CD200 Induces Epithelial-to-Mesenchymal Transition in Head and Neck Squamous Cell Carcinoma via β-Catenin-Mediated Nuclear Translocation
Abstract
:1. Introduction
2. Results
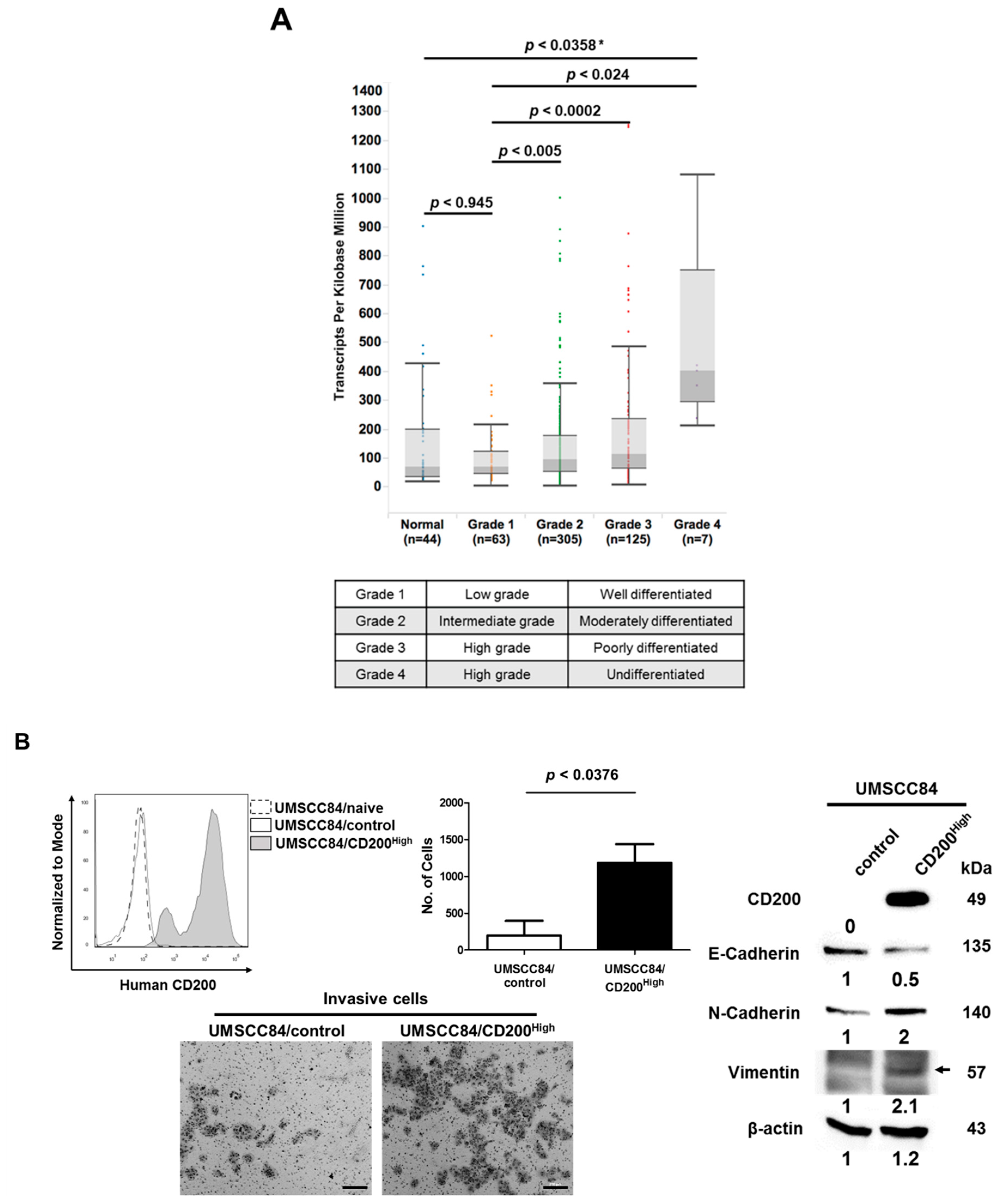
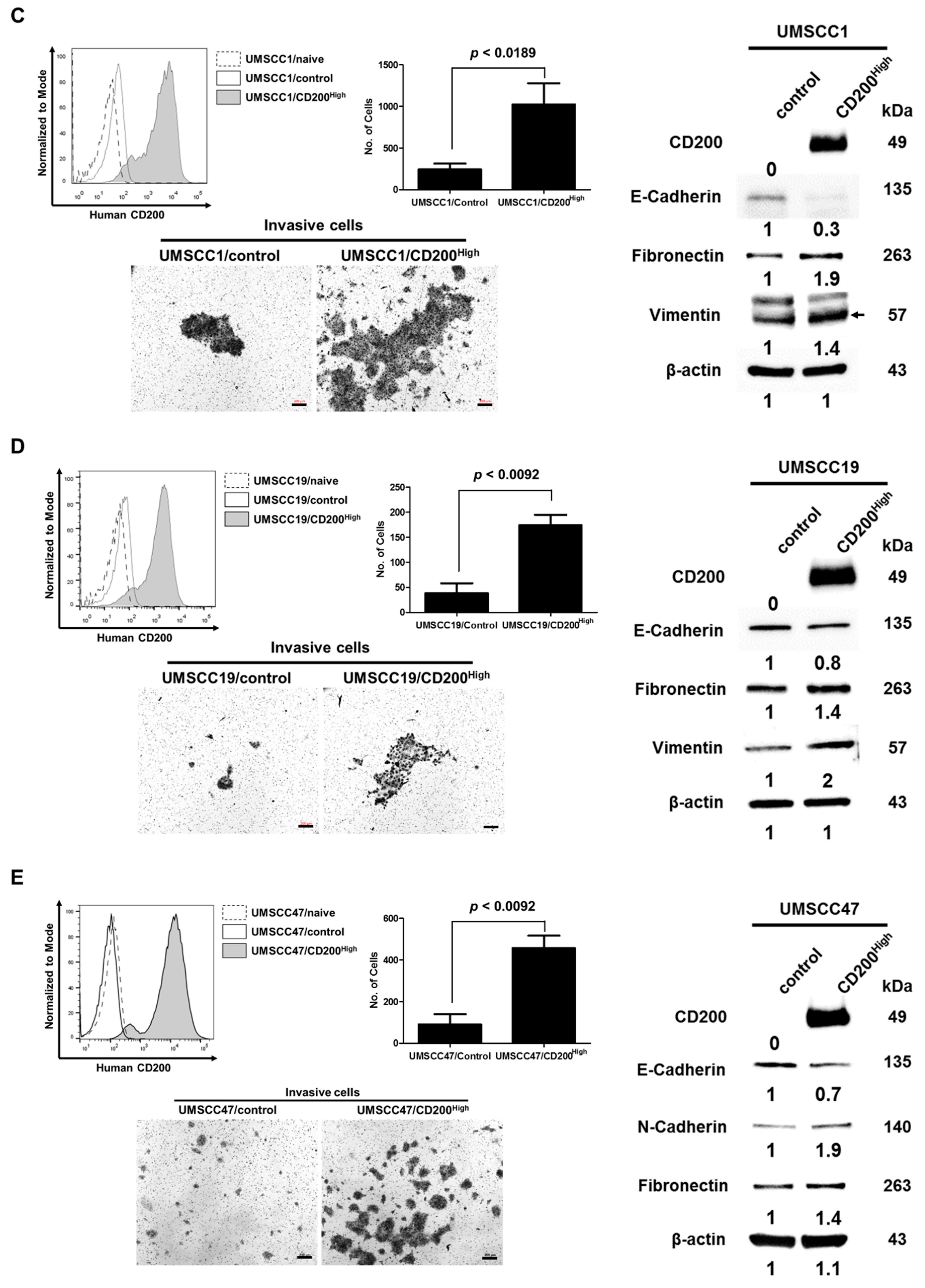
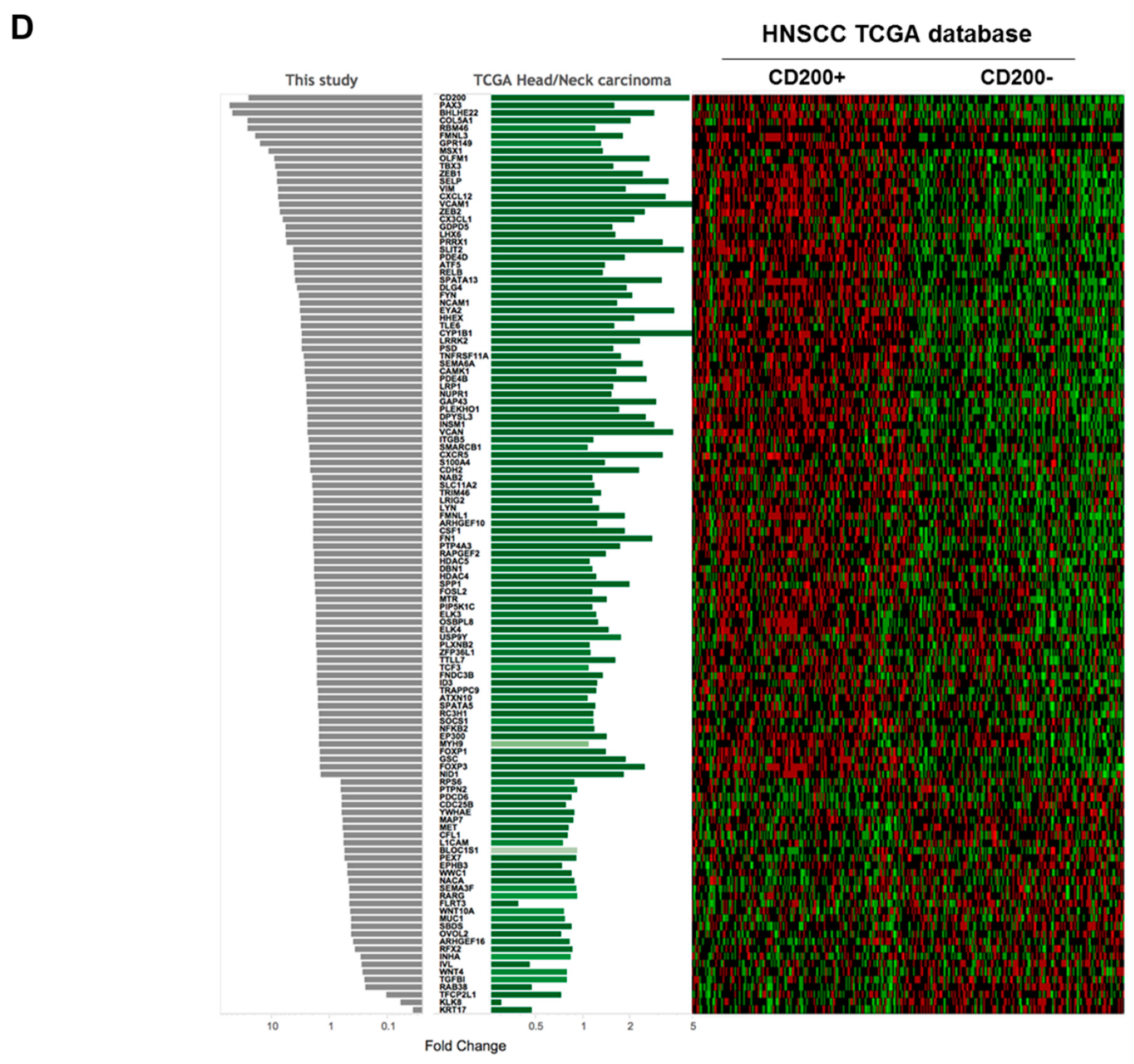
2.1. CD200 Was Associated with EMT Features in HNSCC
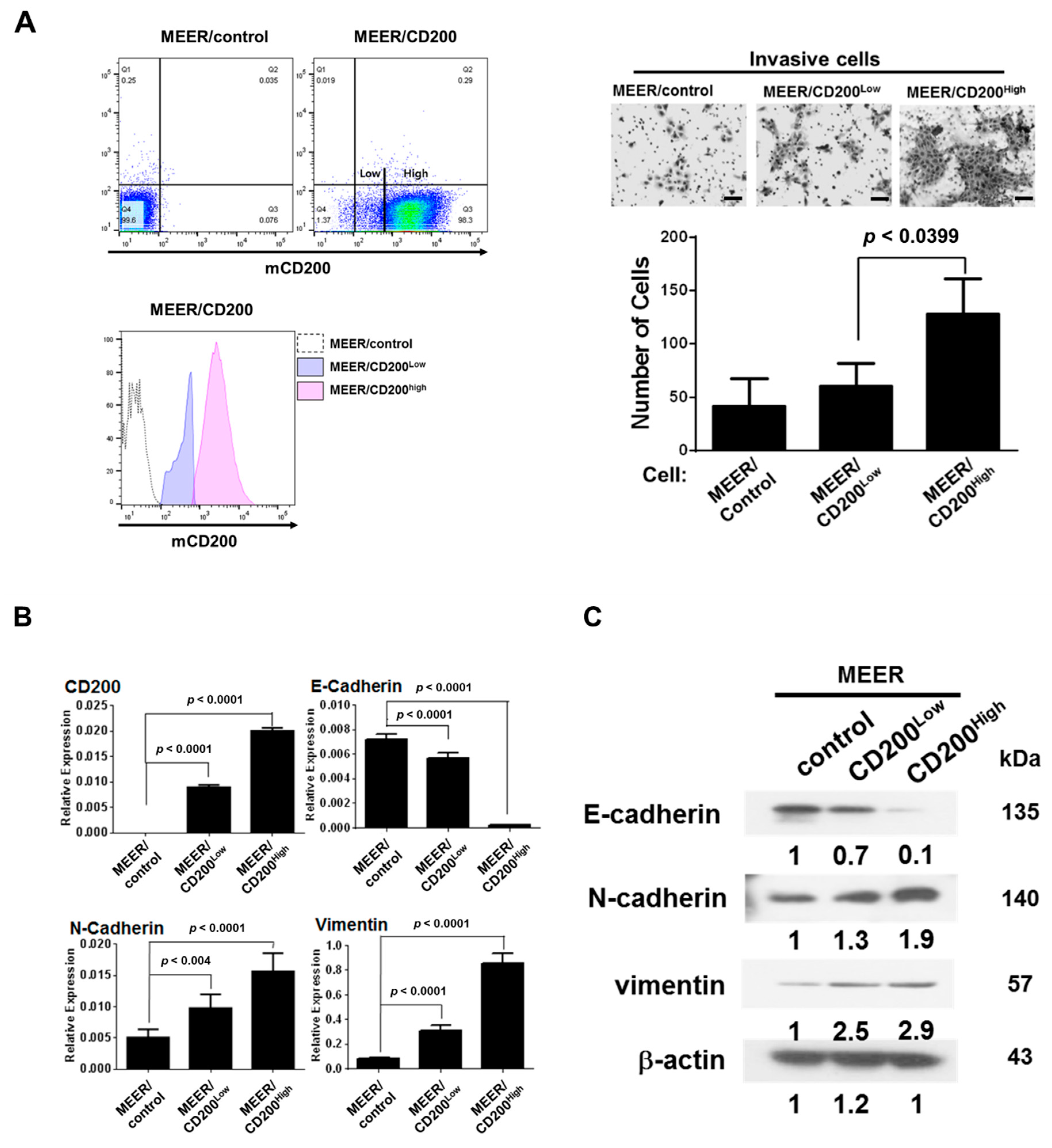
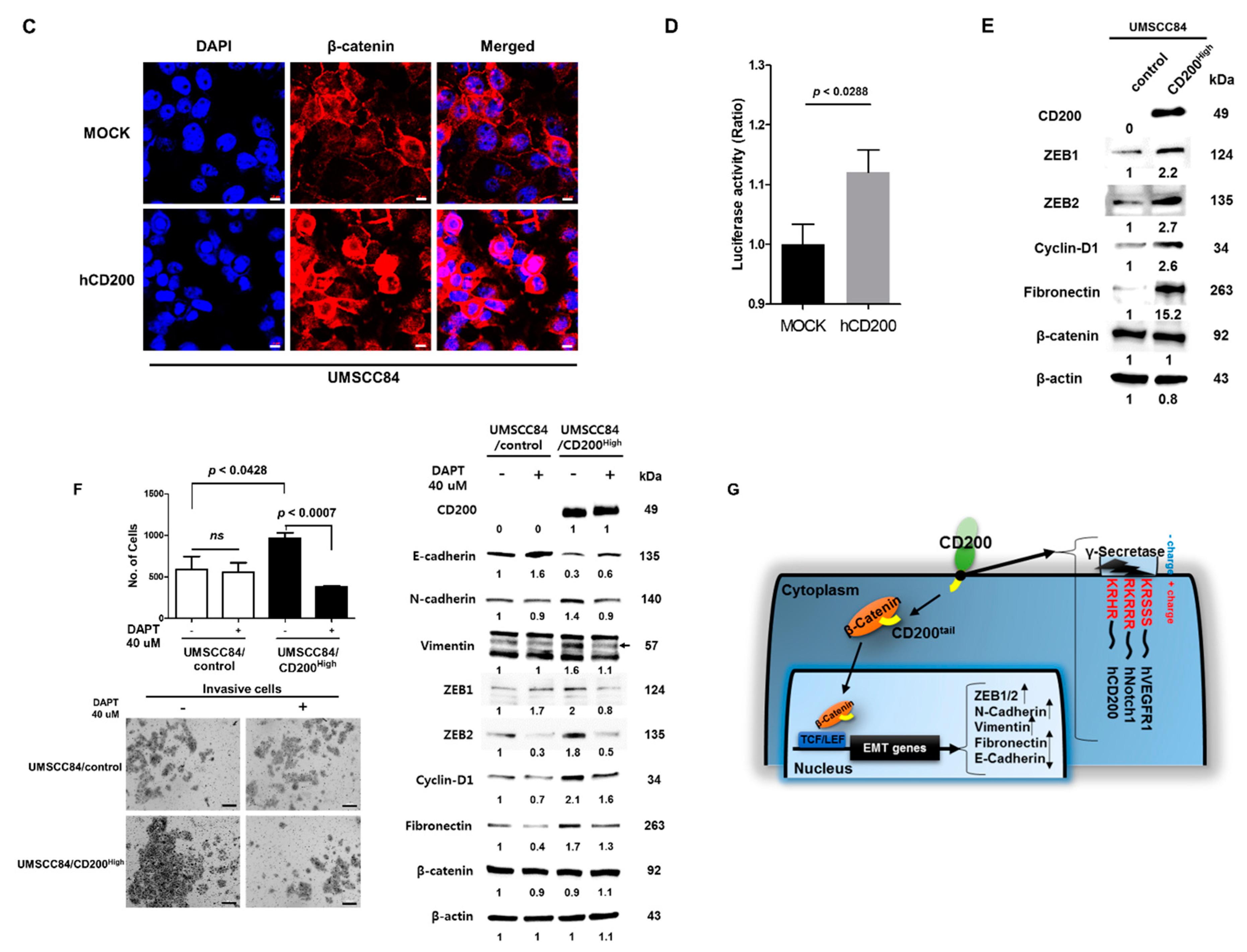
2.2. EMT after CD200 Overexpression in MEER Cells
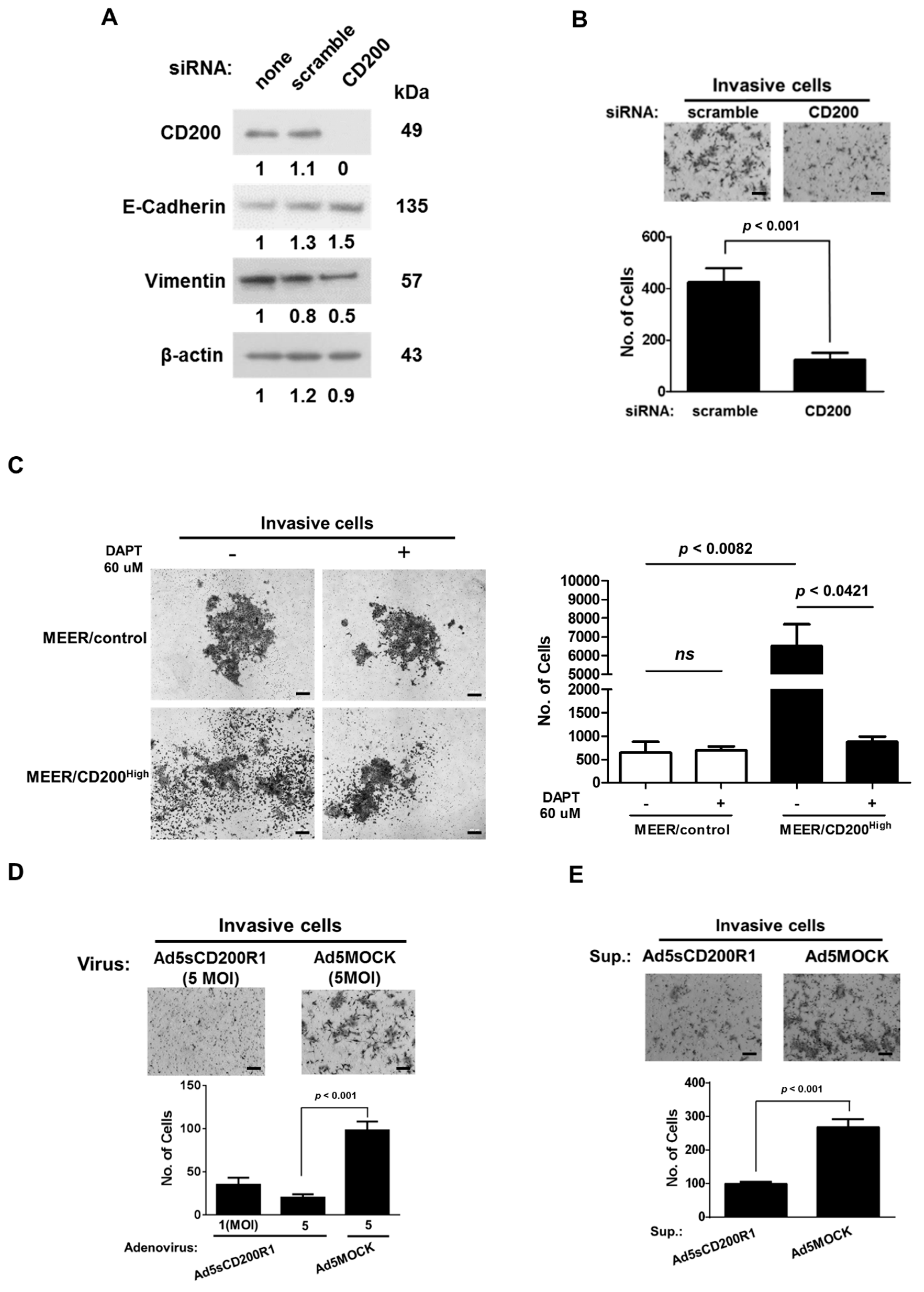
2.3. Changes in the EMT Features after CD200 Inhibition
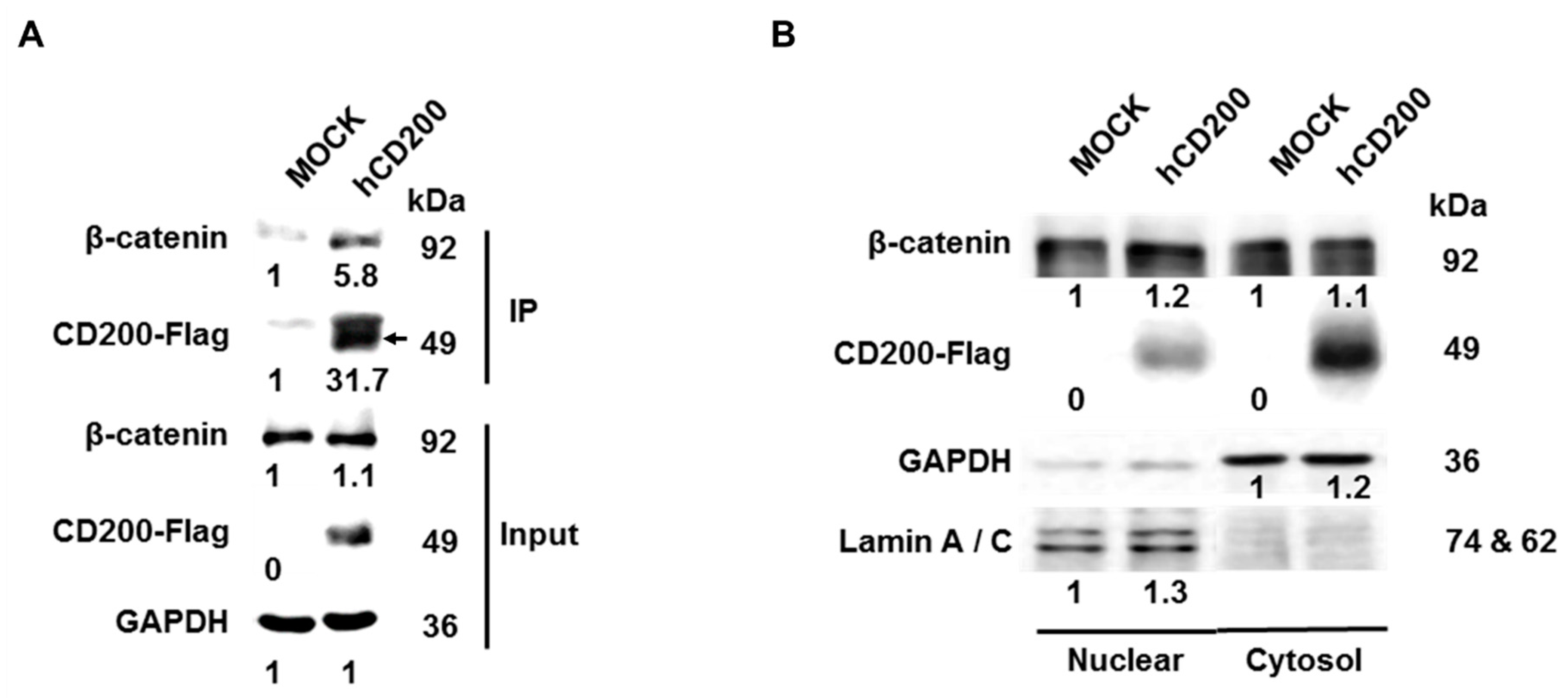
2.4. Interaction of Cleaved CD200 with β-catenin in the Cytoplasm
2.5. CD200-Induced Increase in Metastatic Ability in an Animal Model
3. Discussion
4. Materials and Methods
4.1. Cell Line and Culture
4.2. Analysis of The Cancer Genome Atlas (TCGA) Data
4.3. Western Blot Analysis and Flow Cytometry
4.4. Co-Immunoprecipitation (co-IP)
4.5. Adenovirus Construction
4.6. PCR and Quantitative Reverse Transcription PCR (qRT-PCR)
4.7. Transient Transfection
4.8. Immunocytochemistry (ICC) and Confocal Microscopy
4.9. Invasion Assay
4.10. Animal Experiment
4.11. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Shibuya, K.; Mathers, C.D.; Boschi-Pinto, C.; Lopez, A.D.; Murray, C.J. Global and regional estimates of cancer mortality and incidence by site: II. Results for the global burden of disease 2000. BMC cancer 2002, 2, 37. [Google Scholar] [CrossRef]
- Chaturvedi, A.; Engels, E.; Pfeiffer, R.; Hernandez, B.; Xiao, W.; Kim, E.; Jiang, B.; Goodman, M.; Sibug-Saber, M.; Cozen, W. Human papillomavirus (HPV) and rising oropharyngeal cancer incidence and survival in the United States. J. Clin. Oncol. 2011, 29, 5529. [Google Scholar] [CrossRef]
- Näsman, A.; Attner, P.; Hammarstedt, L.; Du, J.; Eriksson, M.; Giraud, G.; Ährlund-Richter, S.; Marklund, L.; Romanitan, M.; Lindquist, D. Incidence of human papillomavirus (HPV) positive tonsillar carcinoma in Stockholm, Sweden: An epidemic of viral-induced carcinoma? Int. J. Cancer 2009, 125, 362–366. [Google Scholar] [CrossRef] [PubMed]
- Shin, A.; Jung, Y.S.; Jung, K.W.; Kim, K.; Ryu, J.; Won, Y.J. Trends of human papillomavirus-related head and neck cancers in Korea: National cancer registry data. Laryngoscope 2013, 123, E30–E37. [Google Scholar] [CrossRef]
- Vigneswaran, N.; Williams, M.D. Epidemiologic trends in head and neck cancer and aids in diagnosis. Oral Maxillofac. Surg. Clin. N. Am. 2014, 26, 123–141. [Google Scholar] [CrossRef]
- Edge, S.B.; Compton, C.C. The American Joint Committee on Cancer: The 7th edition of the AJCC cancer staging manual and the future of TNM. Ann. Surg. Oncol. 2010, 17, 1471–1474. [Google Scholar] [CrossRef]
- Puram, S.V.; Tirosh, I.; Parikh, A.S.; Patel, A.P.; Yizhak, K.; Gillespie, S.; Rodman, C.; Luo, C.L.; Mroz, E.A.; Emerick, K.S.; et al. Single-Cell Transcriptomic Analysis of Primary and Metastatic Tumor Ecosystems in Head and Neck Cancer. Cell 2017, 171, 1611–1624.e24. [Google Scholar] [CrossRef] [Green Version]
- Clark, D.A.; Arredondo, J.L.; Dhesy-Thind, S. The CD200 tolerance-signaling molecule and its receptor, CD200R1, are expressed in human placental villus trophoblast and in peri-implant decidua by 5 weeks’ gestation. J. Reprod. Immunol. 2015, 112, 20–23. [Google Scholar] [CrossRef]
- Coles, S.J.; Gilmour, M.N.; Reid, R.; Knapper, S.; Burnett, A.; Man, S.; Tonks, A.; Darley, R.L. The immunosuppressive ligands PD-L1 and CD200 are linked in AML T-cell immunosuppression: Identification of a new immunotherapeutic synapse. Leukemia 2015, 29, 1952. [Google Scholar] [CrossRef]
- Aref, S.; Azmy, E.; El-Gilany, A.H. Upregulation of CD200 is associated with regulatory T cell expansion and disease progression in multiple myeloma. Hematol. Oncol. 2017, 35, 51–57. [Google Scholar] [CrossRef]
- Erin, N.; Podnos, A.; Tanriover, G.; Duymuş, Ö.; Cote, E.; Khatri, I.; Gorczynski, R. Bidirectional effect of CD200 on breast cancer development and metastasis, with ultimate outcome determined by tumor aggressiveness and a cancer-induced inflammatory response. Oncogene 2015, 34, 3860. [Google Scholar] [CrossRef] [PubMed]
- Vaine, C.A.; Soberman, R.J. The CD200–CD200R1 Inhibitory Signaling Pathway: Immune Regulation and Host–Pathogen Interactions. Adv. Immunol. 2014, 121, 191–211. [Google Scholar] [PubMed]
- Gorczynski, L.; Chen, Z.; Hu, J.; Kai, Y.; Lei, J.; Ramakrishna, V.; Gorczynski, R.M. Evidence That an OX-2-Positive Cell Can Inhibit the Stimulation of Type 1 Cytokine Production by Bone Marrow-Derived B7-1 (and B7-2)-Positive Dendritic Cells. J. Immunol. 1999, 162, 774–781. [Google Scholar] [PubMed]
- Burger, P.E.; Gupta, R.; Xiong, X.; Ontiveros, C.S.; Salm, S.N.; Moscatelli, D.; Wilson, E.L. High aldehyde dehydrogenase activity: A novel functional marker of murine prostate stem/progenitor cells. Stem Cells 2009, 27, 2220–2228. [Google Scholar] [CrossRef] [PubMed]
- Jung, Y.S.; Vermeer, P.D.; Vermeer, D.W.; Lee, S.J.; Goh, A.R.; Ahn, H.J.; Lee, J.H. CD200: Association with cancer stem cell features and response to chemoradiation in head and neck squamous cell carcinoma. Head Neck 2015, 37, 327–335. [Google Scholar] [CrossRef] [PubMed]
- Kawasaki, B.T.; Farrar, W.L. Cancer stem cells, CD200 and immunoevasion. Trends immunol. 2008, 29, 464–468. [Google Scholar] [CrossRef] [PubMed]
- Kawasaki, B.T.; Mistree, T.; Hurt, E.M.; Kalathur, M.; Farrar, W.L. Co-expression of the toleragenic glycoprotein, CD200, with markers for cancer stem cells. Biochem. Biophys. Res. Commun. 2007, 364, 778–782. [Google Scholar] [CrossRef] [Green Version]
- Colmont, C.S.; BenKetah, A.; Reed, S.H.; Hawk, N.V.; Telford, W.G.; Ohyama, M.; Udey, M.C.; Yee, C.L.; Vogel, J.C.; Patel, G.K. CD200-expressing human basal cell carcinoma cells initiate tumor growth. Proc. Natl. Acad. Sci. USA 2013, 110, 1434–1439. [Google Scholar] [CrossRef] [Green Version]
- Colmont, C.S.; Ketah, A.B.; Errington, R.J.; Reed, S.H.; Udey, M.C.; Patel, G.K. Human Basal Cell Carcinoma Tumor-Initiating Cells Are Resistant to Etoposide. J. Invest. Dermatol. 2014, 134, 867–870. [Google Scholar] [CrossRef] [Green Version]
- Chang, L.; Graham, P.; Hao, J.; Ni, J.; Bucci, J.; Cozzi, P.; Kearsley, J.; Li, Y. Acquisition of epithelial–mesenchymal transition and cancer stem cell phenotypes is associated with activation of the PI3K/Akt/mTOR pathway in prostate cancer radioresistance. Cell Death Dis. 2013, 4, e875. [Google Scholar] [CrossRef]
- Nantajit, D.; Lin, D.; Li, J.J. The network of epithelial–mesenchymal transition: Potential new targets for tumor resistance. J. Cancer Res. Clin. Oncol. 2015, 141, 1697–1713. [Google Scholar] [CrossRef] [PubMed]
- Adhikary, G.; Grun, D.; Kerr, C.; Balasubramanian, S.; Rorke, E.A.; Vemuri, M.; Boucher, S.; Bickenbach, J.R.; Hornyak, T.; Xu, W.; et al. Identification of a Population of Epidermal Squamous Cell Carcinoma Cells with Enhanced Potential for Tumor Formation. PLoS ONE 2013, 8, e84324. [Google Scholar] [CrossRef] [PubMed]
- Chung, J.H.; Jung, H.R.; Jung, A.R.; Lee, Y.C.; Kong, M.; Lee, J.-S.; Eun, Y.-G. SOX2 activation predicts prognosis in patients with head and neck squamous cell carcinoma. Sci. Rep. 2018, 8, 1677. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Kapus, A.; Khatri, I.; Kos, O.; Zhu, F.; Gorczynski, R.M. Cell membrane-bound CD200 signals both via an extracellular domain and following nuclear translocation of a cytoplasmic fragment. Leuk. Res. 2018, 69, 72–80. [Google Scholar] [CrossRef] [PubMed]
- Walker, D.G.; Lue, L.-F. Understanding the neurobiology of CD200 and the CD200 receptor: A therapeutic target for controlling inflammation in human brains? Future Neurol. 2013, 8, 321–332. [Google Scholar] [CrossRef] [PubMed]
- Airoldi, I.; Cocco, C.; Sorrentino, C.; Angelucci, D.; Di Meo, S.; Manzoli, L.; Esposito, S.; Ribatti, D.; Bertolotto, M.; Iezzi, L. Interleukin-30 promotes breast cancer growth and progression. Cancer Res. 2016, 76, 6218–6229. [Google Scholar] [CrossRef] [PubMed]
- Yuan, A.; Hsiao, Y.-J.; Chen, H.-Y.; Chen, H.-W.; Ho, C.-C.; Chen, Y.-Y.; Liu, Y.-C.; Hong, T.-H.; Yu, S.-L.; Chen, J.J. Opposite effects of M1 and M2 macrophage subtypes on lung cancer progression. Sci. Rep. 2015, 5, 14273. [Google Scholar] [CrossRef]
- Huber, M.A.; Kraut, N.; Beug, H. Molecular requirements for epithelial–mesenchymal transition during tumor progression. Curr. Opin. Cell Biol. 2005, 17, 548–558. [Google Scholar] [CrossRef]
- Xu, J.; Lamouille, S.; Derynck, R. TGF-β-induced epithelial to mesenchymal transition. Cell Res. 2009, 19, 156. [Google Scholar] [CrossRef]
- Gato-Cañas, M.; Zuazo, M.; Arasanz, H.; Ibañez-Vea, M.; Lorenzo, L.; Fernandez-Hinojal, G.; Vera, R.; Smerdou, C.; Martisova, E.; Arozarena, I. PDL1 signals through conserved sequence motifs to overcome interferon-mediated cytotoxicity. Cell Rep. 2017, 20, 1818–1829. [Google Scholar] [CrossRef]
- Kretz-Rommel, A.; Qin, F.; Dakappagari, N.; Cofiell, R.; Faas, S.J.; Bowdish, K.S. Blockade of CD200 in the presence or absence of antibody effector function: Implications for anti-CD200 therapy. J. Immunol. 2008, 180, 699–705. [Google Scholar] [CrossRef] [PubMed]
- Hoover, A.C.; Spanos, W.C.; Harris, G.F.; Anderson, M.E.; Klingelhutz, A.J.; Lee, J.H. The role of human papillomavirus 16 E6 in anchorage-independent and invasive growth of mouse tonsil epithelium. Arch. Otolaryngol. Head Neck Surg. 2007, 133, 495–502. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Aksoy, B.A.; Dogrusoz, U.; Dresdner, G.; Gross, B.; Sumer, S.O.; Sun, Y.; Jacobsen, A.; Sinha, R.; Larsson, E.; et al. Integrative Analysis of Complex Cancer Genomics and Clinical Profiles Using the cBioPortal. Sci. Signal. 2013, 6, pl1. [Google Scholar] [CrossRef] [PubMed]
- Cerami, E.; Gao, J.; Dogrusoz, U.; Gross, B.E.; Sumer, S.O.; Aksoy, B.A.; Jacobsen, A.; Byrne, C.J.; Heuer, M.L.; Larsson, E.; et al. The cBio Cancer Genomics Portal: An Open Platform for Exploring Multidimensional Cancer Genomics Data. Cancer Discov. 2012, 2, 401–404. [Google Scholar] [CrossRef] [PubMed]
- Shin, S.P.; Seo, H.-H.; Shin, J.H.; Park, H.B.; Lim, D.P.; Eom, H.S.; Bae, Y.S.; Kim, I.H.; Choi, K.; Lee, S.J. Adenovirus expressing both thymidine kinase and soluble PD1 enhances antitumor immunity by strengthening CD8 T-cell response. Mol. Ther. 2013, 21, 688–695. [Google Scholar] [CrossRef]

© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shin, S.-P.; Goh, A.R.; Kang, H.-G.; Kim, S.-J.; Kim, J.-K.; Kim, K.-T.; Lee, J.H.; Bae, Y.-S.; Jung, Y.-S.; Lee, S.-J. CD200 Induces Epithelial-to-Mesenchymal Transition in Head and Neck Squamous Cell Carcinoma via β-Catenin-Mediated Nuclear Translocation. Cancers 2019, 11, 1583. https://doi.org/10.3390/cancers11101583
Shin S-P, Goh AR, Kang H-G, Kim S-J, Kim J-K, Kim K-T, Lee JH, Bae Y-S, Jung Y-S, Lee S-J. CD200 Induces Epithelial-to-Mesenchymal Transition in Head and Neck Squamous Cell Carcinoma via β-Catenin-Mediated Nuclear Translocation. Cancers. 2019; 11(10):1583. https://doi.org/10.3390/cancers11101583
Chicago/Turabian StyleShin, Seung-Phil, Ah Ra Goh, Hyeon-Gu Kang, Seok-Jun Kim, Jong-Kwang Kim, Kyung-Tae Kim, John H Lee, Yong-Soo Bae, Yuh-Seog Jung, and Sang-Jin Lee. 2019. "CD200 Induces Epithelial-to-Mesenchymal Transition in Head and Neck Squamous Cell Carcinoma via β-Catenin-Mediated Nuclear Translocation" Cancers 11, no. 10: 1583. https://doi.org/10.3390/cancers11101583
APA StyleShin, S. -P., Goh, A. R., Kang, H. -G., Kim, S. -J., Kim, J. -K., Kim, K. -T., Lee, J. H., Bae, Y. -S., Jung, Y. -S., & Lee, S. -J. (2019). CD200 Induces Epithelial-to-Mesenchymal Transition in Head and Neck Squamous Cell Carcinoma via β-Catenin-Mediated Nuclear Translocation. Cancers, 11(10), 1583. https://doi.org/10.3390/cancers11101583





