Proteomic Biomarkers for the Detection of Endometrial Cancer
Abstract
:1. Introduction
2. Search for Endometrial Cancer (EC) Diagnostic Biomarkers Using High-Throughput Technologies
3. Proteomic Approaches for EC Detection
Mass Spectrometry-Based Proteomic Approaches for EC Detection
4. Proteomics Biomarkers for EC Detection
4.1. Proteomic Analysis of Blood for EC Detection
4.2. Proteomic Analysis of Tissue Samples for EC Detection
4.3. Proteomic Analysis of Urine for EC Detection
4.4. The Ideal EC Proteomic Biomarker
4.5. Challenges in Endometrial Cancer Diagnostic Biomarker Validation and Usage
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lortet-Tieulent, J.; Ferlay, J.; Bray, F.; Jemal, A. International patterns and trends in endometrial cancer incidence, 1978-2013. J. Natl. Cancer Inst. 2018, 110, 354–361. [Google Scholar] [CrossRef] [PubMed]
- Cancer Research UK Uterine Cancer Statistics. Available online: http://www.cancerresearchuk.org (accessed on 9 February 2019).
- Sundar, S.; Balega, J.; Crosbie, E.; Drake, A.; Edmondson, R.; Fotopoulou, C.; Gallos, I.; Ganesan, R.; Gupta, J.; Johnson, N. BGCS uterine cancer guidelines: Recommendations for practice. Eur. J. Obstet. Gynecol. Reprod. Biol. 2017, 213, 71–97. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weiderpass, E.; Antoine, J.; Bray, F.I.; Oh, J.-K.; Arbyn, M. Trends in corpus uteri cancer mortality in member states of the European Union. Eur. J. Cancer 2014, 50, 1675–1684. [Google Scholar] [CrossRef]
- Bokhman, J.V. Two pathogenetic types of endometrial carcinoma. Gynecol. Oncol. 1983, 15, 10–17. [Google Scholar] [CrossRef]
- Suarez, A.A.; Felix, A.S.; Cohn, D.E. Bokhman redux: Endometrial cancer “types” in the 21st century. Gynecol. Oncol. 2017, 144, 243–249. [Google Scholar] [CrossRef]
- Talhouk, A.; McConechy, M.K.; Leung, S.; Yang, W.; Lum, A.; Senz, J.; Boyd, N.; Pike, J.; Anglesio, M.; Kwon, J.S. Confirmation of ProMisE: A simple, genomics-based clinical classifier for endometrial cancer. Cancer 2017, 123, 802–813. [Google Scholar] [CrossRef] [Green Version]
- Stelloo, E.; Bosse, T.; Nout, R.A.; MacKay, H.J.; Church, D.N.; Nijman, H.W.; Leary, A.; Edmondson, R.J.; Powell, M.E.; Crosbie, E.J. Refining prognosis and identifying targetable pathways for high-risk endometrial cancer; a TransPORTEC initiative. Mod. Pathol. 2015, 28, 836. [Google Scholar] [CrossRef]
- Colombo, N.; Creutzberg, C.; Amant, F.; Bosse, T.; Gonzalez-Martin, A.; Ledermann, J.; Marth, C.; Nout, R.; Querleu, D.; Mirza, M.R.; et al. ESMO-ESGO-ESTRO Consensus Conference on Endometrial Cancer: Diagnosis, Treatment and Follow-up. Int. J. Gynecol. Cancer 2016, 26, 2–30. [Google Scholar] [CrossRef] [Green Version]
- Smith-Bindman, R.; Weiss, E.; Feldstein, V. How thick is too thick? When endometrial thickness should prompt biopsy in postmenopausal women without vaginal bleeding. Ultrasound Obstet. Gynecol. Off. J. Int. Soc. Ultrasound Obstet. Gynecol. 2004, 24, 558–565. [Google Scholar] [CrossRef]
- Guido, R.; Kanbour-Shakir, A.; Rulin, M.; Christopherson, W. Pipelle endometrial sampling. Sensitivity in the detection of endometrial cancer. J. Reprod. Med. 1995, 40, 553–555. [Google Scholar] [PubMed]
- Clark, T.J.; Voit, D.; Gupta, J.K.; Hyde, C.; Song, F.; Khan, K.S. Accuracy of hysteroscopy in the diagnosis of endometrial cancer and hyperplasia: A systematic quantitative review. JAMA 2002, 288, 1610–1621. [Google Scholar] [CrossRef] [PubMed]
- Guest, P.C.; Gottschalk, M.G.; Bahn, S. Proteomics: Improving biomarker translation to modern medicine? Genome Med. 2013, 5, 17. [Google Scholar] [CrossRef] [PubMed]
- Lockhart, D.J.; Winzeler, E.A. Genomics, gene expression and DNA arrays. Nature 2000, 405, 827. [Google Scholar] [CrossRef] [PubMed]
- Banach, P.; Suchy, W.; Derezinski, P.; Matysiak, J.; Kokot, Z.J.; Nowak-Markwitz, E. Mass spectrometry as a tool for biomarkers searching in gynecological oncology. Biomed. Pharmacother. 2017, 92, 836–842. [Google Scholar] [CrossRef] [PubMed]
- Levine, D.A.; Network, C.G.A.R. Integrated genomic characterization of endometrial carcinoma. Nature 2013, 497, 67. [Google Scholar] [CrossRef] [PubMed]
- Nair, N.; Camacho-Vanegas, O.; Rykunov, D.; Dashkoff, M.; Camacho, S.C.; Schumacher, C.A.; Irish, J.C.; Harkins, T.T.; Freeman, E.; Garcia, I.; et al. Genomic Analysis of Uterine Lavage Fluid Detects Early Endometrial Cancers and Reveals a Prevalent Landscape of Driver Mutations in Women without Histopathologic Evidence of Cancer: A Prospective Cross-Sectional Study. PLoS Med. 2016, 13, e1002206. [Google Scholar] [CrossRef]
- Lim, L.; Yang, Y.C.; Wu, C.C.; Hsu, Y.T.; Chang, C.L. Screening of Endometrial Cancer Using Cervical Swab. J. Obstet. Gynaecol. Res. 2018, 44, 1514. [Google Scholar]
- Shruthi, B.S.; Palani Vinodhkumar, S. Proteomics: A new perspective for cancer. Adv. Biomed. Res. 2016, 5, 67. [Google Scholar] [CrossRef]
- Rakyan, V.K.; Down, T.A.; Balding, D.J.; Beck, S. Epigenome-wide association studies for common human diseases. Nat. Rev. Genet. 2011, 12, 529. [Google Scholar] [CrossRef]
- Lorincz, A.T. The promise and the problems of epigenetic biomarkers in cancer. Expert Opin. Med. Diagn. 2011, 5, 375–379. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ying, J.; Xu, T.; Wang, Q.; Ye, J.; Lyu, J. Exploration of DNA methylation markers for diagnosis and prognosis of patients with endometrial cancer. Epigenetics 2018, 13, 490–504. [Google Scholar] [CrossRef] [PubMed]
- Hagarman, J.A.; Soloway, P.D. Epigenomics: One molecule at a time. Cell Cycle 2013, 12, 3451–3452. [Google Scholar] [CrossRef] [PubMed]
- Jones, A.; Teschendorff, A.E.; Li, Q.; Hayward, J.D.; Kannan, A.; Mould, T.; West, J.; Zikan, M.; Cibula, D.; Fiegl, H. Role of DNA methylation and epigenetic silencing of HAND2 in endometrial cancer development. PLoS Med. 2013, 10, e1001551. [Google Scholar] [CrossRef]
- Wentzensen, N.; Bakkum-Gamez, J.N.; Killian, J.K.; Sampson, J.; Guido, R.; Glass, A.; Adams, L.; Luhn, P.; Brinton, L.A.; Rush, B. Discovery and validation of methylation markers for endometrial cancer. Int. J. Cancer 2014, 135, 1860–1868. [Google Scholar] [CrossRef]
- Huang, R.L.; Su, P.H.; Liao, Y.P.; Wu, T.I.; Hsu, Y.T.; Lin, W.Y.; Wang, H.C.; Weng, Y.C.; Ou, Y.C.; Huang, T.H.M. Integrated epigenomics analysis reveals a DNA methylation panel for endometrial cancer detection using cervical scrapings. Clin. Cancer Res. 2017, 23, 263–272. [Google Scholar] [CrossRef]
- Takenaka, K.; Chen, B.J.; Modesitt, S.C.; Byrne, F.L.; Hoehn, K.L.; Janitz, M. The emerging role of long non-coding RNAs in endometrial cancer. Cancer Genet. 2016, 209, 445–455. [Google Scholar] [CrossRef]
- Cieślik, M.; Chinnaiyan, A.M. Cancer transcriptome profiling at the juncture of clinical translation. Nat. Rev. Genet. 2018, 19, 93. [Google Scholar] [CrossRef]
- Shi, Z.; Li, C.; Tarwater, L.; Li, J.; Li, Y.; Kaliney, W.; Chandrashekar, D.S.; Stack, M.S. RNA-seq Reveals the Overexpression of IGSF9 in Endometrial Cancer. J. Oncol. 2018, 2018, 2439527. [Google Scholar] [CrossRef]
- Jiang, Y.; Wang, N.; Yin, D.; Li, Y.K.; Guo, L.; Shi, L.P.; Huang, X. Changes in the Expression of Serum MiR-887-5p in Patients With Endometrial Cancer. Int. J. Gynecol. Cancer 2016, 26, 1143–1147. [Google Scholar] [CrossRef]
- Lan, J.; Nunez Galindo, A.; Doecke, J.; Fowler, C.; Martins, R.N.; Rainey-Smith, S.R.; Cominetti, O.; Dayon, L. Systematic Evaluation of the Use of Human Plasma and Serum for Mass-Spectrometry-Based Shotgun Proteomics. J. Proteome Res. 2018, 17, 1426–1435. [Google Scholar] [CrossRef] [PubMed]
- Cho, W.C. Proteomics technologies and challenges. Genom. Proteom. Bioinform. 2007, 5, 77–85. [Google Scholar] [CrossRef]
- Penque, D. Two-dimensional gel electrophoresis and mass spectrometry for biomarker discovery. Proteom. Clin. Appl. 2009, 3, 155–172. [Google Scholar] [CrossRef] [PubMed]
- Sutandy, F.R.; Qian, J.; Chen, C.S.; Zhu, H. Overview of protein microarrays. Curr. Protocols Protein Sci. 2013, 72, 27-1. [Google Scholar] [CrossRef]
- Resing, K.A.; Ahn, N.G. Proteomics strategies for protein identification. FEBS Lett. 2005, 579, 885–889. [Google Scholar] [CrossRef]
- Martinez-Garcia, E.; Lopez-Gil, C.; Campoy, I.; Vallve, J.; Coll, E.; Cabrera, S.; Ramon, Y.; Cajal, S.; Matias-Guiu, X.; Van Oostrum, J.; et al. Advances in endometrial cancer protein biomarkers for use in the clinic. Expert Rev. Proteom. 2018, 15, 81–99. [Google Scholar] [CrossRef]
- Lindemann, C.; Thomanek, N.; Hundt, F.; Lerari, T.; Meyer, H.E.; Wolters, D.; Marcus, K. Strategies in relative and absolute quantitative mass spectrometry based proteomics. Biol. Chem. 2017, 398, 687–699. [Google Scholar] [CrossRef]
- Meyer, J.G.; Schilling, B. Clinical applications of quantitative proteomics using targeted and untargeted data-independent acquisition techniques. Expert Rev. Proteom. 2017, 14, 419–429. [Google Scholar] [CrossRef]
- Mittal, P.; Klingler-Hoffmann, M.; Arentz, G.; Zhang, C.; Kaur, G.; Oehler, M.K.; Hoffmann, P. Proteomics of endometrial cancer diagnosis, treatment, and prognosis. Proteom. Clin. Appl. 2016, 10, 217–229. [Google Scholar] [CrossRef]
- Bauer, M.; Ahrné, E.; Baron, A.P.; Glatter, T.; Fava, L.L.; Santamaria, A.; Nigg, E.A.; Schmidt, A. Evaluation of data-dependent and-independent mass spectrometric workflows for sensitive quantification of proteins and phosphorylation sites. J. Proteome Res. 2014, 13, 5973–5988. [Google Scholar] [CrossRef]
- Ludwig, C.; Gillet, L.; Rosenberger, G.; Amon, S.; Collins, B.C.; Aebersold, R. Data-independent acquisition-based SWATH-MS for quantitative proteomics: A tutorial. Mol. Syst. Biol. 2018, 14, e8126. [Google Scholar] [CrossRef] [PubMed]
- Bateson, H.; Saleem, S.; Loadman, P.M.; Sutton, C.W. Use of matrix-assisted laser desorption/ionisation mass spectrometry in cancer research. J. Pharmacol. Toxicol. Methods 2011, 64, 197–206. [Google Scholar] [CrossRef] [PubMed]
- Aichler, M.; Walch, A. MALDI Imaging mass spectrometry: Current frontiers and perspectives in pathology research and practice. Lab. Investig. 2015, 95, 422. [Google Scholar] [CrossRef] [PubMed]
- Muinelo-Romay, L.; Casas-Arozamena, C.; Abal, M. Liquid biopsy in endometrial cancer: New opportunities for personalized oncology. Int. J. Mol. Sci. 2018, 19, 2311. [Google Scholar] [CrossRef]
- Barelli, S.; Crettaz, D.; Thadikkaran, L.; Rubin, O.; Tissot, J.D. Plasma/serum proteomics: Pre-analytical issues. Expert Rev. Proteom. 2007, 4, 363–370. [Google Scholar] [CrossRef]
- Abbott, K.L.; Lim, J.M.; Wells, L.; Benigno, B.B.; McDonald, J.F.; Pierce, M. Identification of candidate biomarkers with cancerspecific glycosylation in the tissue and serum of endometrioid ovarian cancer patients by glycoproteomic analysis. Proteomics 2010, 10, 470–481. [Google Scholar] [CrossRef]
- Feist, P.; Hummon, A. Proteomic challenges: Sample preparation techniques for microgram-quantity protein analysis from biological samples. Int. J. Mol. Sci. 2015, 16, 3537–3563. [Google Scholar] [CrossRef]
- Clair, G.; Piehowski, P.D.; Nicola, T.; Kitzmiller, J.A.; Huang, E.L.; Zink, E.M.; Sontag, R.L.; Orton, D.J.; Moore, R.J.; Carson, J.P. Spatially-resolved proteomics: Rapid quantitative analysis of laser capture microdissected alveolar tissue samples. Sci. Rep. 2016, 6, 39223. [Google Scholar] [CrossRef]
- Wiśniewski, J.R.; Zougman, A.; Nagaraj, N.; Mann, M. Universal sample preparation method for proteome analysis. Nat. Methods 2009, 6, 359. [Google Scholar] [CrossRef]
- Linkov, F.; Goughnour, S.L.; Ma, T.; Xu, Z.; Edwards, R.P.; Lokshin, A.E.; Ramanathan, R.C.; Hamad, G.G.; McCloskey, C.; Bovbjerg, D.H. Changes in inflammatory endometrial cancer risk biomarkers in individuals undergoing surgical weight loss. Gynecol. Oncol. 2017, 147, 133–138. [Google Scholar] [CrossRef]
- Mackintosh, M.; Crosbie, E. Obesity-driven endometrial cancer: Is weight loss the answer? BJOG Int. J. Obstet. Gynaecol. 2013, 120, 791–794. [Google Scholar] [CrossRef] [PubMed]
- Kok, P.; Roelfsema, F.; Frölich, M.; Meinders, A.E.; Pijl, H. Prolactin release is enhanced in proportion to excess visceral fat in obese women. J. Clin. Endocrinol. Metab. 2004, 89, 4445–4449. [Google Scholar] [CrossRef] [PubMed]
- Sanyal, D.; Raychaudhuri, M. Hypothyroidism and obesity: An intriguing link. Indian J. Endocrinol. Metab. 2016, 20, 554. [Google Scholar] [CrossRef] [PubMed]
- Li, L.M.; Zhu, Y.X.; Zhong, Y.; Su, T.; Fan, X.M.; Xi, Q.; Li, M.Y.; Fu, J.; Tan, H.; Liu, S. Human epididymis protein 4 in endometrial cancer: A meta-analysis. Clin. Chim. Acta 2018, 482, 215–223. [Google Scholar] [CrossRef] [PubMed]
- Knific, T.; Osredkar, J.; Smrkolj, Š.; Tonin, I.; Vouk, K.; Blejec, A.; Grazio, S.F.; Rižner, T.L. Novel algorithm including CA-125, HE4 and body mass index in the diagnosis of endometrial cancer. Gynecol. Oncol. 2017, 147, 126–132. [Google Scholar] [CrossRef]
- Yurkovetsky, Z.; Ta’asan, S.; Skates, S.; Rand, A.; Lomakin, A.; Linkov, F.; Marrangoni, A.; Velikokhatnaya, L.; Winans, M.; Gorelik, E. Development of multimarker panel for early detection of endometrial cancer. High diagnostic power of prolactin. Gynecol. Oncol. 2007, 107, 58–65. [Google Scholar] [CrossRef] [Green Version]
- Corbacho, A.; De La Escalera, G.M.; Clapp, C. Roles of prolactin and related members of the prolactin/growth hormone/placental lactogen family in angiogenesis. J. Endocrinol. 2002, 173, 219–238. [Google Scholar] [CrossRef] [Green Version]
- Leiser, A.; Hamid, A.; Blanchard, R. Recurrence of prolactin-producing endometrial stromal sarcoma with sex-cord stromal component treated with progestin and aromatase inhibitor. Gynecol. Oncol. 2004, 94, 567–571. [Google Scholar] [CrossRef]
- Wang, Y.S.; Cao, R.; Jin, H.; Huang, Y.P.; Zhang, X.Y.; Cong, Q.; He, Y.F.; Xu, C.J. Altered protein expression in serum from endometrial hyperplasia and carcinoma patients. J. Hematol. Oncol. 2011, 4, 15. [Google Scholar] [CrossRef]
- Cocco, E.; Bellone, S.; El-Sahwi, K.; Cargnelutti, M.; Buza, N.; Tavassoli, F.A.; Schwartz, P.E.; Rutherford, T.J.; Pecorelli, S.; Santin, A.D. Serum amyloid A: A novel biomarker for endometrial cancer. Cancer Interdiscip. Int. J. Am. Cancer Soc. 2010, 116, 843–851. [Google Scholar] [CrossRef]
- Omer, B.; Genc, S.; Takmaz, O.; Dirican, A.; Kusku-Kiraz, Z.; Berkman, S.; Gurdol, F. The diagnostic role of human epididymis protein 4 and serum amyloid-A in early-stage endometrial cancer patients. Tumor Biol. 2013, 34, 2645–2650. [Google Scholar] [CrossRef] [PubMed]
- Tarney, C.M.; Wang, G.; Bateman, N.W.; Conrads, K.A.; Zhou, M.; Hood, B.L.; Loffredo, J.; Tian, C.; Darcy, K.M.; Hamilton, C.A.; et al. Biomarker Panel for Early Detection of Endometrial Cancer in the Prostate, Lung, Colorectal, and Ovarian Cancer Screening Trial. Am. J. Obstet. Gynecol. 2019. [Google Scholar] [CrossRef] [PubMed]
- Kanat-Pektas, M.; Yenicesu, O.; Gungor, T.; Bilge, U. Predictive power of sexual hormones and tumor markers in endometrial cancer. Arch. Gynecol. Obstet. 2010, 281, 709–715. [Google Scholar] [CrossRef] [PubMed]
- Zanotti, L.; Bignotti, E.; Calza, S.; Bandiera, E.; Ruggeri, G.; Galli, C.; Tognon, G.; Ragnoli, M.; Romani, C.; Tassi, R.A. Human epididymis protein 4 as a serum marker for diagnosis of endometrial carcinoma and prediction of clinical outcome. Clin. Chem. Lab. Med. 2012, 50, 2189–2198. [Google Scholar] [CrossRef]
- Angioli, R.; Plotti, F.; Capriglione, S.; Montera, R.; Damiani, P.; Ricciardi, R.; Aloisi, A.; Luvero, D.; Cafà, E.V.; Dugo, N. The role of novel biomarker HE4 in endometrial cancer: A case control prospective study. Tumor Biol. 2013, 34, 571–576. [Google Scholar] [CrossRef]
- Moore, R.G.; Brown, A.K.; Miller, M.C.; Badgwell, D.; Lu, Z.; Allard, W.J.; Granai, C.; Bast, R.C., Jr.; Lu, K. Utility of a novel serum tumor biomarker HE4 in patients with endometrioid adenocarcinoma of the uterus. Gynecol. Oncol. 2008, 110, 196–201. [Google Scholar] [CrossRef] [Green Version]
- Li, L.; Wan, J.; Cai, G.; Yuan, L.; Liang, J.; Song, J.; Wang, F.; Liu, M. Value of serum human epididymis secretory protein 4 as a marker for differential diagnosis of malignant and benign gynecological diseases of patients in southern China. Clin. Chim. Acta 2016, 459, 170–176. [Google Scholar] [CrossRef]
- Abdul-Rahman, P.S.; Lim, B.K.; Hashim, O.H. Expression of high-abundance proteins in sera of patients with endometrial and cervical cancers: Analysis using 2-DE with silver staining and lectin detection methods. Electrophoresis 2007, 28, 1989–1996. [Google Scholar] [CrossRef] [Green Version]
- Negishi, A.; Ono, M.; Handa, Y.; Kato, H.; Yamashita, K.; Honda, K.; Shitashige, M.; Satow, R.; Sakuma, T.; Kuwabara, H. Large-scale quantitative clinical proteomics by label-free liquid chromatography and mass spectrometry. Cancer Sci. 2009, 100, 514–519. [Google Scholar] [CrossRef]
- Takano, M.; Kikuchi, Y.; Asakawa, T.; Goto, T.; Kita, T.; Kudoh, K.; Kigawa, J.; Sakuragi, N.; Sakamoto, M.; Sugiyama, T. Identification of potential serum markers for endometrial cancer using protein expression profiling. J. Cancer Res. Clin. Oncol. 2010, 136, 475–481. [Google Scholar] [CrossRef]
- Farias-Eisner, G.; Su, F.; Robbins, T.; Kotlerman, J.; Reddy, S.; Farias-Eisner, R. Validation of serum biomarkers for detection of early-and late-stage endometrial cancer. Am. J. Obstet. Gynecol. 2010, 202, 73-e1. [Google Scholar] [CrossRef] [PubMed]
- Diefenbach, C.S.; Shah, Z.; Iasonos, A.; Barakat, R.R.; Levine, D.A.; Aghajanian, C.; Sabbatini, P.; Hensley, M.L.; Konner, J.; Tew, W. Preoperative serum YKL-40 is a marker for detection and prognosis of endometrial cancer. Gynecol. Oncol. 2007, 104, 435–442. [Google Scholar] [CrossRef]
- Kemik, P.; Saatli, B.; Yıldırım, N.; Kemik, V.D.; Deveci, B.; Terek, M.C.; Koçtürk, S.; Koyuncuoğlu, M.; Saygılı, U. Diagnostic and prognostic values of preoperative serum levels of YKL-40, HE-4 and DKK-3 in endometrial cancer. Gynecol. Oncol. 2016, 140, 64–69. [Google Scholar] [CrossRef]
- Jiang, T.; Huang, L.; Wang, S.; Zhang, S. Clinical significance of serum Dkk-3 in patients with gynecological cancer. J. Obstet. Gynaecol. Res. 2010, 36, 769–773. [Google Scholar] [CrossRef] [PubMed]
- Ilhan, T.T.; Kebapcilar, A.; Yilmaz, S.A.; Ilhan, T.; Kerimoglu, O.S.; Pekin, A.T.; Akyurek, F.; Unlu, A.; Celik, C. Relations of serum visfatin and resistin levels with endometrial cancer and factors associated with its prognosis. Asian Pac. J. Cancer Prev. 2015, 16, 4503–4508. [Google Scholar] [CrossRef] [PubMed]
- Peng, X.; Li, J.; Li, M.; Ye, X.; Yan, W. Clinical significance of vascular endothelial growth factor in sera of patients with gynaecological malignant tumors. Ai Zheng Aizheng Chin. J. Cancer 2002, 21, 181–185. [Google Scholar]
- Dobrzycka, B.; Terlikowski, S.J.; Kowalczuk, O.; Kulikowski, M.; Niklinski, J. Serum levels of VEGF and VEGF-C in patients with endometrial cancer. Eur. Cytokine Netw. 2011, 22, 45–51. [Google Scholar] [CrossRef] [Green Version]
- Jóźwik, M.; Okungbowa, O.E.; Lipska, A.; Jóźwik, M.; Smoktunowicz, M.; Semczuk, A.; Jóźwik, M.; Radziwon, P. Surface antigen expression on peripheral blood monocytes in women with gynecologic malignancies. BMC Cancer 2015, 15, 129. [Google Scholar] [CrossRef]
- Li, Z.; Min, W.; Huang, C.; Bai, S.; Tang, M.; Zhao, X. Proteomics-based approach identified differentially expressed proteins with potential roles in endometrial carcinoma. Int. J. Gynecol. Cancer 2010, 20, 9–15. [Google Scholar] [CrossRef]
- Li, Z.; Zhao, X.; Bai, S.; Wang, Z.; Chen, L.; Wei, Y.; Huang, C. Proteomics identification of cyclophilin a as a potential prognostic factor and therapeutic target in endometrial carcinoma. Mol. Cell. Proteom. 2008, 7, 1810–1823. [Google Scholar] [CrossRef]
- Baser, E.; Togrul, C.; Ozgu, E.; Ayhan, S.; Caglar, M.; Erkaya, S.; Gungor, T. Sperm-associated antigen 9 is a promising marker for early diagnosis of endometrial cancer. Asian Pac. J. Cancer Prev. 2013, 14, 7635–7638. [Google Scholar] [CrossRef] [PubMed]
- Nishikawa, R.; Suzumori, N.; Nishiyama, T.; Nishikawa, H.; Arakawa, A.; Sugiura-Ogasawara, M. Clinical significance of serum growth-regulated oncogene alpha (GROalpha) in patients with gynecological cancer. Eur. J. Gynaecol. Oncol. 2012, 33, 138–141. [Google Scholar] [PubMed]
- Staff, A.C.; Trovik, J.; Zahl, E.A.G.; Wik, E.; Wollert, K.C.; Kempf, T.; Salvesen, H.B. Elevated plasma growth differentiation factor-15 correlates with lymph node metastases and poor survival in endometrial cancer. Clin. Cancer Res. 2011, 17, 4825–4833. [Google Scholar] [CrossRef]
- Nowosielski, K.; Pozowski, J.; Ulman-Wlodarz, I.; Romanik, M.; Poreba, R.; Sioma-Markowska, U. Adiponectin to leptin index as a marker of endometrial cancer in postmenopausal women with abnormal vaginal bleeding: An observational study. Neuroendocrinol. Lett. 2012, 33, 217–223. [Google Scholar] [PubMed]
- Konno, R.; Takano, T.; Sato, S.; Yajima, A. Serum soluble fas level as a prognostic factor in patients with gynecological malignancies. Clin. Cancer Res. 2000, 6, 3576–3580. [Google Scholar]
- Graesslin, O.; Cortez, A.; Fauvet, R.; Lorenzato, M.; Birembaut, P.; Daraï, E. Metalloproteinase-2,-7 and-9 and tissue inhibitor of metalloproteinase-1 and-2 expression in normal, hyperplastic and neoplastic endometrium: A clinical-pathological correlation study. Ann. Oncol. 2006, 17, 637–645. [Google Scholar] [CrossRef]
- Byrjalsen, I.; Mose Larsen, P.; Fey, S.; Nilas, L.; Larsen, M.; Christiansen, C. Two-dimensional gel analysis of human endometrial proteins: Characterization of proteins with increased expression in hyperplasia and adenocarcinoma. Mol. Hum. Reprod. 1999, 5, 748–756. [Google Scholar] [CrossRef]
- DeSouza, L.; Diehl, G.; Rodrigues, M.J.; Guo, J.; Romaschin, A.D.; Colgan, T.J.; Siu, K.M. Search for cancer markers from endometrial tissues using differentially labeled tags iTRAQ and cICAT with multidimensional liquid chromatography and tandem mass spectrometry. J. Proteome Res. 2005, 4, 377–386. [Google Scholar] [CrossRef]
- Ławicki, S.; Będkowska, G.E.; Gacuta-Szumarska, E.; Szmitkowski, M. Hematopoietic cytokines as tumor markers in gynecological malignancies: A multivariate analysis with ROC curve in endometrial cancer patients. Growth Factors 2012, 30, 29–36. [Google Scholar] [CrossRef]
- Ciocca, D.R.; Calderwood, S.K. Heat shock proteins in cancer: Diagnostic, prognostic, predictive, and treatment implications. Cell Stress Chaperones 2005, 10, 86. [Google Scholar] [CrossRef]
- DeSouza, L.V.; Krakovska, O.; Darfler, M.M.; Krizman, D.B.; Romaschin, A.D.; Colgan, T.J.; Siu, K.W.M. mTRAQ-based quantification of potential endometrial carcinoma biomarkers from archived formalin-fixed paraffin-embedded tissues. Proteomics 2010, 10, 3108–3116. [Google Scholar] [CrossRef] [PubMed]
- Dube, V.; Grigull, J.; DeSouza, L.V.; Ghanny, S.; Colgan, T.J.; Romaschin, A.D.; Michael Siu, K.W. Verification of endometrial tissue biomarkers previously discovered using mass spectrometry-based proteomics by means of immunohistochemistry in a tissue microarray format. J. Proteome Res. 2007, 6, 2648–2655. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.; Colgan, T.J.; DeSouza, L.V.; Rodrigues, M.J.; Romaschin, A.D.; Siu, K.M. Direct analysis of laser capture microdissected endometrial carcinoma and epithelium by matrix-assisted laser desorption/ionization mass spectrometry. Rapid Commun. Mass Spectrom. Int. J. Devoted Rapid Dissem. Minute Res. Mass Spectrom. 2005, 19, 2762–2766. [Google Scholar] [CrossRef] [PubMed]
- Yang, E.C.; Guo, J.; Diehl, G.; DeSouza, L.; Rodrigues, M.J.; Romaschin, A.D.; Colgan, T.J.; Siu, K.M. Protein expression profiling of endometrial malignancies reveals a new tumor marker: Chaperonin 10. J. Proteome Res. 2004, 3, 636–643. [Google Scholar] [CrossRef] [PubMed]
- DeSouza, L.V.; Grigull, J.; Ghanny, S.; Dubé, V.; Romaschin, A.D.; Colgan, T.J.; Siu, K.M. Endometrial carcinoma biomarker discovery and verification using differentially tagged clinical samples with multidimensional liquid chromatography and tandem mass spectrometry. Mol. Cell. Proteom. 2007, 6, 1170–1182. [Google Scholar] [CrossRef] [PubMed]
- Cappello, F.; Bellafiore, M.; David, S.; Anzalone, R.; Zummo, G. Ten kilodalton heat shock protein (HSP10) is overexpressed during carcinogenesis of large bowel and uterine exocervix. Cancer Lett. 2003, 196, 35–41. [Google Scholar] [CrossRef] [Green Version]
- Xiaoguang, F.; Laixi, Y.; Shunhou, J.; Aiying, M.; Changxin, Q. A study of early pregnancy factor activity in the sera of women with trophoblastic tumor. Am. J. Reprod. Immunol. 1999, 41, 204–208. [Google Scholar] [CrossRef]
- Martinez-Garcia, E.; Lesur, A.; Devis, L.; Campos, A.; Cabrera, S.; van Oostrum, J.; Matias-Guiu, X.; Gil-Moreno, A.; Reventos, J.; Colas, E. Development of a sequential workflow based on LC-PRM for the verification of endometrial cancer protein biomarkers in uterine aspirate samples. Oncotarget 2016, 7, 53102. [Google Scholar] [CrossRef]
- Martinez-Garcia, E.; Lesur, A.; Devis, L.; Cabrera, S.; Matias-Guiu, X.; Hirschfeld, M.; Asberger, J.; Van Oostrum, J.; Casares de Cal, M.D.L.A.; Gomez-Tato, A.; et al. Targeted proteomics identifies proteomic signatures in liquid biopsies of the endometrium to diagnose endometrial cancer and assist in the prediction of the optimal surgical treatment. Clin. Cancer Res. 2017, 23, 6458–6467. [Google Scholar] [CrossRef]
- Ura, B.; Monasta, L.; Arrigoni, G.; Franchin, C.; Radillo, O.; Peterlunger, I.; Ricci, G.; Scrimin, F. A proteomic approach for the identification of biomarkers in endometrial cancer uterine aspirate. Oncotarget 2017, 8, 109536–109545. [Google Scholar] [CrossRef]
- Dombrauckas, J.D.; Santarsiero, B.D.; Mesecar, A.D. Structural basis for tumor pyruvate kinase M2 allosteric regulation and catalysis. Biochemistry 2005, 44, 9417–9429. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.; Yang, E.C.; DeSouza, L.; Diehl, G.; Rodrigues, M.J.; Romaschin, A.D.; Colgan, T.J.; Siu, K.M. A strategy for high-resolution protein identification in surface-enhanced laser desorption/ionization mass spectrometry: Calgranulin A and chaperonin 10 as protein markers for endometrial carcinoma. Proteomics 2005, 5, 1953–1966. [Google Scholar] [CrossRef] [PubMed]
- Voisin, S.N.; Krakovska, O.; Matta, A.; DeSouza, L.V.; Romaschin, A.D.; Colgan, T.J.; Siu, K.M. Identification of novel molecular targets for endometrial cancer using a drill-down LC-MS/MS approach with iTRAQ. PLoS ONE 2011, 6, e16352. [Google Scholar] [CrossRef] [PubMed]
- Monge, M.; Doll, A.; Colas, E.; Gil-Moreno, A.; Castellvi, J.; Garcia, A.; Colome, N.; Perez-Benavente, A.; Pedrola, N.; Lopez-Lopez, R. Subtractive proteomic approach to the endometrial carcinoma invasion front. J. Proteome Res. 2009, 8, 4676–4684. [Google Scholar] [CrossRef]
- Maxwell, G.L.; Hood, B.L.; Day, R.; Chandran, U.; Kirchner, D.; Kolli, V.K.; Bateman, N.W.; Allard, J.; Miller, C.; Sun, M. Proteomic analysis of stage I endometrial cancer tissue: Identification of proteins associated with oxidative processes and inflammation. Gynecol. Oncol. 2011, 121, 586–594. [Google Scholar] [CrossRef]
- Townsend, M.H.; Ence, Z.E.; Felsted, A.M.; Parker, A.C.; Piccolo, S.R.; Robison, R.A.; O’Neill, K.L. Potential new biomarkers for endometrial cancer. Cancer Cell Int.l 2019, 19, 19. [Google Scholar] [CrossRef]
- Morelli, M.; Scumaci, D.; Di Cello, A.; Venturella, R.; Donato, G.; Faniello, M.C.; Quaresima, B.; Cuda, G.; Zullo, F.; Costanzo, F. DJ-1 in endometrial cancer: A possible biomarker to improve differential diagnosis between subtypes. Int. J. Gynecol. Cancer 2014, 24, 649–658. [Google Scholar] [CrossRef]
- Mu, A.K.W.; Lim, B.K.; Hashim, O.H.; Shuib, A.S. Detection of differential levels of proteins in the urine of patients with endometrial cancer: Analysis using two-dimensional gel electrophoresis and O-glycan binding lectin. Int. J. Mol. Sci. 2012, 13, 9489–9501. [Google Scholar] [CrossRef]
- Wu, J.; Chen, Y.D.; Gu, W. Urinary proteomics as a novel tool for biomarker discovery in kidney diseases. J. Zhejiang Univ. Sci. B 2010, 11, 227–237. [Google Scholar] [CrossRef] [Green Version]
- Grayson, K.; Gregory, E.; Khan, G.; Guinn, B.A. Urine Biomarkers for the Early Detection of Ovarian Cancer–Are We There Yet? Biomarke. Cancer 2019, 11, 1179299X19830977. [Google Scholar] [CrossRef]
- Austin, R.M. George Papanicolaou’s efforts to develop novel cytologic methods for the early diagnosis of endometrial carcinoma. Acta Cytol. 2017, 61, 281–298. [Google Scholar] [CrossRef] [PubMed]
- Kinde, I.; Bettegowda, C.; Wang, Y.; Wu, J.; Agrawal, N.; Shih, I.M.; Kurman, R.; Dao, F.; Levine, D.A.; Giuntoli, R. Evaluation of DNA from the Papanicolaou test to detect ovarian and endometrial cancers. Sci. Transl. Med. 2013, 5, 167ra164. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Li, L.; Douville, C.; Cohen, J.D.; Yen, T.T.; Kinde, I.; Sundfelt, K.; Kjær, S.K.; Hruban, R.H.; Shih, I.M. Evaluation of liquid from the Papanicolaou test and other liquid biopsies for the detection of endometrial and ovarian cancers. Sci. Transl. Med. 2018, 10, eaap8793. [Google Scholar] [CrossRef] [PubMed]
- Kim, G.E.; Kweon, S.S.; Lee, J.S.; Lee, J.H.; Nam, J.H.; Choi, C. Quantitative assessment of DNA methylation for the detection of cervical and endometrial adenocarcinomas in liquid-based cytology specimens. Anal. Quant. Cytopathol. Histopathol. 2012, 34, 195–203. [Google Scholar]
- Bakkum-Gamez, J.N.; Wentzensen, N.; Maurer, M.J.; Hawthorne, K.M.; Voss, J.S.; Kroneman, T.N.; Famuyide, A.O.; Clayton, A.C.; Halling, K.C.; Kerr, S.E. Detection of endometrial cancer via molecular analysis of DNA collected with vaginal tampons. Gynecol. Oncol. 2015, 137, 14–22. [Google Scholar] [CrossRef] [Green Version]
- Ransohoff, D.F.; Gourlay, M.L. Sources of bias in specimens for research about molecular markers for cancer. J. Clin. Oncol. 2010, 28, 698. [Google Scholar] [CrossRef]
- Altmae, S.; Esteban, F.J.; Stavreus-Evers, A.; Simon, C.; Giudice, L.; Lessey, B.A.; Horcajadas, J.A.; Macklon, N.S.; D’Hooghe, T.; Campoy, C.; et al. Guidelines for the design, analysis and interpretation of ’omics’ data: Focus on human endometrium. Hum. Reprod. Update 2014, 20, 12–28. [Google Scholar] [CrossRef]
- Henry, N.L.; Hayes, D.F. Cancer biomarkers. Mol. Oncol. 2012, 6, 140–146. [Google Scholar] [CrossRef] [Green Version]
- McLerran, D.; Grizzle, W.E.; Feng, Z.; Thompson, I.M.; Bigbee, W.L.; Cazares, L.H.; Chan, D.W.; Dahlgren, J.; Diaz, J.; Kagan, J. SELDI-TOF MS whole serum proteomic profiling with IMAC surface does not reliably detect prostate cancer. Clin. Chem. 2008, 54, 53–60. [Google Scholar] [CrossRef]
- Skates, S.J.; Gillette, M.A.; LaBaer, J.; Carr, S.A.; Anderson, L.; Liebler, D.C.; Ransohoff, D.; Rifai, N.; Kondratovich, M.; Tezak, Z. Statistical design for biospecimen cohort size in proteomics-based biomarker discovery and verification studies. J. Proteome Res. 2013, 12, 5383–5394. [Google Scholar] [CrossRef]
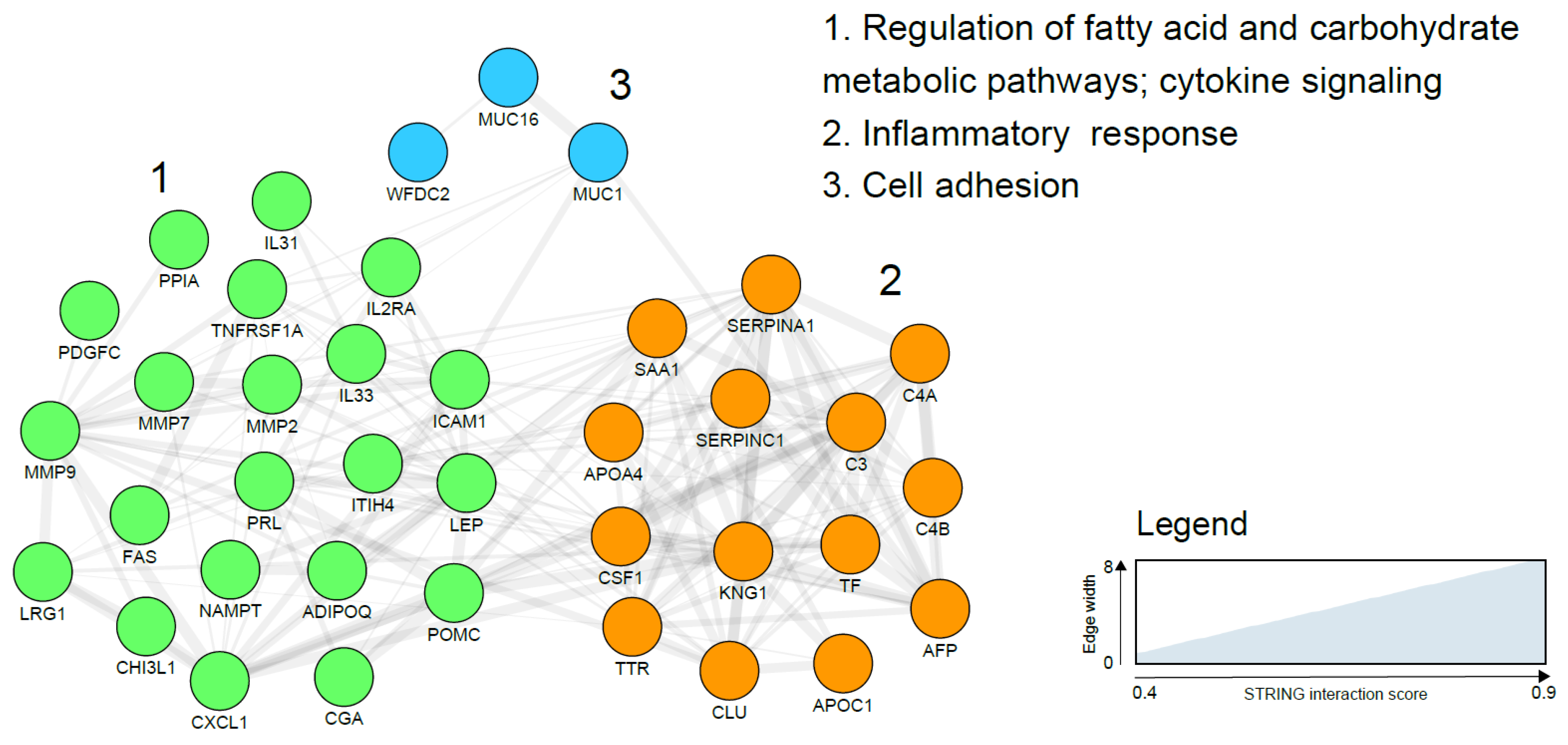
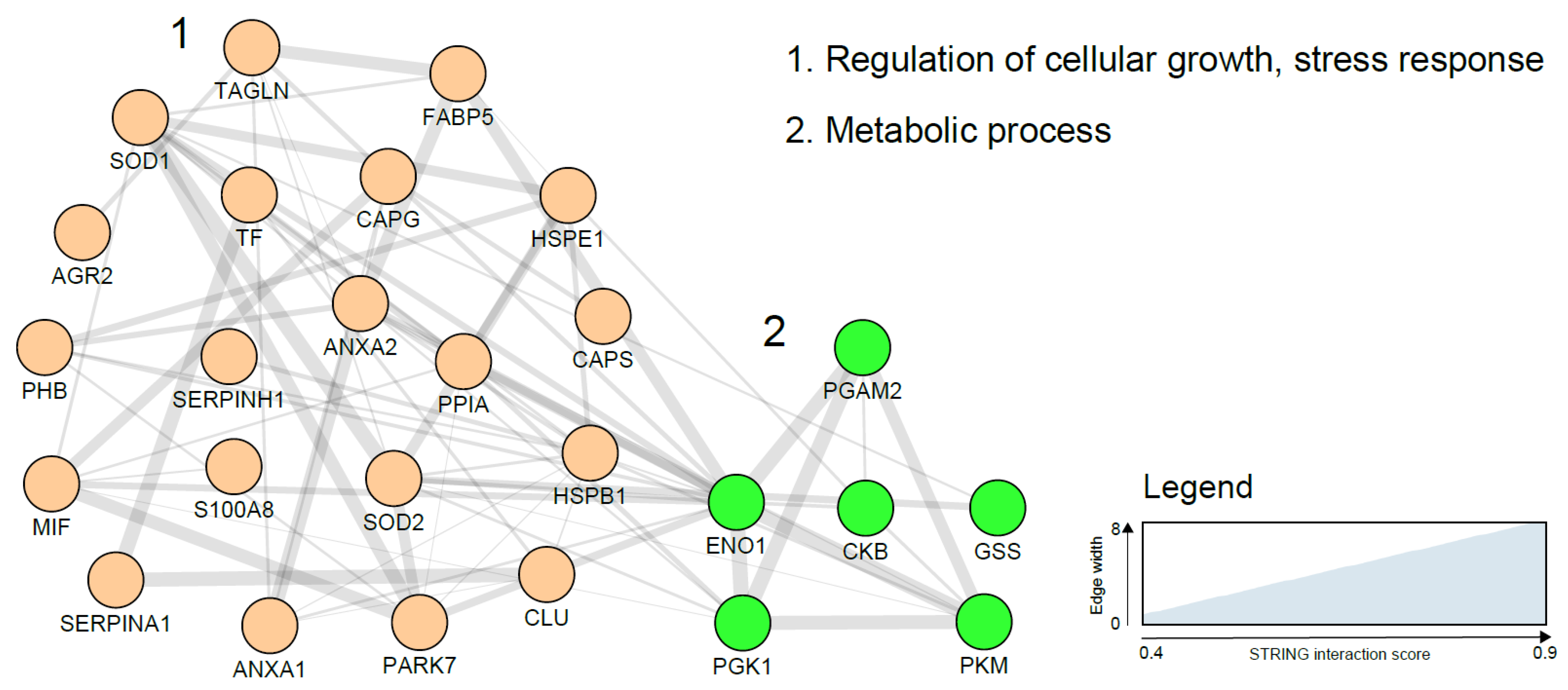
| Potential Sources of EC Biomarkers | Description | Advantages | Disadvantages |
|---|---|---|---|
| Blood (serum/plasma) | Blood drawn into sample collection tubes. | Easily accessible Minimally invasive High acceptability to both clinicians and patients. | Challenging matrix for proteomic analysis High protein dynamic range Low cancer derived proteins in early phases of disease. Poor concordance with tissue-derived proteins |
| Hysterectomy specimens | Tissue specimens obtained following hysterectomy | Viable source of biomarkers Relatively low protein dynamic range Good matrix for proteomic analysis | Highly invasive Low acceptability Not feasible for pre-treatment diagnosis |
| Pipelle biopsy specimens | Endometrial sampling by insertion of the pipelle into the uterine cavity either blindly or at hysteroscopy | Viable source of biomarkers Minimally invasive Relatively low protein dynamic range Relatively good matrix for proteomics Useful in both symptomatic and asymptomatic women. | Severe pain in up to 25% May miss focal pathologies High risk of insertion failure (22% in nulliparous, 8% in parous) Infection, bleeding, uterine perforation |
| Uterine lavage | Saline is introduced into the uterine cavity and returned by aspiration. | Viable source of biomarkers Relatively low protein dynamic range Relatively good matrix for proteomics Useful in both symptomatic and asymptomatic women. | Relatively invasive Discomfort and pain Low acceptability especially in asymptomatic women |
| Pap Smear/cervical scrape | A cervical brush is used to sample the ecto-cervix and the endocervical canal. | Simple and minimally invasive Low cost Widely acceptable Viable source of biomarkers | Discomfort from speculum examination Intimate procedure Less useful in asymptomatic women |
| Tao Brush biopsy specimens | The Tao brush is inserted into the uterine cavity and used to obtain tissue specimens | Less discomfort than pipelle biopsy Viable source of biomarkers Relatively low protein dynamic range | High cost High risk of insertion failure (20% nulliparous, 8% in parous) |
| Vaginal tampons/swabs | Vaginal tampons used for 8–12 h | Minimally invasive Potential source of uterine biomarkers in symptomatic women | Unappealing to postmenopausal women Inadequate for EC detection in women without bleeding symptoms |
| Urine | Usually self-collected | Cheap, simple, non-invasive High level of acceptability Can be collected at home/in privacy Relies on renal excretion of systemic biomarkers or urinary contamination by uterine biomarkers Proteins and peptides are stable in urine and less complex | Biomarkers may not be excreted in urine Urinary contamination by uterine biomarkers may be unreliable especially in asymptomatic or minimally symptomatic women Wide variability in urinary protein concentrations |
| Potential Biomarker | Gene Names | Summary of Evidence | Proteomic Techniques Used | Known Biochemical Function | Limitations | Panels |
|---|---|---|---|---|---|---|
| Serum Amyloid A (SAA) | SAA1 | Inconsistent evidence Upregulated in EC in some studies [60,61]. No difference between cases and controls in others [62,63]. | Isobaric tags for relative and absolute quantification (iTRAQ) technology and 2-dimensional liquid chromatography coupled to tandem mass spectrometry (LC–MS/MS). Particle-enhanced immunonephelometry and LS–MS/MS. | High density lipoprotein. Modulates inflammation. Metabolism and transport of cholesterol. Acute phase reactant | Lacks selectivity as also elevated in lung, colon and other cancers | SAA+HE4 had 73.3% sensitivity and 64% specificity [62] |
| Prolactin (PRL) | PRL | Consistent but limited evidence Sensitivity of 98.3% and specificity of 98% [57]. Upregulated in EC with 16.3% sensitivity and 100.0% specificity at PRL >30 ng/mil [64]. | Multiplex xMAP™ bead-based immunoassay Electrochemoluminescence immunoassay | Single-chain protein closely related to GH Secretion by stromal cells in response to tumour growth and differentiation Cytokine with immune and inflammatory functions. | Elevated in ovarian, pancreatic and lung cancers. | Prolactin, GH, TSH, eotaxin and E-selectin had better accuracy [57] |
| Human Epididymis Protein 4 (HE4) | WFDC2 | Consistent (evidence from meta-analysis) Sensitivity and specificity of 0.65 (95% CI 0.56–0.73) and 0.9 (95% CI (0.8–0.95) respectively [55]. | Enzyme immunoassay, Microparticle immunoassay Electrochemiluminescence | A member of the Whey acidic protein family, located on chromosome 20q 12–13 and acts as a proteinase inhibitor (trypsin inhibitor properties). Possible role in sperm maturation | Expressed in ovarian, renal, lung, colon and breast cancers Suboptimal sensitivity Methodological heterogeneity across studies. | HE4+CA125 57–76% sensitivity and 90–100% specificity [65,66,67,68] |
| Alpha-1-beta-gylycoprotein (AIBG) | AIBG | Limited evidence Upregulated in EC [69] | 2-Dimensional gel electrophoresis | Plasma glycoprotein encoded by A1BG gene with unknown function. | Limited evidence Few studies, obsolete technique | None |
| Complement factors (C3, C4A, C4B) | C3, C4A, C4B | Limited evidence Upregulated in EC [70] | LC–ESI–QTOF(MS1) | Complement proteins involved in immunity and tolerance. | Limited evidence Few studies, small sample size High abundance proteins with low specificity for EC. | None |
| Cancer Antigens CA125 CA72.4, CA15-3 | MUC 16 TAG-72 MUC 1 | Consistent evidence CA125 (MUC 16) sensitivity of 17.8–52.6% and specificity of 33.35% to 95% [37]. | Enzyme-linked immunosorbent assay (ELISA) Electrochemiluminescence Multiplex bead-based immunoassay | Mucin family glycoprotein, a component of the female reproductive tract, respiratory and ocular surfaces. | Sub-optimal diagnostic accuracy. Elevated in several other malignancies such as ovarian and pancreatic cancers. | See above |
| Apolipoproteins (A-IV), C1 | APOA1-4 APOC1 | Limited and inconsistent evidence Apo A-IV Downregulated in EC [60,70,71]. AI showed SEN 78% and SPE 90%. Apo C1 Upregulated (SEN 82%, SPE 86%) [71]. | LC–ESI–QTOF (MS1)/SELDI TOF (MSI) SELDI TOF (MSI) | Lipid transport proteins, stabilise lipoprotein structure and act as enzyme cofactors. | Sub-optimal diagnostic accuracy. High abundance blood proteins with low specificity for EC. | ApoA-1+TTR+TF 71% SEN, 88% SPE [72] |
| Clusterin (CLU) | CLU | Limited Upregulated in EC [69] | 2 DE Electrophoresis | Also known as Apolipoprotein J, Chaperone with anti-apoptotic properties, involved in preventing the aggregation of non-native protein | Limited evidence. Involved in many conditions related to oxidative stress such as ageing, cancers, neuro- degenerative diseases. | None |
| Antithrombin III (SERPINC I) | SERPINC 1 | Limited Upregulated in EC [69] | 2 DE Electrophoresis | Glycoprotein produced by the liver, involved in the coagulation system. May inhibit angiogenesis. | Limited evidence, low specificity for EC. | None |
| Alpha 1 antitrypsin (SERPINA1) | SERPINA1 | Limited Downregulated in EC [69] | 2 DE Electrophoresis | A serine protease inhibitor, inhibits enzymes such as trypsin and neutrophil elastase, produced in the liver and transported to the lungs | Limited evidence, low specificity for EC. | None |
| Human chitinase-3 like protein1 (YKL-40) | CHI3L1 | Consistent, limited Upregulated in EC, 76% sensitivity, 89% specificity [73,74]. | Enzyme-linked immunosorbent assay (ELISA) | Glycoprotein of the chitinase family, involved in degradation of extracellular matrix. | Nonspecific. Elevated in colorectal, breast, leukaemia, lung, melanoma cancers, rheumatoid arthritis etc. | None |
| Dickkopf-related protein 3 precursor (DKK3) | DKK3 | Limited, inconsistent Upregulated in EC [75] No difference between cases and controls [74] | Enzyme-linked immunosorbent assay (ELISA) | Member of the Wnt signalling pathway important in cell division, formation and cell death during embryogenesis. Reported pro-angiogenic effect in tumour growth. | Limited and inconsistent evidence. Implicated in bone disease, cancer and Alzheimers’ disese. | None |
| Visfatin (NAMPT) | NAMPT | Limited Upregulated in EC, 14.9± 10.6 ng/mL and 8.1± 6,9 ng/mL in EC vs. controls respectively (p:0.011) [76] | Enzyme-linked immunosorbent assay (ELISA) | Secreted by visceral fat, mimics insulin. Possible involvement in metabolic pathways, immune response and cancers. | Limited evidence, may be surrogate for EC risk factors. | None |
| VEGFA, VEGFVEGFC | PDGFC | Limited, Inconsistent Upregulated in EC [77,78] in some studies, Down regulated in EC [57] in others. | Enzyme-linked immunosorbent assay (ELISA) | Endothelial cell growth factor involved in physiological and pathological angiogenesis. | Limited and inconsistent evidence, non-specific, elevated in many physiological and pathological states. | None |
| TSH, ACTH, FSH | CGA, POMC, CGA | Limited TSH and ACTH upregulated in EC while FSH is downregulated [57] | Enzyme-linked immunosorbent assay (ELISA), multiplex bead based immunoassay. | TSH and ACTH: communication between immune cells and regulation. FSH: Glycoprotein regulating growth and reproductive processes. | Limited evidence, all non-specific. | Prolactin, GH, TSH, eotaxin and E-selectin had better accuracy [57] |
| ICAM1/CD54 | ICAM 1 | Limited Upregulated in EC [79] | Flow cytometry | Leucocyte-endothelial transmigration | Limited evidence | None |
| Interleukin s/receptors (IL31, 33, IL2R) | IL31, IL33, IL2R | Limited Upregulated in EC [57] | Enzyme-linked immunosorbent assay (ELISA), multiplex bead based immunoassay. | Protein expressed on the surface of immune cells and response to cytokines. | Limited evidence | None |
| Cyclophilin A (CypA) | PPIA | Limited Upregulated in EC [80,81] | Two-dimensional gel electrophoresis and MALDI-Q-TOF MS/MS | Ubiquitous protein, ubiquitous protein, regulates protein folding and trafficking. Plays role in malignant transformation. | Limited evidence, non-specific, high abundance protein, increases with aging and pro-inflammatory conditions such sepsis. | None |
| Sperm associated antigen-9 | SPAG9 | Limited Upregulated in EC vs. controls: 18.3 (12.7–53.8) vs. 14.1(4.3–65.3); SEN 70.4% & SPE 82.5% at SPAG9 > 17 ng/mL [82] | Enzyme-linked immunosorbent assay (ELISA). | Scaffold protein involved in signalling pathways, Expressed in testicular haploid germ cells, implicated in infertility. | Limited evidence, non-specific, elevated in cervical, bladder and lung cancers. | None |
| Growth-regulated oncogene alpha (CXCL1) | CXCL1 | Limited Upregulated in EC vs. controls (145(70–270)/90(45–237), p < 0.001. AUC = 0.80 [83] | Immunoassay | Chemokine involved in inflammation and tumorigenesis. | Non-specific, elevated in colorectal, melanoma, gastric cancer, ovarian cancer etc. | None |
| Growth differentiation factor 15 (GDF-15) | GDF15 | Limited Upregulated in EC vs. controls. AUC 0.86 [84] | Immunoradiometric sandwich assay with polyclonal goat antihuman GDF-15 antibodies. | A transforming growth factor involved in tissue differentiation and maintenance. | Nonspecific, elevated in ovarian thyroid, pancreatic and colon cancers | None |
| Adiponectin, Leptin | ADIPOQ LEP | Consistent Adiponectin: Downregulated in EC vs. control (mean g/mL 11.3 vs. 17.2 (p < 0.0001) [85] Leptin: Upregulated in EC vs. control, mean ng/mL (19.5 vs. 13.4, p = 0.03) [85] | Enzyme-linked immunosorbent assay (ELISA) and RIA | Adipokines with metabolic, inflammatory and immune functions. Leptin is pro-inflammatory and adiponectin is anti-inflammatory. | Markers of obesity and metabolic syndrome. Non-specific. | None |
| FAS (APO1, CD95) | FAS | Limited Upregulated in EC [86]. | Enzyme-linked immunosorbent assay (ELISA) and RIA | Fas-Fas ligand system important in CTL and NK mediated apoptosis. | Limited evidence | None |
| Leucine-rich glycoprotein (LRG1) | LRG1 | Limited Upregulated in EC vs. controls [69]. | 2 DE Electrophoresis | Involved in protein-protein interaction signal transduction, cell adhesion and neovascularization. | Limited evidence, non-specific. | None |
| Matrix metalloproteinase 2,7,9 | MMP2 MMP7 MMP9 | Inconsistent, limited MMP7 upregulated in EC, MMP2 and MMP9 downregulated [87]. | Enzyme-linked immunosorbent assay (ELISA), multiplex bead based immunoassay. | Enzymes involved in the degradation of extracellular matrix proteins during organogenesis, growth and tissue turnover. | Limited evidence | None |
| Transthyretin (TTR)/Transferin (TF) | TTR TF | Limited evidence Upregulated in EC [88] | Immunoassay | TTR: Transport protein that carries thyroid hormone and retinol-binding protein.TF: Iron-binding glycoprotein | Limited evidence, non-specific, associated with amyloidosis, cardiomyopathy etc. | TTR+TF+ ApoA:71%SEN &88% SPE |
| Inter-alpha-trypsin inhibitor family heavy chain-related protein (HRP) | ITIH4 | Limited evidence Upregulated in EC [69] | 2 DE Electrophoresis LC-ESI-QTOF (MS1) | Plasma glycoprotein, Serine protease inhibitors. | Limited evidence, non-specific. Dysregulated in multiple solid tumours. | None |
| Cleaved high molecular weight kininogen | KNG1 | Limited evidence Down-regulated in EC vs. Controls [69,89]. | 2 DE Electrophoresis ITRAQ technology and 2D LC–MS/MS. | Multifunctional plasma proteins involved in the blood coagulation cascade. | Limited evidence, non-specific, high abundance plasma proteins. | None |
| Tumor necrosis factor receptor 1A (TNFRSF1A) | TNFRSF1A | Limited evidence Upregulated in EC [57]. | Enzyme-linked immunosorbent assay (ELISA). | A ubiquitous receptor binding TNF, activating the NF-KB transcription factor, mediating apoptosis and regulating inflammation. | Limited evidence, few studies, not specific elevated in multiple sclerosis, dementia, schizophrenia etc. | None |
| Colony stimulating factor 1(CSF1) | CSF1 | Limited evidence Upregulated in EC vs. Controls [90]. | Enzyme-linked immunosorbent assay (ELISA). | Regulatory cytokine involved in the proliferation and differentiation of haematopoietic stem cells. | Limited evidence, few studies, non-specific. | None |
| Alpha fetoprotein (AFP) | AFP | Limited evidence Downregulated in EC [64] | Electrochemiluminescence | Plasma protein whose function in adult humans is less clear. Prevents transport of estradiol across placenta in rodents. | Limited evidence, non-specific, elevated in hepatic cancers germ cell tumours etc. | None |
| Potential Biomarker | Gene Names | Summary of Evidence | Proteomic Techniques Used | Known Biochemical Function | Limitations | Panels |
|---|---|---|---|---|---|---|
| Chaperonin 10 (CPN 10) | HSPE1 | Consistent Upregulated in EC tissues [89,92,94,95,96,103] | iTRAQ and cleavable isotope coded affinity tags (ciCAT) labelled LC-Tandem MS SELDI QTOF MSI | Chaperones involved in normal protein folding, cell signalling and maintenance of the conformation of transduction complexes | Heat-shock proteins are elevated in many other conditions. | CPN 10, PK and SERPINA1 had SEN, SPE and PPV of 0.95. |
| Calcyphosine (CAPS) | CAPS | Limited Upregulated in EC tissues [80,81,104]. | 2 DE Electrophoresis+MS Immunobloting immunohistochemistry | A phosphorylated substrate for cAMP-dependent protein kinase cross-signalling regulating proliferation and differentiation | Limited evidence | None |
| Pyruvate Kinase (PK) | PKM | Accumulating evidence Upregulated in EC tissues [89,93,96,104] | iTRAQ and ciCAT labelled LC-Tandem MS | Regulatory function in the glycolytic pathway | Non-specific, elevated in other malignant and metabolic conditions. | CPN10, PKM2, SERPINA1 had SEN 0.85, SPE 0.93, PV 0.90. [93] |
| Cyclophilin A (CYP A) | PPIA | Limited evidence Upregulated in EC by up to 27.23 fold [80,81] | 2 DE Electrophoresis+MS Immunobloting immunohistochemistry | Protein folding and immune regulation. Exogenous CYPA may enhance cancer growth via interaction with CD147 and activation of ERK1/2 and MAPK pathways. | Upregulated in lung, pancreatic, hepatocellular and buccal squamous cell carcinomas. | None |
| Calgizzarin (S-100A11) | S-100A11 | Limited evidence Upregulated in EC [89,103] | iTRAQ and ciCAT labelled LC-Tandem MS | Calcium binding protein which has roles in cell growth, apoptosis and low grade inflammation. | Non-specific. | None |
| Epidermal fatty acid binding protein (EFBP) | FABP5 | Limited evidence E-FABP was upregulated by up to 6.56 fold in EC cases compared to controls [80,81]. | 2 DE Electrophoresis+MS Immunobloting immunohistochemistry | Fatty-acid binding protein involved in cellular signalling and influences gene expression, growth regulation and cell differentiation. | Up-regulated in oesophageal squamous cell cancer and down-regulated in less differentiated bladder cancer | None |
| Calgranulin A (S100A8) | S100A8 | Limited evidence Upregulated in EC [94,103] | MALDI-TOF-MS SELDI-QTOF MSI | S-100 calcium binding protein expressed in multiple cell types. Act as calcium sensors and modulate inflammation. | Limited evidence, non-specific. | None |
| Other heat-shock proteins HSP27 HSP47 | HSPB1 SERPINH1 | Limited evidence Upregulated in EC tissues vs. controls [88,105]. Downregulated in EC [81,104] | 2 DE Electrophoresis MALDI Q-TOF MS/MS | Protein folding, cell signalling and maintenance of the conformation of transduction complexes, cell proliferation and differentiation. | Non-specific, limited evidence. | None |
| Prohibitin (PHB) | PHB | Limited evidence Upregulated in EC tissues vs. Controls [88,106]. | 2 DE Electrophoresis LC–MS/MS | Inhibits DNA synthesis and regulates proliferation. | Limited evidence, few studies. | None |
| Transgelin (TAGLN) | TAGLN | Limited evidence Downregulated in EC [89,96] | iTRAQ and ciCAT labelled LC–Tandem MS | Involved in actin cross linking and protein gelling. Found in many fibroblasts and smooth muscle. | Limited evidence, few studies. | None |
| Phosphoglycerate kinase (PGK1) | PGK1 | Limited evidence Upregulated in EC [88,107] | 2 DE Electrophoresis + MS Immunohistochemistry | Regulatory enzyme in the glycolytic pathway. | Limited evidence, non-specific | None |
| Creatine kinase B | CKB | Limited evidence Downregulated in EC [89,96,104] | iTRAQ and ciCAT labelled LC–Tandem MS | Mainly expressed in brain and smooth muscles including vascular and uterine. Major role in energy transduction. | Limited evidence, non-specific | None |
| Serotransferrin precursor/Transferrin | TF | Limited evidence Upregulated in EC [88] Downregulated in EC [81,104]. | 2 DE Electrophoresis MALDI Q–TOF–MS/MS | Iron-binding plasma protein. | Limited evidence | None |
| Heterogeneous nuclear ribonucleoproteins (A2/B1,D0) | HNPRNPA1 | Limited evidence Upregulated in EC [88,89]. | iTRAQ and ciCAT labelled LC-Tandem MS 2 DE Electrophoresis | Protein complexes of RNA important in cell-cycle processes and DNA damage. | Limited evidence | None |
| Macrophage migratory inhibitory factor (MIF) | MIF | Limited evidence Upregulated in EC [89,96]. | iTRAQ and ciCAT labelled LC–Tandem MS | Important regulator of the cell-mediated immunity and inflammation. | Limited evidence | None |
| Polymeric immunoglobulin receptor precursor (PIGR) | PIGR | Limited evidence Upregulated in EC vs. Controls [89,96]. | iTRAQ and ciCAT labelled LC–Tandem MS | A receptor that binds polymeric IgA and IgM on basolateral surface of epithelial cells. Important in signalling and immunoglobulin transcytosis. | Limited evidence | None |
| Alpha-1-antitypsin precursor (AIAT) | SERPINA1 | Limited evidence Down regulated in EC tissues [89,96]. | iTRAQ and ciCAT labelled LC–Tandem MS | A serine protease inhibitor, inhibits enzymes such as trypsin | Limited evidence | None |
| Capping Actin Protein, Gelsolin Like (CAPG) | CAPG | Limited evidence Upregulated in EC tissues [104,108]. Downregulated in EC [81] | 2 DE Electrophoresis + LC–MS/MS | Actin-based motility in non-muscle cells. | Limited and inconsistent evidence | None |
| Protein Deglycase (DJ-1) | PARK7 | Limited evidence Upregulated in EC [108] | 2 DE Electrophoresis + LC–MS/MS | Redox-sensitive chaperone and sensor for oxidative stress. | Limited evidence | None |
| Annexin-1,2 (ANXA 1,2) | ANXA1, ANXA2 | Limited evidence Upregulated in EC [104,106,108] | 2 DE Electrophoresis +LC–MS/MS Western blotting Tissue microarray | Bind to cellular membranes in a calcium-dependent manner, mimic glucocorticoid function and exhibits anti-inflammatory properties. | Limited evidence | None |
| Peroxiredoxin-1,4 (PRD-X1,X4) | PRDX1-4 | Limited evidence Upregulated in EC [81,106] | LC–MS/MS Western blotting Tissue microarray | Scavenging of peroxides, protection from oxidative stress-induced apoptosis. | Limited evidence | None |
| Costars family protein ABRACL | ABRACL | Limited evidence Upregulated in EC [101] | 2 DE Electrophoresis LC–MS/MS Western blotting | ABRACL is an 82 amino acid protein that regulates actin cytoskeleton dynamics and motility. | Limited evidence | None |
| Phosphoglycerate mutase 2 (PGAM2) | PGAM2 | Limited evidence Upregulated in EC [101] | 2 DE Electrophoresis LC–MS/MS Western blotting | Glycolytic enzyme modulating NADPH homeostasis, impacting cell proliferation and tumour growth. | Limited evidence | None |
| Glutathione synthetase (GSS) | GSS | Limited evidence Upregulated in EC [101] | 2 DE Electrophoresis LC–MS/MS Western blotting | Cellular homeostasis and anti-oxidant properties. | Limited evidence | None |
| Desmin (Des) | DES | Limited evidence Downregulated in EC [81,105]. | DIGE MALDI–TOF | Protein marker for muscle tissue | Limited evidence | None |
| Alpha enolase (ENO1) | ENO1 | Limited evidence Upregulated in EC 220 fold [100,101,106] | 2 DE Electrophoresis LC–MS/MS Western blotting | Glycolytic enzyme. Regulates the PI3K/AKT signalling pathway and induces tumorigenesis by activating plasminogen. | Limited evidence | None |
| Superoxide dismutase (SOD1&2) | SOD1 SOD2 | Limited evidence SOD2 Upregulated in EC 5 fold [101]. SOD1 downregulated in EC [105] | 2 DE Electrophoresis LC–MS/MS Western blotting | Has anti-apoptotic effects against oxidative stress, ionizing radiation, and inflammatory cytokines. | Limited evidence | None |
| Fibrinogen beta chain (FBG) | FBG | Limited evidence Upregulated in EC up to 400 fold [101]. | 2 DE Electrophoresis LC–MS/MS Western blotting | Blood-based glycoprotein | Limited evidence | None |
| Anterior Gradient 2 protein | AGR2 | Limited evidence Upregulated in EC [81,104] | 2 DE Electrophoresis MALDI Q–TOF MS/MS | A protein disulphide isomerase involved in protein folding and implicated in various cancers. | Limited evidence | None |
| Clusterin (CLU) | CLU | Limited evidence Upregulated in EC [96] | LC + Tandem MS/MS | Chaperone with anti-apoptotic properties. | Limited evidence | None |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Njoku, K.; Chiasserini, D.; Whetton, A.D.; Crosbie, E.J. Proteomic Biomarkers for the Detection of Endometrial Cancer. Cancers 2019, 11, 1572. https://doi.org/10.3390/cancers11101572
Njoku K, Chiasserini D, Whetton AD, Crosbie EJ. Proteomic Biomarkers for the Detection of Endometrial Cancer. Cancers. 2019; 11(10):1572. https://doi.org/10.3390/cancers11101572
Chicago/Turabian StyleNjoku, Kelechi, Davide Chiasserini, Anthony D. Whetton, and Emma J. Crosbie. 2019. "Proteomic Biomarkers for the Detection of Endometrial Cancer" Cancers 11, no. 10: 1572. https://doi.org/10.3390/cancers11101572
APA StyleNjoku, K., Chiasserini, D., Whetton, A. D., & Crosbie, E. J. (2019). Proteomic Biomarkers for the Detection of Endometrial Cancer. Cancers, 11(10), 1572. https://doi.org/10.3390/cancers11101572





