Amino Acid Deprivation-Induced Autophagy Requires Upregulation of DIRAS3 through Reduction of E2F1 and E2F4 Transcriptional Repression
Abstract
1. Introduction
2. Results
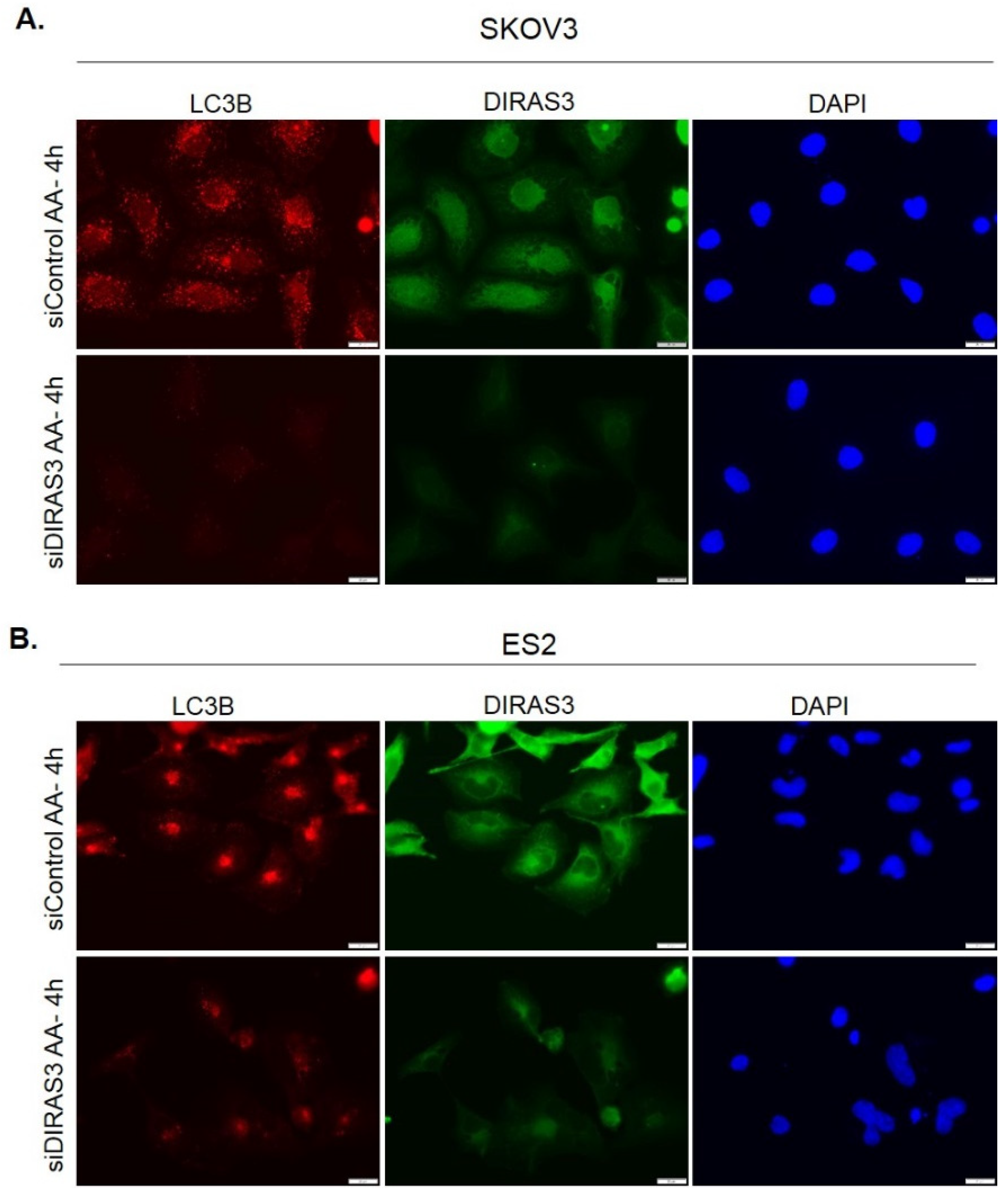
2.1. Amino Acid Deprivation Inhibits mTOR Signaling, Inducing Upregulation of DIRAS3 mRNA Expression and Autophagy
2.2. Knockdown of Sestrin2 Suppresses Amino Acid Starvation-Induced Autophagy
2.3. Acute Amino Acid Starvation Induces Transcriptional Upregulation of DIRAS3 with Decreased Binding of E2F1/E2F4 to the DIRAS3 Promoter and Decreased Levels of E2F1 and E2F4, But Does not Affect Methylation of the DIRAS3 Promoter or miRNA Regulation of DIRAS3 Expression
2.4. Knockdown of E2F1/E2F4 Induces DIRAS3-Mediated Autophagy
2.5. CEBPα Positively Regulates DIRAS3 Expression and Is also Required for Starvation-Induced DIRAS3-Mediated Autophagy
3. Discussion
4. Materials and Methods
4.1. Antibodies and Reagents
4.2. Cell Lines
4.3. Measurement of mRNA Expression
| GAPDH | Forward | ATGGAATCCATCACCATCTT |
| Reverse | CGCCCCACTTGATTTTGG | |
| LC3 | Forward 356F | GAAGCAGCTTCCTGTTCTGG |
| Reverse 588R | TCATCCCGAACGTCTCCTGG | |
| DIRAS3 | Forward | GTACCTGCCGACCATTGAAAA |
| Reverse | GGGTTTCCTTCTTGGTCACTG | |
| E2F1 | Forward 734F | ACGTGACGTGTCAGGACCT |
| Reverse 879R | GATCGGGCCTTGTTTGCTCTT | |
| E2F4 | Forward 202F | ATCGGGCTAATCGAGAAAAAGTC |
| Reverse 350R | TGCTGGTCTAGTTCTTGCTCC |
4.4. Brightfield and Fluorescence Microscopy
4.5. Transmission Electron Microscopy
4.6. Western Blotting
4.7. siRNA Transfection
4.8. Immunofluorescent Staining
4.9. Quantitative Pyrosequencing
| DIRAS3-1-F | GTAAGGGAGAAAGAAGTTAGA |
| DIRAS3-1-R | biotin-TACTATCCTAACAAAACCCTC |
| DIRAS3-1-SF2 | ATTTGGAAAAGGGATTGG |
| DIRAS3-2-F | GTTGGGTTAGTTTTTTATAGTTGGTT |
| DIRAS3-2-R | biotin-AACCAAACAACCTAAAAAACAAATAC |
| DIRAS3-2-SF2 | TTGGGGTGTTTAGTTGGTTG |
4.10. Chromatin Immunoprecipitation (ChIP) Assay
| ddH2O | 3.6 µL |
| iTaqSYBR-Green master Mix (BioRad #172-5124) | 10 µL |
| Primer Mix (10 µM working concentration F+R) | 0.4 µL |
| DNA (20 µL diluted with 38 µL ddH2O) | 6.0 µL |
| Initial Denaturation | 94 °C | 10 min |
| Denature | 94 °C | 20 s |
| Anneal and Extension | 60 °C | 1.0 min |
| DIRAS3-0 | ChIP Forward 524F | TTTACCGGTCTTGCCACTAATG |
| ChIP Reverse 341R | TCCAAAAGCAGTTTAATGCAGG | |
| DIRAS3-1 | ChIP Forward 76F | TTAAGAACCTTTTGCCTAGCC |
| ChIP Reverse 227R | CGGTCCAACTGATTTTAGACGAA |
4.11. miRNA Quantitative Reverse Transcription-Polymerase Chain Reaction (qRT-PCR)
4.12. Statistics
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Appendix A


References
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2018. CA Cancer J. Clin. 2018, 68, 7–30. [Google Scholar] [CrossRef]
- Lu, Z.; Baquero, M.T.; Yang, H.; Yang, M.; Reger, A.S.; Kim, C.; Levine, D.A.; Clarke, C.H.; Liao, W.S.; Bast, R.C., Jr. DIRAS3 regulates the autophagosome initiation complex in dormant ovarian cancer cells. Autophagy 2014, 10, 1071–1092. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Xu, F.; Peng, H.; Fang, X.; Zhao, S.; Li, Y.; Cuevas, B.; Kuo, W.L.; Gray, J.W.; Siciliano, M.; et al. NOEY2 (ARHI), an imprinted putative tumor suppressor gene in ovarian and breast carcinomas. Proc. Natl. Acad. Sci. USA 1999, 96, 214–219. [Google Scholar] [CrossRef] [PubMed]
- Feng, W.; Lu, Z.; Luo, R.Z.; Zhang, X.; Seto, E.; Liao, W.S.; Yu, Y. Multiple histone deacetylases repress tumor suppressor gene ARHI in breast cancer. Int. J. Cancer 2007, 120, 1664–1668. [Google Scholar] [CrossRef] [PubMed]
- Feng, W.; Marquez, R.T.; Lu, Z.; Liu, J.; Lu, K.H.; Issa, J.P.; Fishman, D.M.; Yu, Y.; Bast, R.C., Jr. Imprinted tumor suppressor genes ARHI and PEG3 are the most frequently down-regulated in human ovarian cancers by loss of heterozygosity and promoter methylation. Cancer 2008, 112, 1489–1502. [Google Scholar] [CrossRef] [PubMed]
- Fujii, S. Reactivation of the silenced and imprinted alleles of ARHI is associated with increased histone H3 acetylation and decreased histone H3 lysine 9 methylation. Hum. Mol. Genet. 2003, 12, 1791–1800. [Google Scholar] [CrossRef]
- Yu, Y.; Fujii, S.; Yuan, J.; Luo, R.Z.; Wang, L.; Bao, J.; Kadota, M.; Oshimura, M.; Dent, S.R.; Issa, J.P.; et al. Epigenetic regulation of ARHI in breast and ovarian cancer cells. Ann. N. Y. Acad. Sci. 2003, 983, 268–277. [Google Scholar] [CrossRef] [PubMed]
- Yuan, J.; Luo, R.Z.; Fujii, S.; Wang, L.; Hu, W.; Andreeff, M.; Pan, Y.; Kadota, M.; Oshimura, M.; Sahin, A.A.; et al. Aberrant methylation and silencing of ARHI, an imprinted tumor suppressor gene in which the function is lost in breast cancers. Cancer Res. 2003, 63, 4174–4180. [Google Scholar]
- Lu, Z.; Luo, R.Z.; Peng, H.; Huang, M.; Nishmoto, A.; Hunt, K.K.; Helin, K.; Liao, W.S.; Yu, Y. E2F-HDAC complexes negatively regulate the tumor suppressor gene ARHI in breast cancer. Oncogene 2006, 25, 230–239. [Google Scholar] [CrossRef] [PubMed]
- Lu, Z.; Luo, R.Z.; Peng, H.; Rosen, D.G.; Atkinson, E.N.; Warneke, C.; Huang, M.; Nishmoto, A.; Liu, J.; Liao, W.S.; et al. Transcriptional and posttranscriptional down-regulation of the imprinted tumor suppressor gene ARHI (DRAS3) in ovarian cancer. Clin. Cancer Res. 2006, 12, 2404–2413. [Google Scholar] [CrossRef]
- Lu, Z.; Luo, R.Z.; Lu, Y.; Zhang, X.; Yu, Q.; Khare, S.; Kondo, S.; Kondo, Y.; Yu, Y.; Mills, G.B.; et al. The tumor suppressor gene ARHI regulates autophagy and tumor dormancy in human ovarian cancer cells. J. Clin. Investig. 2008, 118, 3917–3929. [Google Scholar] [CrossRef]
- Sutton, M.N. The Role of the DIRAS Family Members in Regulating Ras Function, Cancer Growth and Autophagy. The University of Texas MD Anderson Cancer Center UTHealth Graduate School of Biomedical Sciences, 2017. Available online: https://digitalcommons.library.tmc.edu/utgsbs_dissertations/741/ (accessed on 30 May 2018).
- Levine, B.; Klionsky, D.J. Development by self-digestion: molecular mechanisms and biological functions of autophagy. Dev. Cell 2004, 6, 463–477. [Google Scholar] [CrossRef]
- Yue, Z.; Jin, S.; Yang, C.; Levine, A.J.; Heintz, N. Beclin 1, an autophagy gene essential for early embryonic development, is a haploinsufficient tumor suppressor. Proc. Natl. Acad. Sci. USA 2003, 100, 15077–15082. [Google Scholar] [CrossRef]
- Lu, Z.; Yang, H.; Sutton, M.N.; Yang, M.; Clarke, C.H.; Liao, W.S.; Bast, R.C., Jr. ARHI (DIRAS3) induces autophagy in ovarian cancer cells by downregulating the epidermal growth factor receptor, inhibiting PI3K and Ras/MAP signaling and activating the FOXo3a-mediated induction of Rab7. Cell Death Differ. 2014, 21, 1275–1289. [Google Scholar] [CrossRef] [PubMed]
- Klionsky, D.J.; Abdelmohsen, K.; Abe, A.; Abedin, M.J.; Abeliovich, H.; Acevedo Arozena, A.; Adachi, H.; Adams, C.M.; Adams, P.D.; Adeli, K.; et al. Guidelines for the use and interpretation of assays for monitoring autophagy (3rd edition). Autophagy 2016, 12, 1–222. [Google Scholar] [CrossRef] [PubMed]
- Khan, N.A.; Nikkanen, J.; Yatsuga, S.; Jackson, C.; Wang, L.; Pradhan, S.; Kivelä, R.; Pessia, A.; Velagapudi, V.; Suomalainen, A. mTORC1 Regulates Mitochondrial Integrated Stress Response and Mitochondrial Myopathy Progression. Cell Metab. 2017, 26, 419–428. [Google Scholar] [CrossRef] [PubMed]
- Jewell, J.L.; Guan, K.L. Nutrient signaling to mTOR and cell growth. Trends Biochem. Sci. 2013, 38, 233–242. [Google Scholar] [CrossRef]
- Wang, C.I.; Chen, Y.Y.; Wang, C.L.; Yu, J.S.; Chang, Y.S.; Yu, C.J. mTOR regulates proteasomal degradation and Dp1/E2F1- mediated transcription of KPNA2 in lung cancer cells. Oncotarget 2016, 7, 25432–25442. [Google Scholar] [CrossRef] [PubMed]
- Saxton, R.A.; Knockenhauer, K.E.; Wolfson, R.L.; Chantranupong, L.; Pacold, M.E.; Wang, T.; Schwartz, T.U.; Sabatini, D.M. Structural basis for leucine sensing by the Sestrin2-mTORC1 pathway. Science 2016, 351, 53–58. [Google Scholar] [CrossRef]
- Wolfson, R.L.; Chantranupong, L.; Saxton, R.A.; Shen, K.; Scaria, S.M.; Cantor, J.R.; Sabatini, D.M. Sestrin2 is a leucine sensor for the mTORC1 pathway. Science 2016, 351, 43–48. [Google Scholar] [CrossRef]
- Neufeld, T.P. Autophagy and cell growth--the yin and yang of nutrient responses. J. Cell Sci. 2012, 125, 2359–2368. [Google Scholar] [CrossRef]
- Sun, C.C.; Zhou, Q.; Hu, W.; Li, S.J.; Zhang, F.; Chen, Z.L.; Li, G.; Bi, Z.Y.; Bi, Y.Y.; Gong, F.Y.; et al. Transcriptional E2F1/2/5/8 as potential targets and transcriptional E2F3/6/7 as new biomarkers for the prognosis of human lung carcinoma. Aging 2018, 10, 973–987. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Kurtyka, C.A.; Boyapalle, S.; Sung, S.S.; Lawrence, H.; Guida, W.; Cress, W.D. A small-molecule E2F inhibitor blocks growth in a melanoma culture model. Cancer Res. 2008, 68, 6292–6299. [Google Scholar] [CrossRef]
- Muller, C.; Calkhoven, C.F.; Sha, X.; Leutz, A. The CCAAT enhancer-binding protein alpha (C/EBPalpha) requires a SWI/SNF complex for proliferation arrest. J. Biol. Chem. 2004, 279, 7353–7358. [Google Scholar] [CrossRef]
- Wang, Y.; Zhu, J.; Zhang, L.; Zhang, Z.; He, L.; Mou, Y.; Deng, Y.; Cao, Y.; Yang, P.; Su, Y.; et al. Role of C/EBP homologous protein and endoplasmic reticulum stress in asthma exacerbation by regulating the IL-4/signal transducer and activator of transcription 6/transcription factor EC/IL-4 receptor alpha positive feedback loop in M2 macrophages. J. Allergy Clin. Immunol. 2017, 140, 1550–1561. [Google Scholar] [CrossRef] [PubMed]
- Slomiany, B.A.; D’Arigo, K.L.; Kelly, M.M.; Kurtz, D.T. C/EBPalpha inhibits cell growth via direct repression of E2F-DP-mediated transcription. Mol. Cell Biol. 2000, 20, 5986–5997. [Google Scholar] [CrossRef]
- Fullgrabe, J.; Ghislat, G.; Cho, D.-H.; Rubinsztein, D.C. Transcriptional regulation of mammalian autophagy at a glance. J. Cell Sci. 2016, 129, 3059–3066. [Google Scholar] [CrossRef] [PubMed]
- White, E. The role for autophagy in cancer. J. Clin. Investig. 2015, 125, 42–46. [Google Scholar] [CrossRef]
- Maycotte, P.; Thorburn, A. Autophagy and cancer therapy. Cancer Biol. Ther. 2011, 11, 127–137. [Google Scholar] [CrossRef] [PubMed]
- Levy, J.M.M.; Towers, C.G.; Thorburn, A. Targeting autophagy in cancer. Nat. Rev. Cancer 2017, 17, 528–542. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.Y.; White, E. Autophagy, Metabolism, and Cancer. Cold Spring Harb Symp Quant. Biol. 2016, 81, 73–78. [Google Scholar] [CrossRef]
- Rabinowitz, J.D.; White, E. Autophagy and metabolism. Science 2010, 330, 1344–1348. [Google Scholar] [CrossRef]
- Qiu, J.; Li, X.; He, Y.; Sun, D.; Li, W.; Xin, Y. Distinct subgroup of the Ras family member 3 (DIRAS3) expression impairs metastasis and induces autophagy of gastric cancer cells in mice. J. Cancer Res. Clin. Oncol. 2018, 144, 1869–1886. [Google Scholar] [CrossRef]
- Wolpin, B.M.; Rubinson, D.A.; Wang, X.; Chan, J.A.; Cleary, J.M.; Enzinger, P.C.; Fuchs, C.S.; McCleary, N.J.; Meyerhardt, J.A.; Ng, K. Phase II and pharmacodynamic study of autophagy inhibition using hydroxychloroquine in patients with metastatic pancreatic adenocarcinoma. Oncologist 2014, 19, 637–638. [Google Scholar] [CrossRef]
- Manic, G.; Obrist, F.; Kroemer, G.; Vitale, I.; Galluzzi, L. Chloroquine and hydroxychloroquine for cancer therapy. Mol. Cell Oncol. 2014, 1, e29911. [Google Scholar] [CrossRef]
- Amaravadi, R.K.; Winkler, J.D. Lys05: a new lysosomal autophagy inhibitor. Autophagy 2012, 8, 1383–1384. [Google Scholar] [CrossRef]
- Mao, W.; Peters, H.L.; Sutton, M.N.; Orozco, A.F.; Pang, L.; Yang, H.; Lu, Z.; Bast, R.C., Jr. The role of vascular endothelial growth factor, interleukin 8, and insulinlike growth factor in sustaining autophagic DIRAS3-induced dormant ovarian cancer xenografts. Cancer 2019, 125, 1267–1280. [Google Scholar] [CrossRef]
- Wang, G.; Luo, X.; Wang, J.; Wan, J.; Xia, S.; Zhu, H.; Qian, J.; Wang, Y. MeDReaders: a database for transcription factors that bind to methylated DNA. Nucleic Acids Res. 2018, 46, D146–D151. [Google Scholar] [CrossRef]
- Zhu, H.; Wang, G.; Qian, J. Transcription factors as readers and effectors of DNA methylation. Nat. Rev. Genet. 2016, 17, 551–565. [Google Scholar] [CrossRef]
- Estecio, M.R.; Yan, P.S.; Ibrahim, A.E.; Tellez, C.S.; Shen, L.; Huang, T.H.; Issa, J.P. High-throughput methylation profiling by MCA coupled to CpG island microarray. Genome Res. 2007, 17, 1529–1536. [Google Scholar] [CrossRef]







© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sutton, M.N.; Huang, G.Y.; Zhou, J.; Mao, W.; Langley, R.; Lu, Z.; Bast, R.C., Jr. Amino Acid Deprivation-Induced Autophagy Requires Upregulation of DIRAS3 through Reduction of E2F1 and E2F4 Transcriptional Repression. Cancers 2019, 11, 603. https://doi.org/10.3390/cancers11050603
Sutton MN, Huang GY, Zhou J, Mao W, Langley R, Lu Z, Bast RC Jr. Amino Acid Deprivation-Induced Autophagy Requires Upregulation of DIRAS3 through Reduction of E2F1 and E2F4 Transcriptional Repression. Cancers. 2019; 11(5):603. https://doi.org/10.3390/cancers11050603
Chicago/Turabian StyleSutton, Margie N., Gilbert Y. Huang, Jinhua Zhou, Weiqun Mao, Robert Langley, Zhen Lu, and Robert C. Bast, Jr. 2019. "Amino Acid Deprivation-Induced Autophagy Requires Upregulation of DIRAS3 through Reduction of E2F1 and E2F4 Transcriptional Repression" Cancers 11, no. 5: 603. https://doi.org/10.3390/cancers11050603
APA StyleSutton, M. N., Huang, G. Y., Zhou, J., Mao, W., Langley, R., Lu, Z., & Bast, R. C., Jr. (2019). Amino Acid Deprivation-Induced Autophagy Requires Upregulation of DIRAS3 through Reduction of E2F1 and E2F4 Transcriptional Repression. Cancers, 11(5), 603. https://doi.org/10.3390/cancers11050603





