: Tools, advances and future approaches. J. Mol. Endocrinol. 2019, 62, R21–R45. [Google Scholar] [CrossRef]
- Filkins, L.M.; Bryson, A.L.; Miller, S.A.; Mitchell, S.L. Navigating clinical utilization of direct-from-specimen metagenomic pathogen detection: Clinical applications, limitations, and testing recommendations. Clin. Chem. 2020, 66, 1381–1395. [Google Scholar] [CrossRef]
- Vojinović, V.; Cabral, J.M.S.; Fonseca, L.P. Real-time bioprocess monitoring: Part I: In situ sensors. Sens. Actuators B Chem. 2006, 114, 1083–1091. [Google Scholar] [CrossRef]
- Teymourian, H.; Tehrani, F.; Longardner, K.; Mahato, K.; Podhajny, T.; Moon, J.-M.; Kotagiri, Y.G.; Sempionatto, J.R.; Litvan, I.; Wang, J. Closing the loop for patients with Parkinson disease: Where are we? Nat. Rev. Neurol. 2022, 18, 497–507. [Google Scholar] [CrossRef] [PubMed]
- Tan, D.S.W.; Thomas, G.V.; Garrett, M.D.; Banerji, U.; De Bono, J.S.; Kaye, S.B.; Workman, P. Biomarker-driven early clinical trials in oncology: A paradigm shift in drug development. Cancer J. 2009, 15, 406–420. [Google Scholar] [CrossRef] [PubMed]
- Oliveira, B.B.; Veigas, B.; Baptista, P.V. Isothermal amplification of nucleic acids: The race for the next “gold standard”. Front. Sens. 2021, 2, 752600. [Google Scholar] [CrossRef]
- Zhu, H.; Zhang, H.; Xu, Y.; Laššáková, S.; Korabečná, M.; Neužil, P. PCR past, present and future. Biotechniques 2020, 69, 317–325. [Google Scholar] [CrossRef]
- Navarro, E.; Serrano-Heras, G.; Castaño, M.J.; Solera, J. Real-time PCR detection chemistry. Clin. Chim. Acta 2015, 439, 231–250. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Xu, J.; Ma, W.; Zheng, W. PCR microfluidic devices for DNA amplification. Biotechnol. Adv. 2006, 24, 243–284. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Xing, D.; Li, Y. Micropumps, microvalves, and micromixers within PCR microfluidic chips: Advances and trends. Biotechnol. Adv. 2007, 25, 483–514. [Google Scholar] [CrossRef]
- Park, S.; Zhang, Y.; Lin, S.; Wang, T.-H.; Yang, S. Advances in microfluidic PCR for point-of-care infectious disease diagnostics. Biotechnol. Adv. 2011, 29, 830–839. [Google Scholar] [CrossRef] [PubMed]
- Mohammed, M.-I.; Desmulliez, M.P.Y. Lab-on-a-chip based immunosensor principles and technologies for the detection of cardiac biomarkers: A review. Lab Chip 2011, 11, 569–595. [Google Scholar] [CrossRef]
- Wu, L.; Qu, X. Cancer biomarker detection: Recent achievements and challenges. Chem. Soc. Rev. 2015, 44, 2963–2997. [Google Scholar] [CrossRef] [PubMed]
- Diao, Z.; Han, Y.; Zhang, R.; Li, J. Circulating tumour DNA: A new biomarker to monitor resistance in NSCLC patients treated with EGFR-TKIs. Biochim. Biophys. Acta (BBA) Rev. Cancer 2020, 1873, 188363. [Google Scholar] [CrossRef] [PubMed]
- Bronkhorst, A.J.; Holdenrieder, S. The changing face of circulating tumor DNA (ctDNA) profiling: Factors that shape the landscape of methodologies, technologies, and commercialization. Med. Genet. 2023, 35, 201–235. [Google Scholar] [CrossRef] [PubMed]
- Behura, S.K. Molecular marker systems in insects: Current trends and future avenues. Mol. Ecol. 2006, 15, 3087–3113. [Google Scholar] [CrossRef] [PubMed]
- Bamodu, O.A.; Chung, C.-C.; Pisanic, T.R. Harnessing liquid biopsies: Exosomes and ctDNA as minimally invasive biomarkers for precision cancer medicine. J. Liq. Biopsy 2023, 2, 100126. [Google Scholar] [CrossRef]
- Van de Sande, B.; Lee, J.S.; Mutasa-Gottgens, E.; Naughton, B.; Bacon, W.; Manning, J.; Wang, Y.; Pollard, J.; Mendez, M.; Hill, J.; et al. Applications of single-cell RNA sequencing in drug discovery and development. Nat. Rev. Drug Discov. 2023, 22, 496–520. [Google Scholar] [CrossRef] [PubMed]
- Brennan, D.; Justice, J.; Corbett, B.; McCarthy, T.; Galvin, P. Emerging optofluidic technologies for point-of-care genetic analysis systems: A review. Anal. Bioanal. Chem. 2009, 395, 621–636. [Google Scholar] [CrossRef]
- Yin, J.; Suo, Y.; Zou, Z.; Sun, J.; Zhang, S.; Wang, B.; Xu, Y.; Darland, D.; Zhao, J.X.; Mu, Y. Integrated microfluidic systems with sample preparation and nucleic acid amplification. Lab Chip 2019, 19, 2769–2785. [Google Scholar] [CrossRef]
- Zhang, C.; Xing, D. Miniaturized PCR chips for nucleic acid amplification and analysis: Latest advances and future trends. Nucleic Acids Res. 2007, 35, 4223–4237. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Chen, Z.; Corstjens, P.L.A.M.; Mauk, M.G.; Bau, H.H. A disposable microfluidic cassette for DNA amplification and detection. Lab Chip 2006, 6, 46–53. [Google Scholar] [CrossRef]
- Cortese, B.; Mowlem, M.C.; Morgan, H. Characterisation of an irreversible bonding process for COC–COC and COC–PDMS–COC sandwich structures and application to microvalves. Sens. Actuators B Chem. 2011, 160, 1473–1480. [Google Scholar] [CrossRef]
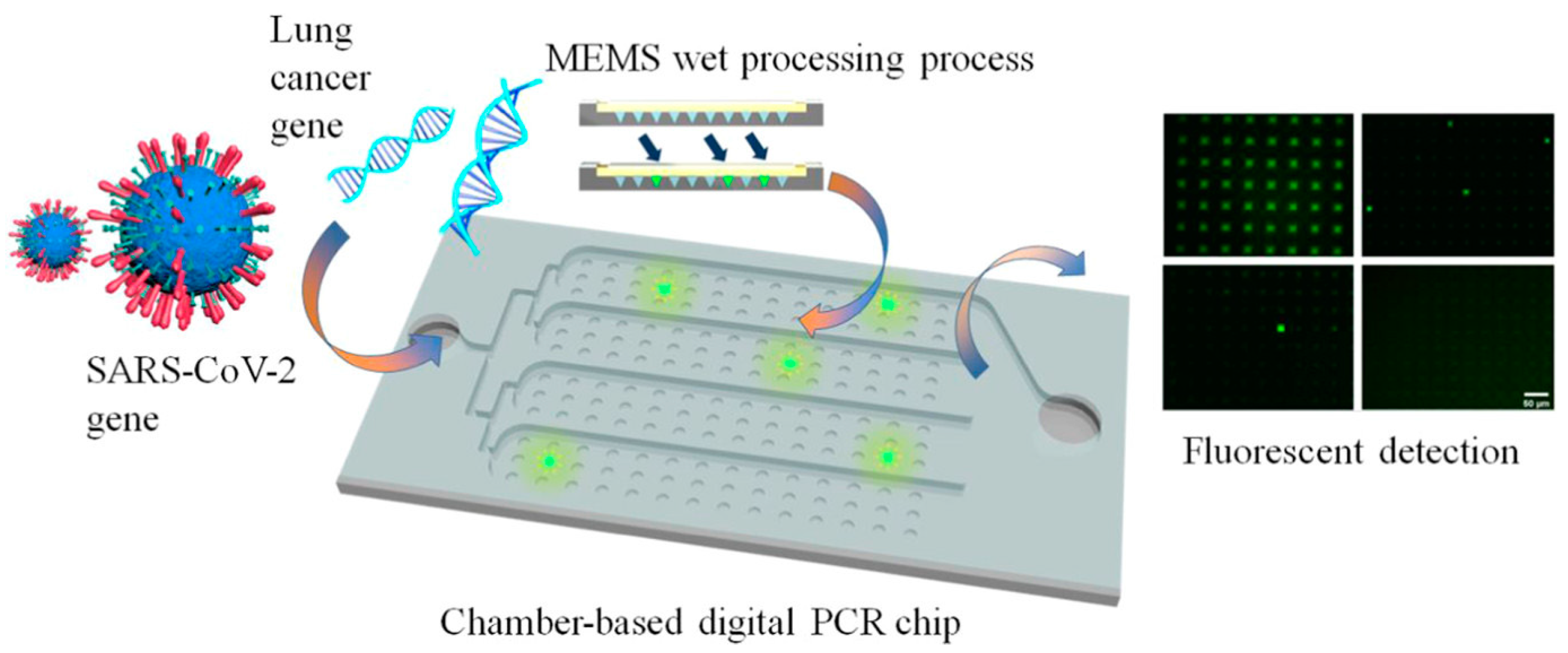
- Sun, Y.; Huang, Y.; Qi, T.; Jin, Q.; Jia, C.; Zhao, J.; Feng, S.; Liang, L. Wet-Etched Microchamber Array Digital PCR Chip for SARS-CoV-2 Virus and Ultra-Early Stage Lung Cancer Quantitative Detection. ACS Omega 2022, 7, 1819–1826. [Google Scholar] [CrossRef] [PubMed]
- Agha, A.; Waheed, W.; Alamoodi, N.; Mathew, B.; Alnaimat, F.; Abu-Nada, E.; Abderrahmane, A.; Alazzam, A. A Review of Cyclic Olefin Copolymer Applications in Microfluidics and Microdevices. Macromol. Mater. Eng. 2022, 307, 2200053. [Google Scholar] [CrossRef]
- Roper, M.G.; Easley, C.J.; Landers, J.P. Advances in Polymerase Chain Reaction on Microfluidic Chips. Anal. Chem. 2005, 77, 3887–3894. [Google Scholar] [CrossRef] [PubMed]
- Arya, M.; Shergill, I.S.; Williamson, M.; Gommersall, L.; Arya, N.; Patel, H.R.H. Basic principles of real-time quantitative PCR. Expert Rev. Mol. Diagn. 2005, 5, 209–219. [Google Scholar] [CrossRef] [PubMed]
- Kulkarni, M.B.; Yashas; Vyas, R. A role of integrated microheaters in a microfluidics based point-of-care-testing and beyond for healthcare applications. Appl. Mater. Today 2024, 38, 102225. [Google Scholar] [CrossRef]
- Moschou, D.; Vourdas, N.; Kokkoris, G.; Papadakis, G.; Parthenios, J.; Chatzandroulis, S.; Tserepi, A. All-plastic, low-power, disposable, continuous-flow PCR chip with integrated microheaters for rapid DNA amplification. Sens. Actuators B Chem. 2014, 199, 470–478. [Google Scholar] [CrossRef]
- Beer, N.R.; Hindson, B.J.; Wheeler, E.K.; Hall, S.B.; Rose, K.A.; Kennedy, I.M.; Colston, B.W. On-Chip, Real-Time, Single-Copy Polymerase Chain Reaction in Picoliter Droplets. Anal. Chem. 2007, 79, 8471–8475. [Google Scholar] [CrossRef] [PubMed]
- Zhu, H.; Fohlerová, Z.; Pekárek, J.; Basova, E.; Neužil, P. Recent advances in lab-on-a-chip technologies for viral diagnosis. Biosens. Bioelectron. 2020, 153, 112041. [Google Scholar] [CrossRef] [PubMed]
- Ranjit Prakash, A.; Adamia, S.; Sieben, V.; Pilarski, P.; Pilarski, L.M.; Backhouse, C.J. Small volume PCR in PDMS biochips with integrated fluid control and vapour barrier. Sens. Actuators B Chem. 2006, 113, 398–409. [Google Scholar] [CrossRef]
- Matsubara, Y.; Kerman, K.; Kobayashi, M.; Yamamura, S.; Morita, Y.; Tamiya, E. Microchamber array based DNA quantification and specific sequence detection from a single copy via PCR in nanoliter volumes. Biosens. Bioelectron. 2005, 20, 1482–1490. [Google Scholar] [CrossRef] [PubMed]
- Yan, Z.; Jin, M.; Li, Z.; Zhou, G.; Shui, L. Droplet-Based Microfluidic Thermal Management Methods for High Performance Electronic Devices. Micromachines 2019, 10, 89. [Google Scholar] [CrossRef] [PubMed]
- Farrar, J.S.; Wittwer, C.T. Extreme PCR: Efficient and Specific DNA Amplification in 15–60 Seconds. Clin. Chem. 2015, 61, 145–153. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Xing, D. Single-Molecule DNA Amplification and Analysis Using Microfluidics. Chem. Rev. 2010, 110, 4910–4947. [Google Scholar] [CrossRef] [PubMed]
- Afshari Babazad, M.; Foroozandeh, A.; Abdouss, M.; SalarAmoli, H.; Babazad, R.A.; Hasanzadeh, M. Recent progress and challenges in biosensing of carcinoembryonic antigen. TrAC Trends Anal. Chem. 2024, 180, 117964. [Google Scholar] [CrossRef]
- de Paz, H.D.; Brotons, P.; Muñoz-Almagro, C. Molecular isothermal techniques for combating infectious diseases: Towards low-cost point-of-care diagnostics. Expert Rev. Mol. Diagn. 2014, 14, 827–843. [Google Scholar] [CrossRef] [PubMed]
- Shen, K.; Chen, X.; Guo, M.; Cheng, J. A microchip-based PCR device using flexible printed circuit technology. Sens. Actuators B Chem. 2005, 105, 251–258. [Google Scholar] [CrossRef]
- Chen, S.; Sun, Y.; Fan, F.; Chen, S.; Zhang, Y.; Zhang, Y.; Meng, X.; Lin, J.-M. Present status of microfluidic PCR chip in nucleic acid detection and future perspective. TrAC Trends Anal. Chem. 2022, 157, 116737. [Google Scholar] [CrossRef]
- Quan, P.-L.; Sauzade, M.; Brouzes, E. dPCR: A Technology Review. Sensors 2018, 18, 1271. [Google Scholar] [CrossRef] [PubMed]
- Gou, T.; Hu, J.; Wu, W.; Ding, X.; Zhou, S.; Fang, W.; Mu, Y. Smartphone-based mobile digital PCR device for DNA quantitative analysis with high accuracy. Biosens. Bioelectron. 2018, 120, 144–152. [Google Scholar] [CrossRef] [PubMed]
- Perkins, G.; Lu, H.; Garlan, F.; Taly, V. Chapter Three–Droplet-Based Digital PCR: Application in Cancer Research. In Advances in Clinical Chemistry; Makowski, G.S., Ed.; Elsevier: Amsterdam, The Netherlands, 2017; Volume 79, pp. 43–91. [Google Scholar]
- Lee, S.H.; Park, S.-m.; Kim, B.N.; Kwon, O.S.; Rho, W.-Y.; Jun, B.-H. Emerging ultrafast nucleic acid amplification technologies for next-generation molecular diagnostics. Biosens. Bioelectron. 2019, 141, 111448. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Liu, M.; Wang, Z.; Li, S.; Deng, Y.; He, N. Point-of-care diagnostics for infectious diseases: From methods to devices. Nano Today 2021, 37, 101092. [Google Scholar] [CrossRef] [PubMed]
- Nasseri, B.; Soleimani, N.; Rabiee, N.; Kalbasi, A.; Karimi, M.; Hamblin, M.R. Point-of-care microfluidic devices for pathogen detection. Biosens. Bioelectron. 2018, 117, 112–128. [Google Scholar] [CrossRef]
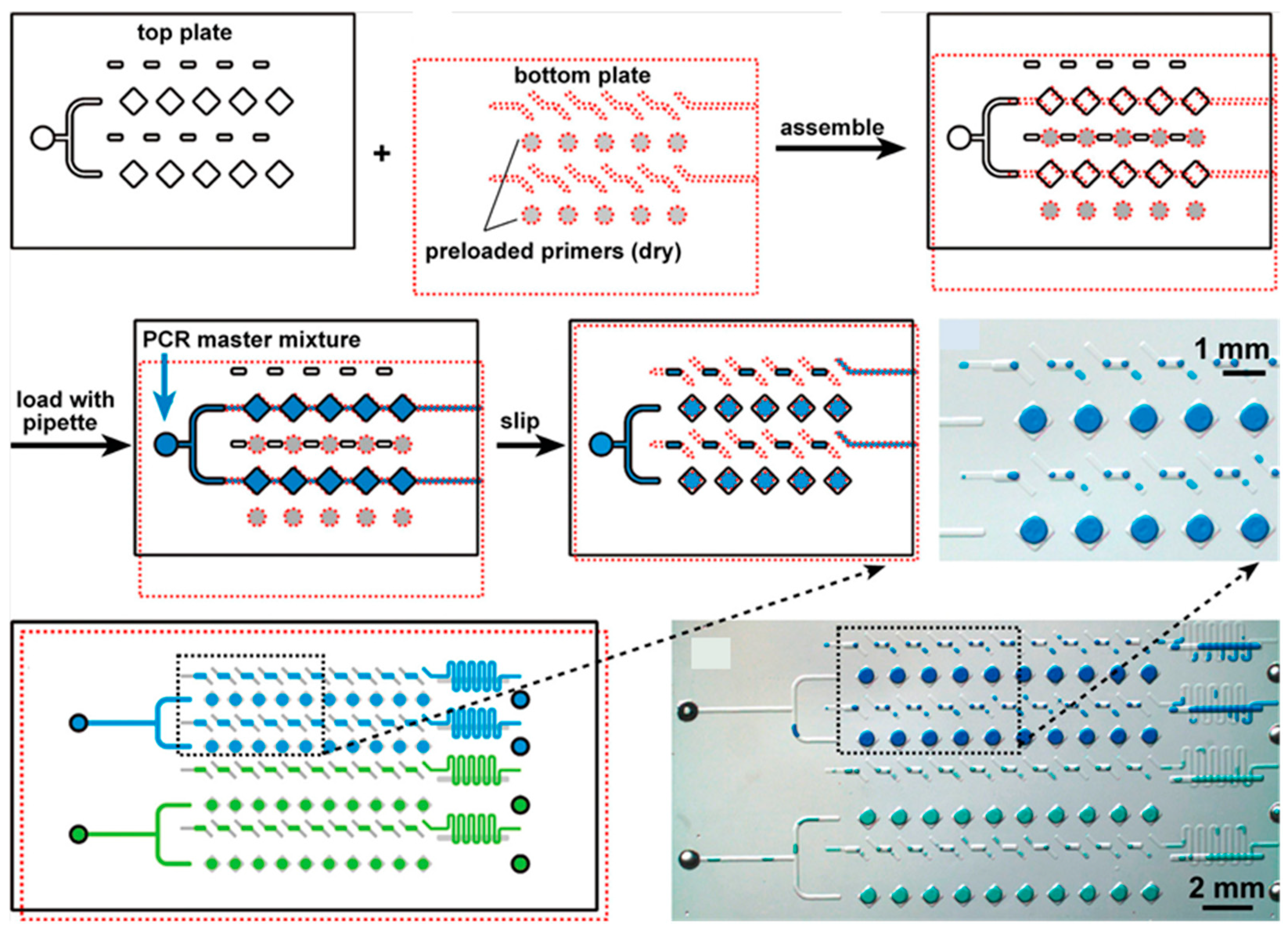
- Shen, F.; Du, W.; Davydova, E.K.; Karymov, M.A.; Pandey, J.; Ismagilov, R.F. Nanoliter Multiplex PCR Arrays on a SlipChip. Anal. Chem. 2010, 82, 4606–4612. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Zhang, Y.-X.; Liu, W.-W.; Ma, Y.; Fang, Q.; Yao, B. Printing 2-Dimentional Droplet Array for Single-Cell Reverse Transcription Quantitative PCR Assay with a Microfluidic Robot. Sci. Rep. 2015, 5, 9551. [Google Scholar] [CrossRef]
- Shao, H.; Chung, J.; Lee, K.; Balaj, L.; Min, C.; Carter, B.S.; Hochberg, F.H.; Breakefield, X.O.; Lee, H.; Weissleder, R. Chip-based analysis of exosomal mRNA mediating drug resistance in glioblastoma. Nat. Commun. 2015, 6, 6999. [Google Scholar] [CrossRef]
- Asiello, P.J.; Baeumner, A.J. Miniaturized isothermal nucleic acid amplification, a review. Lab Chip 2011, 11, 1420–1430. [Google Scholar] [CrossRef] [PubMed]
- Zanoli, L.M.; Spoto, G. Isothermal Amplification Methods for the Detection of Nucleic Acids in Microfluidic Devices. Biosensors 2013, 3, 18–43. [Google Scholar] [CrossRef]
- Zhang, Z.; Zhao, S.; Hu, F.; Yang, G.; Li, J.; Tian, H.; Peng, N. An LED-Driven AuNPs-PDMS Microfluidic Chip and Integrated Device for the Detection of Digital Loop-Mediated Isothermal DNA Amplification. Micromachines 2020, 11, 177. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Kui, L.; Tang, M.; Li, D.; Wei, K.; Chen, W.; Miao, J.; Dong, Y. High-Throughput Transcriptome Profiling in Drug and Biomarker Discovery. Front. Genet. 2020, 11, 19. [Google Scholar] [CrossRef] [PubMed]
- Nero, T.L.; Morton, C.J.; Holien, J.K.; Wielens, J.; Parker, M.W. Oncogenic protein interfaces: Small molecules, big challenges. Nat. Rev. Cancer 2014, 14, 248–262. [Google Scholar] [CrossRef] [PubMed]
- Verma, M.; Manne, U. Genetic and epigenetic biomarkers in cancer diagnosis and identifying high risk populations. Crit. Rev. Oncol./Hematol. 2006, 60, 9–18. [Google Scholar] [CrossRef]
- Islam, M.S.; Gopalan, V.; Lam, A.K.; Shiddiky, M.J.A. Current advances in detecting genetic and epigenetic biomarkers of colorectal cancer. Biosens. Bioelectron. 2023, 239, 115611. [Google Scholar] [CrossRef] [PubMed]
- Sohrabi, H.; Bolandi, N.; Hemmati, A.; Eyvazi, S.; Ghasemzadeh, S.; Baradaran, B.; Oroojalian, F.; Reza Majidi, M.; de la Guardia, M.; Mokhtarzadeh, A. State-of-the-art cancer biomarker detection by portable (Bio) sensing technology: A critical review. Microchem. J. 2022, 177, 107248. [Google Scholar] [CrossRef]
- Khan, N.I.; Song, E. Lab-on-a-Chip Systems for Aptamer-Based Biosensing. Micromachines 2020, 11, 220. [Google Scholar] [CrossRef] [PubMed]
- Chenarani, N.; Emamjomeh, A.; Allahverdi, A.; Mirmostafa, S.; Afsharinia, M.H.; Zahiri, J. Bioinformatic tools for DNA methylation and histone modification: A survey. Genomics 2021, 113, 1098–1113. [Google Scholar] [CrossRef]
- Goodwin, S.; McPherson, J.D.; McCombie, W.R. Coming of age: Ten years of next-generation sequencing technologies. Nat. Rev. Genet. 2016, 17, 333–351. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Lee, H.J.; Corn, R.M. Detection of Protein Biomarkers Using RNA Aptamer Microarrays and Enzymatically Amplified Surface Plasmon Resonance Imaging. Anal. Chem. 2007, 79, 1082–1088. [Google Scholar] [CrossRef] [PubMed]
- Sánchez-Peña, M.L.; Isaza, C.E.; Pérez-Morales, J.; Rodríguez-Padilla, C.; Castro, J.M.; Cabrera-Ríos, M. Identification of potential biomarkers from microarray experiments using multiple criteria optimization. Cancer Med. 2013, 2, 253–265. [Google Scholar] [CrossRef] [PubMed]
- Pös, O.; Radvanszky, J.; Buglyó, G.; Pös, Z.; Rusnakova, D.; Nagy, B.; Szemes, T. DNA copy number variation: Main characteristics, evolutionary significance, and pathological aspects. Biomed. J. 2021, 44, 548–559. [Google Scholar] [CrossRef] [PubMed]
- Reis, A.H.O.; Vargas, F.R.; Lemos, B. Biomarkers of genome instability and cancer epigenetics. Tumor Biol. 2016, 37, 13029–13038. [Google Scholar] [CrossRef] [PubMed]
- Pritchard, C.C.; Salipante, S.J.; Koehler, K.; Smith, C.; Scroggins, S.; Wood, B.; Wu, D.; Lee, M.K.; Dintzis, S.; Adey, A.; et al. Validation and Implementation of Targeted Capture and Sequencing for the Detection of Actionable Mutation, Copy Number Variation, and Gene Rearrangement in Clinical Cancer Specimens. J. Mol. Diagn. 2014, 16, 56–67. [Google Scholar] [CrossRef] [PubMed]
- Kaushik, A.M.; Hsieh, K.; Wang, T.-H. Droplet microfluidics for high-sensitivity and high-throughput detection and screening of disease biomarkers. WIREs Nanomed. Nanobiotechnol. 2018, 10, e1522. [Google Scholar] [CrossRef]
- Lundberg, M.; Thorsen, S.B.; Assarsson, E.; Villablanca, A.; Tran, B.; Gee, N.; Knowles, M.; Nielsen, B.S.; González Couto, E.; Martin, R.; et al. Multiplexed Homogeneous Proximity Ligation Assays for High-throughput Protein Biomarker Research in Serological Material *. Mol. Cell. Proteom. 2011, 10. [Google Scholar] [CrossRef]
- Lianidou, E. Detection and relevance of epigenetic markers on ctDNA: Recent advances and future outlook. Mol. Oncol. 2021, 15, 1683–1700. [Google Scholar] [CrossRef]
- Morselli, M.; Farrell, C.; Rubbi, L.; Fehling, H.L.; Henkhaus, R.; Pellegrini, M. Targeted bisulfite sequencing for biomarker discovery. Methods 2021, 187, 13–27. [Google Scholar] [CrossRef]
- Reinders, J.; Paszkowski, J. Bisulfite Methylation Profiling of Large Genomes. Epigenomics 2010, 2, 209–220. [Google Scholar] [CrossRef] [PubMed]
- Carrigan, P.; Krahn, T. Impact of Biomarkers on Personalized Medicine. In New Approaches to Drug Discovery; Nielsch, U., Fuhrmann, U., Jaroch, S., Eds.; Springer International Publishing: Cham, Switzerland, 2016; pp. 285–311. [Google Scholar]
- Baghel, R.; Maan, K.; Haritwal, T.; Rana, P. Chapter 2–Integration of epigenomics and metabolomics: From biomarkers discovery to personalized medicine. In Epigenetics and Metabolomics; Agrawala, P.K., Rana, P., Eds.; Academic Press: Cambridge, MA, USA, 2021; Volume 28, pp. 31–73. [Google Scholar]
- Uffelmann, E.; Huang, Q.Q.; Munung, N.S.; de Vries, J.; Okada, Y.; Martin, A.R.; Martin, H.C.; Lappalainen, T.; Posthuma, D. Genome-wide association studies. Nat. Rev. Methods Primers 2021, 1, 59. [Google Scholar] [CrossRef]
- Angelakopoulou, A.; Shah, T.; Sofat, R.; Shah, S.; Berry, D.J.; Cooper, J.; Palmen, J.; Tzoulaki, I.; Wong, A.; Jefferis, B.J.; et al. Comparative analysis of genome-wide association studies signals for lipids, diabetes, and coronary heart disease: Cardiovascular Biomarker Genetics Collaboration. Eur. Heart J. 2012, 33, 393–407. [Google Scholar] [CrossRef]
- Deming, Y.; Xia, J.; Cai, Y.; Lord, J.; Del-Aguila, J.L.; Fernandez, M.V.; Carrell, D.; Black, K.; Budde, J.; Ma, S.; et al. Genetic studies of plasma analytes identify novel potential biomarkers for several complex traits. Sci. Rep. 2016, 6, 18092. [Google Scholar] [CrossRef]
- Broza, Y.Y.; Zhou, X.; Yuan, M.; Qu, D.; Zheng, Y.; Vishinkin, R.; Khatib, M.; Wu, W.; Haick, H. Disease Detection with Molecular Biomarkers: From Chemistry of Body Fluids to Nature-Inspired Chemical Sensors. Chem. Rev. 2019, 119, 11761–11817. [Google Scholar] [CrossRef] [PubMed]
- Dhama, K.; Latheef, S.K.; Dadar, M.; Samad, H.A.; Munjal, A.; Khandia, R.; Karthik, K.; Tiwari, R.; Yatoo, M.I.; Bhatt, P. Biomarkers in stress related diseases/disorders: Diagnostic, prognostic, and therapeutic values. Front. Mol. Biosci. 2019, 6, 91. [Google Scholar] [CrossRef] [PubMed]
- La Thangue, N.B.; Kerr, D.J. Predictive biomarkers: A paradigm shift towards personalized cancer medicine. Nat. Rev. Clin. Oncol. 2011, 8, 587–596. [Google Scholar] [CrossRef] [PubMed]
- Pashayan, N.; Antoniou, A.C.; Ivanus, U.; Esserman, L.J.; Easton, D.F.; French, D.; Sroczynski, G.; Hall, P.; Cuzick, J.; Evans, D.G.; et al. Personalized early detection and prevention of breast cancer: ENVISION consensus statement. Nat. Rev. Clin. Oncol. 2020, 17, 687–705. [Google Scholar] [CrossRef] [PubMed]
- Passaro, A.; Al Bakir, M.; Hamilton, E.G.; Diehn, M.; André, F.; Roy-Chowdhuri, S.; Mountzios, G.; Wistuba, I.I.; Swanton, C.; Peters, S. Cancer biomarkers: Emerging trends and clinical implications for personalized treatment. Cell 2024, 187, 1617–1635. [Google Scholar] [CrossRef]
- Li, L.; Zhang, L.; Montgomery, K.C.; Jiang, L.; Lyon, C.J.; Hu, T.Y. Advanced technologies for molecular diagnosis of cancer: State of pre-clinical tumor-derived exosome liquid biopsies. Mater. Today Bio 2023, 18, 100538. [Google Scholar] [CrossRef] [PubMed]
- Sarhadi, V.K.; Armengol, G. Molecular Biomarkers in Cancer. Biomolecules 2022, 12, 1021. [Google Scholar] [CrossRef] [PubMed]
- Nicolini, A.; Ferrari, P.; Duffy, M.J. Prognostic and predictive biomarkers in breast cancer: Past, present and future. Semin. Cancer Biol. 2018, 52, 56–73. [Google Scholar] [CrossRef] [PubMed]
- Markandan, K.; Tiong, Y.W.; Sankaran, R.; Subramanian, S.; Markandan, U.D.; Chaudhary, V.; Numan, A.; Khalid, M.; Walvekar, R. Emergence of infectious diseases and role of advanced nanomaterials in point-of-care diagnostics: A review. Biotechnol. Genet. Eng. Rev. 2024, 40, 3438–3526. [Google Scholar] [CrossRef] [PubMed]
- Rosenheim, J.; Gupta, R.K.; Thakker, C.; Mann, T.; Bell, L.C.K.; Broderick, C.M.; Madon, K.; Papargyris, L.; Dayananda, P.; Kwok, A.J.; et al. SARS-CoV-2 human challenge reveals biomarkers that discriminate early and late phases of respiratory viral infections. Nat. Commun. 2024, 15, 10434. [Google Scholar] [CrossRef] [PubMed]
- Hong, D.; Kim, K.; Jo, E.-J.; Kim, M.-G. Electrochemiluminescence-Incorporated Lateral Flow Immunosensors Using Ru(bpy)32+-Labeled Gold Nanoparticles for the Full-Range Detection of Physiological C-Reactive Protein Levels. Anal. Chem. 2021, 93, 7925–7932. [Google Scholar] [CrossRef] [PubMed]
- Svegliati-Baroni, G.; Pierantonelli, I.; Torquato, P.; Marinelli, R.; Ferreri, C.; Chatgilialoglu, C.; Bartolini, D.; Galli, F. Lipidomic biomarkers and mechanisms of lipotoxicity in non-alcoholic fatty liver disease. Free Radic. Biol. Med. 2019, 144, 293–309. [Google Scholar] [CrossRef]
- Liu, M.; Wen, Y. Point-of-care testing for early-stage liver cancer diagnosis and personalized medicine: Biomarkers, current technologies and perspectives. Heliyon 2024, 10, e38444. [Google Scholar] [CrossRef]
- Stuart, T.; Satija, R. Integrative single-cell analysis. Nat. Rev. Genet. 2019, 20, 257–272. [Google Scholar] [CrossRef] [PubMed]
- Schmid, A.; Kortmann, H.; Dittrich, P.S.; Blank, L.M. Chemical and biological single cell analysis. Curr. Opin. Biotechnol. 2010, 21, 12–20. [Google Scholar] [CrossRef]
- Yin, H.; Marshall, D. Microfluidics for single cell analysis. Curr. Opin. Biotechnol. 2012, 23, 110–119. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Ma, L.; Wu, D.; Chen, G. Advances in bulk and single-cell multi-omics approaches for systems biology and precision medicine. Brief. Bioinform. 2021, 22, bbab024. [Google Scholar] [CrossRef] [PubMed]
- Heath, J.R.; Ribas, A.; Mischel, P.S. Single-cell analysis tools for drug discovery and development. Nat. Rev. Drug Discov. 2016, 15, 204–216. [Google Scholar] [CrossRef] [PubMed]
- Gu, X.; Wei, S.; Lv, X. Circulating tumor cells: From new biological insights to clinical practice. Signal Transduct. Target. Ther. 2024, 9, 226. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Shen, F.; Yang, X.; Han, T.; Long, Z.; Wen, J.; Huang, J.; Shen, J.; Guo, Q. Single-cell sequencing technology applied to epigenetics for the study of tumor heterogeneity. Clin. Epigenetics 2023, 15, 161. [Google Scholar] [CrossRef]
- Zhang, L.; Parvin, R.; Chen, M.; Hu, D.; Fan, Q.; Ye, F. High-throughput microfluidic droplets in biomolecular analytical system: A review. Biosens. Bioelectron. 2023, 228, 115213. [Google Scholar] [CrossRef] [PubMed]
- Salomon, R.; Kaczorowski, D.; Valdes-Mora, F.; Nordon, R.E.; Neild, A.; Farbehi, N.; Bartonicek, N.; Gallego-Ortega, D. Droplet-based single cell RNAseq tools: A practical guide. Lab Chip 2019, 19, 1706–1727. [Google Scholar] [CrossRef]
- Noé, A.; Cargill, T.N.; Nielsen, C.M.; Russell, A.J.C.; Barnes, E. The Application of Single-Cell RNA Sequencing in Vaccinology. J. Immunol. Res. 2020, 2020, 8624963. [Google Scholar] [CrossRef]
- Nath, A.; Bild, A.H. Leveraging Single-Cell Approaches in Cancer Precision Medicine. Trends Cancer 2021, 7, 359–372. [Google Scholar] [CrossRef]
- Chau, C.H.; Rixe, O.; McLeod, H.; Figg, W.D. Validation of Analytic Methods for Biomarkers Used in Drug Development. Clin. Cancer Res. 2008, 14, 5967–5976. [Google Scholar] [CrossRef] [PubMed]
- Kraus, V.B. Biomarkers as drug development tools: Discovery, validation, qualification and use. Nat. Rev. Rheumatol. 2018, 14, 354–362. [Google Scholar] [CrossRef] [PubMed]
- Nakayasu, E.S.; Gritsenko, M.; Piehowski, P.D.; Gao, Y.; Orton, D.J.; Schepmoes, A.A.; Fillmore, T.L.; Frohnert, B.I.; Rewers, M.; Krischer, J.P.; et al. Tutorial: Best practices and considerations for mass-spectrometry-based protein biomarker discovery and validation. Nat. Protoc. 2021, 16, 3737–3760. [Google Scholar] [CrossRef] [PubMed]
- Klyucherev, T.O.; Olszewski, P.; Shalimova, A.A.; Chubarev, V.N.; Tarasov, V.V.; Attwood, M.M.; Syvänen, S.; Schiöth, H.B. Advances in the development of new biomarkers for Alzheimer’s disease. Transl. Neurodegener. 2022, 11, 25. [Google Scholar] [CrossRef] [PubMed]
- Fernández-Metzler, C.; Ackermann, B.; Garofolo, F.; Arnold, M.E.; DeSilva, B.; Gu, H.; Laterza, O.; Mao, Y.; Rose, M.; Vazvaei-Smith, F.; et al. Biomarker Assay Validation by Mass Spectrometry. AAPS J. 2022, 24, 66. [Google Scholar] [CrossRef]
- Vo, D.-K.; Nguyen, T.-T.-L.; Maeng, H.-J. Effects of 1α,25-dihydroxyvitamin D3 on the pharmacokinetics and biodistribution of ergothioneine, an endogenous organic cation/carnitine transporter 1 substrate, in rats. J. Pharm. Investig. 2022, 52, 341–351. [Google Scholar] [CrossRef] [PubMed]
- Masucci, G.V.; Cesano, A.; Hawtin, R.; Janetzki, S.; Zhang, J.; Kirsch, I.; Dobbin, K.K.; Alvarez, J.; Robbins, P.B.; Selvan, S.R.; et al. Validation of biomarkers to predict response to immunotherapy in cancer: Volume I—Pre-analytical and analytical validation. J. ImmunoTherapy Cancer 2016, 4, 76. [Google Scholar] [CrossRef] [PubMed]
- Maïno, N.; Hauling, T.; Cappi, G.; Madaboosi, N.; Dupouy, D.G.; Nilsson, M. A microfluidic platform towards automated multiplexed in situ sequencing. Sci. Rep. 2019, 9, 3542. [Google Scholar] [CrossRef] [PubMed]
- Godfrey, A.; Vandendriessche, B.; Bakker, J.P.; Fitzer-Attas, C.; Gujar, N.; Hobbs, M.; Liu, Q.; Northcott, C.A.; Parks, V.; Wood, W.A.; et al. Fit-for-Purpose Biometric Monitoring Technologies: Leveraging the Laboratory Biomarker Experience. Clin. Transl. Sci. 2021, 14, 62–74. [Google Scholar] [CrossRef] [PubMed]
- Perlis, R.H. Translating biomarkers to clinical practice. Mol. Psychiatry 2011, 16, 1076–1087. [Google Scholar] [CrossRef] [PubMed]
- Chakraborty, S. Democratizing nucleic acid-based molecular diagnostic tests for infectious diseases at resource-limited settings–From point of care to extreme point of care. Sens. Diagn. 2024, 3, 536–561. [Google Scholar] [CrossRef]
- Kulasinghe, A.; Wu, H.; Punyadeera, C.; Warkiani, M.E. The Use of Microfluidic Technology for Cancer Applications and Liquid Biopsy. Micromachines 2018, 9, 397. [Google Scholar] [CrossRef] [PubMed]
- Mohammadniaei, M.; Nguyen, H.V.; Tieu, M.V.; Lee, M.-H. 2D Materials in Development of Electrochemical Point-of-Care Cancer Screening Devices. Micromachines 2019, 10, 662. [Google Scholar] [CrossRef] [PubMed]
- Strom, C.M.; Rivera, S.; Elzinga, C.; Angeloni, T.; Rosenthal, S.H.; Goos-Root, D.; Siaw, M.; Platt, J.; Braastadt, C.; Cheng, L. Development and validation of a next-generation sequencing assay for BRCA1 and BRCA2 variants for the clinical laboratory. PLoS ONE 2015, 10, e0136419. [Google Scholar] [CrossRef] [PubMed]
- Greathouse, K.L.; White, J.R.; Vargas, A.J.; Bliskovsky, V.V.; Beck, J.A.; von Muhlinen, N.; Polley, E.C.; Bowman, E.D.; Khan, M.A.; Robles, A.I.; et al. Interaction between the microbiome and TP53 in human lung cancer. Genome Biol. 2018, 19, 123. [Google Scholar] [CrossRef]
- Brychta, N.; Krahn, T.; von Ahsen, O. Detection of KRAS Mutations in Circulating Tumor DNA by Digital PCR in Early Stages of Pancreatic Cancer. Clin. Chem. 2016, 62, 1482–1491. [Google Scholar] [CrossRef] [PubMed]
- Najjar, D.; Rainbow, J.; Sharma Timilsina, S.; Jolly, P.; de Puig, H.; Yafia, M.; Durr, N.; Sallum, H.; Alter, G.; Li, J.Z.; et al. A lab-on-a-chip for the concurrent electrochemical detection of SARS-CoV-2 RNA and anti-SARS-CoV-2 antibodies in saliva and plasma. Nat. Biomed. Eng. 2022, 6, 968–978. [Google Scholar] [CrossRef] [PubMed]
- Nouri, R.; Jiang, Y.; Politza, A.J.; Liu, T.; Greene, W.H.; Zhu, Y.; Nunez, J.J.; Lian, X.; Guan, W. STAMP-Based Digital CRISPR-Cas13a for Amplification-Free Quantification of HIV-1 Plasma Viral Loads. ACS Nano 2023, 17, 10701–10712. [Google Scholar] [CrossRef] [PubMed]
- Fang, X.; Zheng, Y.; Duan, Y.; Liu, Y.; Zhong, W. Recent Advances in Design of Fluorescence-Based Assays for High-Throughput Screening. Anal. Chem. 2019, 91, 482–504. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Zhao, J.; Wu, X.; Zhu, C.; Liu, Y.; Wang, A.; Deng, G.; Zhu, L. A rapid microfluidic platform with real-time fluorescence detection system for molecular diagnosis. Biotechnol. Biotechnol. Equip. 2019, 33, 223–230. [Google Scholar] [CrossRef]
- Wang, R.C.; Wang, Z. Precision Medicine: Disease Subtyping and Tailored Treatment. Cancers 2023, 15, 3837. [Google Scholar] [CrossRef] [PubMed]
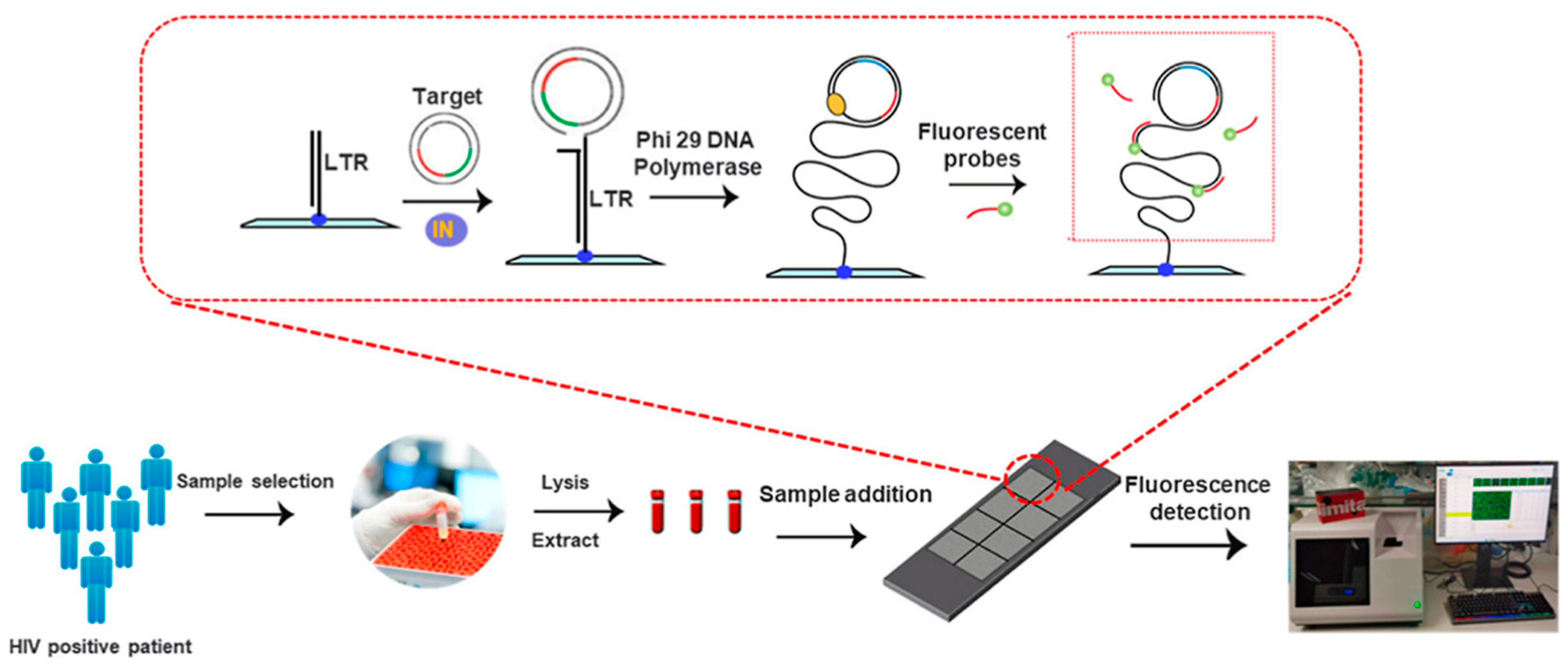
- Chen, F.; Wang, J.; Ma, J.; Song, L.; Yan, H.; Wang, F.; Yang, Z.; Li, F. Novel DNA Biosensing Platform for Detecting HIV Integrase for Highly Sensitive and Quantitative HIV Detection, Diagnosis, and Therapeutic Monitoring. ACS Omega 2024, 9, 25042–25053. [Google Scholar] [CrossRef] [PubMed]
- Sefrioui, D.; Sarafan-Vasseur, N.; Beaussire, L.; Baretti, M.; Gangloff, A.; Blanchard, F.; Clatot, F.; Sabourin, J.-C.; Sesboüé, R.; Frebourg, T.; et al. Clinical value of chip-based digital-PCR platform for the detection of circulating DNA in metastatic colorectal cancer. Dig. Liver Dis. 2015, 47, 884–890. [Google Scholar] [CrossRef]
- Lee, J.-m.; Han, J.J.; Altwerger, G.; Kohn, E.C. Proteomics and biomarkers in clinical trials for drug development. J. Proteom. 2011, 74, 2632–2641. [Google Scholar] [CrossRef] [PubMed]
- Hopkins, A.L.; Groom, C.R. The druggable genome. Nat. Rev. Drug Discov. 2002, 1, 727–730. [Google Scholar] [CrossRef]
- Das, S.; Dey, M.K.; Devireddy, R.; Gartia, M.R. Biomarkers in Cancer Detection, Diagnosis, and Prognosis. Sensors 2024, 24, 37. [Google Scholar] [CrossRef]
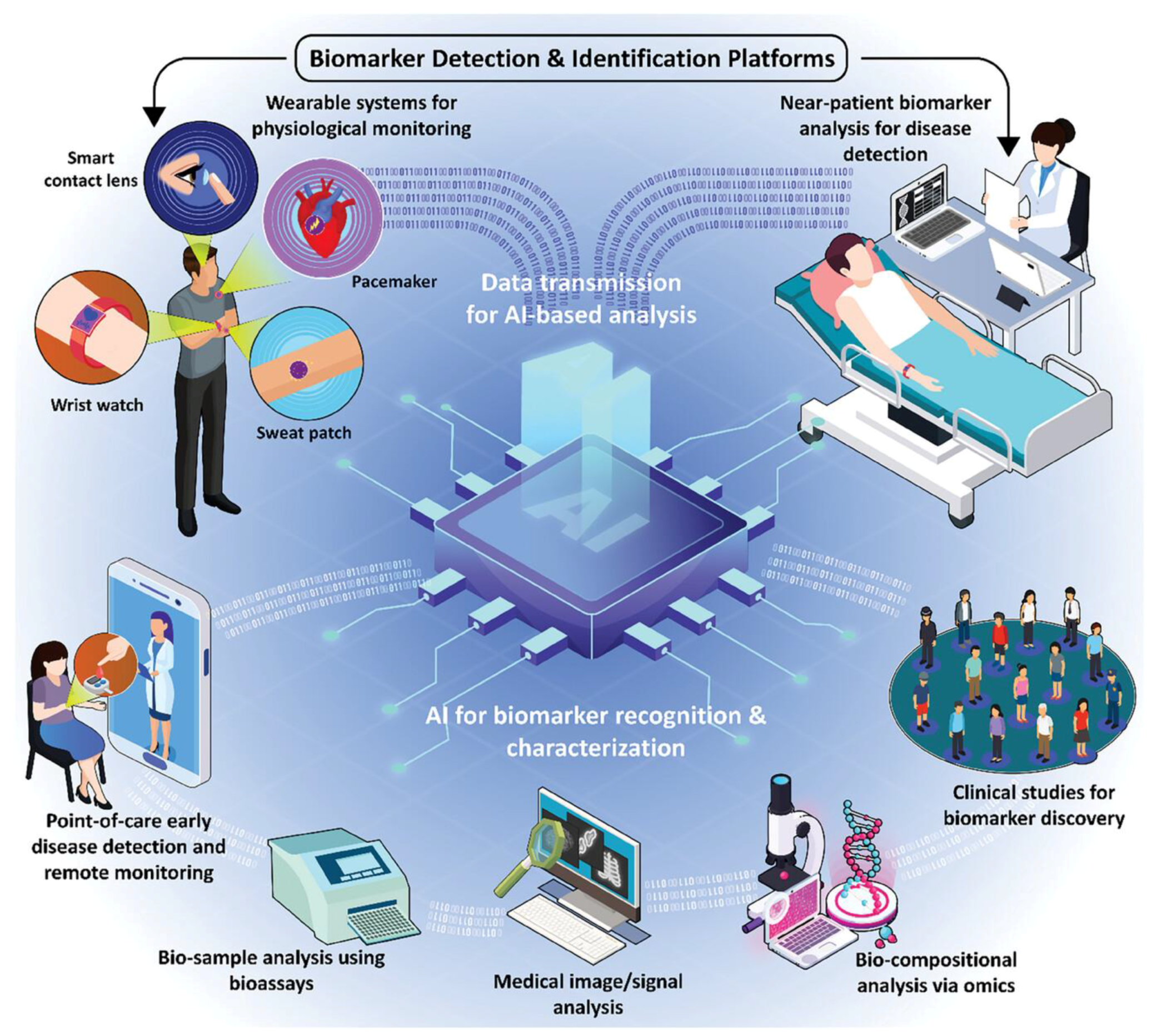
- Haghayegh, F.; Norouziazad, A.; Haghani, E.; Feygin, A.A.; Rahimi, R.H.; Ghavamabadi, H.A.; Sadighbayan, D.; Madhoun, F.; Papagelis, M.; Felfeli, T.; et al. Revolutionary Point-of-Care Wearable Diagnostics for Early Disease Detection and Biomarker Discovery through Intelligent Technologies. Adv. Sci. 2024, 11, 2400595. [Google Scholar] [CrossRef] [PubMed]
- Levantini, E.; Maroni, G.; Del Re, M.; Tenen, D.G. EGFR signaling pathway as therapeutic target in human cancers. Semin. Cancer Biol. 2022, 85, 253–275. [Google Scholar] [CrossRef] [PubMed]
- Rascio, F.; Spadaccino, F.; Rocchetti, M.T.; Castellano, G.; Stallone, G.; Netti, G.S.; Ranieri, E. The Pathogenic Role of PI3K/AKT Pathway in Cancer Onset and Drug Resistance: An Updated Review. Cancers 2021, 13, 3949. [Google Scholar] [CrossRef]
- Li, M.; Chi, X.; Wang, Y.; Setrerrahmane, S.; Xie, W.; Xu, H. Trends in insulin resistance: Insights into mechanisms and therapeutic strategy. Signal Transduct. Target. Ther. 2022, 7, 216. [Google Scholar] [CrossRef]
- Wang, D.-R.; Wu, X.-L.; Sun, Y.-L. Therapeutic targets and biomarkers of tumor immunotherapy: Response versus non-response. Signal Transduct. Target. Ther. 2022, 7, 331. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, Y.; Kawazoe, A.; Lordick, F.; Janjigian, Y.Y.; Shitara, K. Biomarker-targeted therapies for advanced-stage gastric and gastro-oesophageal junction cancers: An emerging paradigm. Nat. Rev. Clin. Oncol. 2021, 18, 473–487. [Google Scholar] [CrossRef]
- Monette, A.; Aguilar-Mahecha, A.; Altinmakas, E.; Angelos, M.G.; Assad, N.; Batist, G.; Bommareddy, P.K.; Bonilla, D.L.; Borchers, C.H.; Church, S.E. The Society for Immunotherapy of Cancer Perspective on Tissue-Based Technologies for Immuno-Oncology Biomarker Discovery and Application. Clin. Cancer Res. 2024, 31, OF1–OF18. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Collet, L.; Rediti, M.; Debien, V.; De Caluwé, A.; Venet, D.; Romano, E.; Rothé, F.; Sotiriou, C.; Buisseret, L. Predictive Biomarkers for Response to Immunotherapy in Triple Negative Breast Cancer: Promises and Challenges. J. Clin. Med. 2023, 12, 953. [Google Scholar] [CrossRef] [PubMed]
- Holmes, M.V.; Richardson, T.G.; Ference, B.A.; Davies, N.M.; Davey Smith, G. Integrating genomics with biomarkers and therapeutic targets to invigorate cardiovascular drug development. Nat. Rev. Cardiol. 2021, 18, 435–453. [Google Scholar] [CrossRef] [PubMed]
- Qiu, S.; Cai, Y.; Yao, H.; Lin, C.; Xie, Y.; Tang, S.; Zhang, A. Small molecule metabolites: Discovery of biomarkers and therapeutic targets. Signal Transduct. Target. Ther. 2023, 8, 132. [Google Scholar] [CrossRef] [PubMed]
- Emmerich, C.H.; Gamboa, L.M.; Hofmann, M.C.J.; Bonin-Andresen, M.; Arbach, O.; Schendel, P.; Gerlach, B.; Hempel, K.; Bespalov, A.; Dirnagl, U.; et al. Improving target assessment in biomedical research: The GOT-IT recommendations. Nat. Rev. Drug Discov. 2021, 20, 64–81. [Google Scholar] [CrossRef] [PubMed]
- Middleton, G.; Robbins, H.; Andre, F.; Swanton, C. A state-of-the-art review of stratified medicine in cancer: Towards a future precision medicine strategy in cancer. Ann. Oncol. 2022, 33, 143–157. [Google Scholar] [CrossRef] [PubMed]
- Lawler, M.; Keeling, P.; Kholmanskikh, O.; Minnaard, W.; Moehlig-Zuttermeister, H.; Normanno, N.; Philip, R.; Popp, C.; Salgado, R.; Santiago-Walker, A.E.; et al. Empowering effective biomarker-driven precision oncology: A call to action. Eur. J. Cancer 2024, 209, 114225. [Google Scholar] [CrossRef]
- Zhou, Z.; Lin, T.; Chen, S.; Zhang, G.; Xu, Y.; Zou, H.; Zhou, A.; Zhang, Y.; Weng, S.; Han, X.; et al. Omics-based molecular classifications empowering in precision oncology. Cell. Oncol. 2024, 47, 759–777. [Google Scholar] [CrossRef] [PubMed]
- Swain, S.M.; Shastry, M.; Hamilton, E. Targeting HER2-positive breast cancer: Advances and future directions. Nat. Rev. Drug Discov. 2023, 22, 101–126. [Google Scholar] [CrossRef] [PubMed]
- Aldea, M.; Andre, F.; Marabelle, A.; Dogan, S.; Barlesi, F.; Soria, J.-C. Overcoming resistance to tumor-targeted and immune-targeted therapies. Cancer Discov. 2021, 11, 874–899. [Google Scholar] [CrossRef] [PubMed]
- Williams, D.M.; Jones, H.; Stephens, J.W. Personalized Type 2 Diabetes Management: An Update on Recent Advances and Recommendations. Diabetes Metab. Syndr. Obes. 2022, 15, 281–295. [Google Scholar] [CrossRef]
- Ponce, D.M.; Alousi, A.M.; Nakamura, R.; Slingerland, J.; Calafiore, M.; Sandhu, K.S.; Barker, J.N.; Devlin, S.; Shia, J.; Giralt, S.; et al. A phase 2 study of interleukin-22 and systemic corticosteroids as initial treatment for acute GVHD of the lower GI tract. Blood 2023, 141, 1389–1401. [Google Scholar] [CrossRef]
- Zanella, E.R.; Grassi, E.; Trusolino, L. Towards precision oncology with patient-derived xenografts. Nat. Rev. Clin. Oncol. 2022, 19, 719–732. [Google Scholar] [CrossRef] [PubMed]
- Di Nicolantonio, F.; Vitiello, P.P.; Marsoni, S.; Siena, S.; Tabernero, J.; Trusolino, L.; Bernards, R.; Bardelli, A. Precision oncology in metastatic colorectal cancer—From biology to medicine. Nat. Rev. Clin. Oncol. 2021, 18, 506–525. [Google Scholar] [CrossRef] [PubMed]
- Muthamilselvan, S.; Ramasami Sundhar Baabu, P.; Palaniappan, A. Microfluidics for Profiling miRNA Biomarker Panels in AI-Assisted Cancer Diagnosis and Prognosis. Technol. Cancer Res. Treat. 2023, 22, 15330338231185284. [Google Scholar] [CrossRef]
- Al-Thani, A.N.; Jan, A.G.; Abbas, M.; Geetha, M.; Sadasivuni, K.K. Nanoparticles in cancer theragnostic and drug delivery: A comprehensive review. Life Sci. 2024, 352, 122899. [Google Scholar] [CrossRef]
- Normanno, N.; Apostolidis, K.; de Lorenzo, F.; Beer, P.A.; Henderson, R.; Sullivan, R.; Biankin, A.V.; Horgan, D.; Lawler, M. Cancer Biomarkers in the era of precision oncology: Addressing the needs of patients and health systems. Semin. Cancer Biol. 2022, 84, 293–301. [Google Scholar] [CrossRef] [PubMed]
- Sanz-Garcia, E.; Zhao, E.; Bratman, S.V.; Siu, L.L. Monitoring and adapting cancer treatment using circulating tumor DNA kinetics: Current research, opportunities, and challenges. Sci. Adv. 2022, 8, eabi8618. [Google Scholar] [CrossRef] [PubMed]
- Pascual, J.; Attard, G.; Bidard, F.C.; Curigliano, G.; De Mattos-Arruda, L.; Diehn, M.; Italiano, A.; Lindberg, J.; Merker, J.D.; Montagut, C.; et al. ESMO recommendations on the use of circulating tumour DNA assays for patients with cancer: A report from the ESMO Precision Medicine Working Group. Ann. Oncol. 2022, 33, 750–768. [Google Scholar] [CrossRef]
- Parkins Michael, D.; Lee Bonita, E.; Acosta, N.; Bautista, M.; Hubert Casey, R.J.; Hrudey Steve, E.; Frankowski, K.; Pang, X.-L. Wastewater-based surveillance as a tool for public health action: SARS-CoV-2 and beyond. Clin. Microbiol. Rev. 2023, 37, e00103-00122. [Google Scholar] [CrossRef]
- Wang, S.; Li, H.; Kou, Z.; Ren, F.; Jin, Y.; Yang, L.; Dong, X.; Yang, M.; Zhao, J.; Liu, H.; et al. Highly sensitive and specific detection of hepatitis B virus DNA and drug resistance mutations utilizing the PCR-based CRISPR-Cas13a system. Clin. Microbiol. Infect. 2021, 27, 443–450. [Google Scholar] [CrossRef]
- Aulin, L.B.S.; de Lange, D.W.; Saleh, M.A.A.; van der Graaf, P.H.; Völler, S.; van Hasselt, J.G.C. Biomarker-Guided Individualization of Antibiotic Therapy. Clin. Pharmacol. Ther. 2021, 110, 346–360. [Google Scholar] [CrossRef]
- Vo, D.-K.; Nguyen, T.-T.-L.; Maeng, H.-J. Impact of 1α,25-dihydroxyvitamin D3 on biodistribution and pharmacokinetics of L-carnitine and creatinine, organic cation/carnitine transporter 2 and organic cation transporter 2 biomarkers. J. Pharm. Investig. 2024, 54, 389–402. [Google Scholar] [CrossRef]
- Xiao, H.; Wang, X.; Li, S.; Liu, Y.; Cui, Y.; Deng, X. Advances in Biomarkers for Detecting Early Cancer Treatment-Related Cardiac Dysfunction. Front. Cardiovasc. Med. 2021, 8, 753313. [Google Scholar] [CrossRef] [PubMed]
- Sharma, R.; Singh, D.; Gaur, P.; Joshi, D. Intelligent automated drug administration and therapy: Future of healthcare. Drug Deliv. Transl. Res. 2021, 11, 1878–1902. [Google Scholar] [CrossRef]
- Valla, V.; Alzabin, S.; Koukoura, A.; Lewis, A.; Nielsen, A.A.; Vassiliadis, E. Companion Diagnostics: State of the Art and New Regulations. Biomark. Insights 2021, 16, 11772719211047763. [Google Scholar] [CrossRef] [PubMed]
- Orellana García, L.P.; Ehmann, F.; Hines, P.A.; Ritzhaupt, A.; Brand, A. Biomarker and Companion Diagnostics—A Review of Medicinal Products Approved by the European Medicines Agency. Front. Med. 2021, 8, 753187. [Google Scholar] [CrossRef] [PubMed]
- Bakker, E.; Starokozhko, V.; Kraaijvanger, J.W.M.; Heerspink, H.J.L.; Mol, P.G.M. Precision medicine in regulatory decision making: Biomarkers used for patient selection in European Public Assessment Reports from 2018 to 2020. Clin. Transl. Sci. 2023, 16, 2394–2412. [Google Scholar] [CrossRef]
- Arafah, A.; Khatoon, S.; Rasool, I.; Khan, A.; Rather, M.A.; Abujabal, K.A.; Faqih, Y.A.; Rashid, H.; Rashid, S.M.; Bilal Ahmad, S.; et al. The Future of Precision Medicine in the Cure of Alzheimer’s Disease. Biomedicines 2023, 11, 335. [Google Scholar] [CrossRef]
- Pal, M.; Muinao, T.; Boruah, H.P.D.; Mahindroo, N. Current advances in prognostic and diagnostic biomarkers for solid cancers: Detection techniques and future challenges. Biomed. Pharmacother. 2022, 146, 112488. [Google Scholar] [CrossRef] [PubMed]
- Marques, L.; Costa, B.; Pereira, M.; Silva, A.; Santos, J.; Saldanha, L.; Silva, I.; Magalhães, P.; Schmidt, S.; Vale, N. Advancing Precision Medicine: A Review of Innovative In Silico Approaches for Drug Development, Clinical Pharmacology and Personalized Healthcare. Pharmaceutics 2024, 16, 332. [Google Scholar] [CrossRef] [PubMed]
- Bartel, P.; Granger, J. Introduction to Companion Diagnostics for Gene Therapy. In Drug Development for Gene Therapy; Wiley: Hoboken, NJ, USA, 2024; pp. 383–392. [Google Scholar]
- Acharya, D.; Mukhopadhyay, A. A comprehensive review of machine learning techniques for multi-omics data integration: Challenges and applications in precision oncology. Brief. Funct. Genom. 2024, 23, 549–560. [Google Scholar] [CrossRef]
- Tolani, P.; Gupta, S.; Yadav, K.; Aggarwal, S.; Yadav, A.K. Chapter Four–Big data, integrative omics and network biology. In Advances in Protein Chemistry and Structural Biology; Donev, R., Karabencheva-Christova, T., Eds.; Academic Press: Cambridge, MA, USA, 2021; Volume 127, pp. 127–160. [Google Scholar]
- Caudai, C.; Galizia, A.; Geraci, F.; Le Pera, L.; Morea, V.; Salerno, E.; Via, A.; Colombo, T. AI applications in functional genomics. Comput. Struct. Biotechnol. J. 2021, 19, 5762–5790. [Google Scholar] [CrossRef] [PubMed]
- Bahl, A.; Ibrahim, C.; Plate, K.; Haase, A.; Dengjel, J.; Nymark, P.; Dumit, V.I. PROTEOMAS: A workflow enabling harmonized proteomic meta-analysis and proteomic signature mapping. J. Cheminformatics 2023, 15, 34. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.; Zhang, R.; Zhou, A.; Lv, J.; Chen, S.; Zou, H.; Zhang, G.; Lin, T.; Wang, Z.; Zhang, Y.; et al. Proteomics appending a complementary dimension to precision oncotherapy. Comput. Struct. Biotechnol. J. 2024, 23, 1725–1739. [Google Scholar] [CrossRef] [PubMed]
- Russell, A.M.; Pack, A.P.; Bailey, S.C.; Weldon, C.B.; Dreyer, M.S.; Kircher, S.M.; Wolf, M.S. A local perspective on internal, external, and reflexive biomarker testing processes for lung cancer in an academic medical center. Cancer 2024, 130, 2085–2090. [Google Scholar] [CrossRef] [PubMed]
- Kaur, P.; Singh, A.; Chana, I. Computational Techniques and Tools for Omics Data Analysis: State-of-the-Art, Challenges, and Future Directions. Arch. Comput. Methods Eng. 2021, 28, 4595–4631. [Google Scholar] [CrossRef]
- Kim, S.; Kang, S.; Choe, J.; Moon, C.; Choi, H.; Kim, J.-Y.; Choi, J.-W. A Microfluidic System for Investigating Anticipatory Medication Effects on Dopamine Homeostasis in Dopaminergic Cells. Anal. Chem. 2023, 95, 3153–3159. [Google Scholar] [CrossRef] [PubMed]
- Batis, N.; Brooks, J.M.; Payne, K.; Sharma, N.; Nankivell, P.; Mehanna, H. Lack of predictive tools for conventional and targeted cancer therapy: Barriers to biomarker development and clinical translation. Adv. Drug Deliv. Rev. 2021, 176, 113854. [Google Scholar] [CrossRef]
- Moqri, M.; Herzog, C.; Poganik, J.R.; Ying, K.; Justice, J.N.; Belsky, D.W.; Higgins-Chen, A.T.; Chen, B.H.; Cohen, A.A.; Fuellen, G.; et al. Validation of biomarkers of aging. Nat. Med. 2024, 30, 360–372. [Google Scholar] [CrossRef] [PubMed]
- Guo, L.; Kong, D.; Liu, J.; Zhan, L.; Luo, L.; Zheng, W.; Zheng, Q.; Chen, C.; Sun, S. Breast cancer heterogeneity and its implication in personalized precision therapy. Exp. Hematol. Oncol. 2023, 12, 3. [Google Scholar] [CrossRef] [PubMed]
- Lenz, G.; Onzi, G.R.; Lenz, L.S.; Buss, J.H.; dos Santos, J.A.; Begnini, K.R. The Origins of Phenotypic Heterogeneity in Cancer. Cancer Res. 2022, 82, 3–11. [Google Scholar] [CrossRef] [PubMed]
- Mani, D.R.; Krug, K.; Zhang, B.; Satpathy, S.; Clauser, K.R.; Ding, L.; Ellis, M.; Gillette, M.A.; Carr, S.A. Cancer proteogenomics: Current impact and future prospects. Nat. Rev. Cancer 2022, 22, 298–313. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, S.; Rajendran, R.L.; Mahajan, A.A.; Chowdhury, A.; Bera, A.; Guha, S.; Chakraborty, K.; Chowdhury, R.; Paul, A.; Jha, S.; et al. Harnessing exosomes as cancer biomarkers in clinical oncology. Cancer Cell Int. 2024, 24, 278. [Google Scholar] [CrossRef] [PubMed]
- del Campo, M.; Zetterberg, H.; Gandy, S.; Onyike, C.U.; Oliveira, F.; Udeh-Momoh, C.; Lleó, A.; Teunissen, C.E.; Pijnenburg, Y. New developments of biofluid-based biomarkers for routine diagnosis and disease trajectories in frontotemporal dementia. Alzheimer’s Dement. 2022, 18, 2292–2307. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, H.T.; Peirsman, A.; Tirpakova, Z.; Mandal, K.; Vanlauwe, F.; Maity, S.; Kawakita, S.; Khorsandi, D.; Herculano, R.; Umemura, C.; et al. Engineered Vasculature for Cancer Research and Regenerative Medicine. Micromachines 2023, 14, 978. [Google Scholar] [CrossRef] [PubMed]
- Füzéry, A.K.; Levin, J.; Chan, M.M.; Chan, D.W. Translation of proteomic biomarkers into FDA approved cancer diagnostics: Issues and challenges. Clin. Proteom. 2013, 10, 13. [Google Scholar] [CrossRef] [PubMed]
- Kang, S.L.; Woo, J.H.; Kim, N.H.; Kwon, J.Y.; Kim, S.M. Necessity of strengthening the current clinical regulatory for companion diagnostics: An institutional comparison of the FDA, EMA, and MFDS. Mol. Ther. Methods Clin. Dev. 2023, 30, 447–458. [Google Scholar] [CrossRef] [PubMed]
- Bakker, E.; Hendrikse, N.M.; Ehmann, F.; van der Meer, D.S.; Llinares Garcia, J.; Vetter, T.; Starokozhko, V.; Mol, P.G.M. Biomarker Qualification at the European Medicines Agency: A Review of Biomarker Qualification Procedures From 2008 to 2020. Clin. Pharmacol. Ther. 2022, 112, 69–80. [Google Scholar] [CrossRef] [PubMed]
- Zwanenburg, A.; Vallières, M.; Abdalah, M.A.; Aerts, H.J.W.L.; Andrearczyk, V.; Apte, A.; Ashrafinia, S.; Bakas, S.; Beukinga, R.J.; Boellaard, R. The image biomarker standardization initiative: Standardized quantitative radiomics for high-throughput image-based phenotyping. Radiology 2020, 295, 328–338. [Google Scholar] [CrossRef] [PubMed]
- Gromova, M.; Vaggelas, A.; Dallmann, G.; Seimetz, D. Biomarkers: Opportunities and challenges for drug development in the current regulatory landscape. Biomark. Insights 2020, 15, 1177271920974652. [Google Scholar] [CrossRef]
- Kingsmore, S.F. Multiplexed protein measurement: Technologies and applications of protein and antibody arrays. Nat. Rev. Drug Discov. 2006, 5, 310–321. [Google Scholar] [CrossRef] [PubMed]
- Hays, A.; Wissel, M.; Colletti, K.; Soon, R.; Azadeh, M.; Smith, J.; Doddareddy, R.; Chalfant, M.; Adamowicz, W.; Ramaswamy, S.S.; et al. Recommendations for Method Development and Validation of qPCR and dPCR Assays in Support of Cell and Gene Therapy Drug Development. AAPS J. 2024, 26, 24. [Google Scholar] [CrossRef] [PubMed]
- Tenchov, R.; Sapra, A.K.; Sasso, J.; Ralhan, K.; Tummala, A.; Azoulay, N.; Zhou, Q.A. Biomarkers for Early Cancer Detection: A Landscape View of Recent Advancements, Spotlighting Pancreatic and Liver Cancers. ACS Pharmacol. Transl. Sci. 2024, 7, 586–613. [Google Scholar] [CrossRef] [PubMed]
- Agarwal, A.; Ressler, D.; Snyder, G. The current and future state of companion diagnostics. Pharmacogenomics Pers. Med. 2015, 8, 99–110. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.Y.H.; Karlawish, J.; Berkman, B.E. Ethics of genetic and biomarker test disclosures in neurodegenerative disease prevention trials. Neurology 2015, 84, 1488–1494. [Google Scholar] [CrossRef] [PubMed]
- Erdmann, A.; Rehmann-Sutter, C.; Bozzaro, C. Patients’ and professionals’ views related to ethical issues in precision medicine: A mixed research synthesis. BMC Med. Ethics 2021, 22, 116. [Google Scholar] [CrossRef]
- Singhal, P.; Tan, A.L.M.; Drivas, T.G.; Johnson, K.B.; Ritchie, M.D.; Beaulieu-Jones, B.K. Opportunities and challenges for biomarker discovery using electronic health record data. Trends Mol. Med. 2023, 29, 765–776. [Google Scholar] [CrossRef] [PubMed]
- Avilés-Santa, M.L.; Heintzman, J.; Lindberg, N.M.; Guerrero-Preston, R.; Ramos, K.; Abraído-Lanza, A.L.; Bull, J.; Falcón, A.; McBurnie, M.A.; Moy, E.; et al. Personalized medicine and Hispanic health: Improving health outcomes and reducing health disparities—A National Heart, Lung, and Blood Institute workshop report. BMC Proc. 2017, 11, 11. [Google Scholar] [CrossRef]
- Jurjako, M.; Malatesti, L.; Brazil, I.A. Some Ethical Considerations About the Use of Biomarkers for the Classification of Adult Antisocial Individuals. Int. J. Forensic Ment. Health 2019, 18, 228–242. [Google Scholar] [CrossRef]
- Walsh, C.G.; Chaudhry, B.; Dua, P.; Goodman, K.W.; Kaplan, B.; Kavuluru, R.; Solomonides, A.; Subbian, V. Stigma, biomarkers, and algorithmic bias: Recommendations for precision behavioral health with artificial intelligence. JAMIA Open 2020, 3, 9–15. [Google Scholar] [CrossRef] [PubMed]
- Brannan, C.; Foulkes, A.L.; Lázaro-Muñoz, G. Preventing discrimination based on psychiatric risk biomarkers. Am. J. Med. Genet. Part B Neuropsychiatr. Genet. 2019, 180, 159–171. [Google Scholar] [CrossRef] [PubMed]
- Khera, R.; Oikonomou, E.K.; Nadkarni, G.N.; Morley, J.R.; Wiens, J.; Butte, A.J.; Topol, E.J. Transforming cardiovascular care with artificial intelligence: From discovery to practice: JACC state-of-the-art review. J. Am. Coll. Cardiol. 2024, 84, 97–114. [Google Scholar] [CrossRef]
- Wang, B.; Li, Y.; Zhou, M.; Han, Y.; Zhang, M.; Gao, Z.; Liu, Z.; Chen, P.; Du, W.; Zhang, X.; et al. Smartphone-based platforms implementing microfluidic detection with image-based artificial intelligence. Nat. Commun. 2023, 14, 1341. [Google Scholar] [CrossRef] [PubMed]
- Gupta, R.; Srivastava, D.; Sahu, M.; Tiwari, S.; Ambasta, R.K.; Kumar, P. Artificial intelligence to deep learning: Machine intelligence approach for drug discovery. Mol. Divers. 2021, 25, 1315–1360. [Google Scholar] [CrossRef] [PubMed]
- Shapiro, M.R.; Tallon, E.M.; Brown, M.E.; Posgai, A.L.; Clements, M.A.; Brusko, T.M. Leveraging artificial intelligence and machine learning to accelerate discovery of disease-modifying therapies in type 1 diabetes. Diabetologia 2024. [Google Scholar] [CrossRef] [PubMed]
- Doherty, T.; Yao, Z.; Khleifat, A.A.l.; Tantiangco, H.; Tamburin, S.; Albertyn, C.; Thakur, L.; Llewellyn, D.J.; Oxtoby, N.P.; Lourida, I.; et al. Artificial intelligence for dementia drug discovery and trials optimization. Alzheimer’s Dement. 2023, 19, 5922–5933. [Google Scholar] [CrossRef] [PubMed]
- Prelaj, A.; Miskovic, V.; Zanitti, M.; Trovo, F.; Genova, C.; Viscardi, G.; Rebuzzi, S.E.; Mazzeo, L.; Provenzano, L.; Kosta, S.; et al. Artificial intelligence for predictive biomarker discovery in immuno-oncology: A systematic review. Ann. Oncol. 2024, 35, 29–65. [Google Scholar] [CrossRef]
- Joshi, R.C.; Srivastava, P.; Mishra, R.; Burget, R.; Dutta, M.K. Biomarker profiling and integrating heterogeneous models for enhanced multi-grade breast cancer prognostication. Comput. Methods Programs Biomed. 2024, 255, 108349. [Google Scholar] [CrossRef]
- Sahiner, B.; Pezeshk, A.; Hadjiiski, L.M.; Wang, X.; Drukker, K.; Cha, K.H.; Summers, R.M.; Giger, M.L. Deep learning in medical imaging and radiation therapy. Med. Phys. 2019, 46, e1–e36. [Google Scholar] [CrossRef] [PubMed]
- Odenkirk, M.T.; Reif, D.M.; Baker, E.S. Multiomic Big Data Analysis Challenges: Increasing Confidence in the Interpretation of Artificial Intelligence Assessments. Anal. Chem. 2021, 93, 7763–7773. [Google Scholar] [CrossRef]
- Chakraborty, S.; Sharma, G.; Karmakar, S.; Banerjee, S. Multi-OMICS approaches in cancer biology: New era in cancer therapy. Biochim. Biophys. Acta (BBA) Mol. Basis Dis. 2024, 1870, 167120. [Google Scholar] [CrossRef]
- Barrat, F.J.; Crow, M.K.; Ivashkiv, L.B. Interferon target-gene expression and epigenomic signatures in health and disease. Nat. Immunol. 2019, 20, 1574–1583. [Google Scholar] [CrossRef] [PubMed]
- Aebersold, R.; Mann, M. Mass-spectrometric exploration of proteome structure and function. Nature 2016, 537, 347–355. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.; Xu, X.; Wang, W.; Tian, T.; Zhu, Z.; Yang, C. Single cell transcriptomics: Moving towards multi-omics. Analyst 2019, 144, 3172–3189. [Google Scholar] [CrossRef] [PubMed]
- Bargahi, N.; Ghasemali, S.; Jahandar-Lashaki, S.; Nazari, A. Recent advances for cancer detection and treatment by microfluidic technology, review and update. Biol. Proced. Online 2022, 24, 5. [Google Scholar] [CrossRef] [PubMed]
- Sanchez-Freire, V.; Ebert, A.D.; Kalisky, T.; Quake, S.R.; Wu, J.C. Microfluidic single-cell real-time PCR for comparative analysis of gene expression patterns. Nat. Protoc. 2012, 7, 829–838. [Google Scholar] [CrossRef]
- Labib, M.; Kelley, S.O. Single-cell analysis targeting the proteome. Nat. Rev. Chem. 2020, 4, 143–158. [Google Scholar] [CrossRef] [PubMed]
- Bennett, H.M.; Stephenson, W.; Rose, C.M.; Darmanis, S. Single-cell proteomics enabled by next-generation sequencing or mass spectrometry. Nat. Methods 2023, 20, 363–374. [Google Scholar] [CrossRef] [PubMed]
- Özyurt, C.; Uludağ, İ.; İnce, B.; Sezgintürk, M.K. Lab-on-a-chip systems for cancer biomarker diagnosis. J. Pharm. Biomed. Anal. 2023, 226, 115266. [Google Scholar] [CrossRef]
- Chan, H.N.; Tan, M.J.A.; Wu, H. Point-of-care testing: Applications of 3D printing. Lab Chip 2017, 17, 2713–2739. [Google Scholar] [CrossRef]
- Prabhakar, P.; Sen, R.K.; Dwivedi, N.; Khan, R.; Solanki, P.R.; Srivastava, A.K.; Dhand, C. 3D-Printed Microfluidics and Potential Biomedical Applications. Front. Nanotechnol. 2021, 3, 609355. [Google Scholar] [CrossRef]
- Mao, M.; He, J.; Li, X.; Zhang, B.; Lei, Q.; Liu, Y.; Li, D. The Emerging Frontiers and Applications of High-Resolution 3D Printing. Micromachines 2017, 8, 113. [Google Scholar] [CrossRef]
- Chavez-Pineda, O.G.; Rodriguez-Moncayo, R.; Cedillo-Alcantar, D.F.; Guevara-Pantoja, P.E.; Amador-Hernandez, J.U.; Garcia-Cordero, J.L. Microfluidic systems for the analysis of blood-derived molecular biomarkers. Electrophoresis 2022, 43, 1667–1700. [Google Scholar] [CrossRef]
- Sharma, S.; Zapatero-Rodríguez, J.; Estrela, P.; Kennedy, R. Point-of-Care Diagnostics in Low Resource Settings: Present Status and Future Role of Microfluidics. Biosensors 2015, 5, 577–601. [Google Scholar] [CrossRef]
- Dkhar, D.S.; Kumari, R.; Malode, S.J.; Shetti, N.P.; Chandra, P. Integrated lab-on-a-chip devices: Fabrication methodologies, transduction system for sensing purposes. J. Pharm. Biomed. Anal. 2023, 223, 115120. [Google Scholar] [CrossRef]
- Ríos, Á.; Zougagh, M.; Avila, M. Miniaturization through lab-on-a-chip: Utopia or reality for routine laboratories? A review. Anal. Chim. Acta 2012, 740, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Clack, K.; Soda, N.; Kasetsirikul, S.; Mahmudunnabi, R.G.; Nguyen, N.-T.; Shiddiky, M.J.A. Toward Personalized Nanomedicine: The Critical Evaluation of Micro and Nanodevices for Cancer Biomarker Analysis in Liquid Biopsy. Small 2023, 19, 2205856. [Google Scholar] [CrossRef]
- Jain, S.; Nehra, M.; Kumar, R.; Dilbaghi, N.; Hu, T.; Kumar, S.; Kaushik, A.; Li, C.-z. Internet of medical things (IoMT)-integrated biosensors for point-of-care testing of infectious diseases. Biosens. Bioelectron. 2021, 179, 113074. [Google Scholar] [CrossRef] [PubMed]
- Chu, H.; Liu, C.; Liu, J.; Yang, J.; Li, Y.; Zhang, X. Recent advances and challenges of biosensing in point-of-care molecular diagnosis. Sens. Actuators B Chem. 2021, 348, 130708. [Google Scholar] [CrossRef] [PubMed]
- Gardy, J.L.; Loman, N.J. Towards a genomics-informed, real-time, global pathogen surveillance system. Nat. Rev. Genet. 2018, 19, 9–20. [Google Scholar] [CrossRef] [PubMed]
- Ielapi, N.; Andreucci, M.; Licastro, N.; Faga, T.; Grande, R.; Buffone, G.; Mellace, S.; Sapienza, P.; Serra, R. Precision Medicine and Precision Nursing: The Era of Biomarkers and Precision Health. Int. J. Gen. Med. 2020, 13, 1705–1711. [Google Scholar] [CrossRef] [PubMed]
- Kelloff, G.J.; Sigman, C.C. Cancer biomarkers: Selecting the right drug for the right patient. Nat. Rev. Drug Discov. 2012, 11, 201–214. [Google Scholar] [CrossRef] [PubMed]
- Herder, C.; Roden, M. A novel diabetes typology: Towards precision diabetology from pathogenesis to treatment. Diabetologia 2022, 65, 1770–1781. [Google Scholar] [CrossRef] [PubMed]
- Walzl, G.; McNerney, R.; du Plessis, N.; Bates, M.; McHugh, T.D.; Chegou, N.N.; Zumla, A. Tuberculosis: Advances and challenges in development of new diagnostics and biomarkers. Lancet Infect. Dis. 2018, 18, e199–e210. [Google Scholar] [CrossRef]
- Zarei, M. Advances in point-of-care technologies for molecular diagnostics. Biosens. Bioelectron. 2017, 98, 494–506. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vo, D.-K.; Trinh, K.T.L. Polymerase Chain Reaction Chips for Biomarker Discovery and Validation in Drug Development. Micromachines 2025, 16, 243. https://doi.org/10.3390/mi16030243
Vo D-K, Trinh KTL. Polymerase Chain Reaction Chips for Biomarker Discovery and Validation in Drug Development. Micromachines. 2025; 16(3):243. https://doi.org/10.3390/mi16030243
Chicago/Turabian StyleVo, Dang-Khoa, and Kieu The Loan Trinh. 2025. "Polymerase Chain Reaction Chips for Biomarker Discovery and Validation in Drug Development" Micromachines 16, no. 3: 243. https://doi.org/10.3390/mi16030243
APA StyleVo, D.-K., & Trinh, K. T. L. (2025). Polymerase Chain Reaction Chips for Biomarker Discovery and Validation in Drug Development. Micromachines, 16(3), 243. https://doi.org/10.3390/mi16030243






