Ion Channel Expression in Human Melanoma Samples: In Silico Identification and Experimental Validation of Molecular Targets
Abstract
1. Introduction
2. Results
2.1. Selection Phase
2.1.1. First-Round Validation Step
2.1.2. Second-Round Validation Step
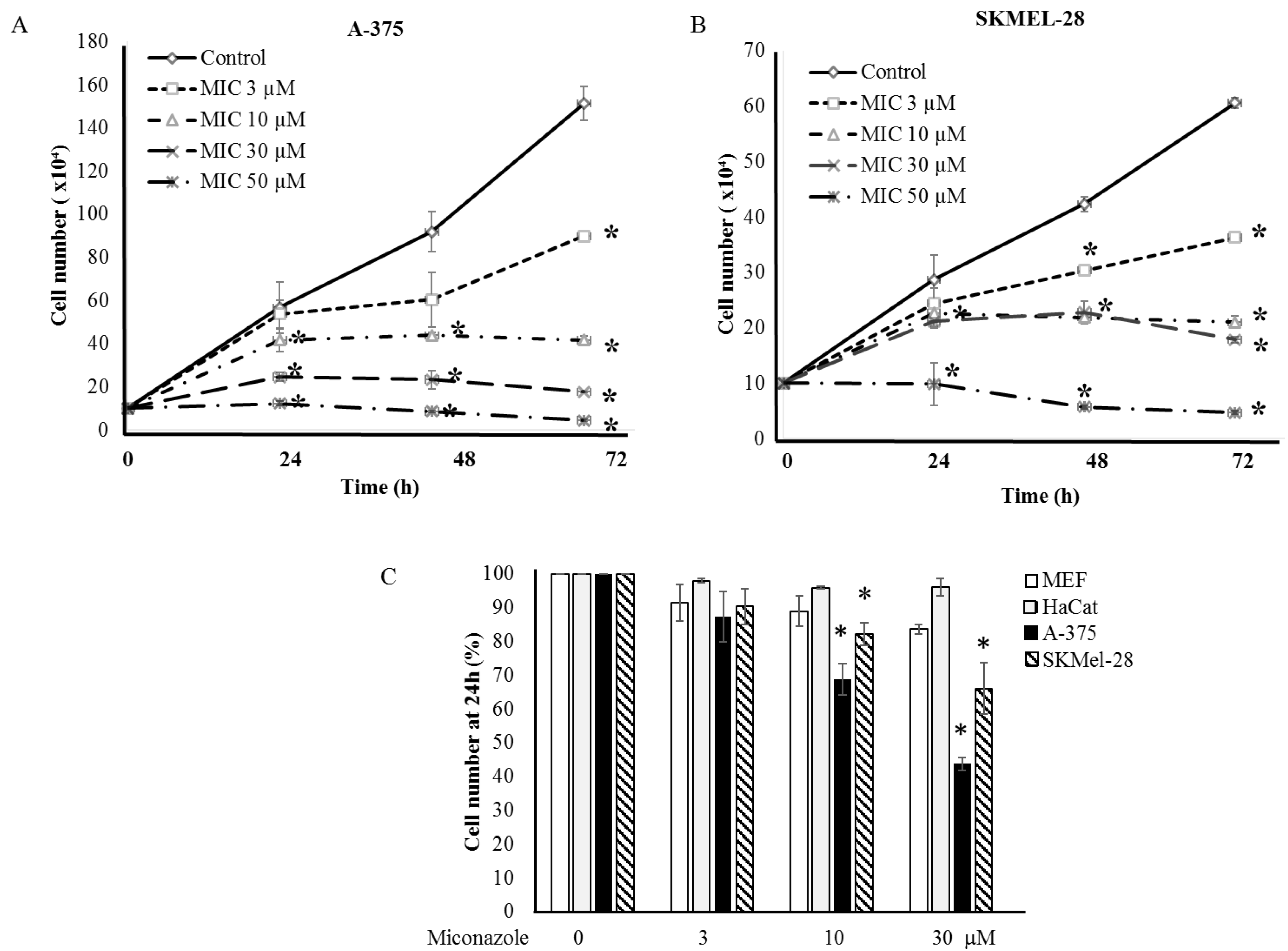
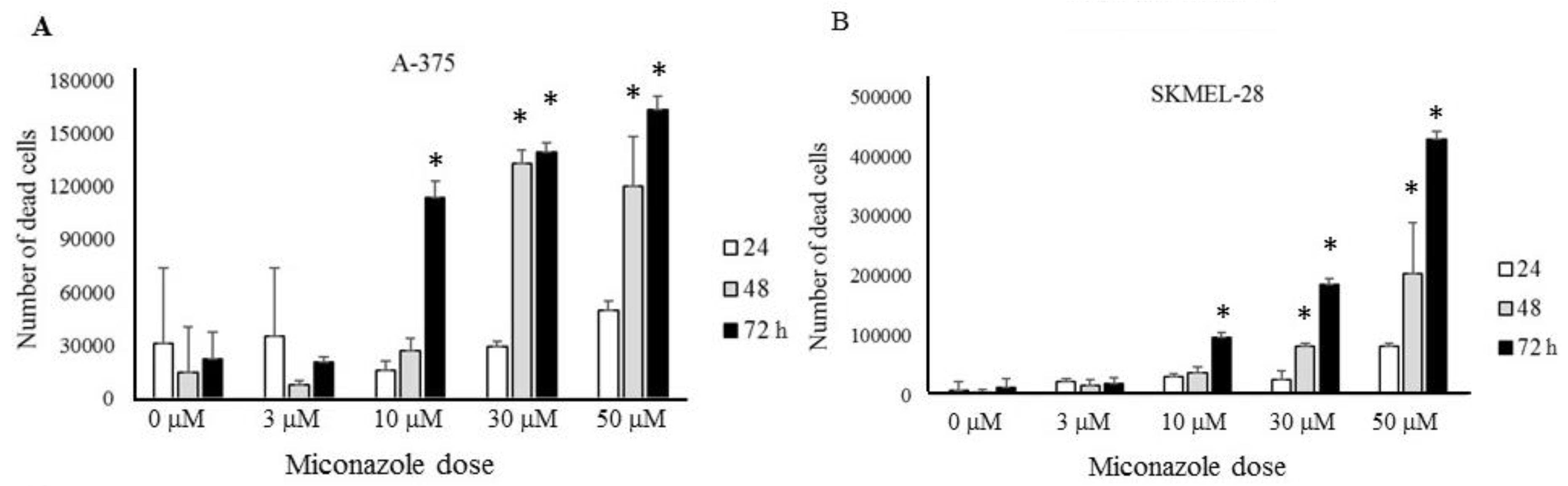
2.2. Experimental Validation
2.3. Mechanisms Underlying Miconazole Action
3. Discussion
4. Materials and Methods
4.1. In Vitro Experimental Validation: Analysis of Potential “Druggability”
4.2. Chilibot Analysis
4.3. Cell Culture and Melanoma Cells In Vitro Proliferation Assay
4.4. KCNN2 Silencing by siRNA
4.5. Western Blot Analysis
4.6. STRING Analysis
4.7. Statistics
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Bagal, S.K.; Brown, A.D.; Cox, P.J.; Omoto, K.; Owen, R.M.; Pryde, D.C.; Sidders, B.; Skerratt, S.E.; Stevens, E.B.; Storer, R.I.; et al. Ion channels as therapeutic targets: A drug discovery perspective. J. Med. Chem. 2013, 56, 593–624. [Google Scholar] [CrossRef] [PubMed]
- Wickenden, A.; Priest, B.; Erdemli, G. Ion channel drug discovery: Challenges and future directions. Future Med. Chem. 2012, 4, 661–679. [Google Scholar] [CrossRef]
- Weyer-Menkhoff, I.; Lötsch, J. Human pharmacological approaches to TRP-ion-channel-based analgesic drug development. Drug Discov. Today 2018, 23, 2003–2012. [Google Scholar] [CrossRef] [PubMed]
- Ledwitch, K.V.; Roberts, A.G. Cardiovascular Ion Channel Inhibitor Drug-Drug Interactions with P-glycoprotein. AAPS J. 2017, 19, 409–420. [Google Scholar] [CrossRef] [PubMed]
- Robertson, G.A. Endocytic control of ion channel density as a target for cardiovascular disease. J. Clin. Investig. 2009, 119, 2531–2534. [Google Scholar] [CrossRef]
- Elliott, W.J.; Bistrika, E.A. Perindopril arginine and amlodipine besylate for hypertension: A safety evaluation. Expert Opin. Drug Saf. 2018, 17, 207–216. [Google Scholar] [CrossRef] [PubMed]
- Kwai, N.C.G.; Arnold, R.; Wickremaarachchi, C.; Lin, C.S.-Y.; Poynten, A.M.; Kiernan, M.C.; Krishnan, A. V Effects of axonal ion channel dysfunction on quality of life in type 2 diabetes. Diabetes Care 2013, 36, 1272–1277. [Google Scholar] [CrossRef]
- Pietschmann, T. Clinically Approved Ion Channel Inhibitors Close Gates for Hepatitis C Virus and Open Doors for Drug Repurposing in Infectious Viral Diseases. J. Virol. 2017, 91, e01914-16. [Google Scholar] [CrossRef]
- Moorthy, N.S.H.N.; Poongavanam, V.; Pratheepa, V. Viral M2 ion channel protein: A promising target for anti-influenza drug discovery. Mini Rev. Med. Chem. 2014, 14, 819–830. [Google Scholar]
- Bhattacharya, A.; Biber, K. The microglial ATP-gated ion channel P2x7 as a CNS drug target. Glia 2016, 64, 1772–1787. [Google Scholar] [CrossRef] [PubMed]
- Wei, F.; Yan, L.-M.; Su, T.; He, N.; Lin, Z.-J.; Wang, J.; Shi, Y.-W.; Yi, Y.-H.; Liao, W.-P. Ion Channel Genes and Epilepsy: Functional Alteration, Pathogenic Potential, and Mechanism of Epilepsy. Neurosci. Bull. 2017, 33, 455–477. [Google Scholar] [CrossRef] [PubMed]
- Seefeld, M.A.; Lin, H.; Holenz, J.; Downie, D.; Donovan, B.; Fu, T.; Pasikanti, K.; Zhen, W.; Cato, M.; Chaudhary, K.W.; et al. Novel KV7 ion channel openers for the treatment of epilepsy and implications for detrusor tissue contraction. Bioorg. Med. Chem. Lett. 2018, 28, 3793–3797. [Google Scholar] [CrossRef]
- Ullah, S.; Ali, N.; Khan, A.; Ali, S.; Nazish, H.R.; Uddin, Z. Epilepsy control with carbamazepine monotherapy from a genetic perspective. BMC Pharmacol. Toxicol. 2018, 19, 73. [Google Scholar] [CrossRef]
- Mayor, S. Cystic fibrosis: Triple drug regimens that target defective ion channel improve lung function, studies show. BMJ 2018, 363, k4377. [Google Scholar] [CrossRef]
- Sala, M.A.; Jain, M. Tezacaftor for the treatment of cystic fibrosis. Expert Rev. Respir. Med. 2018, 12, 725–732. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.; Wang, X.; Xue, Q. Combined targeted ion channel therapy: Can it be an alternative choice for esophageal cancer patients? Med. Hypotheses 2018, 117, 59–62. [Google Scholar] [CrossRef]
- Tuszynski, J.; Tilli, T.M.; Levin, M. Ion Channel and Neurotransmitter Modulators as Electroceutical Approaches to the Control of Cancer. Curr. Pharm. Des. 2017, 23, 4827–4841. [Google Scholar] [CrossRef]
- Vancauwenberghe, E.; Noyer, L.; Derouiche, S.; Lemonnier, L.; Gosset, P.; Sadofsky, L.R.; Mariot, P.; Warnier, M.; Bokhobza, A.; Slomianny, C.; et al. Activation of mutated TRPA1 ion channel by resveratrol in human prostate cancer associated fibroblasts (CAF). Mol. Carcinog. 2017, 56, 1851–1867. [Google Scholar] [CrossRef]
- Biasiotta, A.; D’Arcangelo, D.; Passarelli, F.; Nicodemi, E.M.; Facchiano, A. Ion channels expression and function are strongly modified in solid tumors and vascular malformations. J. Transl. Med. 2016, 14, 285. [Google Scholar] [CrossRef]
- Ko, J.-H.; Ko, E.A.; Gu, W.; Lim, I.; Bang, H.; Zhou, T. Expression profiling of ion channel genes predicts clinical outcome in breast cancer. Mol. Cancer 2013, 12, 106. [Google Scholar] [CrossRef]
- Li, H.; Ma, N.; Wang, J.; Wang, Y.; Yuan, C.; Wu, J.; Luo, M.; Yang, J.; Chen, J.; Shi, J.; et al. Nicotine Induces Progressive Properties of Lung Adenocarcinoma A549 Cells by Inhibiting Cystic Fibrosis Transmembrane Conductance Regulator (CFTR) Expression and Plasma Membrane Localization. Technol. Cancer Res. Treat. 2018, 17, 1533033818809984. [Google Scholar] [CrossRef] [PubMed]
- Berrout, J.; Kyriakopoulou, E.; Moparthi, L.; Hogea, A.S.; Berrout, L.; Ivan, C.; Lorger, M.; Boyle, J.; Peers, C.; Muench, S.; et al. TRPA1-FGFR2 binding event is a regulatory oncogenic driver modulated by miRNA-142-3p. Nat. Commun. 2017, 8, 947. [Google Scholar] [CrossRef] [PubMed]
- Xia, D.; Lai, D.V.; Wu, W.; Webb, Z.D.; Yang, Q.; Zhao, L.; Yu, Z.; Thorpe, J.E.; Disch, B.C.; Ihnat, M.A.; et al. Transition from androgenic to neurosteroidal action of 5α-androstane-3α, 17β-diol through the type A γ-aminobutyric acid receptor in prostate cancer progression. J. Steroid Biochem. Mol. Biol. 2018, 178, 89–98. [Google Scholar] [CrossRef] [PubMed]
- Camacho, N.; Van Loo, P.; Edwards, S.; Kay, J.D.; Matthews, L.; Haase, K.; Clark, J.; Dennis, N.; Thomas, S.; Kremeyer, B.; et al. Appraising the relevance of DNA copy number loss and gain in prostate cancer using whole genome DNA sequence data. PLoS Genet. 2017, 13, e1007001. [Google Scholar] [CrossRef] [PubMed]
- Sagredo, A.I.; Sagredo, E.A.; Cappelli, C.; Báez, P.; Andaur, R.E.; Blanco, C.; Tapia, J.C.; Echeverría, C.; Cerda, O.; Stutzin, A.; et al. TRPM4 regulates Akt/GSK3-β activity and enhances β-catenin signaling and cell proliferation in prostate cancer cells. Mol. Oncol. 2018, 12, 151–165. [Google Scholar] [CrossRef] [PubMed]
- Prickett, T.D.; Gartner, J.J.; Samuels, Y. Genetic and Functional Analysis of GRIN2A in Tumor Samples. Methods Mol. Biol. 2017, 1677, 93–116. [Google Scholar]
- Leanza, L.; Romio, M.; Becker, K.A.; Azzolini, M.; Trentin, L.; Managò, A.; Venturini, E.; Zaccagnino, A.; Mattarei, A.; Carraretto, L.; et al. Direct Pharmacological Targeting of a Mitochondrial Ion Channel Selectively Kills Tumor Cells In Vivo. Cancer Cell 2017, 31, 516–531. [Google Scholar] [CrossRef]
- Kacik, M.; Oliván-Viguera, A.; Köhler, R. Modulation of K(Ca)3.1 channels by eicosanoids, omega-3 fatty acids, and molecular determinants. PLoS ONE 2014, 9, e112081. [Google Scholar] [CrossRef]
- Bauer, D.; Werth, F.; Nguyen, H.A.; Kiecker, F.; Eberle, J. Critical role of reactive oxygen species (ROS) for synergistic enhancement of apoptosis by vemurafenib and the potassium channel inhibitor TRAM-34 in melanoma cells. Cell Death Dis. 2017, 8, e2594. [Google Scholar] [CrossRef]
- Busch, A.E.; Suessbrich, H.; Kunzelmann, K.; Hipper, A.; Greger, R.; Waldegger, S.; Mutschler, E.; Lindemann, B.; Lang, F. Blockade of epithelial Na+ channels by triamterenes—Underlying mechanisms and molecular basis. Pflugers Arch. 1996, 432, 760–766. [Google Scholar] [CrossRef]
- Planer, J.D.; Hulverson, M.A.; Arif, J.A.; Ranade, R.M.; Don, R.; Buckner, F.S. Synergy testing of FDA-approved drugs identifies potent drug combinations against Trypanosoma cruzi. PLoS Negl. Trop. Dis. 2014, 8, e2977. [Google Scholar] [CrossRef]
- Kandel, C.; Schmidt, P.; Perniss, A.; Keshavarz, M.; Scholz, P.; Osterloh, S.; Althaus, M.; Kummer, W.; Deckmann, K. ENaC in Cholinergic Brush Cells. Front. Cell Dev. Biol. 2018, 6, 89. [Google Scholar] [CrossRef]
- Canessa, C.M.; Schild, L.; Buell, G.; Thorens, B.; Gautschi, I.; Horisberger, J.D.; Rossier, B.C. Amiloride-sensitive epithelial Na+ channel is made of three homologous subunits. Nature 1994, 367, 463–467. [Google Scholar] [CrossRef] [PubMed]
- Pawson, A.J.; Sharman, J.L.; Benson, H.E.; Faccenda, E.; Alexander, S.P.H.; Buneman, O.P.; Davenport, A.P.; McGrath, J.C.; Peters, J.A.; Southan, C.; et al. The IUPHAR/BPS Guide to PHARMACOLOGY: An expert-driven knowledgebase of drug targets and their ligands. Nucleic Acids Res. 2014, 42, D1098–D1106. [Google Scholar] [CrossRef]
- Ramzan, I. Phytotherapies: Efficacy, Safety, and Regulation; Wiley: Hoboken, NJ, USA, 2015; ISBN 978-1-118-26806-3. [Google Scholar]
- Wörsdörfer, P.; Maxeiner, S.; Markopoulos, C.; Kirfel, G.; Wulf, V.; Auth, T.; Urschel, S.; von Maltzahn, J.; Willecke, K. Connexin expression and functional analysis of gap junctional communication in mouse embryonic stem cells. Stem Cells 2008, 26, 431–439. [Google Scholar] [CrossRef]
- Vessey, J.P.; Lalonde, M.R.; Mizan, H.A.; Welch, N.C.; Kelly, M.E.M.; Barnes, S. Carbenoxolone inhibition of voltage-gated Ca channels and synaptic transmission in the retina. J. Neurophysiol. 2004, 92, 1252–1256. [Google Scholar] [CrossRef]
- Alvarez, J.; Montero, M.; Garcia-Sancho, J. High affinity inhibition of Ca(2+)-dependent K+ channels by cytochrome P-450 inhibitors. J. Biol. Chem. 1992, 267, 11789–11793. [Google Scholar] [PubMed]
- Chang, H.-T.; Chen, W.-C.; Chen, J.-S.; Lu, Y.-C.; Hsu, S.-S.; Wang, J.-L.; Cheng, H.-H.; Cheng, J.-S.; Jiann, B.-P.; Chiang, A.-J.; et al. Effect of miconazole on intracellular Ca2+ levels and proliferation in human osteosarcoma cells. Life Sci. 2005, 76, 2091–2101. [Google Scholar] [CrossRef] [PubMed]
- Botana, L.M. Seafood and Freshwater Toxins: Pharmacology, Physiology, and Detection; CRC Press: Boca Raton, FL, USA, 2014; ISBN 9781466505148. [Google Scholar]
- Ko, C.; Park, W.-J.; Park, S.; Kim, S.; Windisch, M.P.; Ryu, W.-S. The FDA-approved drug irbesartan inhibits HBV-infection in HepG2 cells stably expressing sodium taurocholate co-transporting polypeptide. Antivir. Ther. 2015, 20, 835–842. [Google Scholar] [CrossRef] [PubMed]
- Comes, N.; Serrano-Albarrás, A.; Capera, J.; Serrano-Novillo, C.; Condom, E.; Ramón, Y.; Cajal, S.; Ferreres, J.C.; Felipe, A. Involvement of potassium channels in the progression of cancer to a more malignant phenotype. Biochim. Biophys. Acta 2015, 1848, 2477–2492. [Google Scholar] [CrossRef] [PubMed]
- Marshall, H.T.; Djamgoz, M.B.A. Immuno-Oncology: Emerging Targets and Combination Therapies. Front. Oncol. 2018, 8, 315. [Google Scholar] [CrossRef] [PubMed]
- D’Arcangelo, D.; Nicodemi, E.M.; Facchiano, A. Letter to the editor: “Ion channels in brain metastasis”—Ion channels in cancer set up and metastatic progression. Int. J. Mol. Sci. 2017, 18, 718. [Google Scholar] [CrossRef]
- He, M.; Liu, S.; Gallolu Kankanamalage, S.; Borromeo, M.D.; Girard, L.; Gazdar, A.F.; Minna, J.D.; Johnson, J.E.; Cobb, M.H. The Epithelial Sodium Channel (αENaC) Is a Downstream Therapeutic Target of ASCL1 in Pulmonary Neuroendocrine Tumors. Transl. Oncol. 2018, 11, 292–299. [Google Scholar] [CrossRef] [PubMed]
- Varley, K.E.; Gertz, J.; Roberts, B.S.; Davis, N.S.; Bowling, K.M.; Kirby, M.K.; Nesmith, A.S.; Oliver, P.G.; Grizzle, W.E.; Forero, A.; et al. Recurrent read-through fusion transcripts in breast cancer. Breast Cancer Res. Treat. 2014, 146, 287–297. [Google Scholar] [CrossRef] [PubMed]
- Jarzab, B.; Wiench, M.; Fujarewicz, K.; Simek, K.; Jarzab, M.; Oczko-Wojciechowska, M.; Wloch, J.; Czarniecka, A.; Chmielik, E.; Lange, D.; et al. Gene expression profile of papillary thyroid cancer: Sources of variability and diagnostic implications. Cancer Res. 2005, 65, 1587–1597. [Google Scholar] [CrossRef]
- Willis, S.; Villalobos, V.M.; Gevaert, O.; Abramovitz, M.; Williams, C.; Sikic, B.I.; Leyland-Jones, B. Single Gene Prognostic Biomarkers in Ovarian Cancer: A Meta-Analysis. PLoS ONE 2016, 11, e0149183. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Chen, J.; He, Y.; Zhan, X.; Zhao, R.; Huang, Y.; Xu, H.; Zhu, Z.; Liu, Q. miR-125b inhibits hepatitis B virus expression in vitro through targeting of the SCNN1A gene. Arch. Virol. 2014, 159, 3335–3343. [Google Scholar] [CrossRef] [PubMed]
- Kappelmann, M.; Kuphal, S.; Meister, G.; Vardimon, L.; Bosserhoff, A.-K. MicroRNA miR-125b controls melanoma progression by direct regulation of c-Jun protein expression. Oncogene 2013, 32, 2984–2991. [Google Scholar] [CrossRef]
- D’Arcangelo, D.; Giampietri, C.; Muscio, M.; Scatozza, F.; Facchiano, F.; Facchiano, A. WIPI1, BAG1, and PEX3 Autophagy-Related Genes Are Relevant Melanoma Markers. Oxid. Med. Cell. Longev. 2018, 2018, 1471682. [Google Scholar] [CrossRef]
- Kondratskyi, A.; Kondratska, K.; Skryma, R.; Klionsky, D.J.; Prevarskaya, N. Ion channels in the regulation of autophagy. Autophagy 2018, 14, 3–21. [Google Scholar] [CrossRef]
- Mandy, S.J.; Garrott, T.C. Miconazole treatment for severe dermatophytoses. JAMA 1974, 230, 72–75. [Google Scholar] [CrossRef]
- Hatton, C.J.; Peers, C. Effects of cytochrome P-450 inhibitors on ionic currents in isolated rat type I carotid body cells. Am. J. Physiol. 1996, 271, C85–C92. [Google Scholar] [CrossRef] [PubMed]
- Verdoliva, V.; Senatore, C.; Polci, M.L.; Rossi, S.; Cordella, M.; Carlucci, G.; Marchetti, P.; Antonini-Cappellini, G.; Facchiano, A.; D’Arcangelo, D.; et al. Differential Denaturation of Serum Proteome Reveals a Significant Amount of Hidden Information in Complex Mixtures of Proteins. PLoS ONE 2013, 8, e57104. [Google Scholar] [CrossRef] [PubMed]
- Rossi, S.; Cordella, M.; Tabolacci, C.; Nassa, G.; D’Arcangelo, D.; Senatore, C.; Pagnotto, P.; Magliozzi, R.; Salvati, A.; Weisz, A.; et al. TNF-alpha and metalloproteases as key players in melanoma cells aggressiveness. J. Exp. Clin. Cancer Res. 2018, 37, 326. [Google Scholar] [CrossRef] [PubMed]
- Haapajärvi, T.; Pitkänen, K.; Laiho, M. Human melanoma cell line UV responses show independency of p53 function. Cell Growth Differ. 1999, 10, 163–171. [Google Scholar] [PubMed]
- Raisova, M.; Hossini, A.M.; Eberle, J.; Riebeling, C.; Wieder, T.; Sturm, I.; Daniel, P.T.; Orfanos, C.E.; Geilen, C.C. The Bax/Bcl-2 ratio determines the susceptibility of human melanoma cells to CD95/Fas-mediated apoptosis. J. Invest. Dermatol. 2001, 117, 333–340. [Google Scholar] [CrossRef] [PubMed]
- Mun, Y.-J.; Lee, S.-W.; Jeong, H.-W.; Lee, K.-G.; Kim, J.-H.; Woo, W.-H. Inhibitory effect of miconazole on melanogenesis. Biol. Pharm. Bull. 2004, 27, 806–809. [Google Scholar] [CrossRef]
- Wu, C.-H.; Jeng, J.-H.; Wang, Y.-J.; Tseng, C.-J.; Liang, Y.-C.; Chen, C.-H.; Lee, H.-M.; Lin, J.-K.; Lin, C.-H.; Lin, S.-Y.; et al. Antitumor effects of miconazole on human colon carcinoma xenografts in nude mice through induction of apoptosis and G0/G1 cell cycle arrest. Toxicol. Appl. Pharmacol. 2002, 180, 22–35. [Google Scholar] [CrossRef]
- Wang, Y.-J.; Jeng, J.-H.; Chen, R.-J.; Tseng, H.; Chen, L.-C.; Liang, Y.-C.; Lin, C.-H.; Chen, C.-H.; Chu, J.-S.; Ho, W.-L.; et al. Ketoconazole potentiates the antitumor effects of nocodazole: In vivo therapy for human tumor xenografts in nude mice. Mol. Carcinog. 2002, 34, 199–210. [Google Scholar] [CrossRef] [PubMed]
- Yuan, S.-Y.; Shiau, M.-Y.; Ou, Y.-C.; Huang, Y.-C.; Chen, C.-C.; Cheng, C.-L.; Chiu, K.-Y.; Wang, S.-S.; Tsai, K.-J. Miconazole induces apoptosis via the death receptor 5-dependent and mitochondrial-mediated pathways in human bladder cancer cells. Oncol. Rep. 2017, 37, 3606–3616. [Google Scholar] [CrossRef] [PubMed]
- Roan, C.-J.; Chou, C.-T.; Liang, W.-Z.; Chang, H.-T.; Kuo, D.-H.; Kuo, C.-C.; Chen, F.-A.; Shieh, P.; Jan, C.-R. Effect of Miconazole on [Ca2+]i and Cytotoxicity in ZR-75-1 Human Breast Cancer Cells. Chin. J. Physiol. 2015, 58, 377–384. [Google Scholar] [CrossRef] [PubMed]
- Torikai, M.; Ibara, S.; Ieiri, S.; Hamada, T.; Noguchi, H.; Sueyoshi, K.; Fukuda, T.; Abeyama, K. Prophylactic efficacy of enteral miconazole administration for neonatal intestinal perforation and its potential mechanism. Pediatr. Surg. Int. 2016, 32, 953–957. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Song, X.; Wu, L.; Zhang, T.; Zhao, C.; Yang, X.; Shan, L.; Yu, P.; Sun, Y.; Wang, Y.; et al. Miconazole stimulates post-ischemic neurogenesis and promotes functional restoration in rats. Neurosci. Lett. 2018, 687, 94–98. [Google Scholar] [CrossRef]
- Singh, M.; Saha, R.K.; Saha, H.; Parhi, J. Effect of miconazole nitrate on immunological response and its preventive efficacy in Labeo rohita fingerlings against oomycetes Saprolegnia parasitica. J. Fish Dis. 2018, 41, 1539–1548. [Google Scholar] [CrossRef] [PubMed]
- Lu, X.; He, S.-Y.; Li, Q.; Yang, H.; Jiang, X.; Lin, H.; Chen, Y.; Qu, W.; Feng, F.; Bian, Y.; et al. Investigation of multi-target-directed ligands (MTDLs) with butyrylcholinesterase (BuChE) and indoleamine 2,3-dioxygenase 1 (IDO1) inhibition: The design, synthesis of miconazole analogues targeting Alzheimer’s disease. Bioorg. Med. Chem. 2018, 26, 1665–1674. [Google Scholar] [CrossRef]
- Pemberton, M.N. Morbidity and mortality associated with the interaction of miconazole oral gel and warfarin. Br. Dent. J. 2018, 225, 129–132. [Google Scholar] [CrossRef]
- Liang, G.; Liu, M.; Wang, Q.; Shen, Y.; Mei, H.; Li, D.; Liu, W. Itraconazole exerts its anti-melanoma effect by suppressing Hedgehog, Wnt, and PI3K/mTOR signaling pathways. Oncotarget 2017, 8, 28510–28525. [Google Scholar] [CrossRef]
- Lee, S.W.; Kim, Y.C.; Kim, D.K.; Yoon, T.Y.; Park, H.J.; Cinn, Y.W. Fungal melanonychia. J. Dermatol. 2004, 31, 904–909. [Google Scholar] [CrossRef] [PubMed]
- Schwartz, R.A. Superficial fungal infections. Lancet 2004, 364, 1173–1182. [Google Scholar] [CrossRef]
- Chung, B.Y.; Kim, S.Y.; Jung, J.M.; Won, C.H.; Choi, J.H.; Lee, M.W.; Chang, S.E. The antimycotic agent clotrimazole inhibits melanogenesis by accelerating ERK and PI3K-/Akt-mediated tyrosinase degradation. Exp. Dermatol. 2015, 24, 386–388. [Google Scholar] [CrossRef] [PubMed]
- Talantov, D.; Mazumder, A.; Yu, J.X.; Briggs, T.; Jiang, Y.; Backus, J.; Atkins, D.; Wang, Y. Novel genes associated with malignant melanoma but not benign melanocytic lesions. Clin. Cancer Res. 2005, 11, 7234–7242. [Google Scholar] [CrossRef] [PubMed]
- Mandrekar, J.N. Receiver operating characteristic curve in diagnostic test assessment. J. Thorac. Oncol. 2010, 5, 1315–1316. [Google Scholar] [CrossRef] [PubMed]
- Raskin, L.; Fullen, D.R.; Giordano, T.J.; Thomas, D.G.; Frohm, M.L.; Cha, K.B.; Ahn, J.; Mukherjee, B.; Johnson, T.M.; Gruber, S.B. Transcriptome profiling identifies HMGA2 as a biomarker of melanoma progression and prognosis. J. Investig. Dermatol. 2013, 133, 2585–2592. [Google Scholar] [CrossRef] [PubMed]
- Riker, A.I.; Enkemann, S.A.; Fodstad, O.; Liu, S.; Ren, S.; Morris, C.; Xi, Y.; Howell, P.; Metge, B.; Samant, R.S.; et al. The gene expression profiles of primary and metastatic melanoma yields a transition point of tumor progression and metastasis. BMC Med. Genomics 2008, 1, 13. [Google Scholar] [CrossRef]
- Chen, H.; Sharp, B.M. Content-rich biological network constructed by mining PubMed abstracts. BMC Bioinform. 2004, 5, 147. [Google Scholar]
- Facchiano, F.; D’Arcangelo, D.; Lentini, A.; Rossi, S.; Senatore, C.; Pannellini, T.; Tabolacci, C.; Facchiano, A.M.; Facchiano, A.; Beninati, S. Tissue transglutaminase activity protects from cutaneous melanoma metastatic dissemination: An in vivo study. Amino Acids 2013, 44, 53–61. [Google Scholar] [CrossRef]
- D’Arcangelo, D.; Facchiano, F.; Nassa, G.; Stancato, A.; Antonini, A.; Rossi, S.; Senatore, C.; Cordella, M.; Tabolacci, C.; Salvati, A.; et al. PDGFR-alpha inhibits melanoma growth via CXCL10/IP-10: A multi-omics approach. Oncotarget 2016, 7, 77257. [Google Scholar] [CrossRef]
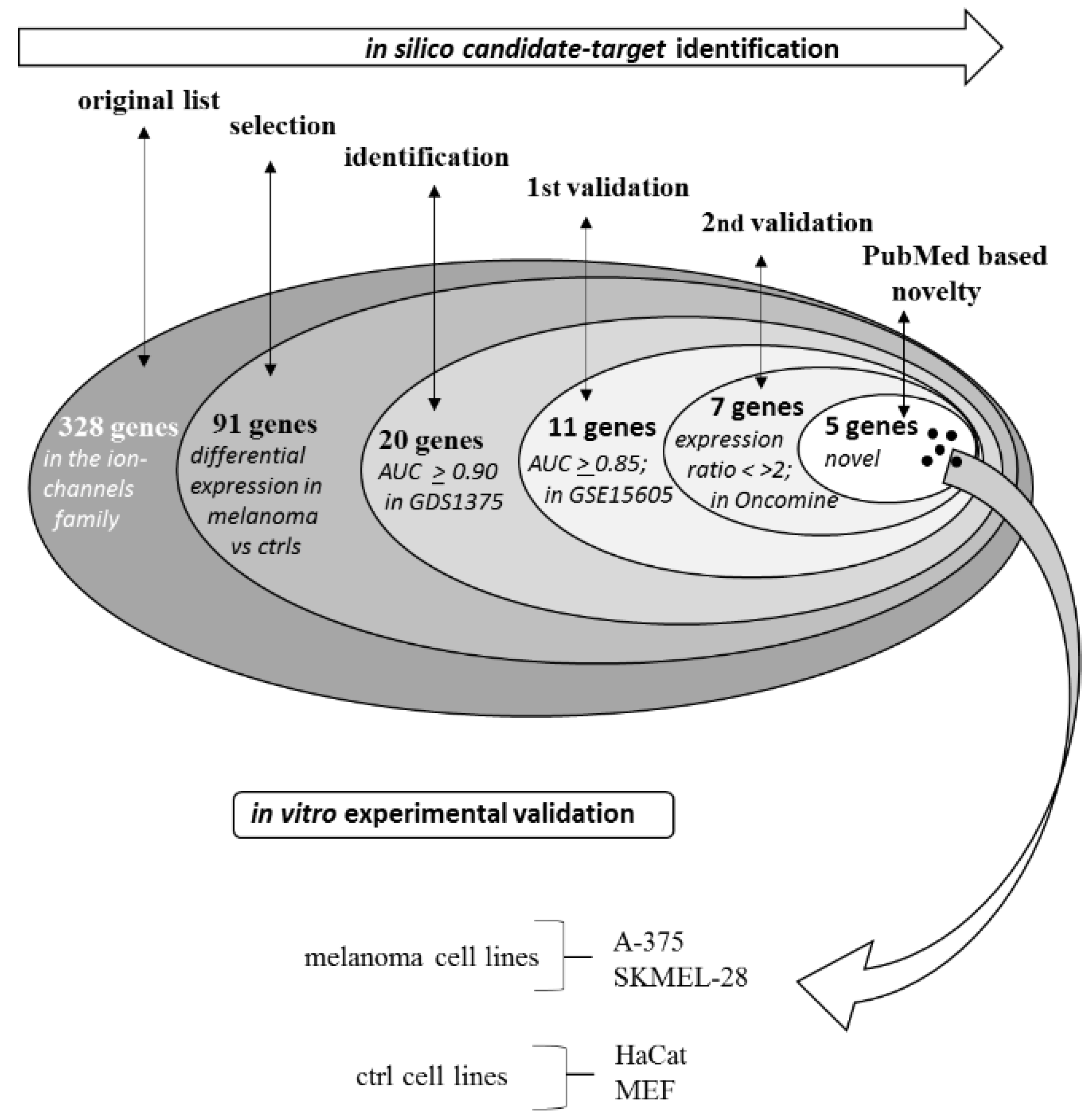



| No. | Gene Name | No. | Gene Name | No. | Gene Name | No. | Gene Name | No. | Gene Name | No. | Gene Name |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | ANO1 | 56 | CACNG5 | 111 | GABRB1 | 166 | GRIN2D | 221 | KCNJ2 | 276 | P2RX6 |
| 2 | ANO10 | 57 | CACNG6 | 112 | GABRB2 | 167 | GRIN3A | 222 | KCNJ3 | 277 | P2RX7 |
| 3 | ANO2 | 58 | CACNG7 | 113 | GABRB3 | 168 | GRIN3B | 223 | KCNJ4 | 278 | PKD2 |
| 4 | ANO3 | 59 | CACNG8 | 114 | GABRD | 169 | HCN1 | 224 | KCNJ5 | 279 | PKD2L1 |
| 5 | ANO4 | 60 | CATSPER1 | 115 | GABRE | 170 | HCN2 | 225 | KCNJ6 | 280 | PKD2L2 |
| 6 | ANO5 | 61 | CATSPER2 | 116 | GABRG1 | 171 | HCN3 | 226 | KCNJ8 | 281 | RYR1 |
| 7 | ANO6 | 62 | CATSPER3 | 117 | GABRG2 | 172 | HCN4 | 227 | KCNJ9 | 282 | RYR2 |
| 8 | ANO7 | 63 | CATSPER4 | 118 | GABRG3 | 173 | HTR3A | 228 | KCNK1 | 283 | RYR3 |
| 9 | ANO8 | 64 | CATSPERB | 119 | GABRP | 174 | HTR3B | 229 | KCNK10 | 284 | SCN10A |
| 10 | ANO9 | 65 | CATSPERD | 120 | GABRQ | 175 | HTR3C | 230 | KCNK12 | 285 | SCN11A |
| 11 | AQP1 | 66 | CATSPERG | 121 | GABRR1 | 176 | HTR3D | 231 | KCNK13 | 286 | SCN1A |
| 12 | AQP10 | 67 | CFTR | 122 | GABRR2 | 177 | HTR3E | 232 | KCNK15 | 287 | SCN1B |
| 13 | AQP11 | 68 | CHRNA1 | 123 | GABRR3 | 178 | HVCN1 | 233 | KCNK16 | 288 | SCN2A |
| 14 | AQP12A | 69 | CHRNA10 | 124 | GJA1 | 179 | ITPR1 | 234 | KCNK17 | 289 | SCN2B |
| 15 | AQP12B | 70 | CHRNA2 | 125 | GJA10 | 180 | ITPR2 | 235 | KCNK18 | 290 | SCN3A |
| 16 | AQP2 | 71 | CHRNA3 | 126 | GJA3 | 181 | ITPR3 | 236 | KCNK2 | 291 | SCN3B |
| 17 | AQP3 | 72 | CHRNA4 | 127 | GJA4 | 182 | KCNA1 | 237 | KCNK3 | 292 | SCN4A |
| 18 | AQP4 | 73 | CHRNA5 | 128 | GJA5 | 183 | KCNA10 | 238 | KCNK4 | 293 | SCN4B |
| 19 | AQP5 | 74 | CHRNA6 | 129 | GJA6P | 184 | KCNA2 | 239 | KCNK5 | 294 | SCN5A |
| 20 | AQP6 | 75 | CHRNA7 | 130 | GJA8 | 185 | KCNA3 | 240 | KCNK6 | 295 | SCN8A |
| 21 | AQP7 | 76 | CHRNA9 | 131 | GJA9 | 186 | KCNA4 | 241 | KCNK7 | 296 | SCN9A |
| 22 | AQP8 | 77 | CHRNB1 | 132 | GJB1 | 187 | KCNA5 | 242 | KCNK9 | 297 | SCNN1A |
| 23 | AQP9 | 78 | CHRNB2 | 133 | GJB2 | 188 | KCNA6 | 243 | KCNMA1 | 298 | SCNN1B |
| 24 | ASIC1 | 79 | CHRNB3 | 134 | GJB3 | 189 | KCNA7 | 244 | KCNN1 | 299 | SCNN1D |
| 25 | ASIC2 | 80 | CHRNB4 | 135 | GJB4 | 190 | KCNB1 | 245 | KCNN2 | 300 | SCNN1G |
| 26 | ASIC3 | 81 | CHRND | 136 | GJB5 | 191 | KCNB2 | 246 | KCNN3 | 301 | TPCN1 |
| 27 | ASIC4 | 82 | CHRNE | 137 | GJB6 | 192 | KCNC1 | 247 | KCNN4 | 302 | TPCN2 |
| 28 | ASIC5 | 83 | CHRNG | 138 | GJB7 | 193 | KCNC2 | 248 | KCNQ1 | 303 | TRPA1 |
| 29 | BEST1 | 84 | CLCN1 | 139 | GJC1 | 194 | KCNC3 | 249 | KCNQ2 | 304 | TRPC1 |
| 30 | BEST2 | 85 | CLCN2 | 140 | GJC2 | 195 | KCNC4 | 250 | KCNQ3 | 305 | TRPC2 |
| 31 | BEST3 | 86 | CLCN3 | 141 | GJC3 | 196 | KCND1 | 251 | KCNQ4 | 306 | TRPC3 |
| 32 | BEST4 | 87 | CLCN4 | 142 | GJD2 | 197 | KCND2 | 252 | KCNQ5 | 307 | TRPC4 |
| 33 | BSND | 88 | CLCN5 | 143 | GJD3 | 198 | KCND3 | 253 | KCNS1 | 308 | TRPC5 |
| 34 | CACNA1A | 89 | CLCN6 | 144 | GJD4 | 199 | KCNF1 | 254 | KCNS2 | 309 | TRPC6 |
| 35 | CACNA1B | 90 | CLCN7 | 145 | GJE1 | 200 | KCNG1 | 255 | KCNS3 | 310 | TRPC7 |
| 36 | CACNA1C | 91 | CLCNKA | 146 | GLRA1 | 201 | KCNG2 | 256 | KCNT1 | 311 | TRPM1 |
| 37 | CACNA1D | 92 | CLCNKB | 147 | GLRA2 | 202 | KCNG3 | 257 | KCNT2 | 312 | TRPM2 |
| 38 | CACNA1E | 93 | CLIC1 | 148 | GLRA3 | 203 | KCNG4 | 258 | KCNU1 | 313 | TRPM3 |
| 39 | CACNA1F | 94 | CLIC2 | 149 | GLRA4 | 204 | KCNH1 | 259 | KCNV1 | 314 | TRPM4 |
| 40 | CACNA1G | 95 | CLIC3 | 150 | GLRB | 205 | KCNH2 | 260 | KCNV2 | 315 | TRPM5 |
| 41 | CACNA1H | 96 | CLIC4 | 151 | GRIA1 | 206 | KCNH3 | 261 | LRRC8A | 316 | TRPM6 |
| 42 | CACNA1I | 97 | CLIC5 | 152 | GRIA2 | 207 | KCNH4 | 262 | LRRC8B | 317 | TRPM7 |
| 43 | CACNA1S | 98 | CLIC6 | 153 | GRIA3 | 208 | KCNH5 | 263 | LRRC8C | 318 | TRPM8 |
| 44 | CACNA2D1 | 99 | CNGA1 | 154 | GRIA4 | 209 | KCNH6 | 264 | LRRC8D | 319 | TRPV1 |
| 45 | CACNA2D2 | 100 | CNGA2 | 155 | GRID1 | 210 | KCNH7 | 265 | LRRC8E | 320 | TRPV2 |
| 46 | CACNA2D3 | 101 | CNGA3 | 156 | GRID2 | 211 | KCNH8 | 266 | MCOLN1 | 321 | TRPV3 |
| 47 | CACNA2D4 | 102 | CNGA4 | 157 | GRIK1 | 212 | KCNJ1 | 267 | MCOLN2 | 322 | TRPV4 |
| 48 | CACNB1 | 103 | CNGB1 | 158 | GRIK2 | 213 | KCNJ10 | 268 | MCOLN3 | 323 | TRPV5 |
| 49 | CACNB2 | 104 | CNGB3 | 159 | GRIK3 | 214 | KCNJ11 | 269 | MIP | 324 | TRPV6 |
| 50 | CACNB3 | 105 | GABRA1 | 160 | GRIK4 | 215 | KCNJ12 | 270 | NALCN | 325 | VDAC1 |
| 51 | CACNB4 | 106 | GABRA2 | 161 | GRIK5 | 216 | KCNJ13 | 271 | P2RX1 | 326 | VDAC2 |
| 52 | CACNG1 | 107 | GABRA3 | 162 | GRIN1 | 217 | KCNJ14 | 272 | P2RX2 | 327 | VDAC3 |
| 53 | CACNG2 | 108 | GABRA4 | 163 | GRIN2A | 218 | KCNJ15 | 273 | P2RX3 | 328 | ZACN |
| 54 | CACNG3 | 109 | GABRA5 | 164 | GRIN2B | 219 | KCNJ16 | 274 | P2RX4 | ||
| 55 | CACNG4 | 110 | GABRA6 | 165 | GRIN2C | 220 | KCNJ18 | 275 | P2RX5 |
| No. | Gene Name | t Test Nevi vs. Melanoma | Ratio Melanoma vs. Nevi | AUC | No. | Gene Name | t Test Nevi vs. Melanoma | Ratio Melanoma vs. Nevi | AUC | ||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | ANO1 | 8.7 × 10−18 | 0.231 |  | 0.98 | 47 | CACNB2 | 1.5 × 10−3 | 0.4388 |  | 0.73 |
| 2 | KCNK7 | 1.8 × 10−17 | 0.081 |  | 0.94 | 48 | TRPV6 | 1.9 × 10−3 | 0.576 |  | 0.65 |
| 3 | SCNN1A | 1.0 × 10−14 | 0.07 |  | 0.98 | 49 | CLIC2 | 2.0 × 10−3 | 0.319 |  | 0.87 |
| 4 | SCNN1B | 1.4 × 10−14 | 0.313 |  | 0.96 | 50 | SCN3A | 2.2 × 10−3 | 0.382 |  | 0.64 |
| 5 | GABRE | 2.3 × 10−13 | 0.203 |  | 0.95 | 51 | GRIN2A | 2.3 × 10−3 | 0.619 |  | 0.69 |
| 6 | GJB5 | 3.4 × 10−13 | 0.1277 |  | 0.97 | 52 | CACNG4 | 2.6 × 10−3 | 1.317 |  | 0.74 |
| 7 | CLIC1 | 1.9 × 10−12 | 1.738 |  | 0.95 | 53 | GABRB1 | 3.0 × 10−3 | 0.698 |  | 0.69 |
| 8 | KCND3 | 2.0 × 10−11 | 0.487 |  | 0.94 | 54 | GJB4 | 3.7 × 10−3 | 0.588 |  | 0.63 |
| 9 | KCNK1 | 8.6 × 10−11 | 0.269 |  | 0.93 | 55 | CACNB4 | 4.3 × 10−3 | 0.474 |  | 0.66 |
| 10 | GRIA1 | 8.7 × 10−11 | 0.1049 |  | 0.93 | 56 | CACNA11 | 5.0 × 10−3 | 0.629 |  | 0.70 |
| 11 | VDAC1 | 2.0 × 10−10 | 1.8 |  | 0.96 | 57 | KCNK2 | 5.0 × 10−3 | 0.512 |  | 0.64 |
| 12 | GJB3 | 2.1 × 10−10 | 0.2453 |  | 0.96 | 58 | TRPC1 | 5.4 × 10−3 | 0.665 |  | 0.70 |
| 13 | KCNN2 | 3.6 × 10−10 | 4.337 |  | 0.91 | 59 | TRPC7 | 5.7 × 10−3 | 1.396 |  | 0.74 |
| 14 | AQP1 | 5.5 × 10−10 | 0.268 |  | 0.84 | 60 | GRIK1 | 6.3 × 10−3 | 1.885 |  | 0.72 |
| 15 | ITPR3 | 1.1 × 10−9 | 2.311 |  | 0.93 | 61 | GRIN2D | 6.7 × 10−3 | 1.922 |  | 0.75 |
| 16 | RYR1 | 1.8 × 10−9 | 0.2414 |  | 0.81 | 62 | CLIC4 | 6.8 × 10−3 | 0.671 |  | 0.69 |
| 17 | GJA1 | 2.1 × 10−9 | 0.086 |  | 0.98 | 63 | CACNA1S | 7.7 × 10−3 | 1.91 |  | 0.77 |
| 18 | KCNJ13 | 1.9 × 10−8 | 0.266 |  | 0.86 | 64 | CATSPER2 | 8.1 × 10−3 | 0.677 |  | 0.75 |
| 19 | TRPV2 | 3.0 × 10−8 | 1.77 |  | 0.88 | 65 | GABRP | 8.1 × 10−3 | 0.3014 |  | 0.69 |
| 20 | GJB1 | 3.4 × 10−8 | 2.643 |  | 0.93 | 66 | GJA4 | 9.0 × 10−3 | 0.5357 |  | 0.68 |
| 21 | CLIC3 | 4.0 × 10−8 | 0.118 |  | 0.85 | 67 | KCNJ15 | 9.0 × 10−3 | 0.506 |  | 0.64 |
| 22 | LRRC8B | 4.4 × 10−8 | 0.459 |  | 0.90 | 68 | CNGB3 | 1.0 × 10−2 | 0.63 |  | 0.69 |
| 23 | HCN2 | 2.1 × 10−7 | 3.88 |  | 0.89 | 69 | SCNN1D | 1.0 × 10−2 | 0.735 |  | 0.69 |
| 24 | TRPM1 | 3.0 × 10−7 | 0.368 |  | 0.77 | 70 | GRIA4 | 1.2 × 10−2 | 0.71 |  | 0.68 |
| 25 | BEST2 | 3.6 × 10−7 | 0.456 |  | 0.86 | 71 | TRPM3 | 1.3 × 10−2 | 0.762 |  | 0.72 |
| 26 | AQP3 | 6.9 × 10−7 | 0.067 |  | 0.97 | 72 | CHRNA10 | 1.5 × 10−2 | 0.759 |  | 0.68 |
| 27 | KCNN4 | 1.5 × 10−6 | 3.79 |  | 0.93 | 73 | AQP9 | 1.9 × 10−2 | 0.56 |  | 0.75 |
| 28 | SCN1B | 1.5 × 10−6 | 0.657 |  | 0.83 | 74 | VDAC3 | 1.9 × 10−2 | 1.27 |  | 0.78 |
| 29 | LRRC8E | 1.5 × 10−6 | 0.563 |  | 0.82 | 75 | KCNV1 | 2.0 × 10−2 | 1.737 |  | 0.70 |
| 30 | CACNB1 | 2.9 × 10−6 | 0.614 |  | 0.82 | 76 | VDAC2 | 2.0 × 10−2 | 1.15 |  | 0.71 |
| 31 | KCNJ12 | 3.5 × 10−6 | 0.388 |  | 0.80 | 77 | CHRNB1 | 2.1 × 10−2 | 0.754 |  | 0.68 |
| 32 | KCNJ18 | 3.5 × 10−6 | 0.388 |  | 0.80 | 78 | ANO2 | 2.4 × 10−2 | 0.67 |  | 0.68 |
| 33 | TRPM4 | 3.6 × 10−6 | 0.154 |  | 0.87 | 79 | CHRNB2 | 2.6 × 10−2 | 1.383 |  | 0.70 |
| 34 | KCNS3 | 7.7 × 10−6 | 0.403 |  | 0.93 | 80 | SCNN1G | 2.8 × 10−2 | 1.45 |  | 0.67 |
| 35 | PKD2L1 | 8.7 × 10−6 | 2.66 |  | 0.86 | 81 | TRPC6 | 2.8 × 10−2 | 0.594 |  | 0.67 |
| 36 | P2RX4 | 1.0 × 10−5 | 1.756 |  | 0.85 | 82 | KCNA1 | 3.0 × 10−2 | 0.79 |  | 0.67 |
| 37 | GRIK2 | 3.9 × 10−5 | 3.922 |  | 0.86 | 83 | PKD2L2 | 3.0 × 10−2 | 0.447 |  | 0.52 |
| 38 | KCNJ4 | 5.0 × 10−5 | 2.87 |  | 0.84 | 84 | GABRA6 | 3.9 × 10−2 | 0.622 |  | 0.65 |
| 39 | TRPM2 | 6.5 × 10−5 | 0.219 |  | 0.93 | 85 | GRINB2 | 3.9 × 10−2 | 1.225 |  | 0.67 |
| 40 | CLCN7 | 1.2 × 10−4 | 1.753 |  | 0.80 | 86 | KCNJ2 | 4.0 × 10−2 | 1.33 |  | 0.72 |
| 41 | MCOLN3 | 2.2 × 10−4 | 0.352 |  | 0.62 | 87 | TRPV5 | 4.0 × 10−2 | 1.29 |  | 0.66 |
| 42 | SCN2B | 2.6 × 10−4 | 0.483 |  | 0.71 | 88 | ASIC1 | 4.1 × 10−2 | 0.71 |  | 0.63 |
| 43 | LRRC8D | 2.8 × 10−4 | 1.39 |  | 0.78 | 89 | AQP5 | 4.1 × 10−2 | 1.498 |  | 0.67 |
| 44 | MCOLN1 | 3.8 × 10−4 | 1.43 |  | 0.72 | 90 | CACNG3 | 4.3 × 10−2 | 1.903 |  | 0.73 |
| 45 | CLCN6 | 5.0 × 10−4 | 0.74 |  | 0.79 | 91 | TRPC4 | 4.3 × 10−2 | 2.097 |  | 0.69 |
| 46 | CNGA1 | 5.0 × 10−4 | 0.58 |  | 0.72 | ||||||
| No. | Gene Name | Screening Phase (in the Talantov Dataset, GEO, GDS1375) | First-Round Validation (in the Raskin Dataset, GEO GSE15605) * | Second-Round Validation (in the Riker Dataset, Oncomine) ** | Novelty (in PubMeds Abstracts) *** | Full in Silico Validation | ||
|---|---|---|---|---|---|---|---|---|
| 63 Patients | 62 Patients | 59 Patients | ||||||
| t Test Melanoma vs. Nevi | Ratio Melan. vs. Nevi | AUC | Validation on AUC Value * | Validation on Ratio Value | Gene Name and Melanoma Words Co-Occurrence | |||
| 1 | SCNN1A | 1.0 × 10−14 | 0.07 | 0.98 | Yes (0.85) | Yes (−4.94) | 0 |  |
| 2 | ANO1 | 8.6 × 10−18 | 0.231 | 0.98 | Yes (0.87) | |||
| 3 | GJA1 | 2.1 × 10−9 | 0.086 | 0.98 | Yes (0.88) | |||
| 4 | GJB5 | 3.4 × 10−13 | 0.1277 | 0.97 | ||||
| 5 | GJB3 | 2.1 × 10−10 | 0.2453 | 0.96 | Yes (0.86) | Yes (−6.662) | 0 |  |
| 6 | AQP3 | 6.9 × 10−7 | 0.067 | 0.97 | ||||
| 7 | SCNN1B | 1.4 × 10−14 | 0.313 | 0.96 | Yes (0.87) | |||
| 8 | VDAC1 | 2.0 × 10−10 | 1.8 | 0.96 | ||||
| 9 | CLIC1 | 1.9 × 10−12 | 1.738 | 0.96 | ||||
| 10 | GABRE | 2.3 × 10−13 | 0.203 | 0.95 | Yes (0.88) | Yes (−3.162) | ≥1 | |
| 11 | KCNK7 | 1.8 × 10−17 | 0.081 | 0.94 | Yes (0.86) | Yes (−2.832) | 0 |  |
| 12 | KCND3 | 2.0 × 10−11 | 0.487 | 0.94 | Yes (0.89) | |||
| 14 | KCNN4 | 1.5 × 10−6 | 3.79 | 0.93 | ||||
| 13 | ITPR3 | 1.1 × 10−9 | 2.311 | 0.93 | ||||
| 15 | KCNK1 | 8.6 × 10−11 | 0.269 | 0.93 | Yes (0.89) | |||
| 16 | KCNS3 | 7.7 × 10−6 | 0.403 | 0.93 | ||||
| 17 | TRPM2 | 6.5 × 10−5 | 0.219 | 0.93 | ||||
| 18 | GRIA1 | 8.7 × 10−11 | 0.1049 | 0.93 | ||||
| 19 | GJB1 | 3.4 × 10−8 | 2.643 | 0.93 | Yes (0.87) | Yes (3.303) | 0 |  |
| 20 | KCNN2 | 3.6 × 10−10 | 4.337 | 0.91 | Yes (0.91) | Yes (2.284) | 0 |  |
| Gene Name | Drug Targeting the Gene, According to www.dgidb.org, and IUPAC Name | Reference (PMID) | Interaction Type with the Gene | Main Target Tissue | Action and Indications | EC50 |
|---|---|---|---|---|---|---|
| SCNN1A | Triamterene 6-phenylpteridine-2,4,7-triamine | [30] | channel blocker | kidney | diuretic, anti-edema | 1660 nM [31] |
| Amiloride 3,5-diamino-6-chloro-N-(diaminomethylidene)pyrazine-2-carboxamide | [32] | channel blocker | kidney | diuretic; congestive heart failure; hypertension. | 0.1 μM [33] | |
| GJB3 | Flufenamic acid 2-{[3-(trifluoromethyl)phenyl]amino}benzoic acid | [34] | inhibitor | muscles joints | anti-cancer | 100/200 μM [35] |
| Carbenoxolone 2S,4aS,6aR,6aS,6bR,8aR,10S,12aS,14bR)-10-(3-carboxypropanoyloxy)-2,4a,6a,6b,9,9,12a-heptamethyl-13-oxo-3,4,5,6,6a,7,8,8a,10,11,12,14b-dodecahydro-1H-picene-2-carboxylic acid | [34,36] | inhibitor | digestive tract | anti-ulcer/neuro protection | 48 μM [37] | |
| KCNK7 | Not found | |||||
| GJB1 | Not found | |||||
| KCNN2 | Miconazole 1-[2-(2,4-dichlorophenyl)-2-[(2,4-dichlorophenyl)methoxy]ethyl]-1H-imidazole | [38] | Inhibitor | systemic, skin | anti-fungal infections | 75 μM [39] |
| Tubocurarine (1S,16R)-9,21-dihydroxy-10,25-dimethoxy-15,15,30-trimethyl-7,23-dioxa-15,30-diazaheptacyclo [22.6.2.23,6.18,12.118,22.027,31.016,34]hexatriaconta-3,5,8(34),9,11,18(33),19,21, 24, 26,31,35-dodecaen-15-ium | [34] | channel blocker | CNS | diagnostic in myastenia gravis; to treat smoking withdrawl syndrom | 1.3 μM [40] | |
| Bendroflumethiazide 3-benzyl-1,1-dioxo-6-(trifluoromethyl)-3,4-dihydro-2H-1λ6,2,4-benzothiadiazine-7-sulfonamide | [34] | channel blocker | kidney smooth muscle cells | Anti HBV, diuretic; anti-edema; hypertension | 53 μM [41] |
| Biological Process | Pathway ID | False Discovery Rate |
|---|---|---|
| Potassium ion transmembrane transport | GO:00781805 | 1.9 × 10−7 |
| Arginine catabolic process | GO:0006527 | 6.4 × 10−6 |
| Synaptic transmission | GO:0007268 | 6.4 × 10−6 |
| Positive regulation of guanylate cyclase activity | GO:0031284 | 4.2 × 10−5 |
| Regulation of system process | CO:0044057 | 4.2 × 10−5 |
| Nitric oxide biosynthetic process | GO:0006809 | 5.3 × 10−5 |
| Regulation of potassium ion transport | GO:0043266 | 5.7 × 10−5 |
| Regulation of blood circulation | GO:1903522 | 5.7 × 10−5 |
| Nitric oxide mediated signal transduction | GO:0007263 | 8.1 × 10−5 |
| Blood circulation | GO:0008015 | 8.3 × 10−5 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
D’Arcangelo, D.; Scatozza, F.; Giampietri, C.; Marchetti, P.; Facchiano, F.; Facchiano, A. Ion Channel Expression in Human Melanoma Samples: In Silico Identification and Experimental Validation of Molecular Targets. Cancers 2019, 11, 446. https://doi.org/10.3390/cancers11040446
D’Arcangelo D, Scatozza F, Giampietri C, Marchetti P, Facchiano F, Facchiano A. Ion Channel Expression in Human Melanoma Samples: In Silico Identification and Experimental Validation of Molecular Targets. Cancers. 2019; 11(4):446. https://doi.org/10.3390/cancers11040446
Chicago/Turabian StyleD’Arcangelo, Daniela, Francesca Scatozza, Claudia Giampietri, Paolo Marchetti, Francesco Facchiano, and Antonio Facchiano. 2019. "Ion Channel Expression in Human Melanoma Samples: In Silico Identification and Experimental Validation of Molecular Targets" Cancers 11, no. 4: 446. https://doi.org/10.3390/cancers11040446
APA StyleD’Arcangelo, D., Scatozza, F., Giampietri, C., Marchetti, P., Facchiano, F., & Facchiano, A. (2019). Ion Channel Expression in Human Melanoma Samples: In Silico Identification and Experimental Validation of Molecular Targets. Cancers, 11(4), 446. https://doi.org/10.3390/cancers11040446





