Secondary Somatic Mutations in G-Protein-Related Pathways and Mutation Signatures in Uveal Melanoma
Abstract
:1. Introduction
2. Results
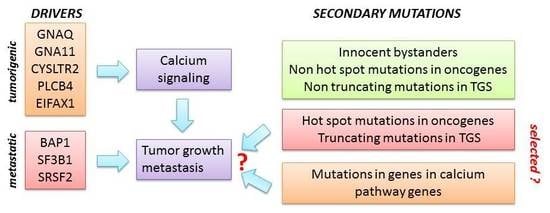
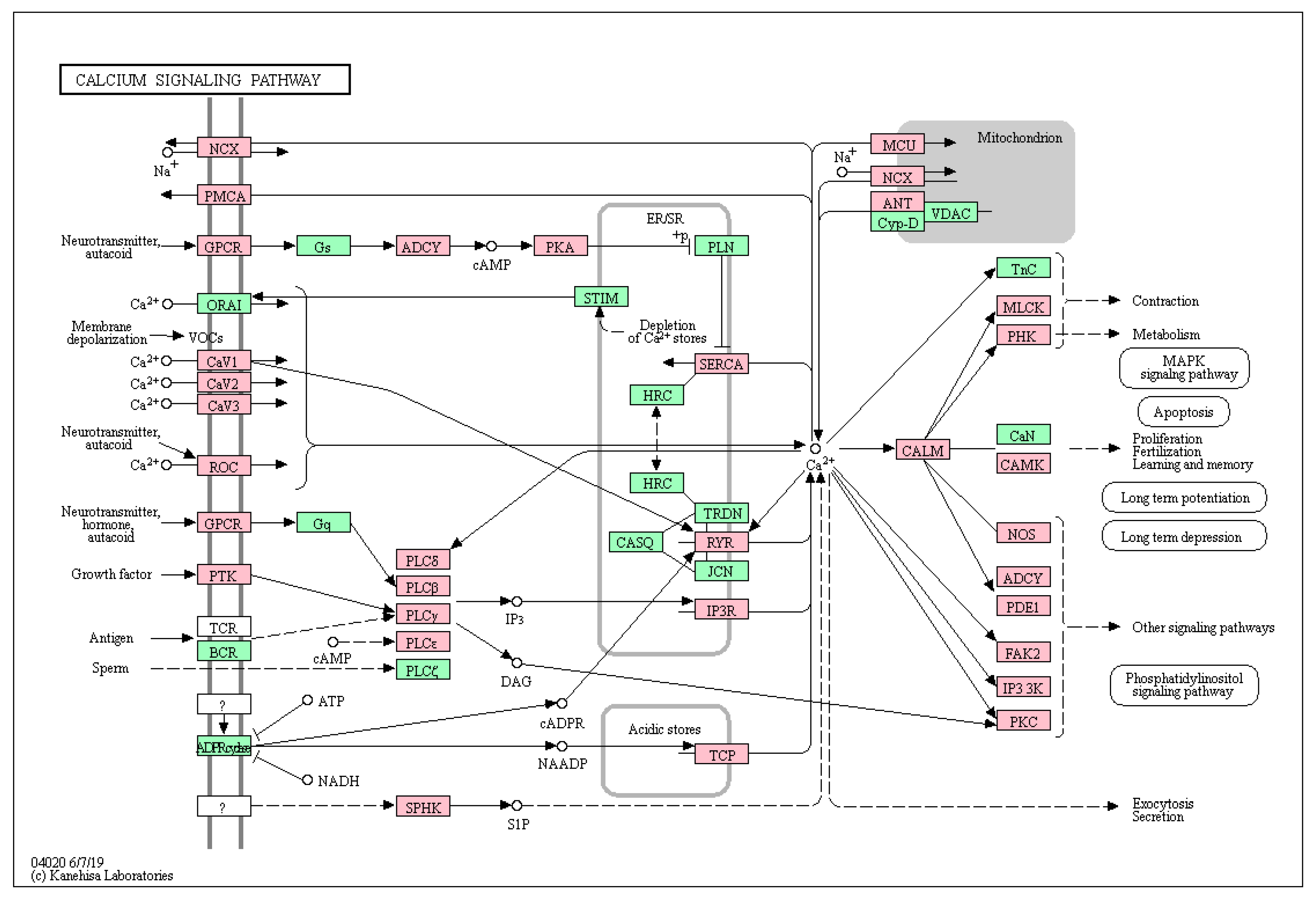
2.1. Enrichment of Secondary Mutations in G-Protein Related Pathways
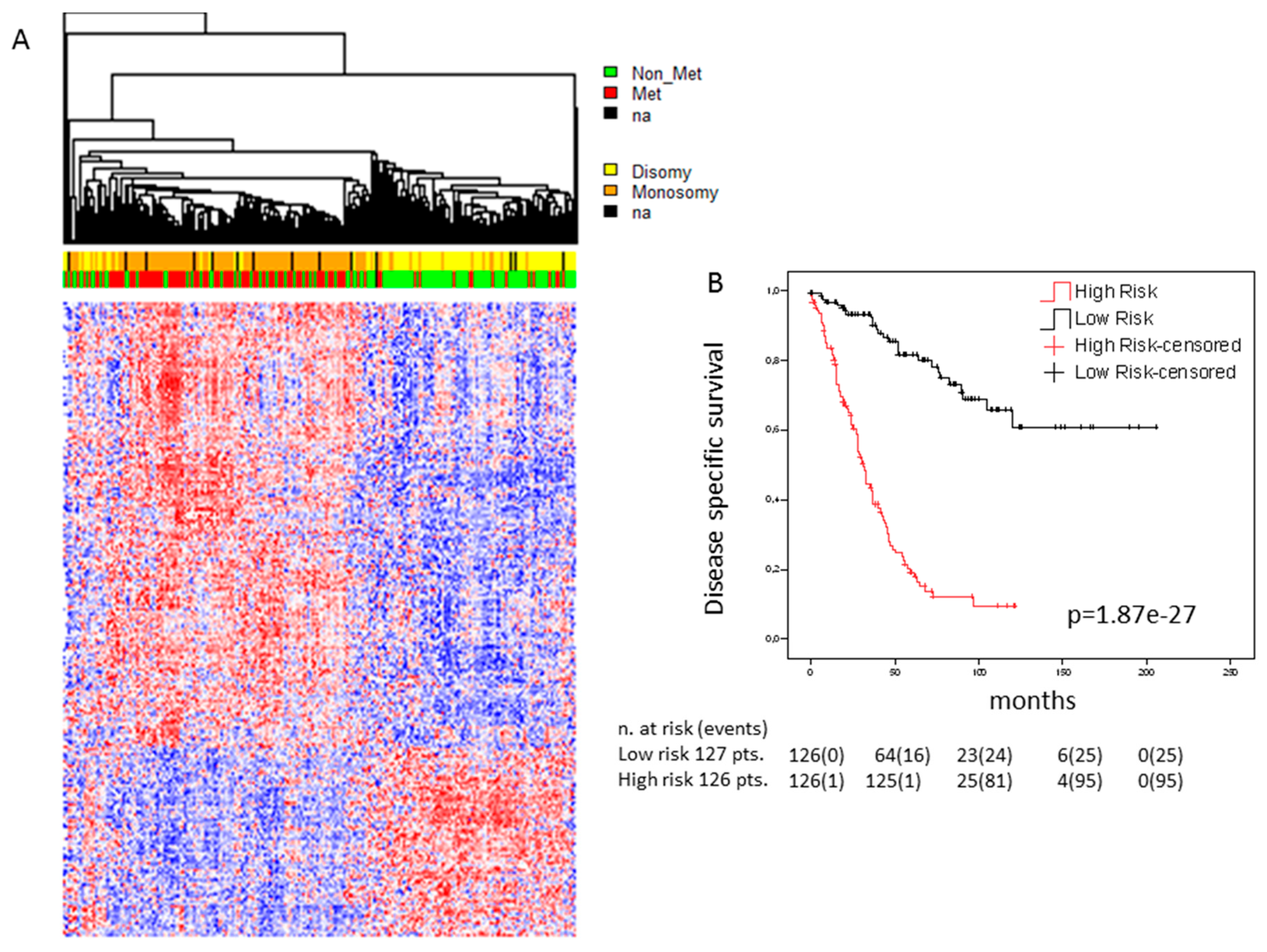
2.2. Expression Analysis
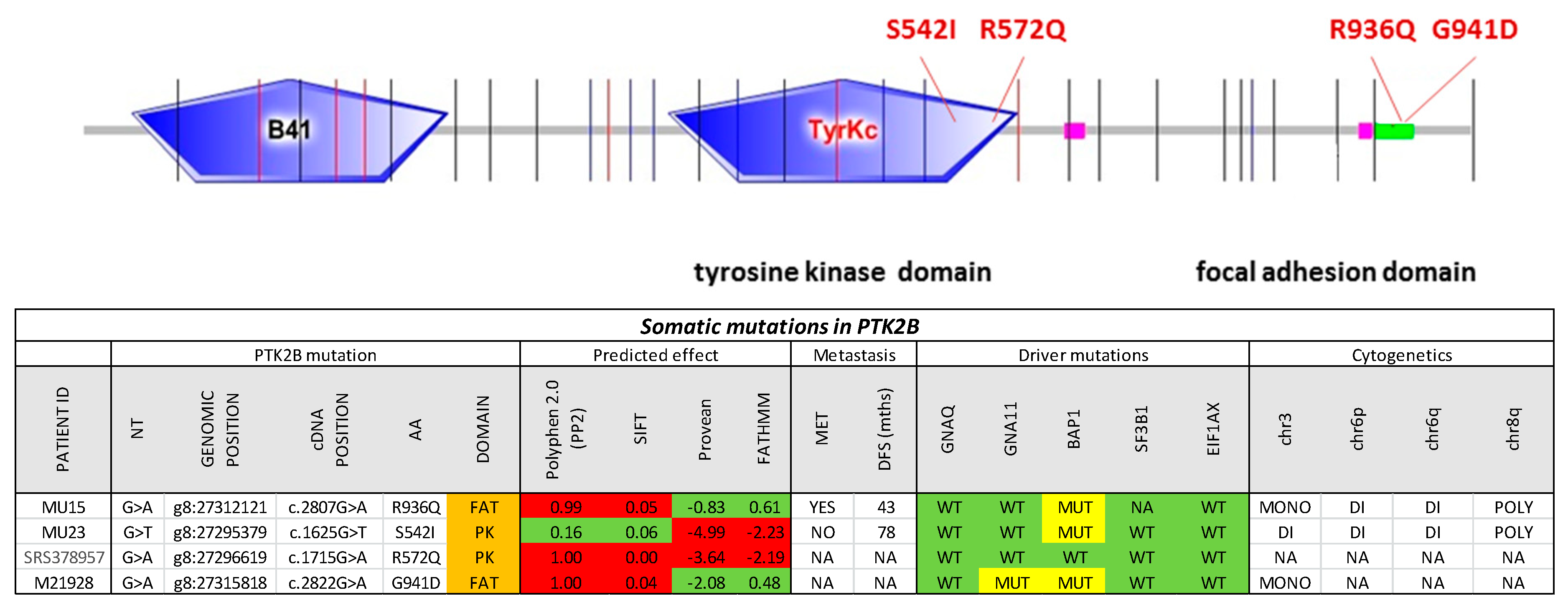
2.3. PTK2B as a Secondary Driver Mutation
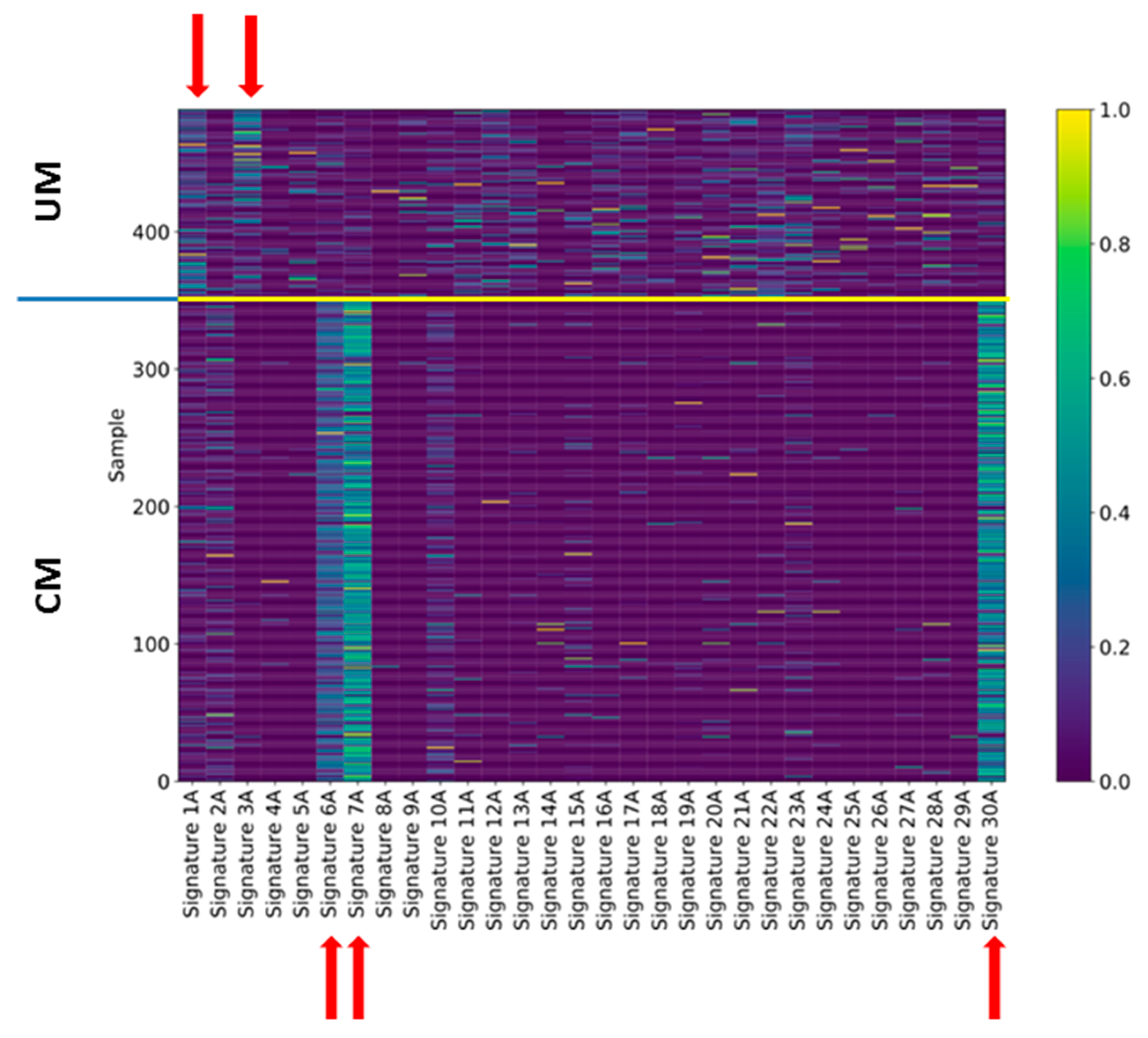
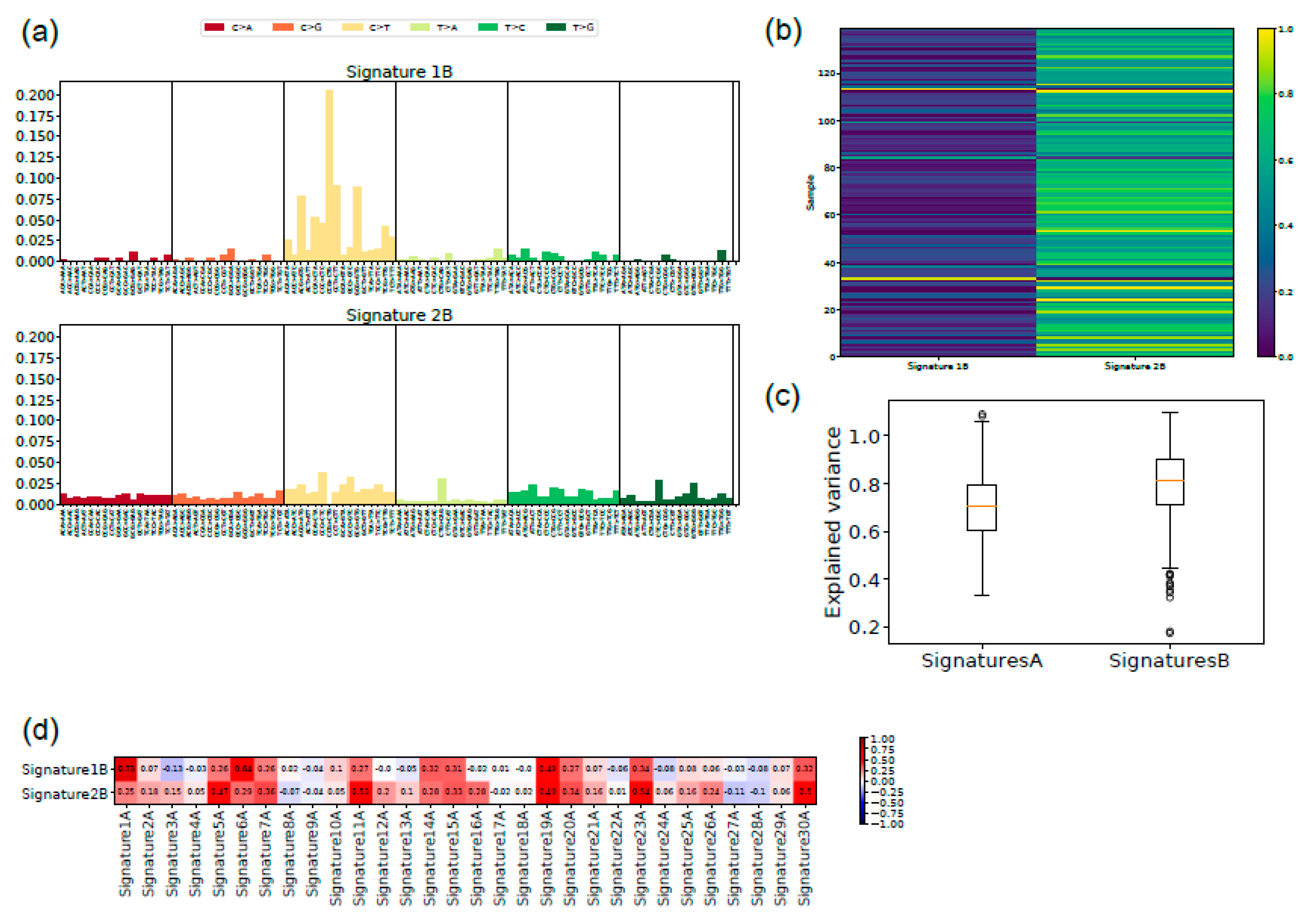
2.4. Signatures of Somatic Mutations in UM
3. Discussion
4. Materials and Methods
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Coupland, S.E.; Lake, S.L.; Zeschnigk, M.; Damato, B.E. Molecular pathology of uveal melanoma. Eye (Lond.) 2013, 27, 230–242. [Google Scholar] [CrossRef]
- Zeschnigk, M.; Lohmann, D.R. Prognostic testing in uveal melanoma. In Cancer Genomics: Molecular Classification, Prognosis and Response Prediction; Pfeffer, U., Ed.; Springer Science and Business Media: Dordrecht, The Netherlands, 2013; pp. 79–96. [Google Scholar]
- Amaro, A.; Gangemi, R.; Piaggio, F.; Angelini, G.; Barisione, G.; Ferrini, S.; Pfeffer, U. The biology of uveal melanoma. Cancer Metastasis Rev. 2017, 36, 109–140. [Google Scholar] [CrossRef]
- Zhang, T.; Dutton-Regester, K.; Brown, K.M.; Hayward, N.K. The genomic landscape of cutaneous melanoma. Pigment Cell Melanoma Res. 2016. [Google Scholar] [CrossRef] [PubMed]
- Van Raamsdonk, C.D.; Bezrookove, V.; Green, G.; Bauer, J.; Gaugler, L.; O’Brien, J.M.; Simpson, E.M.; Barsh, G.S.; Bastian, B.C. Frequent somatic mutations of GNAQ in uveal melanoma and blue naevi. Nature 2009, 457, 599–602. [Google Scholar] [CrossRef] [PubMed]
- Van Raamsdonk, C.D.; Griewank, K.G.; Crosby, M.B.; Garrido, M.C.; Vemula, S.; Wiesner, T.; Obenauf, A.C.; Wackernagel, W.; Green, G.; Bouvier, N.; et al. Mutations in GNA11 in uveal melanoma. N. Engl. J. Med. 2010, 363, 2191–2199. [Google Scholar] [CrossRef] [PubMed]
- Johansson, P.; Aoude, L.G.; Wadt, K.; Glasson, W.J.; Warrier, S.K.; Hewitt, A.W.; Kiilgaard, J.F.; Heegaard, S.; Isaacs, T.; Franchina, M.; et al. Deep sequencing of uveal melanoma identifies a recurrent mutation in PLCB4. Oncotarget 2016, 7, 4624–4631. [Google Scholar] [CrossRef]
- Harbour, J.W.; Onken, M.D.; Roberson, E.D.; Duan, S.; Cao, L.; Worley, L.A.; Council, M.L.; Matatall, K.A.; Helms, C.; Bowcock, A.M. Frequent mutation of BAP1 in metastasizing uveal melanomas. Science 2010, 330, 1410–1413. [Google Scholar] [CrossRef]
- Harbour, J.W.; Roberson, E.D.; Anbunathan, H.; Onken, M.D.; Worley, L.A.; Bowcock, A.M. Recurrent mutations at codon 625 of the splicing factor SF3B1 in uveal melanoma. Nat. Genet. 2013, 45, 133–135. [Google Scholar] [CrossRef]
- Field, M.G.; Durante, M.A.; Anbunathan, H.; Cai, L.Z.; Decatur, C.L.; Bowcock, A.M.; Kurtenbach, S.; Harbour, J.W. Punctuated evolution of canonical genomic aberrations in uveal melanoma. Nat. Commun. 2018, 9, 116. [Google Scholar] [CrossRef]
- Martin, M.; Masshofer, L.; Temming, P.; Rahmann, S.; Metz, C.; Bornfeld, N.; van de Nes, J.; Klein-Hitpass, L.; Hinnebusch, A.G.; Horsthemke, B.; et al. Exome sequencing identifies recurrent somatic mutations in EIF1AX and SF3B1 in uveal melanoma with disomy 3. Nat. Genet. 2013, 45, 933–936. [Google Scholar] [CrossRef] [Green Version]
- Dono, M.; Angelini, G.; Cecconi, M.; Amaro, A.; Esposito, A.I.; Mirisola, V.; Maric, I.; Lanza, F.; Nasciuti, F.; Viaggi, S.; et al. Mutation frequencies of GNAQ, GNA11, BAP1, SF3B1, EIF1AX and TERT in uveal melanoma: Detection of an activating mutation in the TERT gene promoter in a single case of uveal melanoma. Br. J. Cancer 2014, 110, 1058–1065. [Google Scholar] [CrossRef] [PubMed]
- Jager, M.J.; Brouwer, N.J.; Esmaeli, B. The Cancer Genome Atlas Project: An Integrated Molecular View of Uveal Melanoma. Ophthalmology 2018, 125, 1139–1142. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moore, A.R.; Ceraudo, E.; Sher, J.J.; Guan, Y.; Shoushtari, A.N.; Chang, M.T.; Zhang, J.Q.; Walczak, E.G.; Kazmi, M.A.; Taylor, B.S.; et al. Recurrent activating mutations of G-protein-coupled receptor CYSLTR2 in uveal melanoma. Nat. Genet. 2016, 48, 675–680. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Robertson, A.G.; Shih, J.; Yau, C.; Gibb, E.A.; Oba, J.; Mungall, K.L.; Hess, J.M.; Uzunangelov, V.; Walter, V.; Danilova, L.; et al. Integrative Analysis Identifies Four Molecular and Clinical Subsets in Uveal Melanoma. Cancer Cell 2017, 32, 204–220.e15. [Google Scholar] [CrossRef]
- Alexandrov, L.B.; Nik-Zainal, S.; Wedge, D.C.; Aparicio, S.A.; Behjati, S.; Biankin, A.V.; Bignell, G.R.; Bolli, N.; Borg, A.; Borresen-Dale, A.L.; et al. Signatures of mutational processes in human cancer. Nature 2013, 500, 415–421. [Google Scholar] [CrossRef] [Green Version]
- Alexandrov, L.B.; Nik-Zainal, S.; Wedge, D.C.; Campbell, P.J.; Stratton, M.R. Deciphering signatures of mutational processes operative in human cancer. Cell Rep. 2013, 3, 246–259. [Google Scholar] [CrossRef]
- Shah, C.P.; Weis, E.; Lajous, M.; Shields, J.A.; Shields, C.L. Intermittent and chronic ultraviolet light exposure and uveal melanoma: A meta-analysis. Ophthalmology 2005, 112, 1599–1607. [Google Scholar] [CrossRef]
- Dolin, P.J.; Foss, A.J.; Hungerford, J.L. Uveal melanoma: Is solar ultraviolet radiation a risk factor? Ophthalmic Epidemiol. 1994, 1, 27–30. [Google Scholar] [CrossRef]
- Reynaud, C.; Billaud, M. The theory of punctuated equilibrium, a breakthrough in understanding cancer. Med. Sci. M/S 2011, 27, 921–923. [Google Scholar] [CrossRef]
- Amaro, A.; Chiara, S.; Pfeffer, U. Molecular evolution of colorectal cancer: From multistep carcinogenesis to the big bang. Cancer Metastasis Rev. 2016, 35, 63–74. [Google Scholar] [CrossRef]
- Bakhoum, M.F.; Esmaeli, B. Molecular Characteristics of Uveal Melanoma: Insights from the Cancer Genome Atlas (TCGA) Project. Cancers 2019, 11, 1061. [Google Scholar] [CrossRef] [PubMed]
- Vader, M.J.C.; Madigan, M.C.; Versluis, M.; Suleiman, H.M.; Gezgin, G.; Gruis, N.A.; Out-Luiting, J.J.; Bergman, W.; Verdijk, R.M.; Jager, M.J.; et al. GNAQ and GNA11 mutations and downstream YAP activation in choroidal nevi. Br. J. Cancer 2017, 117, 884–887. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Greenman, C.; Stephens, P.; Smith, R.; Dalgliesh, G.L.; Hunter, C.; Bignell, G.; Davies, H.; Teague, J.; Butler, A.; Stevens, C.; et al. Patterns of somatic mutation in human cancer genomes. Nature 2007, 446, 153–158. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vogelstein, B.; Papadopoulos, N.; Velculescu, V.E.; Zhou, S.; Diaz, L.A., Jr.; Kinzler, K.W. Cancer genome landscapes. Science 2013, 339, 1546–1558. [Google Scholar] [CrossRef]
- Nieuwenhuis, B.; Assen-Bolt, A.J.V.; AWH Van Waarde-Verhagen, M.; Sijmons, R.H.; Hout, A.H.V.d.; Bauch, T.; Streffer, C.; Kampinga, H.H. BRCA1 and BRCA2 heterozygosity and repair of X-ray-induced DNA damage. Int. J. Radiat. Biol. 2002, 78, 285–295. [Google Scholar] [CrossRef]
- Cosmic–Catlogue of somatic Mutations in Cancer. Available online: https://cancer.sanger.ac.uk/cosmic (accessed on 9 October 2019).
- Luscan, A.; Just, P.A.; Briand, A.; Burin des Roziers, C.; Goussard, P.; Nitschke, P.; Vidaud, M.; Avril, M.F.; Terris, B.; Pasmant, E. Uveal melanoma hepatic metastases mutation spectrum analysis using targeted next-generation sequencing of 400 cancer genes. Br. J. Ophthalmol. 2015, 99, 437–439. [Google Scholar] [CrossRef]
- McCarthy, C.; Kalirai, H.; Lake, S.L.; Dodson, A.; Damato, B.E.; Coupland, S.E. Insights into genetic alterations of liver metastases from uveal melanoma. Pigment Cell Melanoma Res. 2015, 29, 60–67. [Google Scholar] [CrossRef]
- Drake, J.M.; Graham, N.A.; Stoyanova, T.; Sedghi, A.; Goldstein, A.S.; Cai, H.; Smith, D.A.; Zhang, H.; Komisopoulou, E.; Huang, J.; et al. Oncogene-specific activation of tyrosine kinase networks during prostate cancer progression. Proc. Natl. Acad. Sci. USA 2012. [Google Scholar] [CrossRef]
- Dutton-Regester, K.; Irwin, D.; Hunt, P.; Aoude, L.G.; Tembe, V.; Pupo, G.M.; Lanagan, C.; Carter, C.D.; O’Connor, L.; O’Rourke, M.; et al. A high-throughput panel for identifying clinically relevant mutation profiles in melanoma. Mol. Cancer Ther. 2012, 11, 888–897. [Google Scholar] [CrossRef]
- Prickett, T.D.; Agrawal, N.S.; Wei, X.; Yates, K.E.; Lin, J.C.; Wunderlich, J.R.; Cronin, J.C.; Cruz, P.; Rosenberg, S.A.; Samuels, Y. Analysis of the tyrosine kinome in melanoma reveals recurrent mutations in ERBB4. Nat. Genet. 2009, 41, 1127–1132. [Google Scholar] [CrossRef]
- Jones, A.M.; Ferguson, P.; Gardner, J.; Rooker, S.; Sutton, T.; Ahn, A.; Chatterjee, A.; Bickley, V.M.; Sarwar, M.; Emanuel, P.; et al. NRAS and EPHB6 mutation rates differ in metastatic melanomas of patients in the North Island versus South Island of New Zealand. Oncotarget 2016, 7, 41017–41030. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Adzhubei, I.; Jordan, D.M.; Sunyaev, S.R. Predicting Functional Effect of Human Missense Mutations Using PolyPhen-2. Curr. Protoc. Hum. Genet. 2013. [Google Scholar] [CrossRef] [PubMed]
- Vaser, R.; Adusumalli, S.; Leng, S.N.; Sikic, M.; Ng, P.C. SIFT missense predictions for genomes. Nat. Protoc. 2016, 11, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Choi, Y.; Sims, G.E.; Murphy, S.; Miller, J.R.; Chan, A.P. Predicting the Functional Effect of Amino Acid Substitutions and Indels. PLoS ONE 2012, 7, e46688. [Google Scholar] [CrossRef]
- Shihab, H.A.; Gough, J.; Cooper, D.N.; Day, I.N.M.; Gaunt, T.R. Predicting the Functional Consequences of Cancer-Associated Amino Acid Substitutions. Bioinformatics 2013, 29, 1504–1510. [Google Scholar] [CrossRef] [PubMed]
- Tozzo, V.; Barla, A. Cancer Mutational Signatures Identification with Sparse Dictionary Learning; Springer: Cham, Switzerland, 2017; pp. 32–41. [Google Scholar]
- McFarland, C.D.; Yaglom, J.A.; Wojtkowiak, J.W.; Scott, J.G.; Morse, D.L.; Sherman, M.Y.; Mirny, L.A. The Damaging Effect of Passenger Mutations on Cancer Progression. Cancer Res. 2017, 77, 4763–4772. [Google Scholar] [CrossRef] [Green Version]
- McFarland, C.D.; Korolev, K.S.; Kryukov, G.V.; Sunyaev, S.R.; Mirny, L.A. Impact of deleterious passenger mutations on cancer progression. Proc. Natl. Acad. Sci. USA 2013, 110, 2910–2915. [Google Scholar] [CrossRef] [Green Version]
- Pon, J.R.; Marra, M.A. Driver and passenger mutations in cancer. Annu. Rev. Pathol. 2015, 10, 25–50. [Google Scholar] [CrossRef]
- Bozic, I.; Antal, T.; Ohtsuki, H.; Carter, H.; Kim, D.; Chen, S.; Karchin, R.; Kinzler, K.W.; Vogelstein, B.; Nowak, M.A. Accumulation of driver and passenger mutations during tumor progression. Proc. Natl. Acad. Sci. USA 2010, 107, 18545–18550. [Google Scholar] [CrossRef] [Green Version]
- Knudson, A.G. Mutation and Cancer: Statistical Study of Retinoblastoma. Proc. Natl. Acad. Sci. USA 1971, 68, 820–823. [Google Scholar] [CrossRef] [Green Version]
- Moolgavkar, S.H.; Knudson, A.G. Mutation and Cancer: A Model for Human Carcinogenesis. J. Natl. Cancer Inst. 1981, 66, 1037–1052. [Google Scholar] [CrossRef] [PubMed]
- Vogelstein, B.; Kinzler, K.W. The multistep nature of cancer. Trends Genet. 1993, 9, 138–141. [Google Scholar] [CrossRef]
- Hanahan, D.; Weinberg, R.A. The hallmarks of cancer. Cell 2000, 100, 57–70. [Google Scholar] [CrossRef]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef]
- Sottoriva, A.; Kang, H.; Ma, Z.; Graham, T.A.; Salomon, M.P.; Zhao, J.; Marjoram, P.; Siegmund, K.; Press, M.F.; Shibata, D.; et al. A Big Bang model of human colorectal tumor growth. Nat. Genet. 2015, 47, 209–216. [Google Scholar] [CrossRef]
- Dankort, D.; Curley, D.P.; Cartlidge, R.A.; Nelson, B.; Karnezis, A.N.; Damsky, W.E., Jr.; You, M.J.; DePinho, R.A.; McMahon, M.; Bosenberg, M. Braf(V600E) cooperates with Pten loss to induce metastatic melanoma. Nat. Genet. 2009, 41, 544–552. [Google Scholar] [CrossRef]
- Ben-David, U.; Beroukhim, R.; Golub, T.R. Genomic evolution of cancer models: Perils and opportunities. Nat. Rev. Cancer 2019, 19, 97–109. [Google Scholar] [CrossRef]
- Durante, M.A.; Field, M.G.; Sanchez, M.I.; Covington, K.R.; Decatur, C.L.; Dubovy, S.R.; Harbour, J.W. Genomic evolution of uveal melanoma arising in ocular melanocytosis. Mol. Case Stud. 2019. [Google Scholar] [CrossRef]
- Feng, X.; Arang, N.; Rigiracciolo, D.C.; Lee, J.S.; Yeerna, H.; Wang, Z.; Lubrano, S.; Kishore, A.; Pachter, J.A.; Konig, G.M.; et al. A Platform of Synthetic Lethal Gene Interaction Networks Reveals That the GNAQ Uveal Melanoma Oncogene Controls the Hippo Pathway through FAK. Cancer Cell 2019. [Google Scholar] [CrossRef]
- Naylor, T.L.; Greshock, J.; Wang, Y.; Colligon, T.; Yu, Q.C.; Clemmer, V.; Zaks, T.Z.; Weber, B.L. High resolution genomic analysis of sporadic breast cancer using array-based comparative genomic hybridization. Breast Cancer Res. 2005, 7, R1186–R1198. [Google Scholar] [CrossRef]
- Sun, C.K.; Man, K.; Ng, K.T.; Ho, J.W.; Lim, Z.X.; Cheng, Q.; Lo, C.-M.; Poon, R.T.; Fan, S.-T. Proline-rich tyrosine kinase 2 (Pyk2) promotes proliferation and invasiveness of hepatocellular carcinoma cells through c-Src/ERK activation. Carcinogenesis 2008, 29, 2096–2105. [Google Scholar] [CrossRef] [Green Version]
- Tozzo, V.; D’Amerio, V.; Barla, A. Hey there’s DALILA: A DictionAry LearnIng LibrAry. Dagstuhl Res. Online Publ. Serv. 2017. [Google Scholar] [CrossRef]
- De Lange, M.J.; Razzaq, L.; Versluis, M.; Verlinde, S.; Dogrusoz, M.; Bohringer, S.; Marinkovic, M.; Luyten, G.P.; de Keizer, R.J.; de Gruijl, F.R.; et al. Distribution of GNAQ and GNA11 Mutation Signatures in Uveal Melanoma Points to a Light Dependent Mutation Mechanism. PLoS ONE 2015, 10, e0138002. [Google Scholar] [CrossRef]
- Balazs, E.A. Studies on the structure of the vitreous body. I. The absorption of ultraviolet light. Am. J. Ophthalmol. 1954, 38, 21–28. [Google Scholar] [CrossRef]
- Boru, G.; Cebulla, C.M.; Sample, K.M.; Massengill, J.B.; Davidorf, F.H.; Abdel-Rahman, M.H. Heterogeneity in Mitogen-Activated Protein Kinase (MAPK) Pathway Activation in Uveal Melanoma with Somatic GNAQ and GNA11 Mutations. Investig. Ophthalmol. Vis. Sci. 2019, 60, 2474–2480. [Google Scholar] [CrossRef]
- EnrichR. Available online: http://amp.pharm.mssm.edu/Enrichr/ (accessed on 10 September 2019).
- Amaro, A.; Parodi, F.; Diedrich, K.; Angelini, G.; Gotz, C.; Viaggi, S.; Maric, I.; Coviello, D.; Pistillo, M.P.; Morabito, A.; et al. Analysis of the Expression and Single-Nucleotide Variant Frequencies of the Butyrophilin-Like 2 Gene in Patients with Uveal Melanoma. JAMA Ophthalmol. 2016, 134, 1125–1133. [Google Scholar] [CrossRef]
- Tusher, V.G.; Tibshirani, R.; Chu, G. Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl. Acad. Sci. USA 2001, 98, 5116–5121. [Google Scholar] [CrossRef] [Green Version]
- Amaro, A.; Mirisola, V.; Angelini, G.; Musso, A.; Tosetti, F.; Esposito, A.I.; Perri, P.; Lanza, F.; Nasciuti, F.; Mosci, C.; et al. Evidence of epidermal growth factor receptor expression in uveal melanoma: Inhibition of epidermal growth factor-mediated signalling by Gefitinib and Cetuximab triggered antibody-dependent cellular cytotoxicity. Eur. J. Cancer 2013, 49, 3353–3365. [Google Scholar] [CrossRef]
- Herlihy, N.; Dogrusoz, M.; van Essen, T.H.; Harbour, J.W.; van der Velden, P.A.; van Eggermond, M.C.; Haasnoot, G.W.; van den Elsen, P.J.; Jager, M.J. Skewed expression of the genes encoding epigenetic modifiers in high-risk uveal melanoma. Investig. Ophthalmol. Vis. Sci. 2015, 56, 1447–1458. [Google Scholar] [CrossRef]
- Laurent, C.; Valet, F.; Planque, N.; Silveri, L.; Maacha, S.; Anezo, O.; Hupe, P.; Plancher, C.; Reyes, C.; Albaud, B.; et al. High PTP4A3 phosphatase expression correlates with metastatic risk in uveal melanoma patients. Cancer Res. 2011, 71, 666–674. [Google Scholar] [CrossRef]
- Langfelder, P.; Horvath, S. WGCNA: An R package for weighted correlation network analysis. BMC Bioinform. 2008, 9, 559. [Google Scholar] [CrossRef]
- Johnson, W.E.; Li, C.; Rabinovic, A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics 2007, 8, 118–127. [Google Scholar] [CrossRef]
- Taminau, J.; Meganck, S.; Lazar, C.; Steenhoff, D.; Coletta, A.; Molter, C.; Duque, R.; de Schaetzen, V.; Weiss Solis, D.Y.; Bersini, H.; et al. Unlocking the potential of publicly available microarray data using inSilicoDb and inSilicoMerging R/Bioconductor packages. BMC Bioinform. 2012, 13, 335. [Google Scholar] [CrossRef]
- Nannya, Y.; Sanada, M.; Nakazaki, K.; Hosoya, N.; Wang, L.; Hangaishi, A.; Kurokawa, M.; Chiba, S.; Bailey, D.K.; Kennedy, G.C.; et al. A robust algorithm for copy number detection using high-density oligonucleotide single nucleotide polymorphism genotyping arrays. Cancer Res. 2005, 65, 6071–6079. [Google Scholar] [CrossRef]
- Xie, X.-J.; Whitehurst, A.; White, M. A practical efficient approach in high throughput screening: Using FDR and fold change. Nat. Protoc. 2007. [Google Scholar] [CrossRef]
- Gangemi, R.; Amaro, A.; Gino, A.; Barisione, G.; Fabbi, M.; Pfeffer, U.; Brizzolara, A.; Queirolo, P.; Salvi, S.; Boccardo, S.; et al. ADAM10 correlates with uveal melanoma metastasis and promotes In Vitro invasion. Pigment Cell Melanoma Res. 2014, 27, 1138–1148. [Google Scholar] [CrossRef]
- Krüger, S.; Piro, R. Identification of mutational signatures active in individual tumors. PeerJ 2017, 5. [Google Scholar] [CrossRef]
- Qiao, H. New SVD based initialization strategy for non-negative matrix factorization. Pattern Recognit. Lett. 2015, 63, 71–77. [Google Scholar] [CrossRef] [Green Version]
| KEGG Pathway | # of Query Genes | # Total Annotated | p-Value | Adjusted p-Value | Genes |
|---|---|---|---|---|---|
| Aldosterone synthesis and secretion * | 41 | 98 | 1.38 × 10−6 | 3.79 × 10−4 | SCARB1, CAMK2D, ITPR1, ADCY4, ITPR2, ATP1A3, ADCY3, ATP1A2, CACNA1D, ITPR3, CACNA1C, CALML4, CACNA1F, CACNA1H, LIPE, CACNA1I, MC2R, PRKACG, CACNA1S, CAMK2G, PRKCG, KCNJ5, CAMK1D, HSD3B2, PRKCB, HSD3B1, ATP1B4, ATP2B2, PRKCA, AGT, NR4A2, POMC, PLCB3, AGTR1, PRKD2, CAMK1, PRKD1, PLCB1, DAGLB, PLCB2, ATF4 |
| Calcium signaling pathway * | 66 | 188 | 2.48 × 10−6 | 3.79 × 10−4 | RYR1, RYR2, CHRM1, ATP2A3, ATP2A1, CALML4, RYR3, MYLK3, SLC8A1, MYLK, HTR6, GRM5, CCKAR, EDNRB, PRKACG, BDKRB1, PLCE1, NOS1, PDGFRB, PRKCG, PDGFRA, CAMK1D, PRKCB, SPHK1, PRKCA, ITPKB, PLCB3, ADORA2A, LTB4R2, ADORA2B, AGTR1, PLCB1, SLC25A5, PLCB2, CAMK2D, PDE1C, CACNA1B, ITPR1, ADCY4, ITPR2, ADCY3, CACNA1D, ITPR3, CACNA1C, CACNA1F, CACNA1E, EGFR, CACNA1H, CACNA1I, GRIN2A, ERBB4, PTK2B, CACNA1S, PLCG1, CAMK2G, NTSR1, NOS2, NOS3, ATP2B2, PHKB, TPCN2, TPCN1, GRIN2D, CAMK1, PLCD4, MCU |
| Thyroid hormone signaling pathway | 45 | 116 | 5.08 × 10−6 | 5.17 × 10−4 | NOTCH2, NOTCH3, NOTCH1, HDAC1, ITGB3, SLC2A1, ATP1A3, ATP1A2, MED16, HIF1A, ACTB, SLC9A1, MED17, CASP9, MED12, MED14, MED13, MYC, PRKACG, EP300, PLCE1, PLCG1, SLC16A2, RXRG, PRKCG, NCOA2, CREBBP, PRKCB, ATP1B4, TSC2, SLC16A10, PRKCA, ESR1, MTOR, MED13L, BMP4, KAT2A, PLCB3, PIK3CA, CTNNB1, PLCB1, PLCD4, PLCB2, PFKP, MYH6 |
| Ras-proximate-1 (RAP1) signaling pathway * | 67 | 206 | 3.69 × 10−5 | 2.87 × 10−3 | FLT4, ITGB3, CALML4, SIPA1L3, ACTB, IGF1R, FGF9, KDR, RAC2, RAC3, PLCE1, MAP2K3, MAGI1, PDGFRB, PRKCG, PDGFRA, PRKCB, ARAP3, PRKCA, NGF, VAV2, TIAM1, PLCB3, MRAS, ADORA2A, PIK3CA, ADORA2B, PARD3, KIT, RAPGEF2, PFN4, PRKD2, TLN2, PRKD1, PLCB1, PLCB2, MET, RAPGEF6, RGS14, GNAI3, ADCY4, FPR1, ADCY3, LPAR3, RASGRP2, THBS1, EGFR, GRIN2A, KRIT1, PLCG1, FGF23, NGFR, ANGPT4, EGF, VEGFB, GRIN2B, FGF17, EFNA2, FGF19, CTNNB1, TEK, FGFR4, FGFR2, LAT, SIPA1, FGFR1, FGF10 |
| Protein digestion and absorption | 35 | 90 | 5.15 × 10−5 | 3.14 × 10−3 | COL17A1, COL18A1, PRSS1, CPB2, COL14A1, COL11A2, SLC1A1, ATP1A3, ATP1A2, SLC8A1, PRSS3, SLC36A1, CPA2, SLC6A19, SLC15A1, COL27A1, KCNJ13, COL22A1, ATP1B4, SLC16A10, COL1A1, SLC9A3, COL3A1, SLC7A7, COL2A1, COL5A1, COL4A2, COL4A1, XPNPEP2, MEP1A, COL5A3, COL5A2, COL4A3, COL9A1, COL9A2 |
| Cortisol synthesis and secretion | 26 | 65 | 2.69 × 10−4 | 9.92 × 10−3 | SCARB1, ITPR1, ADCY4, ITPR2, ADCY3, CACNA1D, ITPR3, CACNA1C, CACNA1F, CACNA1H, CYP17A1, CACNA1I, MC2R, PRKACG, CACNA1S, PDE8B, PDE8A, HSD3B2, HSD3B1, AGT, POMC, PLCB3, AGTR1, PLCB1, PLCB2, ATF4 |
| ATP-binding cassette ABC transporters | 20 | 45 | 2.59 × 10−4 | 9.92 × 10−3 | ABCA1, ABCA2, ABCC4, ABCD2, ABCC2, ABCB1, ABCB4, ABCC5, ABCA3, ABCA4, ABCC6, ABCA9, TAP2, ABCA7, ABCA12, ABCB11, ABCA13, ABCA10, ABCB10, ABCD1 |
| Circadian entrainment * | 35 | 97 | 2.93 × 10−4 | 9.92 × 10−3 | RYR1, GRIA1, RYR2, CAMK2D, GNAI3, ITPR1, ADCY4, ADCY3, CACNA1D, ITPR3, CACNA1C, CALML4, RYR3, CACNA1H, CACNA1I, GRIN2A, RASD1, PRKACG, NOS1, CAMK2G, PRKG1, PRKCG, KCNJ5, PRKCB, PRKCA, GRIN2B, GRIN2D, PER1, PLCB3, GNB2, NOS1AP, GNB1, GNB3, PLCB1, PLCB2 |
| Cushing syndrome | 51 | 155 | 2.15 × 10−4 | 9.92 × 10−3 | RB1, SCARB1, PRKACG, PDE8B, PDE8A, MEN1, USP8, AXIN1, AIP, PLCB3, AGTR1, PLCB1, PLCB2, ATF4, KMT2D, CAMK2D, KMT2A, TCF7, GNAI3, ITPR1, ADCY4, ITPR2, ADCY3, CACNA1D, ITPR3, CACNA1C, CACNA1F, EGFR, CACNA1H, CYP17A1, CACNA1I, PDE11A, MC2R, RASD1, DVL2, DVL3, CACNA1S, WNT2, CAMK2G, TCF7L2, WNT10A, TCF7L1, FZD2, HSD3B2, FZD5, WNT3A, HSD3B1, AGT, POMC, APC, CTNNB1 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Piaggio, F.; Tozzo, V.; Bernardi, C.; Croce, M.; Puzone, R.; Viaggi, S.; Patrone, S.; Barla, A.; Coviello, D.; J. Jager, M.; et al. Secondary Somatic Mutations in G-Protein-Related Pathways and Mutation Signatures in Uveal Melanoma. Cancers 2019, 11, 1688. https://doi.org/10.3390/cancers11111688
Piaggio F, Tozzo V, Bernardi C, Croce M, Puzone R, Viaggi S, Patrone S, Barla A, Coviello D, J. Jager M, et al. Secondary Somatic Mutations in G-Protein-Related Pathways and Mutation Signatures in Uveal Melanoma. Cancers. 2019; 11(11):1688. https://doi.org/10.3390/cancers11111688
Chicago/Turabian StylePiaggio, Francesca, Veronica Tozzo, Cinzia Bernardi, Michela Croce, Roberto Puzone, Silvia Viaggi, Serena Patrone, Annalisa Barla, Domenico Coviello, Martine J. Jager, and et al. 2019. "Secondary Somatic Mutations in G-Protein-Related Pathways and Mutation Signatures in Uveal Melanoma" Cancers 11, no. 11: 1688. https://doi.org/10.3390/cancers11111688
APA StylePiaggio, F., Tozzo, V., Bernardi, C., Croce, M., Puzone, R., Viaggi, S., Patrone, S., Barla, A., Coviello, D., J. Jager, M., van der Velden, P. A., Zeschnigk, M., Cangelosi, D., Eva, A., Pfeffer, U., & Amaro, A. (2019). Secondary Somatic Mutations in G-Protein-Related Pathways and Mutation Signatures in Uveal Melanoma. Cancers, 11(11), 1688. https://doi.org/10.3390/cancers11111688