GFP-Fragment Reassembly Screens for the Functional Characterization of Variants of Uncertain Significance in Protein Interaction Domains of the BRCA1 and BRCA2 Genes
Abstract
:1. Introduction
2. Results
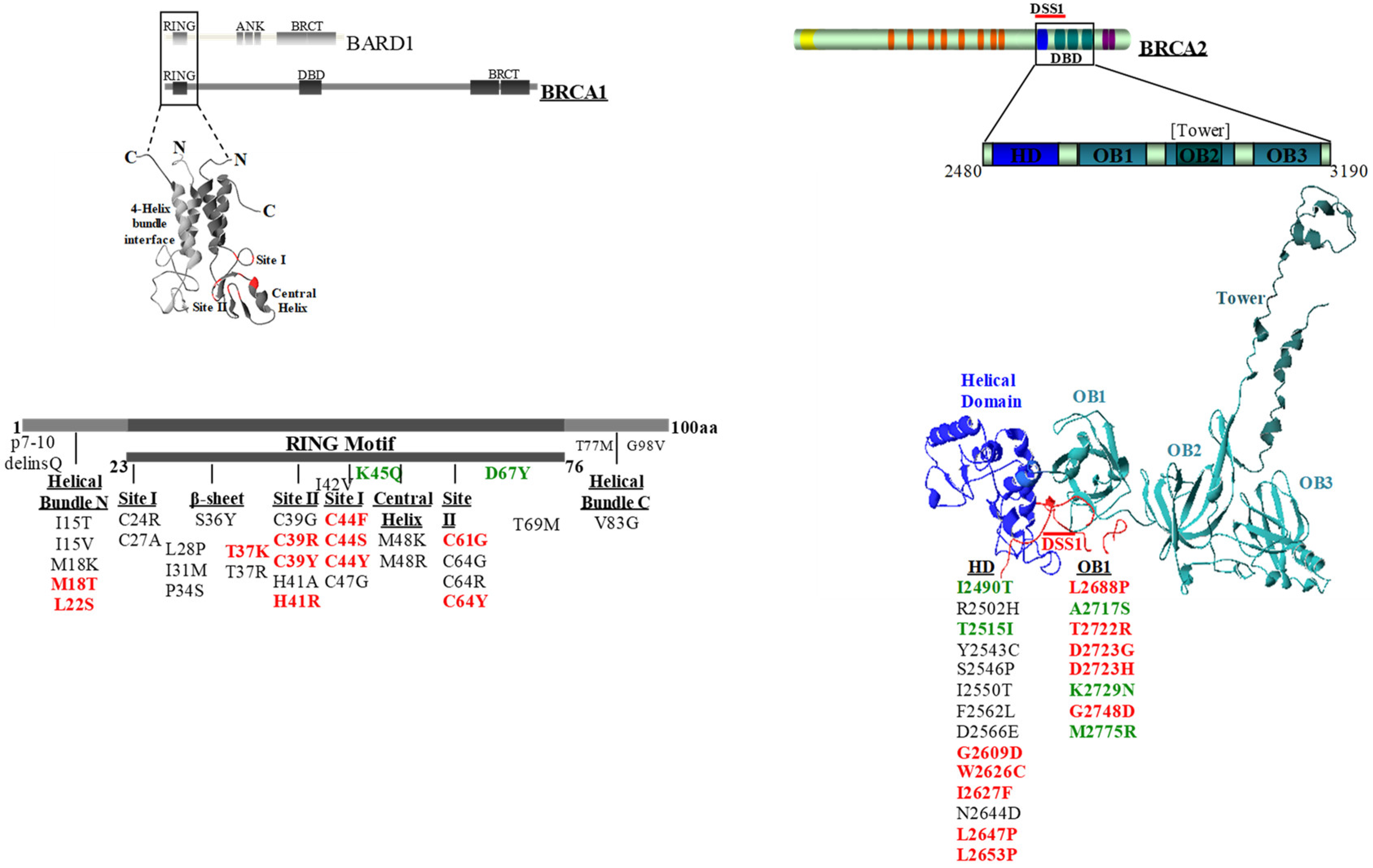
2.1. Selection of the BRCA1 and BRCA2 Variants
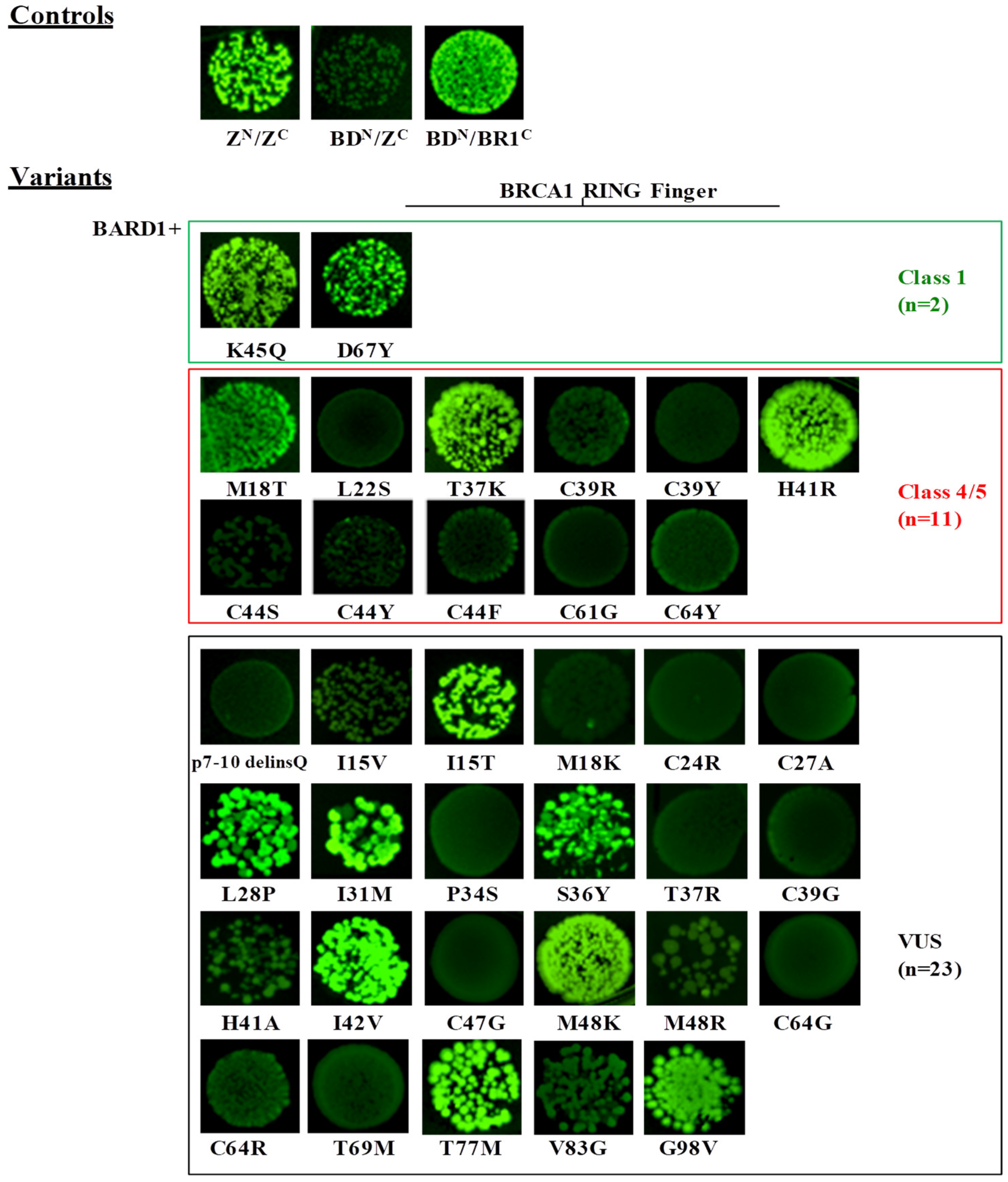
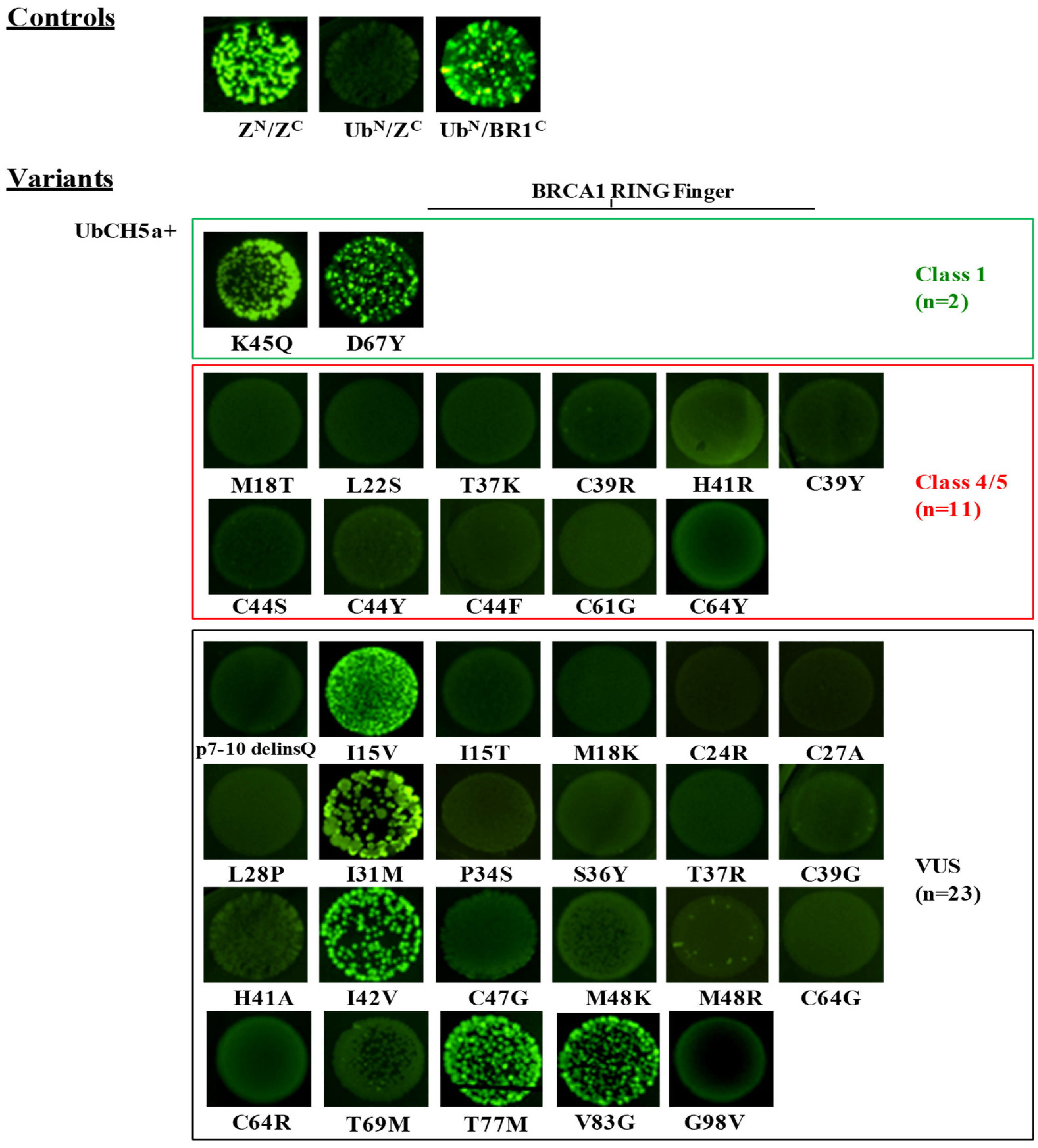
2.2. Evaluation of the Effect of BRCA1 Variants on the Interaction with the BARD1 and the UbcH5a Proteins by the GFP-Reassembly Assay
2.3. Evaluation of the Effect of the BRCA2 Variants on the Interaction with the DSS1 Protein by the GFP-Reassembly Assay
2.4. Validation of the GFP-Reassembly Screenings
2.5. Evaluation of the Effect of VUS on mRNA Splicing
3. Discussion
4. Materials and Methods
4.1. Variant Selection
4.2. Plasmid Construction
4.3. Expression of Recombinant Peptides
4.4. GFP-Fragment Reassembly Screening
4.5. Purification of the Reassembled GFP Complexes
4.6. Protein Structures
4.7. In Silico Analyses
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Mavaddat, N.; Peock, S.; Frost, D.; Ellis, S.; Platte, R.; Fineberg, E.; Evans, D.G.; Izatt, L.; Eeles, R.A.; Adlard, J.; et al. Cancer risks for BRCA1 and BRCA2 mutation carriers: Results from prospective analysis of EMBRACE. J. Natl. Cancer Inst. 2013, 105, 812–822. [Google Scholar] [CrossRef]
- Rebbeck, T.R.; Friebel, T.M.; Friedman, E.; Hamann, U.; Huo, D.; Kwong, A.; Olah, E.; Olopade, O.I.; Solano, A.R.; et al. Mutational spectrum in a worldwide study of 29,700 families with BRCA1 or BRCA2 mutations. Hum. Mutat. 2018, 39, 593–620. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Plon, S.E.; Eccles, D.M.; Easton, D.; Foulkes, W.D.; Genuardi, M.; Greenblatt, M.S.; Hogervorst, F.B.; Hoogerbrugge, N.; Spurdle, A.B.; Tavtigian, S.V.; et al. Sequence variant classification and reporting: Recommendations for improving the interpretation of cancer susceptibility genetic test results. Hum. Mutat. 2008, 29, 1282–1291. [Google Scholar] [CrossRef] [PubMed]
- Tavtigian, S.V.; Byrnes, G.B.; Goldgar, D.E.; Thomas, A. Classification of rare missense substitutions, using risk surfaces, with genetic- and molecular-epidemiology applications. Hum. Mutat. 2008, 29, 1342–1354. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goldgar, D.E.; Easton, D.F.; Deffenbaugh, A.M.; Monteiro, A.N.; Tavtigian, S.V.; Couch, F.J.; Breast Cancer Information Core (BIC) Steering Committee. Integrated Evaluation of DNA Sequence Variants of Unknown Clinical Significance: Application to BRCA1 and BRCA2. Am. J. Hum. Genet. 2004, 75, 535–544. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Easton, D.F.; Deffenbaugh, A.M.; Pruss, D.; Frye, C.; Wenstrup, R.J.; Allen-Brady, K.; Tavtigian, S.V.; Monteiro, A.N.; Iversen, E.S.; Couch, F.J.; et al. A systematic genetic assessment of 1433 sequence variants of unknown clinical significance in the BRCA1 and BRCA2 breast cancer-predisposition genes. Am. J. Hum. Genet. 2007, 81, 873–883. [Google Scholar] [CrossRef]
- Spurdle, A.B. Clinical relevance of rare germline sequence variants in cancer genes: Evolution and application of classification models. Curr. Opin. Genet. 2010, 20, 315–323. [Google Scholar] [CrossRef]
- Lindor, N.M.; Guidugli, L.; Wang, X.; Vallee, M.P.; Monteiro, A.N.; Tavtigian, S.; Goldgar, D.E.; Couch, F.J. A review of a multifactorial probability-based model for classification of BRCA1 and BRCA2 variants of uncertain significance (VUS). Hum. Mutat. 2012, 33, 8–21. [Google Scholar] [CrossRef]
- Millot, G.A.; Carvalho, M.A.; Caputo, S.M.; Vreeswijk, M.P.; Brown, M.A.; Webb, M.; Rouleau, E.; Neuhausen, S.L.; Hansen, T.V.; Galli, A.; et al. A guide for functional analysis of BRCA1 variants of uncertain significance. Hum. Mutat. 2012, 33, 1526–1537. [Google Scholar] [CrossRef]
- Guidugli, L.; Carreira, A.; Caputo, S.M.; Ehlen, A.; Galli, A.; Monteiro, A.N.; Neuhausen, S.L.; Hansen, T.V.; Couch, F.J.; Vreeswijk, M.P.; et al. ENIGMA consortium. Functional assays for analysis of variants of uncertain significance in BRCA2. Hum. Mutat. 2014, 35, 151–164. [Google Scholar] [CrossRef]
- Venkitaraman, A.R. Cancer susceptibility and the functions of BRCA1 and BRCA2. Cell 2002, 108, 171–182. [Google Scholar] [CrossRef]
- Gudmundsdottir, K.; Ashworth, A. The roles of BRCA1 and BRCA2 and associated proteins in themaintenance of genomic stability. Oncogene 2006, 25, 5864–5874. [Google Scholar] [CrossRef] [PubMed]
- Roy, R.; Chun, J.; Powell, S.N. BRCA1 and BRCA2: Different roles in a common pathway of genome protection. Nat. Rev. Cancer 2011, 12, 68–78. [Google Scholar] [CrossRef] [PubMed]
- Sarkar, M.; Magliery, T.J. Re-engineering a split-GFP reassembly screen to examine RING-domain interactions between BARD1 and BRCA1 mutants observed in cancer patients. Mol. Biosyst. 2008, 4, 599–605. [Google Scholar] [CrossRef] [PubMed]
- Caleca, L.; Putignano, A.L.; Colombo, M.; Congregati, C.; Sarkar, M.; Magliery, T.J.; Ripamonti, C.B.; Foglia, C.; Peissel, B.; Zaffaroni, D.; et al. Characterization of an Italian Founder Mutation in the RING-Finger Domain of BRCA1. PLoS ONE 2014, 9, e86924. [Google Scholar] [CrossRef] [PubMed]
- Xia, B.; Sheng, Q.; Nakanishi, K.; Ohashi, A.; Wu, J.; Christ, N.; Liu, X.; Jasin, M.; Couch, F.J.; Livingston, D.M. Control of BRCA2 cellular and clinical functions by a nuclear partner, PALB2. Mol. Cell 2006, 22, 719–729. [Google Scholar] [CrossRef] [PubMed]
- Biswas, K.; Das, R.; Alter, B.P.; Kuznetsov, S.G.; Stauffer, S.M.; North, S.L.; Burkett, S.; Brody, L.C.; Meyer, S.; Byrd, R.A.; et al. A comprehensive functional characterization of BRCA2 variants associated with Fanconi anemia using mouse ES cell based assay. Blood 2011, 118, 2430–2442. [Google Scholar] [CrossRef]
- Brough, R.; Bajrami, I.; Vatcheva, R.; Natrajan, R.; Reis-Filho, J.S.; Lord, C.J.; Ashworth, A. APRIN is a cell cycle specific BRCA2-interacting protein required for genome integrity and a predictor of outcome after chemotherapy in breast cancer. EMBO J. 2012, 31, 1160–1176. [Google Scholar] [CrossRef] [Green Version]
- Brzovic, P.S.; Rajagopal, P.; Hoyt, D.W.; King, M.C.; Klevit, R.E. Structure of a BRCA1-BARD1 heterodimeric RING-RING complex. Nat. Struct. Biol. 2001, 8, 833–837. [Google Scholar] [CrossRef]
- Joukov, V.; Chen, J.; Fox, E.A.; Green, J.B.; Livingston, D.M. Functional communication between endogenous BRCA1 and its partner, BARD1, during Xenopus laevis development. Proc. Natl. Acad. Sci. USA 2001, 98, 12078–12083. [Google Scholar] [CrossRef] [Green Version]
- Thompson, M.E.; Robinson-Benion, C.L.; Holt, J.T. An amino-terminal motif functions as a second nuclear export sequence in BRCA1. J. Biol. Chem. 2005, 280, 21854–21857. [Google Scholar] [CrossRef]
- Wu, L.C.; Wang, Z.W.; Tsan, J.T.; Spillman, M.A.; Phung, A.; Xu, X.L.; Yang, M.C.; Hwang, L.Y.; Bowcock, A.M.; Baer, R. Identification of a RING protein that can interact in vivo with the BRCA1 gene product. Nat. Genet. 1996, 14, 430–440. [Google Scholar] [CrossRef] [PubMed]
- Chen, A.; Kleiman, F.E.; Manley, J.L.; Ouchi, T.; Pan, Z.Q. Autoubiquitination of the BRCA1*BARD1 RING ubiquitin ligase. J. Biol. Chem. 2002, 2772, 2085–2092. [Google Scholar] [CrossRef]
- Mallery, D.L.; Vandenberg, C.J.; Hiom, K. Activation of the E3 ligase function of the BRCA1/BARD1 complex by polyubiquitin chains. EMBO J. 2002, 21, 6755–6762. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu-Baer, F.; Lagrazon, K.; Yuan, W.; Baer, R. The BRCA1/BARD1 heterodimer assembles polyubiquitin chains through an unconventional linkage involving lysine residue K6 of ubiquitin. J. Biol. Chem. 2003, 278, 34743–34746. [Google Scholar] [CrossRef] [PubMed]
- Hongtae, K.; Junjie, C.; Xiaochun, Y. Ubiquitin-Binding Protein RAP80 Mediates BRCA1-Dependent DNA Damage Response. Science 2007, 316, 1202–1205. [Google Scholar] [CrossRef]
- Horwitz, A.A.; Affar, E.B.; Heine, G.F.; Shi, Y.; Parvin, D.J. A mechanism for transcriptional repression dependent on the BRCA1 E3 ubiquitin ligase. PNAS 2007, 104, 6614–6619. [Google Scholar] [CrossRef] [Green Version]
- Wu, W.; Koike, A.; Takeshita, T.; Ohta, T. The ubiquitin E3 ligase activity of BRCA1 and its biological functions. Cell. Div. 2008, 3, 1. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moynahan, M.E.; Cui, T.Y.; Jasin, M. Homology-directed dna repair, mitomycin-c resistance, and chromosome stability is restored with correction of a Brca1 mutation. Cancer Res. 2001, 61, 4842–4850. [Google Scholar]
- Ruffner, H.; Joazeiro, C.A.; Hemmati, D.; Hunter, T.; Verma, I.M. Cancer-predisposing mutations within the RING domain of BRCA1: Loss of ubiquitin protein ligase activity and protection from radiation hypersensitivity. Proc. Natl. Acad. Sci. USA 2001, 98, 5134–5139. [Google Scholar] [CrossRef] [Green Version]
- Nelson, A.C.; Holt, J.T. Impact of RING and BRCT domain mutations on BRCA1 protein stability, localization and recruitment to DNA damage. Radiat. Res. 2010, 174, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Jeffrey, P.D.; Miller, J.; Kinnucan, E.; Sun, Y.; Thoma, N.H.; Zheng, N.; Chen, P.L.; Lee, W.H.; Pavletich, N.P. BRCA2 function in DNA binding and recombination from a BRCA2-DSS1-ssDNA structure. Science 2002, 297, 1837–1848. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Zou, C.; Bai, Y.; Wazer, D.E.; Band, V.; Gao, Q. DSS1 is required for the stability of BRCA2. Oncogene 2006, 25, 1186–1194. [Google Scholar] [CrossRef] [PubMed]
- Gudmundsdottir, K.; Lord, C.J.; Witt, E.; Tutt, A.N.; Ashworth, A. DSS1 is required for RAD51 focus formation and genomic stability in mammalian cells. EMBO Rep. 2004, 5, 989–993. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jeyasekharan, A.D.; Liu, Y.; Hattori, H.; Pisupati, V.; Jonsdottir, A.B.; Rajendra, E.; Lee, M.; Sundaramoorthy, E.; Schlachter, S.; Kaminski, C.F.; et al. A cancer-associated BRCA2 mutation reveals masked nuclear export signals controlling localization. Nat. Struct. Mol. Biol. 2013, 20, 1191–1198. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wilson, C.G.; Magliery, T.J.; Regan, L. Detecting protein-protein interactions with GFP-fragment reassembly. Nat. Methods. 2004, 1, 255–262. [Google Scholar] [CrossRef] [PubMed]
- Morris, J.R.; Pangon, L.; Boutell, C.; Katagiri, T.; Keep, N.H.; Solomon, E. Genetic analysis of BRCA1 ubiquitin ligase activity and its relationship to breast cancer susceptibility. Hum. Mol. Genet. 2006, 15, 599–606. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ransburgh, D.J.; Chiba, N.; Ishioka, C.; Toland, A.E.; Parvin, J.D. Identification of breast tumor mutations in BRCA1 that abolish its function in homologous DNA recombination. Cancer Res. 2010, 70, 988–995. [Google Scholar] [CrossRef] [PubMed]
- Bouwman, P.; van der Gulden, H.; van der Heijden, I.; Drost, R.; Klijn, C.N.; Prasetyanti, P.; Pieterse, M.; Wientjens, E.; Seibler, J.; Hogervorst, F.B.; et al. A high-throughput functional complementation assay for classification of BRCA1 missense variants. Cancer Discov. 2013, 3, 1142–1155. [Google Scholar] [CrossRef] [PubMed]
- Millot, G.A.; Berger, A.; Lejour, V.; Boulé, J.B.; Bobo, C.; Cullin, C.; Lopes, J.; Stoppa-Lyonnet, D.; Nicolas, A. Assessment of human Nter and Cter BRCA1 mutations using growth and localization assays in yeast. Hum. Mutat. 2011, 32, 1470–1480. [Google Scholar] [CrossRef] [PubMed]
- Towler, W.I.; Zhang, J.; Ransburgh, D.J.; Toland, A.E.; Ishioka, C.; Chiba, N.; Parvin, J.D. Analysis of BRCA1 variants in double-strand break repair by homologous recombination and single-strand annealing. Hum. Mutat. 2013, 34, 439–445. [Google Scholar] [CrossRef] [PubMed]
- Kais, Z.; Chiba, N.; Ishioka, C.; Parvin, J.D. Functional differences among BRCA1 missense mutations in the control of centrosome duplication. Oncogene 2012, 31, 799–804. [Google Scholar] [CrossRef]
- Starita, L.M.; Young, D.L.; Islam, M.; Kitzman, J.O.; Gullingsrud, J.; Hause, R.J.; Fowler, D.M.; Parvin, J.D.; Shendure, J.; Fields, S.; et al. Massively Parallel Functional Analysis of BRCA1 RING Domain Variants. Genetics 2015, 200, 413–422. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Findlay, G.M.; Daza, R.M.; Martin, B.; Zhang, M.D.; Leith, A.P.; Gasperini, M.; Janizek, J.D.; Huang, X.; Starita, L.M.; Shendure, J. Accurate classification of BRCA1 variants with saturation genome editing. Nature 2018. [Google Scholar] [CrossRef] [PubMed]
- Wu, K.; Hinson, S.R.; Ohashi, A.; Farrugia, D.; Wendt, P.; Tavtigian, S.V.; Deffenbaugh, A.; Goldgar, D.; Couch, F.J. Functional evaluation and cancer risk assessment of BRCA2 unclassified variants. Cancer Res. 2005, 65, 417–426. [Google Scholar]
- Farrugia, D.J.; Agarwal, M.K.; Pankratz, V.S.; Deffenbaugh, A.M.; Pruss, D.; Frye, C.; Wadum, L.; Johnson, K.; Mentlick, J.; Tavtigian, S.V.; et al. Functional assays for classification of BRCA2 variants of uncertain significance. Cancer Res. 2008, 68, 3523–3531. [Google Scholar] [CrossRef] [PubMed]
- Guidugli, L.; Pankratz, V.S.; Singh, N.; Thompson, J.; Erding, C.A.; Engel, C.; Schmutzler, R.; Domchek, S.; Nathanson, K.; Radice, P.; et al. A classification model for BRCA2 DNA binding domain missense variants based on homology-directed repair activity. Cancer Res. 2013, 73, 265–275. [Google Scholar] [CrossRef] [PubMed]
- Guidugli, L.; Shimelis, H.; Masica, D.L.; Pankratz, V.S.; Lipton, G.B.; Singh, N.; Hu, C.; Monteiro, A.N.A.; Lindor, N.M.; Goldgar, D.E.; et al. Assessment of the Clinical Relevance of BRCA2 Missense Variants by Functional and Computational Approaches. Am. J. Hum. Genet. 2018, 102, 233–248. [Google Scholar] [CrossRef]
- Hendriks, G.; Morolli, B.; Calléja, F.M.; Plomp, A.; Mesman, R.L.; Meijers, M.; Sharan, S.K.; Vreeswijk, M.P.; Vrieling, H. An efficient pipeline for the generation and functional analysis of human BRCA2 variants of uncertain significance. Hum. Mutat. 2014, 35, 1382–1391. [Google Scholar] [CrossRef]
- Mesman, R.L.S.; Calléja, F.M.G.R.; Hendriks, G.; Morolli, B.; Misovic, B.; Devilee, P.; van Asperen, C.J.; Vrieling, H.; Vreeswijk, M.P.G. The functional impact of variants of uncertain significance in BRCA2. Genet. Med. 2018. [Google Scholar] [CrossRef] [PubMed]
- Sweet, K.; Senter, L.; Pilarski, R.; Wei, L.; Toland, A.E. Characterization of BRCA1 ring finger variants of uncertain significance. Breast Cancer Res. Treat. 2010, 119, 737–743. [Google Scholar] [CrossRef] [PubMed]
- Parsons, M.; Tudini, E.; Li, H.; Hahnen, E.; Wappenschmidt, B.; Feliubadalo, L.; Aalfs, C.M.; Agata, S.; Aittomäki, K.; Alducci, E.; et al. Large scale multifactorial likelihood analysis of BRCA1 and BRCA2 variants: An ENIGMA resource to support clinical variant classification. Hum. Mutat. 2019. submitted. [Google Scholar]
- Whiley, P.J.; Parsons, M.T.; Leary, J.; Tucker, K.; Warwick, L.; Dopita, B.; Thorne, H.; Lakhani, S.R.; Goldgar, D.E.; Brown, M.A.; et al. Multifactorial likelihood assessment of BRCA1 and BRCA2 missense variants confirms that BRCA1:c.122A>G(p.His41Arg) is a pathogenic mutation. PLoS ONE 2014, 9, e86836. [Google Scholar] [CrossRef] [PubMed]
- Spearman, A.D.; Sweet, K.; Zhou, X.P.; McLennan, J.; Couch, F.J.; Toland, A.E. Clinically applicable models to characterize BRCA1 and BRCA2 variants of uncertain significance. J. Clin. Oncol. 2008, 26, 5393–5400. [Google Scholar] [CrossRef] [PubMed]
- Spurdle, A.B.; Lakhani, S.R.; Healey, S.; Parry, S.; Da Silva, L.M.; Brinkworth, R.; Hopper, J.L.; Brown, M.A.; Babikyan, D.; Chenevix-Trench, G.; et al. Clinical classification of BRCA1 and BRCA2 DNA sequence variants: The value of cytokeratin profiles and evolutionary analysis-a report from the kConFab Investigators. J. Clin. Oncol. 2008, 26, 1657–1663. [Google Scholar] [CrossRef] [PubMed]
- Walker, L.C.; Whiley, P.J.; Couch, F.J.; Farrugia, D.J.; Healey, S.; Eccles, D.M.; Lin, F.; Butler, S.A.; Goff, S.A.; Thompson, B.A.; et al. Detection of splicing aberrations caused by BRCA1 and BRCA2 sequence variants encoding missense substitutions: Implications for prediction of pathogenicity. Hum. Mutat. 2010, 31, 1484–1505. [Google Scholar] [CrossRef]
- Houdayer, C.; Caux-Moncoutier, V.; Krieger, S.; Barrois, M.; Bonnet, F.; Bourdon, V.; Bronner, M.; Buisson, M.; Coulet, F.; Gaildrat, P.; et al. Guidelines for splicing analysis in molecular diagnosis derived from a set of 327 combined in silico/in vitro studies on BRCA1 and BRCA2 variants. Hum. Mutat. 2012, 33, 1228–1238. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Swaminathan, S.; Martin, B.K.; Sharan, S.K. Aberrant splicing induced by missense mutations in BRCA1: Clues from a humanized mouse model. Hum. Mol. Genet. 2003, 12, 2121–2131. [Google Scholar] [CrossRef] [PubMed]
- Colombo, M.; Blok, M.J.; Whiley, P.; Santamariña, M.; Gutiérrez-Enríquez, S.; Romero, A.; Garre, P.; Becker, A.; Smith, L.D.; De Vecchi, G.; et al. Comprehensive annotation of splice junctions supports pervasive alternative splicing at the BRCA1 locus: A report from the ENIGMA consortium. Hum. Mol. Genet. 2014, 23, 3666–3680. [Google Scholar] [CrossRef] [PubMed]
- Kurian, A.W.; Hare, E.E.; Mills, M.A.; Kingham, K.E.; McPherson, L.; Whittemore, A.S.; McGuire, V.; Ladabaum, U.; Kobayashi, Y.; et al. Clinical evaluation of a multiple-gene sequencing panel for hereditary cancer risk assessment. J. Clin. Oncol. 2014, 32, 2001–2009. [Google Scholar] [CrossRef] [PubMed]
- Hilbers, F.S.; Vreeswijk, M.P.; van Asperen, C.J.; Devilee, P. The impact of next generation sequencing on the analysis of breast cancer susceptibility: A role for extremely rare genetic variation? Clin. Genet. 2013, 84, 407–414. [Google Scholar] [CrossRef] [PubMed]
- Brzovic, P.S.; Meza, J.; King, M.C.; Klevit, R.E. The cancer-predisposing mutation C61G disrupts homodimer formation in the NH2-terminal BRCA1 RING finger domain. J. Biol. Chem. 1998, 273, 7795–7799. [Google Scholar] [CrossRef]
- Drost, T.; Bouwman, P.; Rottemberg, S.; Boon, U.; Schut, S.; Klarenbeek, S.; Klijn, C.; van der Heijden, I.; van der Gulden, H.; Wientjens, E.; et al. BRCA1 RING Function Is Essential for Tumor Suppression but Dispensable for Therapy Resistance. Cancer Cell 2011, 20, 797–809. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shakya, R.; Reid, L.J.; Reczek, C.R.; Cole, F.; Egli, D.; Lin, C.S.; de Rooij, D.G.; Hirsch, S.; Ravi, K.; Hicks, J.B.; et al. BRCA1 Tumor Suppression Depends on BRCT Phosphoprotein Binding, But not Its E3 Ligase Activity. Science 2011, 334, 525–528. [Google Scholar] [CrossRef] [PubMed]
- Cabantous, S.; Nguyen, H.B.; Pedelacq, J.D.; Koraïchi, F.; Chaudhary, A.; Ganguly, K.; Lockard, M.A.; Favre, G.; Terwilliger, T.C.; Waldo, G.S.; et al. A new protein-protein interaction sensor based on tripartite split-GFP association. Sci. Rep. 2013, 3, 2854. [Google Scholar] [CrossRef] [PubMed]
- Thouvenot, P.; Ben, Y.B.; Fourrière, L.; Lescure, A.; Boudier, T.; Del Nery, E.; Chauchereau, A.; Goldgar, D.E.; Houdayer, C.; Stoppa-Lyonnet, D.; et al. Functional Assessment of Genetic Variants with Outcomes Adapted to Clinical Decision-Making. PLoS Genet. 2016, 12, e1006096. [Google Scholar] [CrossRef] [PubMed]
- De Lorenzo, S.B.; Patel, A.G.; Hurley, R.M.; Kaufmann, S.H. The Elephant and the Blind Men: Making Sense of PARP Inhibitors in Homologous Recombination Deficient Tumor. Cells Front. Oncol. 2013, 3, 228. [Google Scholar] [CrossRef]
- Azzollini, J.; Scuvera, G.; Bruno, E.; Pasanisi, P.; Zaffaroni, D.; Calvello, M.; Pasini, B.; Ripamonti, C.B.; Colombo, M.; Pensotti, V.; et al. Mutation detection rates associated with specific selection criteria for BRCA1/2 testing in 1854 high-risk families: A monocentric Italian study. Eur. J. Int. Med. 2016, 32, 65–71. [Google Scholar] [CrossRef] [PubMed]
- Stradella, A.; Del Valle, J.; Rofes, P.; Feliubadaló, L.; Grau Garces, È.; Velasco, À.; González, S.; Vargas, G.; Izquierdo, Á.; Campos, O.; et al. Does multilocus inherited neoplasia alleles syndrome have severe clinical expression? J. Med. Genet. 2018. [Google Scholar] [CrossRef]
- Hansen, T.V.; Jønson, L.; Steffensen, A.Y.; Andersen, M.K.; Kjaergaard, S.; Gerdes, A.M.; Ejlertsen, B.; Nielsen, F.C. Screening of 1331 Danish breast and/or ovarian cancer families identified 40 novel BRCA1 and BRCA2 mutations. Fam. Cancer 2011, 2, 207–212. [Google Scholar] [CrossRef]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Guex, N.; Peitsch, M.C. SWISS-MODEL and the Swiss-PdbViewer: An environment for comparative protein modeling. Electrophoresis 1997, 18, 2714–2723. [Google Scholar] [CrossRef] [PubMed]
- Sussman, J.L.; Lin, D.; Jiang, J.; Manning, N.O.; Prilusky, J.; Ritter, O.; Abola, E.E. Protein Data Bank (PDB): Database of three-dimensional structural information of biological macromolecules. Acta Crystallogr. D Biol. Crystallogr. 1998, 54, 1078–1084. [Google Scholar] [CrossRef] [PubMed]


| Gene | DNA Change | Protein Change | Domain | Motif | IARC Class a | References/Criterion |
|---|---|---|---|---|---|---|
| BRCA1 | c.53T>C | p.Met18Thr | RING finger | Helical bundle N | 4 | [6] |
| c.65T>C | p.Leu22Ser | RING finger | Helical bundle N | 5 | [51] | |
| c.110C>A | p.Thr37Lys | RING finger | β-Sheet | 5 | [51] | |
| c.115T>C | p.Cys39Arg | RING finger | Zn2+/SiteII | 5 | [51] | |
| c.116G>A | p.Cys39Tyr | RING finger | Zn2+/SiteII | 5 | [52] | |
| c.122A>G | p.His41Arg | RING finger | Zn2+/SiteII | 5 | [53] | |
| c.130T>A | p.Cys44Ser | RING finger | Zn2+/SiteI | 5 | [51] | |
| c.131G>T | p.Cys44Phe | RING finger | Zn2+/SiteI | 5 | [52] | |
| c.131G>A | p.Cys44Tyr | RING finger | Zn2+/SiteI | 5 | [51] | |
| c.133A>C | p.Lys45Gln | RING finger | 1 | [51] | ||
| c.181T>G | p.Cys61Gly | RING finger | Zn2+/SiteII | 5 | [54] | |
| c.191G>A | p.Cys64Tyr | RING finger | Zn2+/SiteII | 5 | [52] | |
| c.199G>T | p.Asp67Tyr | RING finger | 1 | [6] | ||
| BRCA2 | c.7469T>C | p.Ile2490Thr | DBD | HD | 1 | Frequency >1% in outbred populations |
| c.7544C>T | p.Thr2515Ile | DBD | HD | 1 | [55] | |
| c.7826G>A | p.Gly2609Asp | DBD | HD | 4 | [8,47] | |
| c.7878G>C | p.Trp2626Cys | DBD | HD | 5 | [6] | |
| c.7879A>T | p.Ile2627Phe | DBD | HD | 5 | [6] | |
| c.7940T>C | p.Leu2647Pro | DBD | HD | 4 | [46] | |
| c.7958T>C | p.Leu2653Pro | DBD | HD | 5 | [6,46] | |
| c.8063T>C | p.Leu2688Pro | DBD | OB1 | 4 | [47] | |
| c.8149G>T | p.Ala2717Ser | DBD | OB1 | 1 | [55] | |
| c.8165C>G | p.Thr2722Arg | DBD | OB1 | 5 | [6,46] | |
| c.8167G>C | p.Asp2723His | DBD | OB1 | 5 | [5,8] | |
| c.8168A>G | p.Asp2723Gly | DBD | OB1 | 5 | [6,47,56] | |
| c.8187G>T | p.Lys2729Asn | DBD | OB1 | 1 | [6,8] | |
| c.8243G>A | p.Gly2748Asp | DBD | OB1 | 5 | [6,8] | |
| c.8324T>G | p.Met2775Arg | DBD | OB1 | 2 | [52] |
| Gene | DNA Change | Protein Change | Domain | Motif | Source | ClinVar |
|---|---|---|---|---|---|---|
| BRCA1 | c.20_28del9 | p.7-10delinsGln | RING finger | INT | VUS (1) | |
| c.43A>G | p.Ile15Val | RING finger | Helical bundle N | INT | NR | |
| c.44T>C | p.Ile15Thr | RING finger | Helical bundle N | INT | VUS (4) | |
| c.53T>A | p.Met18Lys | RING finger | Helical bundle N | ClinVar | VUS (2) | |
| c.70T>C | p.Cys24Arg | RING finger | Zn2+/SiteI | ClinVar | VUS (1); LP (1), P (1) | |
| c.79_80delTGinsGC | p.Cys27Ala | RING finger | Zn2+/SiteI | Synthetica | NR | |
| c.83T>C | p.Leu28Pro | RING finger | INT | VUS (4) | ||
| c.93C>G | p.Ile31Met | RING finger | INT | LB (1), VUS (2) | ||
| c.100C>T | p.Pro34Ser | RING finger | Rigshospitalet | VUS (2) | ||
| c.107C>A | p.Ser36Tyr | RING finger | β-strand | ICO | VUS (5) | |
| c.110C>G | p.Thr37Arg | RING finger | β-strand | ClinVar | LP (1) | |
| c.115T>G | p.Cys39Gly | RING finger | Zn2+/SiteII | Rigshospitalet | VUS (1), LP (1), P (1) | |
| c.121_122delCAinsGC | p.His41Ala | RING finger | Zn2+/SiteII | Synthetic a | NR | |
| c.124A>G | p.Ile42Val | RING finger | Clinvar | 3 LB (1), VUS (3) | ||
| c.139T>G | p.Cys47Gly | RING finger | Zn2+/SiteI | Clinvar | LP (1), P (6) | |
| c.143T>G | p.Met48Arg | RING finger | Central Helix | Rigshospitalet | NR | |
| c.143T>A | p.Met48Lys | RING finger | Central Helix | INT | NR | |
| c.190T>G | p.Cys64Gly | RING finger | Zn2+/SiteII | Clinvar | VUS (1), P (11) | |
| c.190T>C | p.Cys64Arg | RING finger | Zn2+/SiteII | INT | VUS (1), P (4) | |
| c.206_207delCCinsTG | p.Thr69Met | RING finger | INT | NR | ||
| c.230C>T | p.Thr77Met | RING finger | INT | VUS (6) | ||
| c.248T>G | p.Val83Gly | RING finger | Helical bundle C | ICO | NR | |
| c.293G>T | p.Gly98Val | RING finger | INT | NR | ||
| BRCA2 | c.7505G>A | p.Arg2502His | DBD | HD | INT | B (1), LB (5), VUS (3) |
| c.7628A>G | p.Tyr2543Cys | DBD | HD | INT | VUS (7) | |
| c.7636T>C | p.Ser2546Pro | DBD | HD | INT | VUS (1) | |
| c.7649T>C | p.Ile2550Thr | DBD | HD | Clinvar | B (1) | |
| c.7684T>C | p.Phe2562Leu | DBD | HD | INT | VUS (6) | |
| c.7698T>G | p.Asp2566Glu | DBD | HD | INT | NR | |
| c.7930A>G | p.Asn2644Asp | DBD | HD | INT | VUS (3) |
| Variants Group | DNA Change | Protein Change | Motif | Align-GVGD a | IARC Class b | BARD1 Binding d | UbCH5a Binding d | Ubiquitin Ligase Activity e,f | Restoration of Radiation resistance f | HDR g | mESCs-Based Assays h | Cisplatin Response h | Small Colony Phenotype i | Yeast Localization Phenotype i | Single strand Annealing l | Centrosome Number m | HDR Rescue n | Functinal Effect o | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Pred | Func | ||||||||||||||||||
| Validation Panel | c.53T>C | p.Met18Thr | Helical bundle N | C45 | 4 | + (also in c) | − | − | − | − | − | − | − | − | − | − | − | − | LOF |
| c.65T>C | p.Leu22Ser | Helical bundle N | C65 | 5 | − | − | ND | ND | ND | ND | ND | ND | ND | ND | ND | + | − | LOF | |
| c.110C>A | p.Thr37Lys | C65 | 5 | + | − | ND | ND | ND | ND | ND | ND | ND | ND | ND | − | ND | LOF | ||
| c.115T>C | p.Cys39Arg | Zn2+/SiteII | C65 | 5 | − | − | − | ND | ND | ND | ND | ND | ND | ND | ND | − | ND | LOF | |
| c.116G>A | p.Cys39Tyr | Zn2+/SiteII | C65 | 5 | − | − | − | − | − | ND | ND | − | ND | − | − | − | − | LOF | |
| c.122A>G | p.His41Arg | Zn2+/SiteII | C25 | 5 | + | − | − | ND | − | ND | ND | ND | ND | − | − | + | − | LOF | |
| c.130T>A | p.Cys44Ser | Zn2+/SiteI | C65 | 5 | − | − | ND | ND | ND | ND | ND | ND | ND | ND | ND | − | − | LOF | |
| c.131G>A | p.Cys44Tyr | Zn2+/SiteI | C65 | 5 | − | − | ND | ND | ND | ND | ND | − | − | ND | ND | − | ND | LOF | |
| c.131G>T | p.Cys44Phe | Zn2+/SiteI | C65 | 5 | − | − | − | ND | − | ND | ND | ND | ND | − | − | − | − | LOF | |
| c.133A>C | p.Lys45Gln | C0 | 1 | + | + | ND | ND | ND | + | ND | ND | ND | ND | ND | + | + | FUNC | ||
| c.181T>G | p.Cys61Gly | Zn2+/SiteII | C65 | 5 | − | − | − | − | − | − | − | + | − | − | ND | − | − | LOF | |
| c.191G>A | p.Cys64Tyr | Zn2+/SiteII | C65 | 5 | − | − | − | ND | ND | ND | ND | ND | ND | ND | ND | − | ND | LOF | |
| c.199G>T | p.Asp67Tyr | C0 | 1 | + | + | + | ND | + | + | + | ND | ND | + | ND | + | + | FUNC | ||
| VUS | c.20_28del9 | p.7-10delinsGln | NA | NA | − | − | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | |
| c.43A>G | p.Ile15Val | Helical bundle N | C15 | NA | − | + | ND | ND | ND | ND | ND | ND | ND | ND | ND | + | ND | FUNC | |
| c.44T>C | p.Ile15Thr | Helical bundle N | C65 | NA | + (also in a) | − | − | ND | ND | ND | ND | ND | ND | ND | ND | − | − | LOF | |
| c.53T>A | p.Met18Lys | Helical bundle N | C55 | NA | − (also in a) | − | − | ND | ND | ND | ND | ND | ND | ND | ND | − | ND | ND | |
| c.70T>C | p.Cys24Arg | Zn2+/SiteI | C65 | NA | − | − | − | − | − | ND | ND | ND | ND | − | − | − | − | LOF | |
| c.79_80delTGinsGC | p.Cys27Ala | Zn2+/SiteI | C65 | NA | − | − | ND | ND | − | ND | ND | ND | ND | ND | − | − | − | ND | |
| c.83T>C | p.Leu28Pro | C65 | NA | + | − | +/− | ND | ND | ND | ND | ND | ND | ND | ND | + | − | LOF | ||
| c.93C>G | p.Ile31Met | C0 | NA | + | + | + | + | + | ND | ND | ND | ND | + | ND | + | + | FUNC | ||
| c.100C>T | p.Pro34Ser | C65 | NA | − | − | ND | ND | ND | ND | ND | ND | ND | ND | ND | + | ND | LOF | ||
| c.107C>A | p.Ser36Tyr | β-strand | C15 | NA | + | − | ND | ND | ND | ND | ND | ND | ND | ND | ND | + | ND | ND | |
| c.110C>G | p.Thr37Arg | C65 | NA | − | − | − | − | − | ND | ND | ND | ND | − | + | − | − | LOF | ||
| p c.115T>G | p.Cys39Gly | Zn2+/SiteII | C65 | NA | − | − | ND | ND | ND | ND | ND | ND | ND | ND | ND | − | − | LOF | |
| c.121_122delCAinsGC | p.His41Ala | Zn2+/SiteII | C65 | NA | − | − | ND | ND | ND | ND | ND | ND | ND | ND | ND | + | ND | ND | |
| c.124A>G | p.Ile42Val | C0 | NA | + | + | + | + | + | ND | ND | ND | ND | + | − | + | + | FUNC | ||
| c.139T>G | p.Cys47Gly | Zn2+/SiteI | C65 | NA | − | − | − | ND | ND | ND | ND | ND | ND | ND | ND | − | − | LOF | |
| c.143T>G | p.Met48Arg | Central Helix | C35 | NA | − | − | ND | ND | ND | ND | ND | ND | ND | ND | ND | − | − | LOF | |
| c.143T>A | p.Met48Lys | Central Helix | C45 | NA | + | − | ND | ND | ND | ND | ND | ND | ND | ND | ND | + | ND | LOF | |
| q c.190T>G | p.Cys64Gly | Zn2+/SiteII | C65 | NA | − | − | − | ND | − | − | ND | ND | ND | − | ND | − | ND | LOF | |
| r c.190T>C | p.Cys64Arg | Zn2+/SiteII | C65 | NA | − | − | ND | ND | ND | ND | ND | ND | ND | ND | ND | − | ND | LOF | |
| c.206_207delinsTG | p.Thr69Met | C65 | NA | − | − | ND | ND | ND | ND | ND | ND | ND | ND | ND | + | ND | ND | ||
| c.230C>T | p.Thr77Met | C0 | NA | + | + | − | ND | ND | ND | ND | ND | ND | ND | ND | + | ND | FUNC | ||
| c.248T>G | p.Val83Gly | Helical bundle C | C0 | NA | − | + | ND | ND | ND | ND | ND | ND | ND | ND | ND | + | ND | FUNC | |
| c.293G>T | p.Gly98Val | C65 | NA | + | − | ND | ND | ND | ND | ND | ND | ND | ND | ND | + | ND | LOF | ||
| Variants Group | DNA Change | Protein Change | Domain | Motif | Align-GVGD a | IARC Class b | BRCA2/DSS1 Binding c | HDR d | Centrosome Amplification e | mESCs-Based Cell-Viability f,g | mESCs-Based HDR g |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Validation panel | c.7469T>C | p.Ile2490Thr | DBD | HD | C0 | 1 | + | ND | ND | + | ND |
| c.7544C>T | p.Thr2515Ile | DBD | HD | C0 | 1 | + | +/− | +/− | + | + | |
| c.7826G>A | p.Gly2609Asp | DBD | HD | C65 | 4 | − | − | ND | + | − | |
| c.7878G>C | p.Trp2626Cys | DBD | HD | C65 | 5 | − | − | ND | − | ND | |
| c.7879A>T | p.Ile2627Phe | DBD | HD | C15 | 5 | − | − | − | − | ND | |
| c.7940T>C | p.Leu2647Pro | DBD | HD | C65 | 4 | − | − | − | ND | ND | |
| c.7958T>C | p.Leu2653Pro | DBD | HD | C65 | 5 | − | − | − | ND | ND | |
| c.8063T>C | p.Leu2688Pro | DBD | OB1 | C65 | 4 | − | − | ND | − | ND | |
| c.8149G>T | p.Ala2717Ser | DBD | OB1 | C0 | 1 | + | ND | ND | + | + | |
| c.8165C>G | p.Thr2722Arg | DBD | OB1 | C65 | 5 | − | − | − | − | ND | |
| c.8167G>C | p.Asp2723His | DBD | OB1 | C65 | 5 | − | − | − | − | ND | |
| c.8168A>G | p.Asp2723Gly | DBD | OB1 | C65 | 5 | − | − | − | − | ND | |
| c.8187G>T | p.Lys2729Asn | DBD | OB1 | C0 | 1 | + | + | + | + | + | |
| c.8243G>A | p.Gly2748Asp | DBD | OB1 | C65 | 5 | − | − | − | − | ND | |
| c.8324T>G | p.Met2775Arg | DBD | OB1 | C0 | 2 | + | ND | ND | ND | ND | |
| VUS | c.7505G>A | p.Arg2502His | DBD | HD | C0 | NA | + | ND | ND | ND | ND |
| c.7628A>G | p.Tyr2543Cys | DBD | HD | C15 | NA | + | ND | ND | ND | ND | |
| c.7636T>C | p.Ser2546Pro | DBD | HD | C0 | NA | + | ND | ND | ND | ND | |
| c.7649T>C | p.Ile2550Thr | DBD | HD | C25 | NA | + | ND | ND | ND | ND | |
| c.7684T>C | p.Phe2562Leu | DBD | HD | C15 | NA | − | − | ND | ND | ND | |
| c.7698T>G | p.Asp2566Glu | DBD | HD | C0 | NA | + | ND | ND | ND | ND | |
| c.7930A>G | p.Asn2644Asp | DBD | HD | C0 | NA | + | ND | ND | ND | ND |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Caleca, L.; Colombo, M.; van Overeem Hansen, T.; Lázaro, C.; Manoukian, S.; Parsons, M.T.; Spurdle, A.B.; Radice, P. GFP-Fragment Reassembly Screens for the Functional Characterization of Variants of Uncertain Significance in Protein Interaction Domains of the BRCA1 and BRCA2 Genes. Cancers 2019, 11, 151. https://doi.org/10.3390/cancers11020151
Caleca L, Colombo M, van Overeem Hansen T, Lázaro C, Manoukian S, Parsons MT, Spurdle AB, Radice P. GFP-Fragment Reassembly Screens for the Functional Characterization of Variants of Uncertain Significance in Protein Interaction Domains of the BRCA1 and BRCA2 Genes. Cancers. 2019; 11(2):151. https://doi.org/10.3390/cancers11020151
Chicago/Turabian StyleCaleca, Laura, Mara Colombo, Thomas van Overeem Hansen, Conxi Lázaro, Siranoush Manoukian, Michael T. Parsons, Amanda B. Spurdle, and Paolo Radice. 2019. "GFP-Fragment Reassembly Screens for the Functional Characterization of Variants of Uncertain Significance in Protein Interaction Domains of the BRCA1 and BRCA2 Genes" Cancers 11, no. 2: 151. https://doi.org/10.3390/cancers11020151




