Beyond PD-1/PD-L1 Inhibition: What the Future Holds for Breast Cancer Immunotherapy
Abstract
1. Introduction
2. Markers Predominantly Expressed on T-lymphocytes
2.1. LAG-3
2.2. TIM-3
2.3. VISTA
2.4. TIGIT
2.5. GITR
2.6. B7-H3
2.7. ICOS
2.8. 4-1BB (CD137)
2.9. CD27 and CD70
2.10. OX40 and OX40L
2.11. BTLA
2.12. TLR9
2.13. The Adenosine Pathway in Breast Cancer
3. Tumor-Associated Macrophages and Related Markers
3.1. CSF-1/CSF-1R
3.2. CCR2/CCL2
3.3. CD47 and SIRPa
3.4. TLR7
3.5. CD40
4. Natural-Killer Cells and Related Markers
4.1. Killer Immunoglobin Receptors (KIR)
4.2. CD94/NKG2A
4.3. NK-Cell Activating Receptors
5. IDO
6. Myeloid-Derived Suppressor Cells
7. Implementing Combination Immunotherapy in the Clinic
8. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: the next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef]
- Nobelprizemedicine.org. Available online: http://www.nobelprizemedicine.org/wp-content/uploads/2018/10/Adv_info_2018.pdf (accessed on 24 March 2019).
- Denkert, C.; von Minckwitz, G.; Darb-Esfahani, S.; Lederer, B.; Heppner, B.I.; Weber, K.E.; Budczies, J.; Huober, J.; Klauschen, F.; Furlanetto, J.; et al. Tumour-infiltrating lymphocytes and prognosis in different subtypes of breast cancer: a pooled analysis of 3771 patients treated with neoadjuvant therapy. Lancet Oncol. 2018, 19, 40–50. [Google Scholar] [CrossRef]
- Matikas, A.; Lövrot, J.; Ramberg, A.; Eriksson, M.; Lindsten, T.; Lekberg, T.; Hedenfalk, I.; Loman, N.; Bergh, J.; Hatschek, T.; et al. Dynamic evaluation of the immune infiltrate and immune function genes as predictive markers for neoadjuvant chemotherapy in hormone receptor positive, HER2 negative breast cancer. Oncoimmunology 2018, 7, e1466017. [Google Scholar] [CrossRef] [PubMed]
- Foukakis, T.; Lövrot, J.; Matikas, A.; Zerdes, I.; Lorent, J.; Tobin, N.; Suzuki, C.; Brage, S.E.; Carlsson, L.; Einbeigi, Z.; et al. Immune gene expression and response to chemotherapy in advanced breast cancer. Br. J. Cancer 2018, 118, 480–488. [Google Scholar] [CrossRef]
- Ogiya, R.; Niikura, N.; Kumaki, N.; Bianchini, G.; Kitano, S.; Iwamoto, T.; Hayashi, N.; Yokoyama, K.; Oshitanai, R.; Terao, M.; et al. Comparison of tumor-infiltrating lymphocytes between primary and metastatic tumors in breast cancer patients. Cancer Sci 2016, 107, 1730–1735. [Google Scholar] [CrossRef]
- Zerdes, I.; Matikas, A.; Bergh, J.; Rassidakis, G.Z.; Foukakis, T. Genetic, transcriptional and post-translational regulation of the programmed death protein ligand 1 in cancer: biology and clinical correlations. Oncogene 2018, 37, 4639–4661. [Google Scholar] [CrossRef]
- Kassardjian, A.; Shintaku, P.I.; Moatamed, N.A. Expression of immune checkpoint regulators, cytotoxic T lymphocyte antigen 4 (CTLA-4) and programmed death-ligand 1 (PD-L1), in female breast carcinomas. PLoS ONE 2018, 13, e0195958. [Google Scholar] [CrossRef]
- Schmid, P.; Adams, S.; Rugo, H.S.; Schneeweiss, A.; Barrios, C.H.; Iwata, H.; Diéras, V.; Hegg, R.; Im, S.-A.; Shaw Wright, G.; et al. Atezolizumab and Nab-Paclitaxel in Advanced Triple-Negative Breast Cancer. N. Engl. J. Med. 2018, 379, 2108–2121. [Google Scholar] [CrossRef]
- Adams, S.; Gatti-Mays, M.E.; Kalinsky, K.; Korde, L.A.; Sharon, E.; Amiri-Kordestani, L.; Bear, H.; McArthur, H.L.; Frank, E.; Perlmutter, J.; et al. Current Landscape of Immunotherapy in Breast Cancer: A Review. JAMA Oncol. 2019. [Google Scholar] [CrossRef]
- Yarchoan, M.; Hopkins, A.; Jaffee, E.M. Tumor Mutational Burden and Response Rate to PD-1 Inhibition. N. Engl. J. Med. 2017, 377, 2500–2501. [Google Scholar] [CrossRef]
- Huang, C.-T.; Workman, C.J.; Flies, D.; Pan, X.; Marson, A.L.; Zhou, G.; Hipkiss, E.L.; Ravi, S.; Kowalski, J.; Levitsky, H.I.; et al. Role of LAG-3 in regulatory T cells. Immunity 2004, 21, 503–513. [Google Scholar] [CrossRef] [PubMed]
- Triebel, F.; Jitsukawa, S.; Baixeras, E.; Roman-Roman, S.; Genevee, C.; Viegas-Pequignot, E.; Hercend, T. LAG-3, a novel lymphocyte activation gene closely related to CD4. J. Exp. Med. 1990, 171, 1393–1405. [Google Scholar] [CrossRef] [PubMed]
- Huard, B.; Tournier, M.; Triebel, F. LAG-3 does not define a specific mode of natural killing in human. Immunol. Lett. 1998, 61, 109–112. [Google Scholar] [CrossRef]
- Kisielow, M.; Kisielow, J.; Capoferri-Sollami, G.; Karjalainen, K. Expression of lymphocyte activation gene 3 (LAG-3) on B cells is induced by T cells. Eur. J. Immunol. 2005, 35, 2081–2088. [Google Scholar] [CrossRef] [PubMed]
- Buisson, S.; Triebel, F. LAG-3 (CD223) reduces macrophage and dendritic cell differentiation from monocyte precursors. Immunology 2005, 114, 369–374. [Google Scholar] [CrossRef]
- Xu, F.; Liu, J.; Liu, D.; Liu, B.; Wang, M.; Hu, Z.; Du, X.; Tang, L.; He, F. LSECtin expressed on melanoma cells promotes tumor progression by inhibiting antitumor T-cell responses. Cancer Res. 2014, 74, 3418–3428. [Google Scholar] [CrossRef]
- Kouo, T.; Huang, L.; Pucsek, A.B.; Cao, M.; Solt, S.; Armstrong, T.; Jaffee, E. Galectin-3 Shapes Antitumor Immune Responses by Suppressing CD8+ T Cells via LAG-3 and Inhibiting Expansion of Plasmacytoid Dendritic Cells. Cancer Immunol. Res. 2015, 3, 412–423. [Google Scholar] [CrossRef]
- Maçon-Lemaître, L.; Triebel, F. The negative regulatory function of the lymphocyte-activation gene-3 co-receptor (CD223) on human T cells. Immunology 2005, 115, 170–178. [Google Scholar] [CrossRef]
- Workman, C.J.; Dugger, K.J.; Vignali, D.A.A. Cutting edge: molecular analysis of the negative regulatory function of lymphocyte activation gene-3. J. Immunol. 2002, 169, 5392–5395. [Google Scholar] [CrossRef]
- Workman, C.J.; Vignali, D.A.A. Negative regulation of T cell homeostasis by lymphocyte activation gene-3 (CD223). J. Immunol. 2005, 174, 688–695. [Google Scholar] [CrossRef] [PubMed]
- Gandhi, M.K.; Lambley, E.; Duraiswamy, J.; Dua, U.; Smith, C.; Elliott, S.; Gill, D.; Marlton, P.; Seymour, J.; Khanna, R. Expression of LAG-3 by tumor-infiltrating lymphocytes is coincident with the suppression of latent membrane antigen-specific CD8+ T-cell function in Hodgkin lymphoma patients. Blood 2006, 108, 2280–2289. [Google Scholar] [CrossRef]
- Sasidharan Nair, V.; El Salhat, H.; Taha, R.Z.; John, A.; Ali, B.R.; Elkord, E. DNA methylation and repressive H3K9 and H3K27 trimethylation in the promoter regions of PD-1, CTLA-4, TIM-3, LAG-3, TIGIT, and PD-L1 genes in human primary breast cancer. Clin. Epigenetics 2018, 10, 78. [Google Scholar] [CrossRef]
- Kok, M. LAG-3: Another brake to release in breast cancer? Ann. Oncol. 2017, 28, 2907–2908. [Google Scholar] [CrossRef] [PubMed]
- Bottai, G.; Raschioni, C.; Losurdo, A.; Di Tommaso, L.; Tinterri, C.; Torrisi, R.; Reis-Filho, J.S.; Roncalli, M.; Sotiriou, C.; Santoro, A.; et al. An immune stratification reveals a subset of PD-1/LAG-3 double-positive triple-negative breast cancers. Breast Cancer Res. 2016, 18. [Google Scholar] [CrossRef]
- Burugu, S.; Gao, D.; Leung, S.; Chia, S.K.; Nielsen, T.O. LAG-3+ tumor infiltrating lymphocytes in breast cancer: clinical correlates and association with PD-1/PD-L1+ tumors. Ann. Oncol. 2017, 28, 2977–2984. [Google Scholar] [CrossRef]
- Brignone, C.; Gutierrez, M.; Mefti, F.; Brain, E.; Jarcau, R.; Cvitkovic, F.; Bousetta, N.; Medioni, J.; Gligorov, J.; Grygar, C.; et al. First-line chemoimmunotherapy in metastatic breast carcinoma: combination of paclitaxel and IMP321 (LAG-3Ig) enhances immune responses and antitumor activity. J. Transl. Med. 2010, 8, 71. [Google Scholar] [CrossRef]
- Wang, Y.; Dong, T.; Xuan, Q.; Zhao, H.; Qin, L.; Zhang, Q. Lymphocyte-Activation Gene-3 Expression and Prognostic Value in Neoadjuvant-Treated Triple-Negative Breast Cancer. J. Breast Cancer 2018, 21, 124–133. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Xiang, R.; Wu, B.; Li, J.; Luo, G. T-cell immunoglobulin mucin-3 expression in invasive ductal breast carcinoma: Clinicopathological correlations and association with tumor infiltration by cytotoxic lymphocytes. Mol. Clin. Oncol. 2017, 7, 557–563. [Google Scholar] [CrossRef]
- Zhu, S.; Lin, J.; Qiao, G.; Wang, X.; Xu, Y. Tim-3 identifies exhausted follicular helper T cells in breast cancer patients. Immunobiology 2016, 221, 986–993. [Google Scholar] [CrossRef]
- Cari, L.; Nocentini, G.; Migliorati, G.; Riccardi, C. Potential effect of tumor-specific Treg-targeted antibodies in the treatment of human cancers: A bioinformatics analysis. Oncoimmunology 2018, 7, e1387705. [Google Scholar] [CrossRef] [PubMed]
- Burugu, S.; Gao, D.; Leung, S.; Chia, S.K.; Nielsen, T.O. TIM-3 expression in breast cancer. Oncoimmunology 2018, 7. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Canales, S.; Cifuentes, F.; Gregorio, M.L.D.R.; Serrano-Oviedo, L.; Galán-Moya, E.M.; Amir, E.; Pandiella, A.; Győrffy, B.; Ocaña, A. Transcriptomic immunologic signature associated with favorable clinical outcome in basal-like breast tumors. PLoS ONE 2017, 12, e0175128. [Google Scholar] [CrossRef]
- Li, C.-H.; Kuo, W.-H.; Chang, W.-C.; Huang, S.-C.; Chang, K.-J.; Sheu, B.-C. Activation of regulatory T cells instigates functional down-regulation of cytotoxic T lymphocytes in human breast cancer. Immunol. Res. 2011, 51, 71–79. [Google Scholar] [CrossRef]
- Benevides, L.; Cardoso, C.R.B.; Tiezzi, D.G.; Marana, H.R.C.; Andrade, J.M.; Silva, J.S. Enrichment of regulatory T cells in invasive breast tumor correlates with the upregulation of IL-17A expression and invasiveness of the tumor. Eur. J. Immunol. 2013, 43, 1518–1528. [Google Scholar] [CrossRef] [PubMed]
- Krausz, L.T.; Fischer-Fodor, E.; Major, Z.Z.; Fetica, B. GITR-expressing regulatory T-cell subsets are increased in tumor-positive lymph nodes from advanced breast cancer patients as compared to tumor-negative lymph nodes. Int. J. Immunopathol. Pharmacol. 2012, 25, 59–66. [Google Scholar] [CrossRef] [PubMed]
- Ostapchuk, Y.O.; Perfilyeva, Y.V.; Kustova, E.A.; Urazalieva, N.T.; Omarbaeva, N.A.; Talaeva, S.G.; Belyaev, N.N. Functional heterogeneity of circulating T regulatory cell subsets in breast cancer patients. Breast Cancer 2018. [Google Scholar] [CrossRef]
- Sun, J.; Guo, Y.-D.; Li, X.-N.; Zhang, Y.-Q.; Gu, L.; Wu, P.-P.; Bai, G.-H.; Xiao, Y. B7-H3 expression in breast cancer and upregulation of VEGF through gene silence. Onco. Targets Ther. 2014, 7, 1979–1986. [Google Scholar] [CrossRef] [PubMed]
- Arigami, T.; Narita, N.; Mizuno, R.; Nguyen, L.; Ye, X.; Chung, A.; Giuliano, A.E.; Hoon, D.S.B. B7-h3 ligand expression by primary breast cancer and associated with regional nodal metastasis. Ann. Surg. 2010, 252, 1044–1051. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Liu, J.; Wang, J.; Liu, Y.; Zhang, F.; Lin, W.; Gao, A.; Sun, M.; Wang, Y.; Sun, Y. B7-H3 expression in ductal and lobular breast cancer and its association with IL-10. Mol. Med. Rep. 2013, 7, 134–138. [Google Scholar] [CrossRef] [PubMed]
- Maeda, N.; Yoshimura, K.; Yamamoto, S.; Kuramasu, A.; Inoue, M.; Suzuki, N.; Watanabe, Y.; Maeda, Y.; Kamei, R.; Tsunedomi, R.; et al. Expression of B7-H3, a Potential Factor of Tumor Immune Evasion in Combination with the Number of Regulatory T Cells, Affects Against Recurrence-Free Survival in Breast Cancer Patients. Ann. Surg. Oncol. 2014, 21, 546–554. [Google Scholar] [CrossRef]
- Cong, F.; Yu, H.; Gao, X. Expression of CD24 and B7-H3 in breast cancer and the clinical significance. Oncol. Lett. 2017, 14, 7185–7190. [Google Scholar] [CrossRef][Green Version]
- Wilson, K.E.; Bachawal, S.V.; Abou-Elkacem, L.; Jensen, K.; Machtaler, S.; Tian, L.; Willmann, J.K. Spectroscopic Photoacoustic Molecular Imaging of Breast Cancer using a B7-H3-targeted ICG Contrast Agent. Theranostics 2017, 7, 1463–1476. [Google Scholar] [CrossRef]
- Seaman, S.; Zhu, Z.; Saha, S.; Zhang, X.M.; Yang, M.Y.; Hilton, M.B.; Morris, K.; Szot, C.; Morris, H.; Swing, D.A.; et al. Eradication of Tumors through Simultaneous Ablation of CD276/B7-H3-Positive Tumor Cells and Tumor Vasculature. Cancer Cell 2017, 31, 501–515. [Google Scholar] [CrossRef] [PubMed]
- Bachawal, S.V.; Jensen, K.C.; Wilson, K.E.; Tian, L.; Lutz, A.M.; Willmann, J.K. Breast Cancer Detection by B7-H3 Targeted Ultrasound Molecular Imaging. Cancer Res. 2015, 75, 2501–2509. [Google Scholar] [CrossRef] [PubMed]
- Faget, J.; Bendriss-Vermare, N.; Gobert, M.; Durand, I.; Olive, D.; Biota, C.; Bachelot, T.; Treilleux, I.; Goddard-Leon, S.; Lavergne, E.; et al. ICOS-ligand expression on plasmacytoid dendritic cells supports breast cancer progression by promoting the accumulation of immunosuppressive CD4+ T cells. Cancer Res. 2012, 72, 6130–6141. [Google Scholar] [CrossRef]
- Alizadeh, A.A.; Gentles, A.J.; Alencar, A.J.; Liu, C.L.; Kohrt, H.E.; Houot, R.; Goldstein, M.J.; Zhao, S.; Natkunam, Y.; Advani, R.H.; et al. Prediction of survival in diffuse large B-cell lymphoma based on the expression of 2 genes reflecting tumor and microenvironment. Blood 2011, 118, 1350–1358. [Google Scholar] [CrossRef]
- Wang, Q.; Zhang, P.; Zhang, Q.; Wang, X.; Li, J.; Ma, C.; Sun, W.; Zhang, L. Analysis of CD137 and CD137L Expression in Human Primary Tumor Tissues. Croat Med. J. 2008, 49, 192–200. [Google Scholar] [CrossRef] [PubMed]
- Ryan, M.C.; Kostner, H.; Gordon, K.A.; Duniho, S.; Sutherland, M.K.; Yu, C.; Kim, K.M.; Nesterova, A.; Anderson, M.; McEarchern, J.A.; et al. Targeting pancreatic and ovarian carcinomas using the auristatin-based anti-CD70 antibody–drug conjugate SGN-75. Br. J. Cancer 2010, 103, 676–684. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Yin, B.; Yi, Z.; Liu, X.; Hu, Z.; Gao, W.; Yu, H.; Li, Q. Breast cancer stem cells characterized by CD70 expression preferentially metastasize to the lungs. Breast Cancer 2018. [Google Scholar] [CrossRef]
- Tvrdík, D.; Skálová, H.; Dundr, P.; Povýšil, C.; Velenská, Z.; Berková, A.; Staněk, L.; Petruželka, L. Apoptosis – associated genes and their role in predicting responses to neoadjuvant breast cancer treatment. Med. Sci. Monit. 2012, 18, BR60–BR67. [Google Scholar] [CrossRef]
- Xie, F.; Wang, Q.; Chen, Y.; Gu, Y.; Mao, H.; Zeng, W.; Zhang, X. Costimulatory molecule OX40/OX40L expression in ductal carcinoma in situ and invasive ductal carcinoma of breast: an immunohistochemistry-based pilot study. Pathol. Res. Pract. 2010, 206, 735–739. [Google Scholar] [CrossRef] [PubMed]
- Xie, F.; Wang, Q.; Chen, Y.; Gu, Y.; Shi, Q.; Ge, Y.; Yu, G.; Wu, H.; Mao, Y.; Wang, X.; et al. Characterization and application of two novel monoclonal antibodies against human OX40: costimulation of T cells and expression on tumor as well as normal gland tissues. Tissue Antigens 2006, 67, 307–317. [Google Scholar] [CrossRef]
- Morris, A.; Vetto, J.T.; Ramstad, T.; Funatake, C.J.; Choolun, E.; Entwisle, C.; Weinberg, A.D. Induction of anti-mammary cancer immunity by engaging the OX-40 receptor in vivo. Breast Cancer Res. Treat. 2001, 67, 71–80. [Google Scholar] [CrossRef] [PubMed]
- Ramstad, T.; Lawnicki, L.; Vetto, J.; Weinberg, A. Immunohistochemical analysis of primary breast tumors and tumor-draining lymph nodes by means of the T-cell costimulatory molecule OX-40. Am. J. Surg. 2000, 179, 400–406. [Google Scholar] [CrossRef]
- Weinberg, A.D.; Rivera, M.M.; Prell, R.; Morris, A.; Ramstad, T.; Vetto, J.T.; Urba, W.J.; Alvord, G.; Bunce, C.; Shields, J. Engagement of the OX-40 receptor in vivo enhances antitumor immunity. J. Immunol. 2000, 164, 2160–2169. [Google Scholar] [CrossRef]
- Liu, Z.; Li, M.; Jiang, Z.; Wang, X. A Comprehensive Immunologic Portrait of Triple-Negative Breast Cancer. Transl. Oncol. 2018, 11, 311–329. [Google Scholar] [CrossRef] [PubMed]
- Muenst, S.; Soysal, S.D.; Gao, F.; Obermann, E.C.; Oertli, D.; EGillanders, W. The presence of programmed death 1 (PD-1)-positive tumor-infiltrating lymphocytes is associated with poor prognosis in human breast cancer. Breast Cancer Res. Treat. 2013, 139. [Google Scholar] [CrossRef]
- Chandler, M.R.; Keene, K.S.; Tuomela, J.M.; Forero-Torres, A.; Desmond, R.; Vuopala, K.S.; Harris, K.W.; Merner, N.D.; Selander, K.S. Lower frequency of TLR9 variant associated with protection from breast cancer among African Americans. PLoS ONE 2017, 12, e0183832. [Google Scholar] [CrossRef]
- Meseure, D.; Vacher, S.; Drak Alsibai, K.; Trassard, M.; Nicolas, A.; Leclere, R.; Lerebours, F.; Guinebretiere, J.M.; Marangoni, E.; Lidereau, R.; et al. Biopathological Significance of TLR9 Expression in Cancer Cells and Tumor Microenvironment Across Invasive Breast Carcinomas Subtypes. Cancer Microenviron 2016, 9, 107–118. [Google Scholar] [CrossRef]
- Tuomela, J.; Sandholm, J.; Karihtala, P.; Ilvesaro, J.; Vuopala, K.S.; Kauppila, J.H.; Kauppila, S.; Chen, D.; Pressey, C.; Härkönen, P.; et al. Low TLR9 expression defines an aggressive subtype of triple-negative breast cancer. Breast Cancer Res. Treat. 2012, 135, 481–493. [Google Scholar] [CrossRef]
- Sandholm, J.; Kauppila, J.H.; Pressey, C.; Tuomela, J.; Jukkola-Vuorinen, A.; Vaarala, M.; Johnson, M.R.; Harris, K.W.; Selander, K.S. Estrogen receptor-α and sex steroid hormones regulate Toll-like receptor-9 expression and invasive function in human breast cancer cells. Breast Cancer Res. Treat. 2012, 132, 411–419. [Google Scholar] [CrossRef]
- Qiu, J.; Shao, S.; Yang, G.; Shen, Z.; Zhang, Y. Association of Toll like receptor 9 expression with lymph node metastasis in human breast cancer. Neoplasma 2011, 58, 251–255. [Google Scholar] [CrossRef]
- González-Reyes, S.; Marín, L.; González, L.; González, L.O.; del Casar, J.M.; Lamelas, M.L.; González-Quintana, J.M.; Vizoso, F.J. Study of TLR3, TLR4 and TLR9 in breast carcinomas and their association with metastasis. BMC Cancer 2010, 10, 665. [Google Scholar] [CrossRef] [PubMed]
- Berger, R.; Fiegl, H.; Goebel, G.; Obexer, P.; Ausserlechner, M.; Doppler, W.; Hauser-Kronberger, C.; Reitsamer, R.; Egle, D.; Reimer, D.; et al. Toll-Like Receptor 9 expression in breast and ovarian cancer is associated with poorly differentiated tumors. Cancer Sci. 2010, 101, 1059–1066. [Google Scholar] [CrossRef]
- Jukkola-Vuorinen, A.; Rahko, E.; Vuopala, K.S.; Desmond, R.; Lehenkari, P.P.; Harris, K.W.; Selander, K.S. Toll-like receptor-9 expression is inversely correlated with estrogen receptor status in breast cancer. J. Innate Immun. 2009, 1, 59–68. [Google Scholar] [CrossRef] [PubMed]
- Zhi, X.; Wang, Y.; Yu, J.; Yu, J.; Zhang, L.; Yin, L.; Zhou, P. Potential prognostic biomarker CD73 regulates epidermal growth factor receptor expression in human breast cancer. IUBMB Life 2012, 64, 911–920. [Google Scholar] [CrossRef]
- Supernat, A.; Markiewicz, A.; Welnicka-Jaskiewicz, M.; Seroczynska, B.; Skokowski, J.; Sejda, A.; Szade, J.; Czapiewski, P.; Biernat, W.; Zaczek, A. CD73 expression as a potential marker of good prognosis in breast carcinoma. Appl. Immunohistochem. Mol. Morphol. 2012, 20, 103–107. [Google Scholar] [CrossRef]
- Krüger, K.H.; Thompson, L.F.; Kaufmann, M.; Möller, P. Expression of ecto-5’-nucleotidase (CD73) in normal mammary gland and in breast carcinoma. Br. J. Cancer 1991, 63, 114–118. [Google Scholar] [CrossRef]
- Samanta, D.; Park, Y.; Ni, X.; Li, H.; Zahnow, C.A.; Gabrielson, E.; Pan, F.; Semenza, G.L. Chemotherapy induces enrichment of CD47+/CD73+/PDL1+ immune evasive triple-negative breast cancer cells. Proc. Natl. Acad. Sci. USA 2018, 115, E1239–E1248. [Google Scholar] [CrossRef]
- Buisseret, L.; Pommey, S.; Allard, B.; Garaud, S.; Bergeron, M.; Cousineau, I.; Ameye, L.; Bareche, Y.; Paesmans, M.; Crown, J.P.A.; et al. Clinical significance of CD73 in triple-negative breast cancer: multiplex analysis of a phase III clinical trial. Ann. Oncol. 2018, 29, 1056–1062. [Google Scholar] [CrossRef]
- Yu, J.; Wang, X.; Lu, Q.; Wang, J.; Li, L.; Liao, X.; Zhu, W.; Lv, L.; Zhi, X.; Yu, J.; et al. Extracellular 5’-nucleotidase (CD73) promotes human breast cancer cells growth through AKT/GSK-3β/β-catenin/cyclinD1 signaling pathway. Int. J. Cancer 2018, 142, 959–967. [Google Scholar] [CrossRef] [PubMed]
- Turcotte, M.; Allard, D.; Mittal, D.; Bareche, Y.; Buisseret, L.; José, V.; Pommey, S.; Delisle, V.; Loi, S.; Joensuu, H.; et al. CD73 Promotes Resistance to HER2/ErbB2 Antibody Therapy. Cancer Res. 2017, 77, 5652–5663. [Google Scholar] [CrossRef] [PubMed]
- Loi, S.; Pommey, S.; Haibe-Kains, B.; Beavis, P.A.; Darcy, P.K.; Smyth, M.J.; Stagg, J. CD73 promotes anthracycline resistance and poor prognosis in triple negative breast cancer. Proc. Natl. Acad. Sci. USA 2013, 110, 11091–11096. [Google Scholar] [CrossRef]
- Canale, F.P.; Ramello, M.C.; Núñez, N.; Araujo Furlan, C.L.; Bossio, S.N.; Gorosito Serrán, M.; Tosello Boari, J.; Del Castillo, A.; Ledesma, M.; Sedlik, C.; et al. CD39 Expression Defines Cell Exhaustion in Tumor-Infiltrating CD8+ T Cells. Cancer Res. 2018, 78, 115–128. [Google Scholar] [CrossRef] [PubMed]
- Syed Khaja, A.S.; Toor, S.M.; El Salhat, H.; Faour, I.; Ul Haq, N.; Ali, B.R.; Elkord, E. Preferential accumulation of regulatory T cells with highly immunosuppressive characteristics in breast tumor microenvironment. Oncotarget 2017, 8, 33159–33171. [Google Scholar]
- Thibaudin, M.; Chaix, M.; Boidot, R.; Végran, F.; Derangère, V.; Limagne, E.; Berger, H.; Ladoire, S.; Apetoh, L.; Ghiringhelli, F. Human ectonucleotidase-expressing CD25high Th17 cells accumulate in breast cancer tumors and exert immunosuppressive functions. Oncoimmunology 2016, 5, e1055444. [Google Scholar] [CrossRef] [PubMed]
- Bastid, J.; Cottalorda-Regairaz, A.; Alberici, G.; Bonnefoy, N.; Eliaou, J.-F.; Bensussan, A. ENTPD1/CD39 is a promising therapeutic target in oncology. Oncogene 2013, 32, 1743–1751. [Google Scholar] [CrossRef] [PubMed]
- Duhoux, F.P.; Jager, A.; Dirix, L.Y.; Huizing, M.T.; Jerusalem, G.H.M.; Vuylsteke, P.; De Cuypere, E.; Breiner, D.; Mueller, C.; Brignone, C.; et al. Combination of paclitaxel and a LAG-3 fusion protein (eftilagimod alpha), as a first-line chemoimmunotherapy in patients with metastatic breast carcinoma (MBC): Final results from the run-in phase of a placebo-controlled randomized phase II. JCO 2018, 36, 1050. [Google Scholar] [CrossRef]
- Hong, D.S.; Schoffski, P.; Calvo, A.; Sarantopoulos, J.; Ochoa De Olza, M.; Carvajal, R.D.; Prawira, A.; Kyi, C.; Esaki, T.; Akerley, W.L.; et al. Phase I/II study of LAG525 ± spartalizumab (PDR001) in patients (pts) with advanced malignancies. JCO 2018, 36, 3012. [Google Scholar] [CrossRef]
- Koon, H.B.; Shepard, D.R.; Merghoub, T.; Schaer, D.A.; Sirard, C.A.; Wolchok, J.D. First-in-human phase 1 single-dose study of TRX-518, an anti-human glucocorticoid-induced tumor necrosis factor receptor (GITR) monoclonal antibody in adults with advanced solid tumors. JCO 2016, 34, 3017. [Google Scholar] [CrossRef]
- Siu, L.L.; Steeghs, N.; Meniawy, T.; Joerger, M.; Spratlin, J.L.; Rottey, S.; Nagrial, A.; Cooper, A.; Meier, R.; Guan, X.; et al. Preliminary results of a phase I/IIa study of BMS-986156 (glucocorticoid-induced tumor necrosis factor receptor–related gene [GITR] agonist), alone and in combination with nivolumab in pts with advanced solid tumors. JCO 2017, 35, 104. [Google Scholar] [CrossRef]
- Burris, H.A.; Callahan, M.K.; Tolcher, A.W.; Kummar, S.; Falchook, G.S.; Pachynski, R.K.; Tykodi, S.S.; Gibney, G.T.; Seiwert, T.Y.; Gainor, J.F.; et al. Phase 1 safety of ICOS agonist antibody JTX-2011 alone and with nivolumab (nivo) in advanced solid tumors; predicted vs observed pharmacokinetics (PK) in ICONIC. JCO 2017, 35, 3033. [Google Scholar] [CrossRef]
- Segal, N.H.; Logan, T.F.; Hodi, F.S.; McDermott, D.; Melero, I.; Hamid, O.; Schmidt, H.; Robert, C.; Chiarion-Sileni, V.; Ascierto, P.A.; et al. Results from an Integrated Safety Analysis of Urelumab, an Agonist Anti-CD137 Monoclonal Antibody. Clin. Cancer Res. 2017, 23, 1929–1936. [Google Scholar] [CrossRef] [PubMed]
- Awada, A.; Rolfo, C.D.; Rottey, S.; Ysebrant de Lendonck, L.; Schroyens, W.A.; Offner, F.; Silence, K.; Dreier, T.; Moshir, M.; de Haard, H.; et al. A phase I, first-in-human study of ARGX-110, a monoclonal antibody targeting CD70, a receptor involved in immune escape and tumor growth in patients with solid and hematologic malignancies. JCO 2014, 32, 3023. [Google Scholar] [CrossRef]
- Infante, J.R.; Hansen, A.R.; Pishvaian, M.J.; Chow, L.Q.M.; McArthur, G.A.; Bauer, T.M.; Liu, S.V.; Sandhu, S.K.; Tsai, F.Y.-C.; Kim, J.; et al. A phase Ib dose escalation study of the OX40 agonist MOXR0916 and the PD-L1 inhibitor atezolizumab in patients with advanced solid tumors. JCO 2016, 34, 101. [Google Scholar] [CrossRef]
- Babiker, H.M.; Borazanci, E.H.; Subbiah, V.; Diab, A.; Woodhead, G.; Hennemeyer, C.; Shah, A.H.; Hultsch, R.; Murthy, R.; Miller, C.; et al. Preliminary safety of deep/visceral (D/V) image guided (IG) intratumoral injection (ITI) of IMO-2125. JCO 2018, 36, e15150. [Google Scholar] [CrossRef]
- Siu, L.L.; Burris, H.; Le, D.T.; Hollebecque, A.; Steeghs, N.; Delord, J.-P.; Hilton, J.; Barnhart, B.; Sega, E.; Sanghavi, K.; et al. Abstract CT180: Preliminary phase 1 profile of BMS-986179, an anti-CD73 antibody, in combination with nivolumab in patients with advanced solid tumors. Cancer Res. 2018, 78, CT180. [Google Scholar] [CrossRef]
- Monney, L.; Sabatos, C.A.; Gaglia, J.L.; Ryu, A.; Waldner, H.; Chernova, T.; Manning, S.; Greenfield, E.A.; Coyle, A.J.; Sobel, R.A.; et al. Th1-specific cell surface protein Tim-3 regulates macrophage activation and severity of an autoimmune disease. Nature 2002, 415, 536–541. [Google Scholar] [CrossRef]
- Gao, X.; Zhu, Y.; Li, G.; Huang, H.; Zhang, G.; Wang, F.; Sun, J.; Yang, Q.; Zhang, X.; Lu, B. TIM-3 Expression Characterizes Regulatory T Cells in Tumor Tissues and Is Associated with Lung Cancer Progression. PLoS ONE 2012, 7. [Google Scholar] [CrossRef] [PubMed]
- Anderson, A.C.; Anderson, D.E.; Bregoli, L.; Hastings, W.D.; Kassam, N.; Lei, C.; Chandwaskar, R.; Karman, J.; Su, E.W.; Hirashima, M.; et al. Promotion of tissue inflammation by the immune receptor Tim-3 expressed on innate immune cells. Science 2007, 318, 1141–1143. [Google Scholar] [CrossRef] [PubMed]
- Gleason, M.K.; Lenvik, T.R.; McCullar, V.; Felices, M.; O’Brien, M.S.; Cooley, S.A.; Verneris, M.R.; Cichocki, F.; Holman, C.J.; Panoskaltsis-Mortari, A.; et al. Tim-3 is an inducible human natural killer cell receptor that enhances interferon gamma production in response to galectin-9. Blood 2012, 119, 3064–3072. [Google Scholar] [CrossRef] [PubMed]
- Zhu, C.; Anderson, A.C.; Schubart, A.; Xiong, H.; Imitola, J.; Khoury, S.J.; Zheng, X.X.; Strom, T.B.; Kuchroo, V.K. The Tim-3 ligand galectin-9 negatively regulates T helper type 1 immunity. Nat. Immunol. 2005, 6, 1245–1252. [Google Scholar] [CrossRef]
- Huang, Y.-H.; Zhu, C.; Kondo, Y.; Anderson, A.C.; Gandhi, A.; Russell, A.; Dougan, S.K.; Petersen, B.-S.; Melum, E.; Pertel, T.; et al. CEACAM1 regulates TIM-3-mediated tolerance and exhaustion. Nature 2015, 517, 386–390. [Google Scholar] [CrossRef]
- Chiba, S.; Baghdadi, M.; Akiba, H.; Yoshiyama, H.; Kinoshita, I.; Dosaka-Akita, H.; Fujioka, Y.; Ohba, Y.; Gorman, J.V.; Colgan, J.D.; et al. Tumor-infiltrating DCs suppress nucleic acid-mediated innate immune responses through interactions between the receptor TIM-3 and the alarmin HMGB1. Nat. Immunol. 2012, 13, 832–842. [Google Scholar] [CrossRef]
- DeKruyff, R.H.; Bu, X.; Ballesteros, A.; Santiago, C.; Chim, Y.-L.E.; Lee, H.-H.; Karisola, P.; Pichavant, M.; Kaplan, G.G.; Umetsu, D.T.; et al. T cell/transmembrane, Ig, and mucin-3 allelic variants differentially recognize phosphatidylserine and mediate phagocytosis of apoptotic cells. J. Immunol. 2010, 184, 1918–1930. [Google Scholar] [CrossRef]
- Fourcade, J.; Sun, Z.; Benallaoua, M.; Guillaume, P.; Luescher, I.F.; Sander, C.; Kirkwood, J.M.; Kuchroo, V.; Zarour, H.M. Upregulation of Tim-3 and PD-1 expression is associated with tumor antigen-specific CD8+ T cell dysfunction in melanoma patients. J. Exp. Med. 2010, 207, 2175–2186. [Google Scholar] [CrossRef]
- Sehrawat, S.; Suryawanshi, A.; Hirashima, M.; Rouse, B.T. Role of Tim-3/Galectin-9 Inhibitory Interaction In Viral Induced Immunopathology: Shifting The Balance Towards Regulators. J. Immunol. 2009, 182, 3191–3201. [Google Scholar] [CrossRef] [PubMed]
- Dardalhon, V.; Anderson, A.C.; Karman, J.; Apetoh, L.; Chandwaskar, R.; Lee, D.H.; Cornejo, M.; Nishi, N.; Yamauchi, A.; Quintana, F.J.; et al. Tim-3/galectin-9 pathway: regulation of Th1 immunity through promotion of CD11b+Ly-6G+ myeloid cells. J. Immunol. 2010, 185, 1383–1392. [Google Scholar] [CrossRef] [PubMed]
- De Mingo Pulido, Á.; Gardner, A.; Hiebler, S.; Soliman, H.; Rugo, H.S.; Krummel, M.F.; Coussens, L.M.; Ruffell, B. TIM-3 Regulates CD103+ Dendritic Cell Function and Response to Chemotherapy in Breast Cancer. Cancer Cell 2018, 33, 60–74. [Google Scholar] [CrossRef] [PubMed]
- Cheng, S.; Ju, Y.; Han, F.; Wang, Y.; Xu, Y.; Qu, T.; Lu, Z. T Cell Immunoglobulin- and Mucin-Domain-Containing Molecule 3 Gene Polymorphisms and Susceptibility to Invasive Breast Cancer. Ann. Clin. Lab. Sci. 2017, 47, 668–675. [Google Scholar] [PubMed]
- Gao, X.; Yang, J.; He, Y.; Zhang, J. Quantitative assessment of TIM-3 polymorphisms and cancer risk in Chinese Han population. Oncotarget 2016, 7, 35768–35775. [Google Scholar] [CrossRef]
- Wang, Z.; Liu, X.; Wang, X.; Chong, T.; Lin, S.; Wang, M.; Ma, X.; Liu, K.; Xu, P.; Feng, Y.; et al. Polymorphisms in TIM-3 and breast cancer susceptibility in Chinese women: A case-control study. Oncotarget 2016, 7, 43703–43712. [Google Scholar] [CrossRef]
- Nowak, E.C.; Lines, J.L.; Varn, F.S.; Deng, J.; Sarde, A.; Mabaera, R.; Kuta, A.; Le Mercier, I.; Cheng, C.; Noelle, R.J. Immunoregulatory functions of VISTA. Immunol. Rev. 2017, 276, 66–79. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Rubinstein, R.; Lines, J.L.; Wasiuk, A.; Ahonen, C.; Guo, Y.; Lu, L.-F.; Gondek, D.; Wang, Y.; Fava, R.A.; et al. VISTA, a novel mouse Ig superfamily ligand that negatively regulates T cell responses. J. Exp. Med. 2011, 208, 577–592. [Google Scholar] [CrossRef]
- Lines, J.L.; Sempere, L.F.; Wang, L.; Pantazi, E.; Mak, J.; O’Connell, S.; Ceeraz, S.; Suriawinata, A.A.; Yan, S.; Ernstoff, M.S.; et al. VISTA is an immune checkpoint molecule for human T cells. Cancer Res. 2014, 74, 1924–1932. [Google Scholar] [CrossRef] [PubMed]
- Böger, C.; Behrens, H.-M.; Krüger, S.; Röcken, C. The novel negative checkpoint regulator VISTA is expressed in gastric carcinoma and associated with PD-L1/PD-1: A future perspective for a combined gastric cancer therapy? Oncoimmunology 2017, 6. [Google Scholar] [CrossRef]
- Loos, M.; Hedderich, D.M.; Ottenhausen, M.; Giese, N.A.; Laschinger, M.; Esposito, I.; Kleeff, J.; Friess, H. Expression of the costimulatory molecule B7-H3 is associated with prolonged survival in human pancreatic cancer. BMC Cancer 2009, 9, 463. [Google Scholar] [CrossRef]
- Sakr, M.A.; Takino, T.; Domoto, T.; Nakano, H.; Wong, R.W.; Sasaki, M.; Nakanuma, Y.; Sato, H. GI24 enhances tumor invasiveness by regulating cell surface membrane-type 1 matrix metalloproteinase. Cancer Sci. 2010, 101, 2368–2374. [Google Scholar] [CrossRef]
- Boles, K.S.; Vermi, W.; Facchetti, F.; Fuchs, A.; Wilson, T.J.; Diacovo, T.; Cella, M.; Colonna, M. A novel molecular interaction for the adhesion of follicular CD4 T cells to follicular dendritic cells. Eur. J. Immunol. 2009, 39, 695–703. [Google Scholar] [CrossRef]
- Yu, X.; Harden, K.; Gonzalez, L.C.; Francesco, M.; Chiang, E.; Irving, B.; Tom, I.; Ivelja, S.; Refino, C.J.; Clark, H.; et al. The surface protein TIGIT suppresses T cell activation by promoting the generation of mature immunoregulatory dendritic cells. Nat. Immunol. 2009, 10, 48–57. [Google Scholar] [CrossRef] [PubMed]
- Blockade of CD112R and TIGIT signaling sensitizes human natural killer cell functions. Available online: https://www-ncbi-nlm-nih-gov.doc-distant.univ-lille2.fr/pubmed/28623459 (accessed on 2 September 2018).
- Stanietsky, N.; Simic, H.; Arapovic, J.; Toporik, A.; Levy, O.; Novik, A.; Levine, Z.; Beiman, M.; Dassa, L.; Achdout, H.; et al. The interaction of TIGIT with PVR and PVRL2 inhibits human NK cell cytotoxicity. Proc. Natl. Acad. Sci. USA 2009, 106, 17858–17863. [Google Scholar] [CrossRef]
- Joller, N.; Hafler, J.P.; Brynedal, B.; Kassam, N.; Spoerl, S.; Levin, S.D.; Sharpe, A.H.; Kuchroo, V.K. TIGIT has T cell intrinsic inhibitory functions. J. Immunol. 2011, 186, 1338–1342. [Google Scholar] [CrossRef]
- Zhang, Y.; Maksimovic, J.; Naselli, G.; Qian, J.; Chopin, M.; Blewitt, M.E.; Oshlack, A.; Harrison, L.C. Genome-wide DNA methylation analysis identifies hypomethylated genes regulated by FOXP3 in human regulatory T cells. Blood 2013, 122, 2823–2836. [Google Scholar] [CrossRef]
- Joller, N.; Lozano, E.; Burkett, P.R.; Patel, B.; Xiao, S.; Zhu, C.; Xia, J.; Tan, T.G.; Sefik, E.; Yajnik, V.; et al. Treg cells expressing the coinhibitory molecule TIGIT selectively inhibit proinflammatory Th1 and Th17 cell responses. Immunity 2014, 40, 569–581. [Google Scholar] [CrossRef] [PubMed]
- Johnston, R.J.; Comps-Agrar, L.; Hackney, J.; Yu, X.; Huseni, M.; Yang, Y.; Park, S.; Javinal, V.; Chiu, H.; Irving, B.; et al. The immunoreceptor TIGIT regulates antitumor and antiviral CD8(+) T cell effector function. Cancer Cell 2014, 26, 923–937. [Google Scholar] [CrossRef]
- Shimizu, J.; Yamazaki, S.; Takahashi, T.; Ishida, Y.; Sakaguchi, S. Stimulation of CD25(+)CD4(+) regulatory T cells through GITR breaks immunological self-tolerance. Nat. Immunol. 2002, 3, 135–142. [Google Scholar] [CrossRef]
- McHugh, R.S.; Whitters, M.J.; Piccirillo, C.A.; Young, D.A.; Shevach, E.M.; Collins, M.; Byrne, M.C. CD4(+)CD25(+) immunoregulatory T cells: gene expression analysis reveals a functional role for the glucocorticoid-induced TNF receptor. Immunity 2002, 16, 311–323. [Google Scholar] [CrossRef]
- Hanabuchi, S.; Watanabe, N.; Wang, Y.-H.; Wang, Y.-H.; Ito, T.; Shaw, J.; Cao, W.; Qin, F.X.-F.; Liu, Y.-J. Human plasmacytoid predendritic cells activate NK cells through glucocorticoid-induced tumor necrosis factor receptor-ligand (GITRL). Blood 2006, 107, 3617–3623. [Google Scholar] [CrossRef]
- Tone, M.; Tone, Y.; Adams, E.; Yates, S.F.; Frewin, M.R.; Cobbold, S.P.; Waldmann, H. Mouse glucocorticoid-induced tumor necrosis factor receptor ligand is costimulatory for T cells. Proc. Natl. Acad. Sci. USA 2003, 100, 15059–15064. [Google Scholar] [CrossRef]
- Kim, J.D.; Choi, B.K.; Bae, J.S.; Lee, U.H.; Han, I.S.; Lee, H.W.; Youn, B.S.; Vinay, D.S.; Kwon, B.S. Cloning and characterization of GITR ligand. Genes Immun. 2003, 4, 564–569. [Google Scholar] [CrossRef]
- Ronchetti, S.; Zollo, O.; Bruscoli, S.; Agostini, M.; Bianchini, R.; Nocentini, G.; Ayroldi, E.; Riccardi, C. GITR, a member of the TNF receptor superfamily, is costimulatory to mouse T lymphocyte subpopulations. Eur. J. Immunol. 2004, 34, 613–622. [Google Scholar] [CrossRef]
- Kanamaru, F.; Youngnak, P.; Hashiguchi, M.; Nishioka, T.; Takahashi, T.; Sakaguchi, S.; Ishikawa, I.; Azuma, M. Costimulation via Glucocorticoid-Induced TNF Receptor in Both Conventional and CD25+ Regulatory CD4+ T Cells. J. Immunol. 2004, 172, 7306–7314. [Google Scholar] [CrossRef] [PubMed]
- Ji, H.; Liao, G.; Faubion, W.A.; Abadía-Molina, A.C.; Cozzo, C.; Laroux, F.S.; Caton, A.; Terhorst, C. Cutting edge: the natural ligand for glucocorticoid-induced TNF receptor-related protein abrogates regulatory T cell suppression. J. Immunol. 2004, 172, 5823–5827. [Google Scholar] [CrossRef]
- Coe, D.; Begom, S.; Addey, C.; White, M.; Dyson, J.; Chai, J.-G. Depletion of regulatory T cells by anti-GITR mAb as a novel mechanism for cancer immunotherapy. Cancer Immunol. Immunother. 2010, 59, 1367–1377. [Google Scholar] [CrossRef]
- Nocentini, G.; Giunchi, L.; Ronchetti, S.; Krausz, L.T.; Bartoli, A.; Moraca, R.; Migliorati, G.; Riccardi, C. A new member of the tumor necrosis factor/nerve growth factor receptor family inhibits T cell receptor-induced apoptosis. Proc. Natl. Acad. Sci. USA 1997, 94, 6216–6221. [Google Scholar] [CrossRef]
- Steinberger, P.; Majdic, O.; Derdak, S.V.; Pfistershammer, K.; Kirchberger, S.; Klauser, C.; Zlabinger, G.; Pickl, W.F.; Stöckl, J.; Knapp, W. Molecular Characterization of Human 4Ig-B7-H3, a Member of the B7 Family with Four Ig-Like Domains. J. Immunol. 2004, 172, 2352–2359. [Google Scholar] [CrossRef]
- Chapoval, A.I.; Ni, J.; Lau, J.S.; Wilcox, R.A.; Flies, D.B.; Liu, D.; Dong, H.; Sica, G.L.; Zhu, G.; Tamada, K.; et al. B7-H3: a costimulatory molecule for T cell activation and IFN-gamma production. Nat. Immunol. 2001, 2, 269–274. [Google Scholar] [CrossRef]
- Luo, L.; Chapoval, A.I.; Flies, D.B.; Zhu, G.; Hirano, F.; Wang, S.; Lau, J.S.; Dong, H.; Tamada, K.; Flies, A.S.; et al. B7-H3 Enhances Tumor Immunity In Vivo by Costimulating Rapid Clonal Expansion of Antigen-Specific CD8+ Cytolytic T Cells. J. Immunol. 2004, 173, 5445–5450. [Google Scholar] [CrossRef] [PubMed]
- Suh, W.-K.; Gajewska, B.U.; Okada, H.; Gronski, M.A.; Bertram, E.M.; Dawicki, W.; Duncan, G.S.; Bukczynski, J.; Plyte, S.; Elia, A.; et al. The B7 family member B7-H3 preferentially down-regulates T helper type 1-mediated immune responses. Nat. Immunol. 2003, 4, 899–906. [Google Scholar] [CrossRef] [PubMed]
- Fukushima, A.; Sumi, T.; Fukuda, K.; Kumagai, N.; Nishida, T.; Yamazaki, T.; Akiba, H.; Okumura, K.; Yagita, H.; Ueno, H. B7-H3 regulates the development of experimental allergic conjunctivitis in mice. Immunol. Lett. 2007, 113, 52–57. [Google Scholar] [CrossRef]
- Castriconi, R.; Dondero, A.; Augugliaro, R.; Cantoni, C.; Carnemolla, B.; Sementa, A.R.; Negri, F.; Conte, R.; Corrias, M.V.; Moretta, L.; et al. Identification of 4Ig-B7-H3 as a neuroblastoma-associated molecule that exerts a protective role from an NK cell-mediated lysis. Proc. Natl. Acad. Sci. USA 2004, 101, 12640–12645. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.-W.; Tekle, C.; Fodstad, O. The immunoregulatory protein human B7H3 is a tumor-associated antigen that regulates tumor cell migration and invasion. Curr. Cancer Drug Targets 2008, 8, 404–413. [Google Scholar] [CrossRef]
- Xie, C.; Liu, D.; Chen, Q.; Yang, C.; Wang, B.; Wu, H. Soluble B7-H3 promotes the invasion and metastasis of pancreatic carcinoma cells through the TLR4/NF-κB pathway. Sci. Rep. 2016, 6, 27528. [Google Scholar] [CrossRef] [PubMed]
- Hutloff, A.; Dittrich, A.M.; Beier, K.C.; Eljaschewitsch, B.; Kraft, R.; Anagnostopoulos, I.; Kroczek, R.A. ICOS is an inducible T-cell co-stimulator structurally and functionally related to CD28. Nature 1999, 397, 263–266. [Google Scholar] [CrossRef] [PubMed]
- McAdam, A.J.; Chang, T.T.; Lumelsky, A.E.; Greenfield, E.A.; Boussiotis, V.A.; Duke-Cohan, J.S.; Chernova, T.; Malenkovich, N.; Jabs, C.; Kuchroo, V.K.; et al. Mouse Inducible Costimulatory Molecule (ICOS) Expression Is Enhanced by CD28 Costimulation and Regulates Differentiation of CD4+ T Cells. J. Immunol. 2000, 165, 5035–5040. [Google Scholar] [CrossRef] [PubMed]
- Swallow, M.M.; Wallin, J.J.; Sha, W.C. B7h, a novel costimulatory homolog of B7.1 and B7.2, is induced by TNFalpha. Immunity 1999, 11, 423–432. [Google Scholar] [CrossRef]
- Yoshinaga, S.K.; Whoriskey, J.S.; Khare, S.D.; Sarmiento, U.; Guo, J.; Horan, T.; Shih, G.; Zhang, M.; Coccia, M.A.; Kohno, T.; et al. T-cell co-stimulation through B7RP-1 and ICOS. Nature 1999, 402, 827–832. [Google Scholar] [CrossRef] [PubMed]
- Aicher, A.; Hayden-Ledbetter, M.; Brady, W.A.; Pezzutto, A.; Richter, G.; Magaletti, D.; Buckwalter, S.; Ledbetter, J.A.; Clark, E.A. Characterization of Human Inducible Costimulator Ligand Expression and Function. J. Immunol. 2000, 164, 4689–4696. [Google Scholar] [CrossRef]
- Qian, X.; Agematsu, K.; Freeman, G.J.; Tagawa, Y.; Sugane, K.; Hayashi, T. The ICOS-ligand B7-H2, expressed on human type II alveolar epithelial cells, plays a role in the pulmonary host defense system. Eur. J. Immunol. 2006, 36, 906–918. [Google Scholar] [CrossRef]
- Khayyamian, S.; Hutloff, A.; Büchner, K.; Gräfe, M.; Henn, V.; Kroczek, R.A.; Mages, H.W. ICOS-ligand, expressed on human endothelial cells, costimulates Th1 and Th2 cytokine secretion by memory CD4+ T cells. Proc. Natl. Acad. Sci. USA 2002, 99, 6198–6203. [Google Scholar] [CrossRef]
- Gigoux, M.; Lovato, A.; Leconte, J.; Leung, J.; Sonenberg, N.; Suh, W.-K. Inducible costimulator facilitates T-dependent B cell activation by augmenting IL-4 translation. Mol. Immunol. 2014, 59, 46–54. [Google Scholar] [CrossRef]
- Van Berkel, M.E.A.T.; Oosterwegel, M.A. CD28 and ICOS: similar or separate costimulators of T cells? Immunol. Lett. 2006, 105, 115–122. [Google Scholar] [CrossRef] [PubMed]
- Zhifu, Y.; Mingli, J.; Shuang, C.; Fan, W.; Zhenkun, F.; Wangyang, C.; Lin, Z.; Guangxiao, L.; Yashuang, Z.; Dianjun, L. SNP-SNP interactions of immunity related genes involved in the CD28/B7 pathway with susceptibility to invasive ductal carcinoma of the breast. Gene 2015, 566, 217–222. [Google Scholar] [CrossRef]
- Xu, F.; Li, D.; Zhang, Q.; Fu, Z.; Zhang, J.; Yuan, W.; Chen, S.; Pang, D.; Li, D. ICOS gene polymorphisms are associated with sporadic breast cancer: a case-control study. BMC Cancer 2011, 11, 392. [Google Scholar] [CrossRef]
- Schoenbrunn, A.; Frentsch, M.; Kohler, S.; Keye, J.; Dooms, H.; Moewes, B.; Dong, J.; Loddenkemper, C.; Sieper, J.; Wu, P.; et al. A converse 4-1BB and CD40 ligand expression pattern delineates activated regulatory T cells (Treg) and conventional T cells enabling direct isolation of alloantigen-reactive natural Foxp3+ Treg. J. Immunol. 2012, 189, 5985–5994. [Google Scholar] [CrossRef]
- Kwon, B.S.; Weissman, S.M. cDNA sequences of two inducible T-cell genes. Proc. Natl. Acad. Sci. USA 1989, 86, 1963–1967. [Google Scholar] [CrossRef] [PubMed]
- Vinay, D.S.; Lee, S.J.; Kim, C.H.; Oh, H.S.; Kwon, B.S. Exposure of a Distinct PDCA-1+ (CD317) B Cell Population to Agonistic Anti-4-1BB (CD137) Inhibits T and B Cell Responses Both In Vitro and In Vivo. PLoS ONE 2012, 7. [Google Scholar] [CrossRef][Green Version]
- Melero, I.; Johnston, J.V.; Shufford, W.W.; Mittler, R.S.; Chen, L. NK1.1 cells express 4-1BB (CDw137) costimulatory molecule and are required for tumor immunity elicited by anti-4-1BB monoclonal antibodies. Cell. Immunol. 1998, 190, 167–172. [Google Scholar] [CrossRef]
- Kim, D.-H.; Chang, W.-S.; Lee, Y.-S.; Lee, K.-A.; Kim, Y.-K.; Kwon, B.S.; Kang, C.-Y. 4-1BB engagement costimulates NKT cell activation and exacerbates NKT cell ligand-induced airway hyperresponsiveness and inflammation. J. Immunol. 2008, 180, 2062–2068. [Google Scholar] [CrossRef]
- Pauly, S.; Broll, K.; Wittmann, M.; Giegerich, G.; Schwarz, H. CD137 is expressed by follicular dendritic cells and costimulates B lymphocyte activation in germinal centers. J. Leukoc. Biol. 2002, 72, 35–42. [Google Scholar]
- Lee, S.-W.; Park, Y.; So, T.; Kwon, B.S.; Cheroutre, H.; Mittler, R.S.; Croft, M. Identification of regulatory functions for 4-1BB and 4-1BBL in myelopoiesis and the development of dendritic cells. Nat. Immunol. 2008, 9, 917–926. [Google Scholar] [CrossRef]
- Bartkowiak, T.; Curran, M.A. 4-1BB Agonists: Multi-Potent Potentiators of Tumor Immunity. Front. Oncol. 2015, 5. [Google Scholar] [CrossRef]
- Lee, H.-W.; Park, S.-J.; Choi, B.K.; Kim, H.H.; Nam, K.-O.; Kwon, B.S. 4-1BB promotes the survival of CD8+ T lymphocytes by increasing expression of Bcl-xL and Bfl-1. J. Immunol. 2002, 169, 4882–4888. [Google Scholar] [CrossRef]
- Shuford, W.W.; Klussman, K.; Tritchler, D.D.; Loo, D.T.; Chalupny, J.; Siadak, A.W.; Brown, T.J.; Emswiler, J.; Raecho, H.; Larsen, C.P.; et al. 4-1BB costimulatory signals preferentially induce CD8+ T cell proliferation and lead to the amplification in vivo of cytotoxic T cell responses. J. Exp. Med. 1997, 186, 47–55. [Google Scholar] [CrossRef]
- Hurtado, J.C.; Kim, Y.J.; Kwon, B.S. Signals through 4-1BB are costimulatory to previously activated splenic T cells and inhibit activation-induced cell death. J. Immunol. 1997, 158, 2600–2609. [Google Scholar] [PubMed]
- Melero, I.; Gangadhar, T.C.; Kohrt, H.E.; Segal, N.H.; Logan, T.; Urba, W.J.; Hodi, F.S.; Ott, P.A.; Perez-Gracia, J.L.; Wolchok, J.D.; et al. A phase I study of the safety, tolerability, pharmacokinetics, and immunoregulatory activity of urelumab (BMS-663513) in subjects with advanced and/or metastatic solid tumors and relapsed/refractory B-cell non-Hodgkin’s lymphoma (B-NHL). JCO 2013, 31, TPS3107. [Google Scholar]
- Hintzen, R.Q.; Lens, S.M.; Beckmann, M.P.; Goodwin, R.G.; Lynch, D.; Lier, R.A. van Characterization of the human CD27 ligand, a novel member of the TNF gene family. J. Immunol. 1994, 152, 1762–1773. [Google Scholar] [PubMed]
- Agematsu, K. Memory B cells and CD27. Histol. Histopathol. 2000, 15, 573–576. [Google Scholar] [PubMed]
- Jung, J.; Choe, J.; Li, L.; Choi, Y.S. Regulation of CD27 expression in the course of germinal center B cell differentiation: the pivotal role of IL-10. Eur. J. Immunol. 2000, 30, 2437–2443. [Google Scholar] [CrossRef]
- Hayakawa, Y.; Smyth, M.J. CD27 Dissects Mature NK Cells into Two Subsets with Distinct Responsiveness and Migratory Capacity. J. Immunol. 2006, 176, 1517–1524. [Google Scholar] [CrossRef] [PubMed]
- Bowman, M.R.; Crimmins, M.A.; Yetz-Aldape, J.; Kriz, R.; Kelleher, K.; Herrmann, S. The cloning of CD70 and its identification as the ligand for CD27. J. Immunol. 1994, 152, 1756–1761. [Google Scholar] [PubMed]
- Lens, S.M.; de Jong, R.; Hooibrink, B.; Koopman, G.; Pals, S.T.; van Oers, M.H.; van Lier, R.A. Phenotype and function of human B cells expressing CD70 (CD27 ligand). Eur. J. Immunol. 1996, 26, 2964–2971. [Google Scholar] [CrossRef] [PubMed]
- Hashimoto-Okada, M.; Kitawaki, T.; Kadowaki, N.; Iwata, S.; Morimoto, C.; Hori, T.; Uchiyama, T. The CD70–CD27 interaction during the stimulation with dendritic cells promotes naive CD4+ T cells to develop into T cells producing a broad array of immunostimulatory cytokines in humans. Int Immunol 2009, 21, 891–904. [Google Scholar] [CrossRef] [PubMed]
- Titanji, K.; Sammicheli, S.; De Milito, A.; Mantegani, P.; Fortis, C.; Berg, L.; Kärre, K.; Travi, G.; Tassandin, C.; Lopalco, L.; et al. Altered distribution of natural killer cell subsets identified by CD56, CD27 and CD70 in primary and chronic human immunodeficiency virus-1 infection. Immunology 2008, 123, 164–170. [Google Scholar] [CrossRef]
- Wajant, H. Therapeutic targeting of CD70 and CD27. Expert Opin. Ther. Targets 2016, 20, 959–973. [Google Scholar] [CrossRef]
- Van Oosterwijk, M.F.; Juwana, H.; Arens, R.; Tesselaar, K.; van Oers, M.H.J.; Eldering, E.; van Lier, R.A.W. CD27-CD70 interactions sensitise naive CD4+ T cells for IL-12-induced Th1 cell development. Int. Immunol. 2007, 19, 713–718. [Google Scholar] [CrossRef] [PubMed]
- Peperzak, V.; Veraar, E.A.M.; Keller, A.M.; Xiao, Y.; Borst, J. The Pim kinase pathway contributes to survival signaling in primed CD8+ T cells upon CD27 costimulation. J. Immunol. 2010, 185, 6670–6678. [Google Scholar] [CrossRef] [PubMed]
- Hendriks, J.; Xiao, Y.; Borst, J. CD27 Promotes Survival of Activated T Cells and Complements CD28 in Generation and Establishment of the Effector T Cell Pool. J. Exp. Med. 2003, 198, 1369–1380. [Google Scholar] [CrossRef]
- Dolfi, D.V.; Boesteanu, A.C.; Petrovas, C.; Xia, D.; Butz, E.A.; Katsikis, P.D. Late signals from CD27 prevent Fas dependent apoptosis of primary CD8+ T cells. J. Immunol. 2008, 180, 2912–2921. [Google Scholar] [CrossRef] [PubMed]
- Carr, J.M.; Carrasco, M.J.; Thaventhiran, J.E.D.; Bambrough, P.J.; Kraman, M.; Edwards, A.D.; Al-Shamkhani, A.; Fearon, D.T. CD27 mediates interleukin-2-independent clonal expansion of the CD8+ T cell without effector differentiation. Proc. Natl. Acad. Sci. USA 2006, 103, 19454–19459. [Google Scholar] [CrossRef]
- Xiao, Y.; Peperzak, V.; Keller, A.M.; Borst, J. CD27 instructs CD4+ T cells to provide help for the memory CD8+ T cell response after protein immunization. J. Immunol. 2008, 181, 1071–1082. [Google Scholar] [CrossRef]
- Hendriks, J.; Gravestein, L.A.; Tesselaar, K.; van Lier, R.A.; Schumacher, T.N.; Borst, J. CD27 is required for generation and long-term maintenance of T cell immunity. Nat. Immunol. 2000, 1, 433–440. [Google Scholar] [CrossRef]
- Peperzak, V.; Veraar, E.A.M.; Xiao, Y.; Babala, N.; Thiadens, K.; Brugmans, M.; Borst, J. CD8+ T cells produce the chemokine CXCL10 in response to CD27/CD70 costimulation to promote generation of the CD8+ effector T cell pool. J. Immunol. 2013, 191, 3025–3036. [Google Scholar] [CrossRef] [PubMed]
- Kelly, J.M.; Darcy, P.K.; Markby, J.L.; Godfrey, D.I.; Takeda, K.; Yagita, H.; Smyth, M.J. Induction of tumor-specific T cell memory by NK cell-mediated tumor rejection. Nat. Immunol. 2002, 3, 83–90. [Google Scholar] [CrossRef] [PubMed]
- Takeda, K.; Oshima, H.; Hayakawa, Y.; Akiba, H.; Atsuta, M.; Kobata, T.; Kobayashi, K.; Ito, M.; Yagita, H.; Okumura, K. CD27-mediated activation of murine NK cells. J. Immunol. 2000, 164, 1741–1745. [Google Scholar] [CrossRef]
- Agematsu, K.; Kobata, T.; Yang, F.C.; Nakazawa, T.; Fukushima, K.; Kitahara, M.; Mori, T.; Sugita, K.; Morimoto, C.; Komiyama, A. CD27/CD70 interaction directly drives B cell IgG and IgM synthesis. Eur. J. Immunol. 1995, 25, 2825–2829. [Google Scholar] [CrossRef]
- Agematsu, K.; Nagumo, H.; Oguchi, Y.; Nakazawa, T.; Fukushima, K.; Yasui, K.; Ito, S.; Kobata, T.; Morimoto, C.; Komiyama, A. Generation of plasma cells from peripheral blood memory B cells: synergistic effect of interleukin-10 and CD27/CD70 interaction. Blood 1998, 91, 173–180. [Google Scholar]
- Kobata, T.; Jacquot, S.; Kozlowski, S.; Agematsu, K.; Schlossman, S.F.; Morimoto, C. CD27-CD70 interactions regulate B-cell activation by T cells. Proc. Natl. Acad. Sci. USA 1995, 92, 11249–11253. [Google Scholar] [CrossRef] [PubMed]
- Petrau, C.; Cornic, M.; Bertrand, P.; Maingonnat, C.; Marchand, V.; Picquenot, J.-M.; Jardin, F.; Clatot, F. CD70: A Potential Target in Breast Cancer? J. Cancer 2014, 5, 761–764. [Google Scholar] [CrossRef] [PubMed]
- Fujita, T.; Ukyo, N.; Hori, T.; Uchiyama, T. Functional characterization of OX40 expressed on human CD8+ T cells. Immunol. Lett. 2006, 106, 27–33. [Google Scholar] [CrossRef] [PubMed]
- Montler, R.; Bell, R.B.; Thalhofer, C.; Leidner, R.; Feng, Z.; Fox, B.A.; Cheng, A.C.; Bui, T.G.; Tucker, C.; Hoen, H.; et al. OX40, PD-1 and CTLA-4 are selectively expressed on tumor-infiltrating T cells in head and neck cancer. Clin. Transl. Immunol. 2016, 5, e70. [Google Scholar] [CrossRef] [PubMed]
- Lai, C.; August, S.; Albibas, A.; Behar, R.; Cho, S.-Y.; Polak, M.E.; Theaker, J.; MacLeod, A.S.; French, R.R.; Glennie, M.J.; et al. OX40+ Regulatory T Cells in Cutaneous Squamous Cell Carcinoma Suppress Effector T-Cell Responses and Associate with Metastatic Potential. Clin. Cancer Res. 2016, 22, 4236–4248. [Google Scholar] [CrossRef] [PubMed]
- Baumann, R.; Yousefi, S.; Simon, D.; Russmann, S.; Mueller, C.; Simon, H.-U. Functional expression of CD134 by neutrophils. Eur. J. Immunol. 2004, 34, 2268–2275. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Lou, Y.; Lizée, G.; Qin, H.; Liu, S.; Rabinovich, B.; Kim, G.J.; Wang, Y.-H.; Ye, Y.; Sikora, A.G.; et al. Plasmacytoid dendritic cells induce NK cell–dependent, tumor antigen–specific T cell cross-priming and tumor regression in mice. J. Clin. Invest 2008, 118, 1165–1175. [Google Scholar] [CrossRef]
- Zaini, J.; Andarini, S.; Tahara, M.; Saijo, Y.; Ishii, N.; Kawakami, K.; Taniguchi, M.; Sugamura, K.; Nukiwa, T.; Kikuchi, T. OX40 ligand expressed by DCs costimulates NKT and CD4+ Th cell antitumor immunity in mice. J. Clin. Invest 2007, 117, 3330–3338. [Google Scholar] [CrossRef] [PubMed]
- Ohshima, Y.; Tanaka, Y.; Tozawa, H.; Takahashi, Y.; Maliszewski, C.; Delespesse, G. Expression and function of OX40 ligand on human dendritic cells. J. Immunol. 1997, 159, 3838–3848. [Google Scholar] [PubMed]
- Karulf, M.; Kelly, A.; Weinberg, A.D.; Gold, J.A. OX40 ligand regulates inflammation and mortality in the innate immune response to sepsis. J. Immunol. 2010, 185, 4856–4862. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Chen, Y.; Ge, Y.; Sun, J.; Shi, Q.; Ju, S.; Dai, J.; Yu, G.; Zhang, X. Characterization and functional study of five novel monoclonal antibodies against human OX40L highlight reverse signalling: enhancement of IgG production of B cells and promotion of maturation of DCs. Tissue Antigens 2004, 64, 566–574. [Google Scholar] [CrossRef]
- Fujita, T.; Kambe, N.; Uchiyama, T.; Hori, T. Type I interferons attenuate T cell activating functions of human mast cells by decreasing TNF-alpha production and OX40 ligand expression while increasing IL-10 production. J. Clin. Immunol. 2006, 26, 512–518. [Google Scholar] [CrossRef]
- Souza, H.; Elia, C.; Spencer, J.; MacDonald, T. Expression of lymphocyte-endothelial receptor-ligand pairs, α4β7/MAdCAM-1 and OX40/OX40 ligand in the colon and jejunum of patients with inflammatory bowel disease. Gut 1999, 45, 856–863. [Google Scholar] [CrossRef]
- Sato, T.; Ishii, N.; Murata, K.; Kikuchi, K.; Nakagawa, S.; Ndhlovu, L.C.; Sugamura, K. Consequences of OX40-OX40 ligand interactions in langerhans cell function: enhanced contact hypersensitivity responses in OX40L-transgenic mice. Eur. J. Immunol. 2002, 32, 3326–3335. [Google Scholar] [CrossRef]
- Weinberg, A.D.; Wegmann, K.W.; Funatake, C.; Whitham, R.H. Blocking OX-40/OX-40 ligand interaction in vitro and in vivo leads to decreased T cell function and amelioration of experimental allergic encephalomyelitis. J. Immunol. 1999, 162, 1818–1826. [Google Scholar]
- Zingoni, A.; Sornasse, T.; Cocks, B.G.; Tanaka, Y.; Santoni, A.; Lanier, L.L. Cross-talk between activated human NK cells and CD4+ T cells via OX40-OX40 ligand interactions. J. Immunol. 2004, 173, 3716–3724. [Google Scholar] [CrossRef]
- Maxwell, J.R.; Weinberg, A.; Prell, R.A.; Vella, A.T. Danger and OX40 Receptor Signaling Synergize to Enhance Memory T Cell Survival by Inhibiting Peripheral Deletion. J. Immunol. 2000, 164, 107–112. [Google Scholar] [CrossRef] [PubMed]
- Gramaglia, I.; Jember, A.; Pippig, S.D.; Weinberg, A.D.; Killeen, N.; Croft, M. The OX40 costimulatory receptor determines the development of CD4 memory by regulating primary clonal expansion. J. Immunol. 2000, 165, 3043–3050. [Google Scholar] [CrossRef]
- Gramaglia, I.; Weinberg, A.D.; Lemon, M.; Croft, M. Ox-40 Ligand: A Potent Costimulatory Molecule for Sustaining Primary CD4 T Cell Responses. J. Immunol. 1998, 161, 6510–6517. [Google Scholar]
- Ito, T.; Wang, Y.-H.; Duramad, O.; Hori, T.; Delespesse, G.J.; Watanabe, N.; Qin, F.X.-F.; Yao, Z.; Cao, W.; Liu, Y.-J. TSLP-activated dendritic cells induce an inflammatory T helper type 2 cell response through OX40 ligand. J. Exp. Med. 2005, 202, 1213–1223. [Google Scholar] [CrossRef]
- Zhang, Z.; Zhong, W.; Hinrichs, D.; Wu, X.; Weinberg, A.; Hall, M.; Spencer, D.; Wegmann, K.; Rosenbaum, J.T. Activation of OX40 Augments Th17 Cytokine Expression and Antigen-Specific Uveitis. Am. J. Pathol. 2010, 177, 2912–2920. [Google Scholar] [CrossRef] [PubMed]
- Baum, P.R.; Gayle, R.B.; Ramsdell, F.; Srinivasan, S.; Sorensen, R.A.; Watson, M.L.; Seldin, M.F.; Baker, E.; Sutherland, G.R.; Clifford, K.N. Molecular characterization of murine and human OX40/OX40 ligand systems: identification of a human OX40 ligand as the HTLV-1-regulated protein gp34. EMBO J. 1994, 13, 3992–4001. [Google Scholar] [CrossRef]
- Bansal-Pakala, P.; Halteman, B.S.; Cheng, M.H.-Y.; Croft, M. Costimulation of CD8 T cell responses by OX40. J. Immunol. 2004, 172, 4821–4825. [Google Scholar] [CrossRef]
- Lee, S.-W.; Park, Y.; Song, A.; Cheroutre, H.; Kwon, B.S.; Croft, M. Functional dichotomy between OX40 and 4-1BB in modulating effector CD8 T cell responses. J. Immunol. 2006, 177, 4464–4472. [Google Scholar] [CrossRef] [PubMed]
- Han, P.; Goularte, O.D.; Rufner, K.; Wilkinson, B.; Kaye, J. An Inhibitory Ig Superfamily Protein Expressed by Lymphocytes and APCs Is Also an Early Marker of Thymocyte Positive Selection. J. Immunol. 2004, 172, 5931–5939. [Google Scholar] [CrossRef]
- Iwata, A.; Watanabe, N.; Oya, Y.; Owada, T.; Ikeda, K.; Suto, A.; Kagami, S.; Hirose, K.; Kanari, H.; Kawashima, S.; et al. Protective roles of B and T lymphocyte attenuator in NKT cell-mediated experimental hepatitis. J. Immunol. 2010, 184, 127–133. [Google Scholar] [CrossRef]
- Šedý, J.R.; Bjordahl, R.L.; Bekiaris, V.; Macauley, M.G.; Ware, B.C.; Norris, P.S.; Lurain, N.S.; Benedict, C.A.; Ware, C.F. CD160 activation by herpesvirus entry mediator augments inflammatory cytokine production and cytolytic function by NK cells. J. Immunol. 2013, 191, 828–836. [Google Scholar] [CrossRef]
- Del Rio, M.-L.; Kaye, J.; Rodriguez-Barbosa, J.-I. Detection of protein on BTLAlow cells and in vivo antibody-mediated down-modulation of BTLA on lymphoid and myeloid cells of C57BL/6 and BALB/c BTLA allelic variants. Immunobiology 2010, 215, 570–578. [Google Scholar] [CrossRef] [PubMed]
- M’Hidi, H.; Thibult, M.-L.; Chetaille, B.; Rey, F.; Bouadallah, R.; Nicollas, R.; Olive, D.; Xerri, L. High expression of the inhibitory receptor BTLA in T-follicular helper cells and in B-cell small lymphocytic lymphoma/chronic lymphocytic leukemia. Am. J. Clin. Pathol. 2009, 132, 589–596. [Google Scholar] [CrossRef] [PubMed]
- Kwon, B.S.; Tan, K.B.; Ni, J.; Kwi-Ok-Oh, Z.H.L.; Kim, K.K.; Kim, Y.-J.; Wang, S.; Gentz, R.; Yu, G.-L.; Harrop, J.; et al. A Newly Identified Member of the Tumor Necrosis Factor Receptor Superfamily with a Wide Tissue Distribution and Involvement in Lymphocyte Activation. J. Biol. Chem. 1997, 272, 14272–14276. [Google Scholar] [CrossRef] [PubMed]
- Duhen, T.; Pasero, C.; Mallet, F.; Barbarat, B.; Olive, D.; Costello, R.T. LIGHT costimulates CD40 triggering and induces immunoglobulin secretion; a novel key partner in T cell-dependent B cell terminal differentiation. Eur. J. Immunol. 2004, 34, 3534–3541. [Google Scholar] [CrossRef]
- Morel, Y.; Truneh, A.; Sweet, R.W.; Olive, D.; Costello, R.T. The TNF superfamily members LIGHT and CD154 (CD40 ligand) costimulate induction of dendritic cell maturation and elicit specific CTL activity. J. Immunol. 2001, 167, 2479–2486. [Google Scholar] [CrossRef]
- Zhai, Y.; Guo, R.; Hsu, T.L.; Yu, G.L.; Ni, J.; Kwon, B.S.; Jiang, G.W.; Lu, J.; Tan, J.; Ugustus, M.; et al. LIGHT, a novel ligand for lymphotoxin beta receptor and TR2/HVEM induces apoptosis and suppresses in vivo tumor formation via gene transfer. J. Clin. Invest 1998, 102, 1142–1151. [Google Scholar] [CrossRef]
- Xu, Z.; Shen, J.; Wang, M.H.; Yi, T.; Yu, Y.; Zhu, Y.; Chen, B.; Chen, J.; Li, L.; Li, M.; et al. Comprehensive molecular profiling of the B7 family of immune-regulatory ligands in breast cancer. Oncoimmunology 2016, 5. [Google Scholar] [CrossRef] [PubMed]
- Chemnitz, J.M.; Lanfranco, A.R.; Braunstein, I.; Riley, J.L. B and T Lymphocyte Attenuator-Mediated Signal Transduction Provides a Potent Inhibitory Signal to Primary Human CD4 T Cells That Can Be Initiated by Multiple Phosphotyrosine Motifs. J. Immunol. 2006, 176, 6603–6614. [Google Scholar] [CrossRef] [PubMed]
- Krieg, C.; Han, P.; Stone, R.; Goularte, O.D.; Kaye, J. Functional analysis of B and T lymphocyte attenuator engagement on CD4+ and CD8+ T cells. J. Immunol. 2005, 175, 6420–6427. [Google Scholar] [CrossRef]
- Otsuki, N.; Kamimura, Y.; Hashiguchi, M.; Azuma, M. Expression and function of the B and T lymphocyte attenuator (BTLA/CD272) on human T cells. Biochem. Biophys. Res. Commun. 2006, 344, 1121–1127. [Google Scholar] [CrossRef] [PubMed]
- Murphy, T.L.; Murphy, K.M. Slow down and survive: Enigmatic immunoregulation by BTLA and HVEM. Annu. Rev. Immunol. 2010, 28, 389–411. [Google Scholar] [CrossRef] [PubMed]
- Derré, L.; Rivals, J.-P.; Jandus, C.; Pastor, S.; Rimoldi, D.; Romero, P.; Michielin, O.; Olive, D.; Speiser, D.E. BTLA mediates inhibition of human tumor-specific CD8+ T cells that can be partially reversed by vaccination. J. Clin. Invest. 2010, 120, 157–167. [Google Scholar] [CrossRef]
- Watanabe, N.; Gavrieli, M.; Sedy, J.R.; Yang, J.; Fallarino, F.; Loftin, S.K.; Hurchla, M.A.; Zimmerman, N.; Sim, J.; Zang, X.; et al. BTLA is a lymphocyte inhibitory receptor with similarities to CTLA-4 and PD-1. Nat. Immunol. 2003, 4, 670–679. [Google Scholar] [CrossRef]
- Vendel, A.C.; Calemine-Fenaux, J.; Izrael-Tomasevic, A.; Chauhan, V.; Arnott, D.; Eaton, D.L. B and T Lymphocyte Attenuator Regulates B Cell Receptor Signaling by Targeting Syk and BLNK. J. Immunol. 2009, 182, 1509–1517. [Google Scholar] [CrossRef]
- Leifer, C.A.; Kennedy, M.N.; Mazzoni, A.; Lee, C.; Kruhlak, M.J.; Segal, D.M. TLR9 is localized in the endoplasmic reticulum prior to stimulation. J. Immunol. 2004, 173, 1179–1183. [Google Scholar] [CrossRef]
- Latz, E.; Schoenemeyer, A.; Visintin, A.; Fitzgerald, K.A.; Monks, B.G.; Knetter, C.F.; Lien, E.; Nilsen, N.J.; Espevik, T.; Golenbock, D.T. TLR9 signals after translocating from the ER to CpG DNA in the lysosome. Nat. Immunol. 2004, 5, 190–198. [Google Scholar] [CrossRef]
- Kawagoe, T.; Sato, S.; Jung, A.; Yamamoto, M.; Matsui, K.; Kato, H.; Uematsu, S.; Takeuchi, O.; Akira, S. Essential role of IRAK-4 protein and its kinase activity in Toll-like receptor-mediated immune responses but not in TCR signaling. J. Exp. Med. 2007, 204, 1013–1024. [Google Scholar] [CrossRef] [PubMed]
- Sato, S.; Sanjo, H.; Takeda, K.; Ninomiya-Tsuji, J.; Yamamoto, M.; Kawai, T.; Matsumoto, K.; Takeuchi, O.; Akira, S. Essential function for the kinase TAK1 in innate and adaptive immune responses. Nat. Immunol. 2005, 6, 1087–1095. [Google Scholar] [CrossRef]
- Adachi, O.; Kawai, T.; Takeda, K.; Matsumoto, M.; Tsutsui, H.; Sakagami, M.; Nakanishi, K.; Akira, S. Targeted disruption of the MyD88 gene results in loss of IL-1- and IL-18-mediated function. Immunity 1998, 9, 143–150. [Google Scholar] [CrossRef]
- Kawai, T.; Sato, S.; Ishii, K.J.; Coban, C.; Hemmi, H.; Yamamoto, M.; Terai, K.; Matsuda, M.; Inoue, J.; Uematsu, S.; et al. Interferon-alpha induction through Toll-like receptors involves a direct interaction of IRF7 with MyD88 and TRAF6. Nat. Immunol. 2004, 5, 1061–1068. [Google Scholar] [CrossRef]
- Hemmi, H.; Takeuchi, O.; Kawai, T.; Kaisho, T.; Sato, S.; Sanjo, H.; Matsumoto, M.; Hoshino, K.; Wagner, H.; Takeda, K.; et al. A Toll-like receptor recognizes bacterial DNA. Nature 2000, 408, 740–745. [Google Scholar] [CrossRef]
- Lipford, G.B.; Sparwasser, T.; Zimmermann, S.; Heeg, K.; Wagner, H. CpG-DNA-mediated transient lymphadenopathy is associated with a state of Th1 predisposition to antigen-driven responses. J. Immunol. 2000, 165, 1228–1235. [Google Scholar] [CrossRef] [PubMed]
- Karki, K.; Pande, D.; Negi, R.; Khanna, S.; Khanna, R.S.; Khanna, H.D. Correlation of serum toll like receptor 9 and trace elements with lipid peroxidation in the patients of breast diseases. J. Trace Elem. Med. Biol. 2015, 30, 11–16. [Google Scholar] [CrossRef] [PubMed]
- AL-HARRAS, M.F.; HOUSSEN, M.E.; SHAKER, M.E.; FARAG, K.; FAROUK, O.; MONIR, R.; EL-MAHDY, R.; ABO-HASHEM, E.M. Polymorphisms of glutathione S-transferase π 1 and toll-like receptors 2 and 9: Association with breast cancer susceptibility. Oncol. Lett. 2016, 11, 2182–2188. [Google Scholar] [CrossRef]
- Wan, G.-X.; Cao, Y.-W.; Li, W.-Q.; Li, Y.-C.; Zhang, W.-J.; Li, F. Associations between TLR9 polymorphisms and cancer risk: evidence from an updated meta-analysis of 25,685 subjects. Asian Pac. J. Cancer Prev. 2014, 15, 8279–8285. [Google Scholar] [CrossRef]
- Resler, A.J.; Malone, K.E.; Johnson, L.G.; Malkki, M.; Petersdorf, E.W.; McKnight, B.; Madeleine, M.M. Genetic variation in TLR or NFkappaB pathways and the risk of breast cancer: a case-control study. BMC Cancer 2013, 13, 219. [Google Scholar] [CrossRef]
- Etokebe, G.E.; Knezević, J.; Petricević, B.; Pavelić, J.; Vrbanec, D.; Dembić, Z. Single-nucleotide polymorphisms in genes encoding toll-like receptor -2, -3, -4, and -9 in case-control study with breast cancer. Genet. Test Mol. Biomarkers 2009, 13, 729–734. [Google Scholar] [CrossRef]
- Cekic, C.; Linden, J. Purinergic regulation of the immune system. Nat. Rev. Immunol. 2016, 16, 177–192. [Google Scholar] [CrossRef]
- Allard, B.; Longhi, M.S.; Robson, S.C.; Stagg, J. The ectonucleotidases CD39 and CD73: novel checkpoint inhibitor targets. Immunol. Rev. 2017, 276, 121–144. [Google Scholar] [CrossRef] [PubMed]
- Antonioli, L.; Pacher, P.; Vizi, E.S.; Haskó, G. CD39 and CD73 in immunity and inflammation. Trends Mol. Med. 2013, 19, 355–367. [Google Scholar] [CrossRef]
- Resta, R.; Yamashita, Y.; Thompson, L.F. Ecto-enzyme and signaling functions of lymphocyte CD73. Immunol. Rev. 1998, 161, 95–109. [Google Scholar] [CrossRef] [PubMed]
- Kansas, G.S.; Wood, G.S.; Tedder, T.F. Expression, distribution, and biochemistry of human CD39. Role in activation-associated homotypic adhesion of lymphocytes. J. Immunol. 1991, 146, 2235–2244. [Google Scholar] [PubMed]
- Koziak, K.; Sévigny, J.; Robson, S.C.; Siegel, J.B.; Kaczmarek, E. Analysis of CD39/ATP diphosphohydrolase (ATPDase) expression in endothelial cells, platelets and leukocytes. Thromb. Haemost. 1999, 82, 1538–1544. [Google Scholar] [CrossRef] [PubMed]
- Borsellino, G.; Kleinewietfeld, M.; Di Mitri, D.; Sternjak, A.; Diamantini, A.; Giometto, R.; Höpner, S.; Centonze, D.; Bernardi, G.; Dell’Acqua, M.L.; et al. Expression of ectonucleotidase CD39 by Foxp3+ Treg cells: hydrolysis of extracellular ATP and immune suppression. Blood 2007, 110, 1225–1232. [Google Scholar] [CrossRef] [PubMed]
- Kas-Deelen, A.M.; Bakker, W.W.; Olinga, P.; Visser, J.; de Maar, E.F.; van Son, W.J.; The, T.H.; Harmsen, M.C. Cytomegalovirus infection increases the expression and activity of ecto-ATPase (CD39) and ecto-5’nucleotidase (CD73) on endothelial cells. FEBS Lett. 2001, 491, 21–25. [Google Scholar] [CrossRef]
- Do Carmo Araújo, M.; Rocha, J.B.T.; Morsch, A.; Zanin, R.; Bauchspiess, R.; Morsch, V.M.; Schetinger, M.R.C. Enzymes that hydrolyze adenine nucleotides in platelets from breast cancer patients. Biochim. Biophys. Acta 2005, 1740, 421–426. [Google Scholar] [CrossRef] [PubMed]
- Leone, R.D.; Emens, L.A. Targeting adenosine for cancer immunotherapy. J Immunother Cancer 2018, 6. [Google Scholar] [CrossRef] [PubMed]
- Yegutkin, G.G.; Wieringa, B.; Robson, S.C.; Jalkanen, S. Metabolism of circulating ADP in the bloodstream is mediated via integrated actions of soluble adenylate kinase-1 and NTPDase1/CD39 activities. FASEB J. 2012, 26, 3875–3883. [Google Scholar] [CrossRef]
- MacKenzie, W.M.; Hoskin, D.W.; Blay, J. Adenosine suppresses alpha(4)beta(7) integrin-mediated adhesion of T lymphocytes to colon adenocarcinoma cells. Exp. Cell Res. 2002, 276, 90–100. [Google Scholar] [CrossRef]
- Zarek, P.E.; Huang, C.-T.; Lutz, E.R.; Kowalski, J.; Horton, M.R.; Linden, J.; Drake, C.G.; Powell, J.D. A2A receptor signaling promotes peripheral tolerance by inducing T-cell anergy and the generation of adaptive regulatory T cells. Blood 2008, 111, 251–259. [Google Scholar] [CrossRef] [PubMed]
- Raskovalova, T.; Lokshin, A.; Huang, X.; Su, Y.; Mandic, M.; Zarour, H.M.; Jackson, E.K.; Gorelik, E. Inhibition of cytokine production and cytotoxic activity of human antimelanoma specific CD8+ and CD4+ T lymphocytes by adenosine-protein kinase A type I signaling. Cancer Res. 2007, 67, 5949–5956. [Google Scholar] [CrossRef]
- Williams, B.A.; Manzer, A.; Blay, J.; Hoskin, D.W. Adenosine acts through a novel extracellular receptor to inhibit granule exocytosis by natural killer cells. Biochem. Biophys. Res. Commun. 1997, 231, 264–269. [Google Scholar] [CrossRef]
- Nowak, M.; Lynch, L.; Yue, S.; Ohta, A.; Sitkovsky, M.; Balk, S.P.; Exley, M.A. The A2aR adenosine receptor controls cytokine production in iNKT cells. Eur. J. Immunol. 2010, 40, 682–687. [Google Scholar] [CrossRef]
- Xaus, J.; Valledor, A.F.; Cardó, M.; Marquès, L.; Beleta, J.; Palacios, J.M.; Celada, A. Adenosine inhibits macrophage colony-stimulating factor-dependent proliferation of macrophages through the induction of p27kip-1 expression. J. Immunol. 1999, 163, 4140–4149. [Google Scholar] [PubMed]
- Wilson, J.M.; Ross, W.G.; Agbai, O.N.; Frazier, R.; Figler, R.A.; Rieger, J.; Linden, J.; Ernst, P.B. The A2B Adenosine Receptor Impairs the Maturation and Immunogenicity of Dendritic Cells. J. Immunol. 2009, 182, 4616–4623. [Google Scholar] [CrossRef]
- Sevigny, C.P.; Li, L.; Awad, A.S.; Huang, L.; McDuffie, M.; Linden, J.; Lobo, P.I.; Okusa, M.D. Activation of Adenosine 2A Receptors Attenuates Allograft Rejection and Alloantigen Recognition. J. Immunol. 2007, 178, 4240–4249. [Google Scholar] [CrossRef]
- Williams, C.B.; Yeh, E.S.; Soloff, A.C. Tumor-associated macrophages: unwitting accomplices in breast cancer malignancy. NPJ Breast Cancer 2016, 2. [Google Scholar] [CrossRef]
- Aras, S.; Zaidi, M.R. TAMeless traitors: macrophages in cancer progression and metastasis. Br. J. Cancer 2017, 117, 1583–1591. [Google Scholar] [CrossRef]
- Zhang, Q.; Liu, L.; Gong, C.; Shi, H.; Zeng, Y.; Wang, X.; Zhao, Y.; Wei, Y. Prognostic significance of tumor-associated macrophages in solid tumor: a meta-analysis of the literature. PLoS ONE 2012, 7, e50946. [Google Scholar] [CrossRef]
- Zhao, X.; Qu, J.; Sun, Y.; Wang, J.; Liu, X.; Wang, F.; Zhang, H.; Wang, W.; Ma, X.; Gao, X.; et al. Prognostic significance of tumor-associated macrophages in breast cancer: a meta-analysis of the literature. Oncotarget 2017, 8, 30576–30586. [Google Scholar] [CrossRef] [PubMed]
- Mantovani, A.; Marchesi, F.; Malesci, A.; Laghi, L.; Allavena, P. Tumour-associated macrophages as treatment targets in oncology. Nat. Rev. Clin. Oncol. 2017, 14, 399–416. [Google Scholar] [CrossRef] [PubMed]
- DeNardo, D.G.; Ruffell, B. Macrophages as regulators of tumour immunity and immunotherapy. Nat. Rev. Immunol. 2019. [Google Scholar] [CrossRef] [PubMed]
- Thomas, J.K.; Mir, H.; Kapur, N.; Bae, S.; Singh, S. CC chemokines are differentially expressed in Breast Cancer and are associated with disparity in overall survival. Sci. Rep. 2019, 9, 4014. [Google Scholar] [CrossRef]
- Achkova, D.; Maher, J. Role of the colony-stimulating factor (CSF)/CSF-1 receptor axis in cancer. Biochem. Soc. Trans. 2016, 44, 333–341. [Google Scholar] [CrossRef] [PubMed]
- Richardsen, E.; Uglehus, R.D.; Johnsen, S.H.; Busund, L.-T. Macrophage-colony stimulating factor (CSF1) predicts breast cancer progression and mortality. Anticancer Res. 2015, 35, 865–874. [Google Scholar]
- Aharinejad, S.; Salama, M.; Paulus, P.; Zins, K.; Berger, A.; Singer, C.F. Elevated CSF1 serum concentration predicts poor overall survival in women with early breast cancer. Endocr. Relat. Cancer 2013, 20, 777–783. [Google Scholar] [CrossRef] [PubMed]
- Scholl, S.M.; Pallud, C.; Beuvon, F.; Hacene, K.; Stanley, E.R.; Rohrschneider, L.; Tang, R.; Pouillart, P.; Lidereau, R. Anti-colony-stimulating factor-1 antibody staining in primary breast adenocarcinomas correlates with marked inflammatory cell infiltrates and prognosis. J. Natl. Cancer Inst. 1994, 86, 120–126. [Google Scholar] [CrossRef]
- Kluger, H.M.; Dolled-Filhart, M.; Rodov, S.; Kacinski, B.M.; Camp, R.L.; Rimm, D.L. Macrophage colony-stimulating factor-1 receptor expression is associated with poor outcome in breast cancer by large cohort tissue microarray analysis. Clin. Cancer Res. 2004, 10, 173–177. [Google Scholar] [CrossRef]
- Bonapace, L.; Coissieux, M.-M.; Wyckoff, J.; Mertz, K.D.; Varga, Z.; Junt, T.; Bentires-Alj, M. Cessation of CCL2 inhibition accelerates breast cancer metastasis by promoting angiogenesis. Nature 2014, 515, 130–133. [Google Scholar] [CrossRef] [PubMed]
- Qian, B.-Z.; Li, J.; Zhang, H.; Kitamura, T.; Zhang, J.; Campion, L.R.; Kaiser, E.A.; Snyder, L.A.; Pollard, J.W. CCL2 recruits inflammatory monocytes to facilitate breast-tumour metastasis. Nature 2011, 475, 222–225. [Google Scholar] [CrossRef]
- Heiskala, M.; Leidenius, M.; Joensuu, K.; Heikkilä, P. High expression of CCL2 in tumor cells and abundant infiltration with CD14 positive macrophages predict early relapse in breast cancer. Virchows Arch. 2019, 474, 3–12. [Google Scholar] [CrossRef] [PubMed]
- Yao, M.; Yu, E.; Staggs, V.; Fan, F.; Cheng, N. Elevated expression of chemokine C-C ligand 2 in stroma is associated with recurrent basal-like breast cancers. Mod. Pathol. 2016, 29, 810–823. [Google Scholar] [CrossRef][Green Version]
- Labovsky, V.; Martinez, L.M.; Davies, K.M.; de Luján Calcagno, M.; García-Rivello, H.; Wernicke, A.; Feldman, L.; Matas, A.; Giorello, M.B.; Borzone, F.R.; et al. Prognostic significance of TRAIL-R3 and CCR-2 expression in tumor epithelial cells of patients with early breast cancer. BMC Cancer 2017, 17, 280. [Google Scholar] [CrossRef]
- Ueno, T.; Toi, M.; Saji, H.; Muta, M.; Bando, H.; Kuroi, K.; Koike, M.; Inadera, H.; Matsushima, K. Significance of macrophage chemoattractant protein-1 in macrophage recruitment, angiogenesis, and survival in human breast cancer. Clin. Cancer Res. 2000, 6, 3282–3289. [Google Scholar] [PubMed]
- Lavender, N.; Yang, J.; Chen, S.-C.; Sai, J.; Johnson, C.A.; Owens, P.; Ayers, G.D.; Richmond, A. The Yin/Yan of CCL2: a minor role in neutrophil anti-tumor activity in vitro but a major role on the outgrowth of metastatic breast cancer lesions in the lung in vivo. BMC Cancer 2017, 17, 88. [Google Scholar] [CrossRef]
- Slobodova, Z.; Ehrmann, J.; Krejci, V.; Zapletalova, J.; Melichar, B. Analysis of CD40 expression in breast cancer and its relation to clinicopathological characteristics. Neoplasma 2011, 58, 189–197. [Google Scholar] [CrossRef] [PubMed]
- Frazao, A.; Messaoudene, M.; Nunez, N.; Dulphy, N.; Roussin, F.; Sedlik, C.; Zitvogel, L.; Piaggio, E.; Toubert, A.; Caignard, A. CD16+NKG2Ahigh Natural Killer Cells Infiltrate Breast Cancer-Draining Lymph Nodes. Cancer Immunol. Res. 2019, 7, 208–218. [Google Scholar] [CrossRef]
- De Kruijf, E.M.; Sajet, A.; van Nes, J.G.H.; Putter, H.; Smit, V.T.H.B.M.; Eagle, R.A.; Jafferji, I.; Trowsdale, J.; Liefers, G.J.; van de Velde, C.J.H.; et al. NKG2D ligand tumor expression and association with clinical outcome in early breast cancer patients: an observational study. BMC Cancer 2012, 12, 24. [Google Scholar] [CrossRef] [PubMed]
- Asghar, K.; Loya, A.; Rana, I.A.; Tahseen, M.; Ishaq, M.; Farooq, A.; Bakar, M.A.; Masood, I. Indoleamine 2,3-dioxygenase expression and overall survival in patients diagnosed with breast cancer in Pakistan. Cancer Manag. Res. 2019, 11, 475–481. [Google Scholar] [CrossRef] [PubMed]
- Soliman, H.; Rawal, B.; Fulp, J.; Lee, J.-H.; Lopez, A.; Bui, M.M.; Khalil, F.; Antonia, S.; Yfantis, H.G.; Lee, D.H.; et al. Analysis of indoleamine 2-3 dioxygenase (IDO1) expression in breast cancer tissue by immunohistochemistry. Cancer Immunol. Immunother. 2013, 62, 829–837. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Sun, J.; Wang, S.E.; Li, H.; Cao, S.; Cong, Y.; Liu, J.; Ren, X. Upregulated expression of indoleamine 2, 3-dioxygenase in primary breast cancer correlates with increase of infiltrated regulatory T cells in situ and lymph node metastasis. Clin. Dev. Immunol. 2011, 2011, 469135. [Google Scholar] [CrossRef] [PubMed]
- Jacquemier, J.; Bertucci, F.; Finetti, P.; Esterni, B.; Charafe-Jauffret, E.; Thibult, M.-L.; Houvenaeghel, G.; Van den Eynde, B.; Birnbaum, D.; Olive, D.; et al. High expression of indoleamine 2,3-dioxygenase in the tumour is associated with medullary features and favourable outcome in basal-like breast carcinoma. Int. J. Cancer 2012, 130, 96–104. [Google Scholar] [CrossRef]
- Dill, E.A.; Dillon, P.M.; Bullock, T.N.; Mills, A.M. IDO expression in breast cancer: an assessment of 281 primary and metastatic cases with comparison to PD-L1. Mod. Pathol. 2018, 31, 1513–1522. [Google Scholar] [CrossRef] [PubMed]
- Wei, L.; Zhu, S.; Li, M.; Li, F.; Wei, F.; Liu, J.; Ren, X. High Indoleamine 2,3-Dioxygenase Is Correlated With Microvessel Density and Worse Prognosis in Breast Cancer. Front. Immunol. 2018, 9. [Google Scholar] [CrossRef]
- Li, F.; Zhao, Y.; Wei, L.; Li, S.; Liu, J. Tumor-infiltrating Treg, MDSC, and IDO expression associated with outcomes of neoadjuvant chemotherapy of breast cancer. Cancer Biol. Ther. 2018, 19, 695–705. [Google Scholar] [CrossRef] [PubMed]
- Ye, Q.; Wang, C.; Xian, J.; Zhang, M.; Cao, Y.; Cao, Y. Expression of programmed cell death protein 1 (PD-1) and indoleamine 2,3-dioxygenase (IDO) in the tumor microenvironment and in tumor-draining lymph nodes of breast cancer. Hum. Pathol. 2018, 75, 81–90. [Google Scholar] [CrossRef]
- Eftekhari, R.; Esmaeili, R.; Mirzaei, R.; Bidad, K.; de Lima, S.; Ajami, M.; Shirzad, H.; Hadjati, J.; Majidzadeh-A, K. Study of the tumor microenvironment during breast cancer progression. Cancer Cell Int. 2017, 17, 123. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Li, F.; Wei, L.; Li, S.; Liu, J. Indoleamine-2,3-dioxygenase and Interleukin-6 associated with tumor response to neoadjuvant chemotherapy in breast cancer. Oncotarget 2017, 8, 107844–107858. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Carvajal-Hausdorf, D.E.; Mani, N.; Velcheti, V.; Schalper, K.A.; Rimm, D.L. Objective measurement and clinical significance of IDO1 protein in hormone receptor-positive breast cancer. J. Immunother. Cancer 2017, 5, 81. [Google Scholar] [CrossRef]
- Chen, J.-Y.; Li, C.-F.; Kuo, C.-C.; Tsai, K.K.; Hou, M.-F.; Hung, W.-C. Cancer/stroma interplay via cyclooxygenase-2 and indoleamine 2,3-dioxygenase promotes breast cancer progression. Breast Cancer Res. 2014, 16, 410. [Google Scholar] [CrossRef] [PubMed]
- Isla Larrain, M.T.; Rabassa, M.E.; Lacunza, E.; Barbera, A.; Cretón, A.; Segal-Eiras, A.; Croce, M.V. IDO is highly expressed in breast cancer and breast cancer-derived circulating microvesicles and associated to aggressive types of tumors by in silico analysis. Tumour Biol. 2014, 35, 6511–6519. [Google Scholar] [CrossRef]
- Yu, J.; Du, W.; Yan, F.; Wang, Y.; Li, H.; Cao, S.; Yu, W.; Shen, C.; Liu, J.; Ren, X. Myeloid-derived suppressor cells suppress antitumor immune responses through IDO expression and correlate with lymph node metastasis in patients with breast cancer. J. Immunol. 2013, 190, 3783–3797. [Google Scholar] [CrossRef]
- Juhász, C.; Nahleh, Z.; Zitron, I.; Chugani, D.C.; Janabi, M.Z.; Bandyopadhyay, S.; Ali-Fehmi, R.; Mangner, T.J.; Chakraborty, P.K.; Mittal, S.; et al. Tryptophan metabolism in breast cancers: molecular imaging and immunohistochemistry studies. Nucl. Med. Biol. 2012, 39, 926–932. [Google Scholar] [CrossRef] [PubMed]
- Dowlati, A.; Rugo, H.S.; Harvey, R.D.; Kudchadkar, R.R.; Carvajal, R.D.; Manji, G.A.; Hamid, O.; Klempner, S.J.; Tang, S.; Yu, D.; et al. A phase I study of LY3022855, a colony-stimulating factor-1 receptor (CSF-1R) inhibitor, in patients (pts) with advanced solid tumors. JCO 2017, 35, 2523. [Google Scholar] [CrossRef]
- Gomez-Roca, C.A.; Cassier, P.A.; Italiano, A.; Cannarile, M.; Ries, C.; Brillouet, A.; Mueller, C.; Jegg, A.-M.; Meneses-Lorente, G.; Baehner, M.; et al. Phase I study of RG7155, a novel anti-CSF1R antibody, in patients with advanced/metastatic solid tumors. JCO 2015, 33, 3005. [Google Scholar] [CrossRef]
- Lakhani, N.J.; LoRusso, P.; Hafez, N.; Krishnamurthy, A.; O’Rourke, T.J.; Kamdar, M.K.; Fanning, P.; Zhao, Y.; Jin, F.; Wan, H.; et al. A phase 1 study of ALX148, a CD47 blocker, alone and in combination with established anticancer antibodies in patients with advanced malignancy and non-Hodgkin lymphoma. JCO 2018, 36, 3068. [Google Scholar] [CrossRef]
- Gangadhar, T.C.; Hamid, O.; Smith, D.C.; Bauer, T.M.; Wasser, J.S.; Luke, J.J.; Balmanoukian, A.S.; Kaufman, D.R.; Zhao, Y.; Maleski, J.; et al. Preliminary results from a Phase I/II study of epacadostat (incb024360) in combination with pembrolizumab in patients with selected advanced cancers. J. Immunother. Cancer 2015, 3, O7. [Google Scholar] [CrossRef]
- Fujiwara, Y.; Shitara, K.; Shimizu, T.; Yonemori, K.; Matsubara, N.; Ohno, I.; Kogawa, T.; Naito, Y.; Leopold, L.; Sasahara, K.; et al. Abstract A204: INCB024360 (Epacadostat) monotherapy and in combination with pembrolizumab in patients with advanced solid tumors: primary results from first-in-Japanese phase I study (KEYNOTE-434). Mol. Cancer Ther. 2018, 17, A204. [Google Scholar]
- Beatty, G.L.; O’Dwyer, P.J.; Clark, J.; Shi, J.G.; Bowman, K.J.; Scherle, P.A.; Newton, R.C.; Schaub, R.; Maleski, J.; Leopold, L.; et al. First-in-Human Phase I Study of the Oral Inhibitor of Indoleamine 2,3-Dioxygenase-1 Epacadostat (INCB024360) in Patients with Advanced Solid Malignancies. Clin. Cancer Res. 2017. [Google Scholar] [CrossRef] [PubMed]
- Brown, E.J.; Frazier, W.A. Integrin-associated protein (CD47) and its ligands. Trends Cell Biol. 2001, 11, 130–135. [Google Scholar] [CrossRef]
- Willingham, S.B.; Volkmer, J.-P.; Gentles, A.J.; Sahoo, D.; Dalerba, P.; Mitra, S.S.; Wang, J.; Contreras-Trujillo, H.; Martin, R.; Cohen, J.D.; et al. The CD47-signal regulatory protein alpha (SIRPa) interaction is a therapeutic target for human solid tumors. Proc. Natl. Acad. Sci. USA 2012, 109, 6662–6667. [Google Scholar] [CrossRef] [PubMed]
- Advani, R.; Flinn, I.; Popplewell, L.; Forero, A.; Bartlett, N.L.; Ghosh, N.; Kline, J.; Roschewski, M.; LaCasce, A.; Collins, G.P.; et al. CD47 Blockade by Hu5F9-G4 and Rituximab in Non-Hodgkin’s Lymphoma. N. Engl. J. Med. 2018, 379, 1711–1721. [Google Scholar] [CrossRef] [PubMed]
- Le Mercier, I.; Poujol, D.; Sanlaville, A.; Sisirak, V.; Gobert, M.; Durand, I.; Dubois, B.; Treilleux, I.; Marvel, J.; Vlach, J.; et al. Tumor promotion by intratumoral plasmacytoid dendritic cells is reversed by TLR7 ligand treatment. Cancer Res. 2013, 73, 4629–4640. [Google Scholar] [CrossRef] [PubMed]
- Yusuf, N. Toll-like receptor mediated regulation of breast cancer: a case of mixed blessings. Front. Immunol. 2014, 5, 224. [Google Scholar] [PubMed]
- Salazar, L.G.; Lu, H.; Reichow, J.L.; Childs, J.S.; Coveler, A.L.; Higgins, D.M.; Waisman, J.; Allison, K.H.; Dang, Y.; Disis, M.L. Topical Imiquimod Plus Nab-paclitaxel for Breast Cancer Cutaneous Metastases: A Phase 2 Clinical Trial. JAMA Oncol. 2017, 3, 969–973. [Google Scholar] [CrossRef] [PubMed]
- Adams, S.; Kozhaya, L.; Martiniuk, F.; Meng, T.-C.; Chiriboga, L.; Liebes, L.; Hochman, T.; Shuman, N.; Axelrod, D.; Speyer, J.; et al. Topical TLR7 agonist imiquimod can induce immune-mediated rejection of skin metastases in patients with breast cancer. Clin. Cancer Res. 2012, 18, 6748–6757. [Google Scholar] [CrossRef]
- Vonderheide, R.H.; Glennie, M.J. Agonistic CD40 antibodies and cancer therapy. Clin. Cancer Res. 2013, 19, 1035–1043. [Google Scholar] [CrossRef] [PubMed]
- Ma, H.S.; Poudel, B.; Torres, E.R.; Sidhom, J.-W.; Robinson, T.M.; Christmas, B.; Scott, B.; Cruz, K.; Woolman, S.; Wall, V.Z.; et al. A CD40 Agonist and PD-1 Antagonist Antibody Reprogram the Microenvironment of Nonimmunogenic Tumors to Allow T-cell-Mediated Anticancer Activity. Cancer Immunol. Res. 2019, 7, 428–442. [Google Scholar] [CrossRef] [PubMed]
- Winograd, R.; Byrne, K.T.; Evans, R.A.; Odorizzi, P.M.; Meyer, A.R.L.; Bajor, D.L.; Clendenin, C.; Stanger, B.Z.; Furth, E.E.; Wherry, E.J.; et al. Induction of T-cell Immunity Overcomes Complete Resistance to PD-1 and CTLA-4 Blockade and Improves Survival in Pancreatic Carcinoma. Cancer Immunol. Res. 2015, 3, 399–411. [Google Scholar] [CrossRef] [PubMed]
- Vonderheide, R.H.; Flaherty, K.T.; Khalil, M.; Stumacher, M.S.; Bajor, D.L.; Hutnick, N.A.; Sullivan, P.; Mahany, J.J.; Gallagher, M.; Kramer, A.; et al. Clinical activity and immune modulation in cancer patients treated with CP-870,893, a novel CD40 agonist monoclonal antibody. J. Clin. Oncol. 2007, 25, 876–883. [Google Scholar] [CrossRef] [PubMed]
- Guillerey, C.; Smyth, M.J. NK Cells and Cancer Immunoediting. Curr. Top. Microbiol. Immunol. 2016, 395, 115–145. [Google Scholar] [PubMed]
- Muntasell, A.; Ochoa, M.C.; Cordeiro, L.; Berraondo, P.; López-Díaz de Cerio, A.; Cabo, M.; López-Botet, M.; Melero, I. Targeting NK-cell checkpoints for cancer immunotherapy. Curr. Opin. Immunol. 2017, 45, 73–81. [Google Scholar] [CrossRef]
- Jobim, M.R.; Jobim, M.; Salim, P.H.; Portela, P.; Jobim, L.F.; Leistner-Segal, S.; Bittelbrunn, A.C.; Menke, C.H.; Biazús, J.V.; Roesler, R.; et al. Analysis of KIR gene frequencies and HLA class I genotypes in breast cancer and control group. Hum. Immunol. 2013, 74, 1130–1133. [Google Scholar] [CrossRef] [PubMed]
- Beksac, K.; Beksac, M.; Dalva, K.; Karaagaoglu, E.; Tirnaksiz, M.B. Impact of “Killer Immunoglobulin-Like Receptor /Ligand” Genotypes on Outcome following Surgery among Patients with Colorectal Cancer: Activating KIRs Are Associated with Long-Term Disease Free Survival. PLoS ONE 2015, 10, e0132526. [Google Scholar] [CrossRef] [PubMed]
- Braud, V.M.; Allan, D.S.; O’Callaghan, C.A.; Söderström, K.; D’Andrea, A.; Ogg, G.S.; Lazetic, S.; Young, N.T.; Bell, J.I.; Phillips, J.H.; et al. HLA-E binds to natural killer cell receptors CD94/NKG2A, B and C. Nature 1998, 391, 795–799. [Google Scholar] [CrossRef]
- André, P.; Denis, C.; Soulas, C.; Bourbon-Caillet, C.; Lopez, J.; Arnoux, T.; Bléry, M.; Bonnafous, C.; Gauthier, L.; Morel, A.; et al. Anti-NKG2A mAb Is a Checkpoint Inhibitor that Promotes Anti-tumor Immunity by Unleashing Both T and NK Cells. Cell 2018, 175, 1731–1743. [Google Scholar] [CrossRef]
- Van Montfoort, N.; Borst, L.; Korrer, M.J.; Sluijter, M.; Marijt, K.A.; Santegoets, S.J.; van Ham, V.J.; Ehsan, I.; Charoentong, P.; André, P.; et al. NKG2A Blockade Potentiates CD8 T Cell Immunity Induced by Cancer Vaccines. Cell 2018, 175, 1744–1755. [Google Scholar] [CrossRef]
- Raulet, D.H. Roles of the NKG2D immunoreceptor and its ligands. Nat. Rev. Immunol. 2003, 3, 781–790. [Google Scholar] [CrossRef]
- Moretta, A.; Bottino, C.; Vitale, M.; Pende, D.; Cantoni, C.; Mingari, M.C.; Biassoni, R.; Moretta, L. Activating receptors and coreceptors involved in human natural killer cell-mediated cytolysis. Annu. Rev. Immunol. 2001, 19, 197–223. [Google Scholar] [CrossRef] [PubMed]
- Ascierto, M.L.; Idowu, M.O.; Zhao, Y.; Khalak, H.; Payne, K.K.; Wang, X.-Y.; Dumur, C.I.; Bedognetti, D.; Tomei, S.; Ascierto, P.A.; et al. Molecular signatures mostly associated with NK cells are predictive of relapse free survival in breast cancer patients. J. Transl. Med. 2013, 11, 145. [Google Scholar] [CrossRef] [PubMed]
- Abouelghar, A.; Hasnah, R.; Taouk, G.; Saad, M.; Karam, M. Prognostic values of the mRNA expression of natural killer receptor ligands and their association with clinicopathological features in breast cancer patients. Oncotarget 2018, 9, 27171–27196. [Google Scholar] [CrossRef][Green Version]
- Brochez, L.; Chevolet, I.; Kruse, V. The rationale of indoleamine 2,3-dioxygenase inhibition for cancer therapy. Eur. J. Cancer 2017, 76, 167–182. [Google Scholar] [CrossRef] [PubMed]
- Théate, I.; van Baren, N.; Pilotte, L.; Moulin, P.; Larrieu, P.; Renauld, J.-C.; Hervé, C.; Gutierrez-Roelens, I.; Marbaix, E.; Sempoux, C.; et al. Extensive profiling of the expression of the indoleamine 2,3-dioxygenase 1 protein in normal and tumoral human tissues. Cancer Immunol. Res. 2015, 3, 161–172. [Google Scholar] [CrossRef]
- Mellor, A.L.; Keskin, D.B.; Johnson, T.; Chandler, P.; Munn, D.H. Cells expressing indoleamine 2,3-dioxygenase inhibit T cell responses. J. Immunol. 2002, 168, 3771–3776. [Google Scholar] [CrossRef]
- Jung, K.H.; LoRusso, P.M.; Burris, H.A.; Gordon, M.S.; Bang, Y.-J.; Hellmann, M.D.; Cervantes, A.; Ochoa de Olza, M.; Marabelle, A.; Hodi, F.S.; et al. Phase I Study of the Indoleamine 2,3-Dioxygenase 1 (IDO1) Inhibitor Navoximod (GDC-0919) Administered with PD-L1 Inhibitor (Atezolizumab) in Advanced Solid Tumors. Clin. Cancer Res. 2019. [Google Scholar] [CrossRef]
- Corzo, C.A.; Cotter, M.J.; Cheng, P.; Cheng, F.; Kusmartsev, S.; Sotomayor, E.; Padhya, T.; McCaffrey, T.V.; McCaffrey, J.C.; Gabrilovich, D.I. Mechanism regulating reactive oxygen species in tumor-induced myeloid-derived suppressor cells. J. Immunol. 2009, 182, 5693–5701. [Google Scholar] [CrossRef]
- Raber, P.L.; Thevenot, P.; Sierra, R.; Wyczechowska, D.; Halle, D.; Ramirez, M.E.; Ochoa, A.C.; Fletcher, M.; Velasco, C.; Wilk, A.; et al. Subpopulations of myeloid-derived suppressor cells impair T cell responses through independent nitric oxide-related pathways. Int. J. Cancer 2014, 134, 2853–2864. [Google Scholar] [CrossRef] [PubMed]
- Ku, A.W.; Muhitch, J.B.; Powers, C.A.; Diehl, M.; Kim, M.; Fisher, D.T.; Sharda, A.P.; Clements, V.K.; O’Loughlin, K.; Minderman, H.; et al. Tumor-induced MDSC act via remote control to inhibit L-selectin-dependent adaptive immunity in lymph nodes. Elife 2016, 5. [Google Scholar] [CrossRef] [PubMed]
- Huang, B.; Pan, P.-Y.; Li, Q.; Sato, A.I.; Levy, D.E.; Bromberg, J.; Divino, C.M.; Chen, S.-H. Gr-1+CD115+ immature myeloid suppressor cells mediate the development of tumor-induced T regulatory cells and T-cell anergy in tumor-bearing host. Cancer Res. 2006, 66, 1123–1131. [Google Scholar] [CrossRef]
- Beury, D.W.; Parker, K.H.; Nyandjo, M.; Sinha, P.; Carter, K.A.; Ostrand-Rosenberg, S. Cross-talk among myeloid-derived suppressor cells, macrophages, and tumor cells impacts the inflammatory milieu of solid tumors. J. Leukoc. Biol. 2014, 96, 1109–1118. [Google Scholar] [CrossRef]
- Rodriguez, P.C.; Quiceno, D.G.; Zabaleta, J.; Ortiz, B.; Zea, A.H.; Piazuelo, M.B.; Delgado, A.; Correa, P.; Brayer, J.; Sotomayor, E.M.; et al. Arginase I production in the tumor microenvironment by mature myeloid cells inhibits T-cell receptor expression and antigen-specific T-cell responses. Cancer Res. 2004, 64, 5839–5849. [Google Scholar] [CrossRef]
- Srivastava, M.K.; Sinha, P.; Clements, V.K.; Rodriguez, P.; Ostrand-Rosenberg, S. Myeloid-derived suppressor cells inhibit T-cell activation by depleting cystine and cysteine. Cancer Res. 2010, 70, 68–77. [Google Scholar] [CrossRef]
- Lu, C.; Redd, P.S.; Lee, J.R.; Savage, N.; Liu, K. The expression profiles and regulation of PD-L1 in tumor-induced myeloid-derived suppressor cells. Oncoimmunology 2016, 5, e1247135. [Google Scholar] [CrossRef]
- Li, J.; Wang, L.; Chen, X.; Li, L.; Li, Y.; Ping, Y.; Huang, L.; Yue, D.; Zhang, Z.; Wang, F.; et al. CD39/CD73 upregulation on myeloid-derived suppressor cells via TGF-β-mTOR-HIF-1 signaling in patients with non-small cell lung cancer. Oncoimmunology 2017, 6, e1320011. [Google Scholar] [CrossRef]
- Toh, B.; Wang, X.; Keeble, J.; Sim, W.J.; Khoo, K.; Wong, W.-C.; Kato, M.; Prevost-Blondel, A.; Thiery, J.-P.; Abastado, J.-P. Mesenchymal transition and dissemination of cancer cells is driven by myeloid-derived suppressor cells infiltrating the primary tumor. PLoS Biol. 2011, 9, e1001162. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Huang, J.; Ren, X.; Gorska, A.E.; Chytil, A.; Aakre, M.; Carbone, D.P.; Matrisian, L.M.; Richmond, A.; Lin, P.C.; et al. Abrogation of TGF beta signaling in mammary carcinomas recruits Gr-1+CD11b+ myeloid cells that promote metastasis. Cancer Cell 2008, 13, 23–35. [Google Scholar] [CrossRef] [PubMed]
- Cui, T.X.; Kryczek, I.; Zhao, L.; Zhao, E.; Kuick, R.; Roh, M.H.; Vatan, L.; Szeliga, W.; Mao, Y.; Thomas, D.G.; et al. Myeloid-derived suppressor cells enhance stemness of cancer cells by inducing microRNA101 and suppressing the corepressor CtBP2. Immunity 2013, 39, 611–621. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; DeBusk, L.M.; Fukuda, K.; Fingleton, B.; Green-Jarvis, B.; Shyr, Y.; Matrisian, L.M.; Carbone, D.P.; Lin, P.C. Expansion of myeloid immune suppressor Gr+CD11b+ cells in tumor-bearing host directly promotes tumor angiogenesis. Cancer Cell 2004, 6, 409–421. [Google Scholar] [CrossRef] [PubMed]
- Yan, H.H.; Pickup, M.; Pang, Y.; Gorska, A.E.; Li, Z.; Chytil, A.; Geng, Y.; Gray, J.W.; Moses, H.L.; Yang, L. Gr-1+CD11b+ myeloid cells tip the balance of immune protection to tumor promotion in the premetastatic lung. Cancer Res. 2010, 70, 6139–6149. [Google Scholar] [CrossRef] [PubMed]
- Jin, G.; Zhang, Y.; Chang, X.; Zhang, Y.; Xu, J.; Wei, M.; Zeng, X. Increased Percentage of mo-MDSCs in Human Peripheral Blood May Be a Potential Indicator in the Diagnosis of Breast Cancer. Oncol. Res. Treat. 2017, 40, 603–608. [Google Scholar] [CrossRef]
- Diaz-Montero, C.M.; Salem, M.L.; Nishimura, M.I.; Garrett-Mayer, E.; Cole, D.J.; Montero, A.J. Increased circulating myeloid-derived suppressor cells correlate with clinical cancer stage, metastatic tumor burden, and doxorubicin-cyclophosphamide chemotherapy. Cancer Immunol. Immunother. 2009, 58, 49–59. [Google Scholar] [CrossRef]
- Toor, S.M.; Syed Khaja, A.S.; El Salhat, H.; Faour, I.; Kanbar, J.; Quadri, A.A.; Albashir, M.; Elkord, E. Myeloid cells in circulation and tumor microenvironment of breast cancer patients. Cancer Immunol. Immunother. 2017, 66, 753–764. [Google Scholar] [CrossRef]
- Kumar, S.; Wilkes, D.W.; Samuel, N.; Blanco, M.A.; Nayak, A.; Alicea-Torres, K.; Gluck, C.; Sinha, S.; Gabrilovich, D.; Chakrabarti, R. ΔNp63-driven recruitment of myeloid-derived suppressor cells promotes metastasis in triple-negative breast cancer. J. Clin. Invest. 2018, 128, 5095–5109. [Google Scholar] [CrossRef]
- Peng, D.; Tanikawa, T.; Li, W.; Zhao, L.; Vatan, L.; Szeliga, W.; Wan, S.; Wei, S.; Wang, Y.; Liu, Y.; et al. Myeloid-derived suppressor cells endow stem-like qualities to breast cancer cells through IL-6/STAT3 and NO/NOTCH cross-talk signaling. Cancer Res. 2016, 76, 3156–3165. [Google Scholar] [CrossRef]
- Gonda, K.; Shibata, M.; Ohtake, T.; Matsumoto, Y.; Tachibana, K.; Abe, N.; Ohto, H.; Sakurai, K.; Takenoshita, S. Myeloid-derived suppressor cells are increased and correlated with type 2 immune responses, malnutrition, inflammation, and poor prognosis in patients with breast cancer. Oncol. Lett. 2017, 14, 1766–1774. [Google Scholar] [CrossRef]
- Wesolowski, R.; Duggan, M.C.; Stiff, A.; Markowitz, J.; Trikha, P.; Levine, K.M.; Schoenfield, L.; Abdel-Rasoul, M.; Layman, R.; Ramaswamy, B.; et al. Circulating myeloid derived suppressor cells increase in patients undergoing neo-adjuvant chemotherapy for breast cancer. Cancer Immunol. Immunother. 2017, 66, 1437–1447. [Google Scholar] [CrossRef] [PubMed]
- Montero, A.J.; Diaz-Montero, C.M.; Deutsch, Y.E.; Hurley, J.; Koniaris, L.G.; Rumboldt, T.; Yasir, S.; Jorda, M.; Garret-Mayer, E.; Avisar, E.; et al. Phase 2 study of neoadjuvant treatment with NOV-002 in combination with doxorubicin and cyclophosphamide followed by docetaxel in patients with HER-2 negative clinical stage II-IIIc breast cancer. Breast Cancer Res. Treat. 2012, 132, 215–223. [Google Scholar] [CrossRef]
- Su, Z.; Ni, P.; She, P.; Liu, Y.; Richard, S.A.; Xu, W.; Zhu, H.; Wang, J. Bio-HMGB1 from breast cancer contributes to M-MDSC differentiation from bone marrow progenitor cells and facilitates conversion of monocytes into MDSC-like cells. Cancer Immunol. Immunother. 2017, 66, 391–401. [Google Scholar] [CrossRef]
- Hong, H.-J.; Lim, H.X.; Song, J.H.; Lee, A.; Kim, E.; Cho, D.; Cohen, E.P.; Kim, T.S. Aminoacyl-tRNA synthetase-interacting multifunctional protein 1 suppresses tumor growth in breast cancer-bearing mice by negatively regulating myeloid-derived suppressor cell functions. Cancer Immunol. Immunother. 2016, 65, 61–72. [Google Scholar] [CrossRef]
- Qian, X.; Zhang, Q.; Shao, N.; Shan, Z.; Cheang, T.; Zhang, Z.; Su, Q.; Wang, S.; Lin, Y. Respiratory hyperoxia reverses immunosuppression by regulating myeloid-derived suppressor cells and PD-L1 expression in a triple-negative breast cancer mouse model. Am. J. Cancer Res. 2019, 9, 529–545. [Google Scholar]
- Yin, T.; Zhao, Z.-B.; Guo, J.; Wang, T.; Yang, J.-B.; Wang, C.; Long, J.; Ma, S.; Huang, Q.; Zhang, K.; et al. Aurora-A inhibition eliminates myeloid cell-mediated immunosuppression and enhances the efficacy of anti-PD-L1 therapy in breast cancer. Cancer Res. 2019. [Google Scholar] [CrossRef]
- Ugel, S.; Delpozzo, F.; Desantis, G.; Papalini, F.; Simonato, F.; Sonda, N.; Zilio, S.; Bronte, V. Therapeutic targeting of myeloid-derived suppressor cells. Curr. Opin. Pharmacol. 2009, 9, 470–481. [Google Scholar] [CrossRef] [PubMed]
- Shen, X.; Zhao, B. Efficacy of PD-1 or PD-L1 inhibitors and PD-L1 expression status in cancer: meta-analysis. BMJ 2018, 362, k3529. [Google Scholar] [CrossRef] [PubMed]
- Wolchok, J.D.; Chiarion-Sileni, V.; Gonzalez, R.; Rutkowski, P.; Grob, J.-J.; Cowey, C.L.; Lao, C.D.; Wagstaff, J.; Schadendorf, D.; Ferrucci, P.F.; et al. Overall Survival with Combined Nivolumab and Ipilimumab in Advanced Melanoma. N. Engl. J. Med. 2017, 377, 1345–1356. [Google Scholar] [CrossRef]
- Wei, S.C.; Duffy, C.R.; Allison, J.P. Fundamental Mechanisms of Immune Checkpoint Blockade Therapy. Cancer Discov. 2018, 8, 1069–1086. [Google Scholar] [CrossRef]
- Epacadostat Combined with Pembrolizumab in Patients with Unresectable or Metastatic Melanoma - The ASCO Post. Available online: http://www.ascopost.com/News/58726 (accessed on 17 March 2019).
- Freeman-Keller, M.; Kim, Y.; Cronin, H.; Richards, A.; Gibney, G.; Weber, J.S. Nivolumab in Resected and Unresectable Metastatic Melanoma: Characteristics of Immune-Related Adverse Events and Association with Outcomes. Clin. Cancer Res. 2016, 22, 886–894. [Google Scholar] [CrossRef]
- Teraoka, S.; Fujimoto, D.; Morimoto, T.; Kawachi, H.; Ito, M.; Sato, Y.; Nagata, K.; Nakagawa, A.; Otsuka, K.; Uehara, K.; et al. Early Immune-Related Adverse Events and Association with Outcome in Advanced Non-Small Cell Lung Cancer Patients Treated with Nivolumab: A Prospective Cohort Study. J. Thorac. Oncol. 2017, 12, 1798–1805. [Google Scholar] [CrossRef] [PubMed]
- Oh, A.; Tran, D.M.; McDowell, L.C.; Keyvani, D.; Barcelon, J.A.; Merino, O.; Wilson, L. Cost-Effectiveness of Nivolumab-Ipilimumab Combination Therapy Compared with Monotherapy for First-Line Treatment of Metastatic Melanoma in the United States. J Manag Care Spec. Pharm. 2017, 23, 653–664. [Google Scholar] [CrossRef] [PubMed]
- Hellmann, M.D.; Ciuleanu, T.-E.; Pluzanski, A.; Lee, J.S.; Otterson, G.A.; Audigier-Valette, C.; Minenza, E.; Linardou, H.; Burgers, S.; Salman, P.; et al. Nivolumab plus Ipilimumab in Lung Cancer with a High Tumor Mutational Burden. N. Engl. J. Med. 2018, 378, 2093–2104. [Google Scholar] [CrossRef] [PubMed]
- Motzer, R.J.; Tannir, N.M.; McDermott, D.F.; Arén Frontera, O.; Melichar, B.; Choueiri, T.K.; Plimack, E.R.; Barthélémy, P.; Porta, C.; George, S.; et al. Nivolumab plus Ipilimumab versus Sunitinib in Advanced Renal-Cell Carcinoma. N. Engl. J. Med. 2018, 378, 1277–1290. [Google Scholar] [CrossRef] [PubMed]
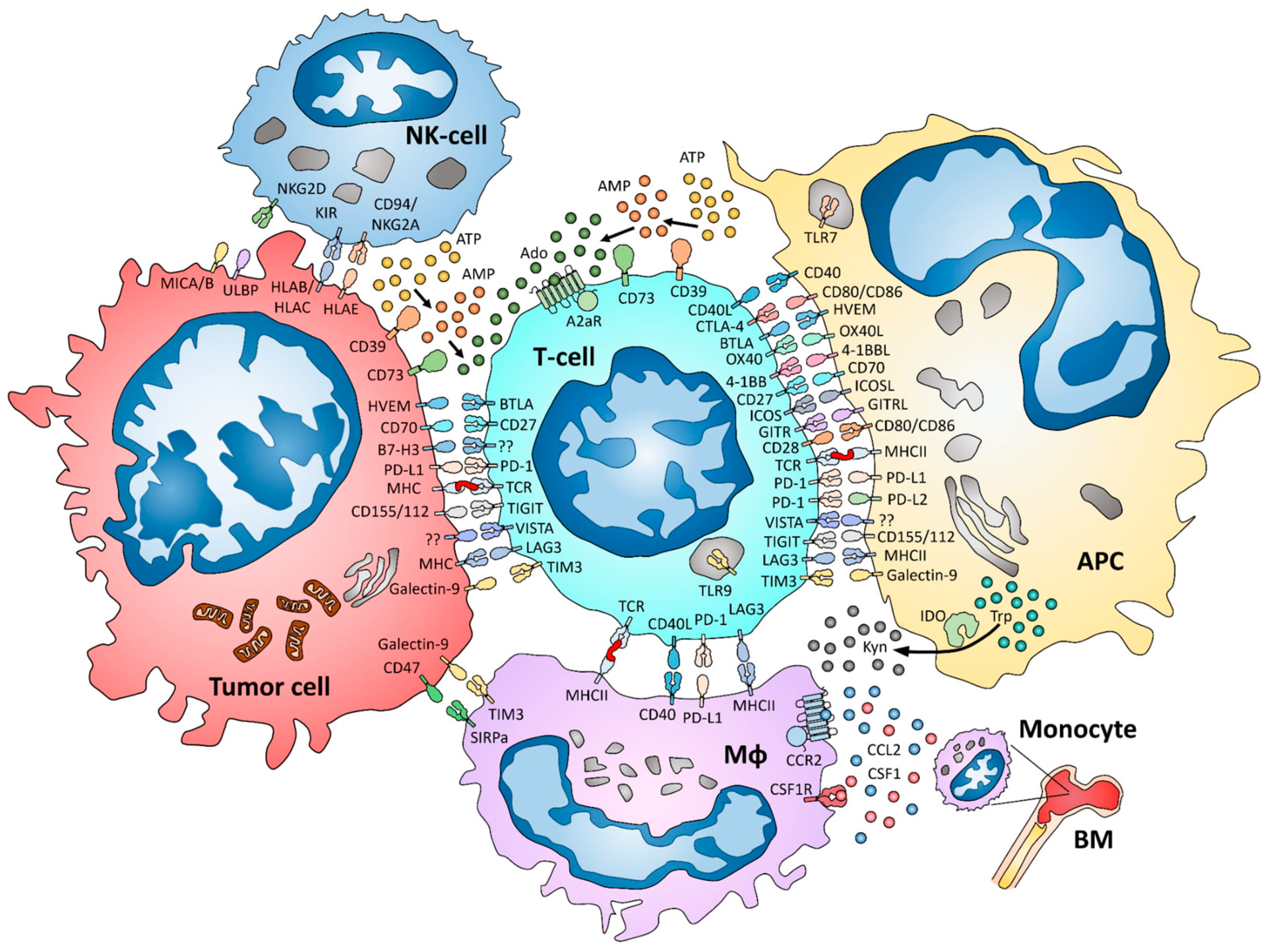
| Marker | BC Subtype | Number of Patients | Method | Positive/ Overexpressing Cases | Prognostic/Predictive Value | Comments | Reference |
|---|---|---|---|---|---|---|---|
| LAG-3 | All | 8 | RT-PCR | LAG-3 expression: 8/8 (100%) | NA | LAG-3 overexpression in BC compared to adjacent healthy tissue | [23] |
| All | 148 pre-NACT 114 post-NACT | IHC | LAG-3 positivity: Pre-NACT: 33/148 (22.3%) Post-NACT: 38/114 (33.3%) | LAG-3 expression: pedictive for pCR in UA but not MA | Positive case cut-off: expression ≥ 5% | [28] | |
| TNBC | 259 (training set) 104 (validation set) | IHC | LAG-3 positivity: 65/363 (18%) | LAG-3 positivity: trend to better RFS and OS in UA | Positive case cut-off: expression ≥ 5% | [25] | |
| All | 330 (training set) 3992 (validation set) | IHC | LAG-3 positivity: 327/2921 (11%) | LAG-3 positivity: better BCSS and RFS in MA but not when considering CD8, PD-1 and PD-L1 | Positive case cut-off: ≥ 1 TILs per TMA core 2921 evaluable in validation set | [26] | |
| TIM-3 | All | 150 | IHC | BC cases: TIM-3 + tumor cells 147/150 (98%) TIM-3+CD8+ T cells 135/150 (90%)Healthy controls: TIM-3+ epithelial cells 13/100 (13%) TIM-3+CD8+ T-cells 23/100 (23%) | NA | [29] | |
| All | 20 | FC | NA | NA | Peripheral blood: overexpression of TIM-3 in CD4+CXCR5+ICOS+ T cells compared to healthy controls TILs: overexpression of TIM-3 in CD4+CXCR5+ICOS+ T cells compared to peripheral blood of same patients | [30] | |
| All | 8 | RT-PCR | TIM-3 expression: 8/8 (100%) | NA | Overexpression in BC compared to healthy adjacent tissue | [23] | |
| All | 3169 | Gene expression dataset | No overexpression | NA | Use of gene expression dataset Genevestigator v3 | [31] | |
| All | 3992 (3148 evaluable) | IHC | TIM-3+ iTILs: 332/3148 (11%) TIM-3+ sTILs: 630/3148 | TIM-3+ iTILs: better BCSS TIM-3+ sTILs: statistically not significant better BCSS | TIM-3 iTILs cut-off: expression ≥ 1 iTIL TIM-3 sTILs cut-off: expression ≥ 2 sTILs TIM-3+ iTILs correlated to basal-like subtype | [32] | |
| VISTA | NA | NA | NA | NA | NA | NA | NA |
| TIGIT | All | 3169 | Gene expression dataset | TIGIT overexpression in 72% | NA | Use of gene expression dataset Genevestigator v3 | [31] |
| TNBC | 47 | Gene expression dataset | NA | TIGIT overexpression: better RFS and OS | Use of gene expression dataset from GEO datasets (GDS2250 and GSE3744) | [33] | |
| All | 8 | RT-PCR | TIGIT expression: 8/8 (100%) | NA | No overexpression of TIGIT in BC compared to adjacent healthy tissue | [23] | |
| GITR | All | 33 | FC | NA | NA | PT Tregs: 80.5% expression of GITR Circulating Tregs: 28.9% expression of GITR | [34] |
| All | 39 | FC | NA | NA | PT CD4+h T cells: higher GITR expression than healthy control CD4+ T cells | [35] | |
| All | 3169 | Gene expression dataset | GITR overexpression in 42% | NA | Use of gene expression dataset Genevestigator v3 | [31] | |
| Not specified | 3 | FC | NA | NA | [36] | ||
| Not specified | 17 | FC | NA | NA | More T regs expressing GITR in BC patients than healthy donors (n = 10) | [37] | |
| B7-H3 | All | 221 | IHC | B7-H3 high expression: Healthy controls: 14/85 (16.48%) BC: 178/221 (80.55%) | NA | [38] | |
| All | 82 | RT-PCR | B7-H3 overexpression: 32/82 (39%) | NA | [39] | ||
| All | 117 | IHC | B7-H3 positivity: 106/117 (90.6%) | NA | Positive case cut-off: expression > 10% | [40] | |
| All | 90 | IHC | B7-H3 high: 83/90 (92%) | B7-H3 high: worse RFS but no association with OS | [41] | ||
| All | 74 | IHC | B7-H3 IHC positivity: BC 42/74 (56.8%) healthy controls 32/74 (43.2%) | B7-H3 positivity: worse OS | [42] | ||
| All | 97 | IHC | NA | NA | B7-H3 expression significantly higher in BC (n=97) compared to normal tissue (n = 53), benign, and precursor lesion (n = 182) | [43] | |
| All | 208 | IHC | B7-H3 positivity: BC: 154/208 (74%) Healthy controls: 3/7 (43%) | NA | [44] | ||
| All | 101 | IHC | B7-H3 positivity: BC: 88/101 (88%) Healthy controls: 6/47 (12.8%) | NA | [45] | ||
| ICOS | All | 120 | IHC FC | NA | ICOS positivity: UA: worse PFS and OS MA: not significant | Positive case cut-off: expression ≥1.7 positive cells Tumoral Treg ICOS+: 69.9% BC circulating Treg ICOS+: 16.6% Healthy circulating Treg ICOS+: 21.3% | [46] |
| 4-1BB | All | 3169 | Gene expression dataset | 4-1BB overexpression in 42% | NA | Use of gene expression dataset Genevestigator v3 | [31] |
| All | 286 | Gene expression dataset | NA | 4-1BB expression: better DFMS | [47] | ||
| Not specified | 4 | IHC | 4-1BB positivity: 2/4 (50%) | NA | Positive case cut-off: expression > 10% | [48] | |
| CD70 | All | 204 | IHC | CD70 positivity: 5/204 (2.45%) | NA | [49] | |
| All | 139 (110/139 with metastasis) 233 (stage I – III) | IHC | CD70 expression: 81/139 (58.3%) | CD70 expression: worse lung MFS | [50] | ||
| All | 16 (pre and post-NACT) | RT-PCR | NA | CD70 overexpression after NACT: better PFS | [51] | ||
| OX40 and OX40L | All | 107 9 DICS | IHC | Positivity in PT: OX40 91/107 (85%) OX40L 89/107 (83.2%) Positivity in DCIS: OX40 6/9 (66.7%) OX40L 7/9 (77.8%) | NA | Positive case cut-off: expression on > 10% tumor cells OX40 associated with advanced stage | [52] |
| Not specified | 19 | IHC | OX40 positivity: 10/19 (52.6%) | NA | Positive case cut-off: expression on > 10% cells | [53] | |
| Not specified | Not specified | IHC FC | OX40+CD4+ TILs in 43% of the BC cases | NA | No OX40 expression on circulation CD4 T cells | [54] | |
| Not specified | 45 | IHC | OX40 positivity: 7/45 (15.55%) | OX40 expressed on TILs OX40 expression also found on positive LN | [55] | ||
| Not specified | 44 | IHC | OX40 positivity: 7/18 (30%) of theCD4+ cases | NA | [56] | ||
| BTLA | All | 3080 | Gene-expression dataset | BTLA overexpressed in TNBC compared to non-TNBC | BTLA overexpression in TNBC: better OS and DFS | Use of gene expression profiles of breast invasive carcinoma from TCGA and METABRIC | [57] |
| All | 660 | IHC FC | BTLA positivity: 15/660 (2.3%) | NA | Positive case cut-off: ≥ 1 BTLA+ TIL All BTLA+ TILs also expressed PD-1 According to FC, CD4 and CD8+TILs don’t express BTLA | [58] | |
| TLR9 | TNBC (Afro-American population) | 51 | IHC | TLR9 ”low” expression: 27/51 (52.9%) TLR9 ”high” expression: 22/51 (43.1%) | TLR9 high: no association with recurrence or BCSS | Variants of TLR9 gene associated with protection from breast cancer | [59] |
| All and TNBC | 84 of all subtypes 80 TNBC 350 of all subtypes | RT- PCRIHC | mRNA expression in cohort of 84 cases of all subtypes: overexpression in TNBC IHC expression in sub-group analyses of 38/84 cases of all subtypes: overexpression in 8/38 (21%) and 5/13 (38.5%) TNBC IHC expression in 80 TNBC cases: 32/80 (40%) mRNA expression in 350 cases of all subtypes: overexpression in 50/350 (14.3%) and 19/64 (29.7%) TNBC | High mRNA expression: trend to better MFS High protein expression in 80 TNBC: better MFS | TLR9 also expressed in pre-invasive lesions | [60] | |
| All | 196 | IHC | TLR9 high expression in TNBC: 51/99 (51.5%) | TLR9 high expression:
| [61] | ||
| All | 12 | RT-PCR | TLR9 expression: 12/12 (100%) | NA | [62] | ||
| All | 124 | IHC | TLR9 positivity: 78/124 (63%) | TLR9 positivity:
| Positive case cut-off: expression > 10% cells Expression significantly higher in tumors with positive axillary LN metastasis, ER- and advanced stage | [63] | |
| All | 74 | IHC | TLR9 expression: By tumor cells: 73/74 (98.6%) By fibroblasts 42/74 (58%) | TLR9 positive expression by fibroblasts: better DMFS | [64] | ||
| All | 124 116 post-menopausal | RT-PCR IHC | TLR9 mRNA: overexpression in ER- TLR9 IHC expression in 116 post-menopausal: 103/116 (88.8%) | IHC expression higher in ER and PR- | [65] | ||
| All | 141 | IHC | TLR9 positivity: 136/141 (98%) | TLR9 positivity: worse DMFS | Higher expression in ER- and high grade tumors | [66] | |
| A2aR | NA | NA | NA | NA | NA | NA | NA |
| CD73 | All | 80 | IHC | NA | CD73 expresion in ER+ cases: no prognostic value CD73 expression in ER- cases: worse OS | CD73 expression associated with EGFR expression | [67] |
| All | 136 | IHC | CD73 positivity: 101/136 (74%) | CD73 positivity: UA: better DFS and OS MA: better DFS, trend to better OS | [68] | ||
| All (Her2 status NA) | 102 | IHC | CD73 positivity: 9/102 (9%) | NA | Positive case cut-off: any expression by tumor cells | [69] | |
| Not specified | 74 | IHC | CD73 positivity: 60/74 (81%) | NA | Positive case cut-off: expression >5% cells | [70] | |
| TNBC | 122 | IF | NA | Tumor cells CD73 expression:
| [71] | ||
| All | 119 | IHC | CD73 positivity: 100/119 (84%) | NA | [72] | ||
| All | 202 | Gene expression dataset | NA | Gene-expression database of 1128 cases of all subtypes: worse DFS Gene-expression of 417 Her2+ cases: worse DFS Gene-expression of 784 ER+ and 211 TNBC cases: statistically NS trend to worse DFS METABRIC cohort of 1981 cases of all subtypes: worse DSS | [73] | ||
| All and TNBC | 6209 all subtypes 59 TNBC | Gene expression dataset | NA | 6209 cases of all subtypes: worse OS for TNBC, no prognostic value for ER+ and Her2+ cases 59 TNBC: worse response to NACT | [74] | ||
| CD39 | Not specified | 33 | FC | PT CD39+CD8+ TILs mean frequency: 18.5% +/− 4.3% Circulating CD8+ T cells: no CD39 expression | NA | [75] | |
| All (Her2 NA) | 11 | FC | NA | NA | CD39+CD4+ TILs 28.7+/−5.8% vs 8.2+/−5.9% in normal adjacent tissue CD39+CD8+ TILs 9+/−3.5% vs 0.4+/−0.3% in normal adjacent tissue | [76] | |
| All | 50 | FC IF RT-PCR | NA | NA | CD39 +Th17 TILs 93.6% CD39 + TILs Tregs 50.9% CD39 overexpressed among IL-17Hi tumors | [77] | |
| All | 3169 | Gene expression dataset | No CD39 overexpression | NA | Use of gene expression dataset Genevestigator v3 | [31] | |
| Not specified | 10 | Gene expression dataset | CD39 overexpressed in BC compared to healthy tissue | NA | Micro-array dataset from Turashvili et al. (BMC Cancer. 2007 Mar 27;7:55.) | [78] |
| Target | Drug | Other Agent(s) | Phase | Disease | Line | NCT Identifier | Trial Status |
|---|---|---|---|---|---|---|---|
| LAG-3 | IMP 321 (Eftilagimod) | + Paclitaxel | I/II | Advanced BC | 1st line | NCT00349934 | Completed, published results [27] |
| + Paclitaxel | Iib | Hormone positive advanced BC | 1st line | NCT02614833 | Recruiting, safety results published [79] | ||
| + Paclitaxel | I | Advanced BC (chinese population) | 1st line | NCT03600090 | Not yet recruiting | ||
| + standard therapy | I | Advanced solid tumors | Any line | NCT03252938 | Recruiting | ||
| MK-4280 | +/− Pembrolizumab (anti-PD1) | I | Advanced solid tumors | No standard therapy available | NCT02720068 | Recruiting | |
| BMS-986016 (Relatlimab) | +/− Nivolumab (anti-PD1) | I | Advanced solid tumors | No standard therapy available | NCT02966548 | Recruiting | |
| + Nivolumab (anti-PD1) and BMS-986205 (IDO1 inhibitor) Or + Nivolumab (anti-PD1) and Ipilimumab (anti-CTLA4) | I/II | Advanced solid tumors | Any line | NCT03459222 | Recruiting | ||
| REGN3767 | +/− REGN2810 (anti-PD1) | I | Advanced solid tumors | No standard therapy available | NCT03005782 | Recruiting | |
| LAG525 (IMP701) | +/− PDR001 (anti-PD1) | I/II | Advanced solid tumors including TNBC | ≥ 1 line | NCT02460224 | Active, not recruiting Preliminary results published [80] | |
| +/− PDR001 (anti-PD1) +/− Carboplatin | II | Advanced TNBC | 1st or 2nd line | NCT03499899 | Suspended | ||
| + PDR001 (anti-PD1) + NIR178 (A2aR antagonist) or Capmantinib (C-MET inhibitor) or MCS110 (anti-M-CSF) or Canakinumab (anti-IL1) | I/Ib | TNBC | ≤ 2 lines | NCT03742349 | Recruiting | ||
| TSR-033 | + anti-PD1 | I | Advanced solid tumors | No standard therapy available | NCT03250832 | Recruiting | |
| INCAGN02385 | No | I | Advanced solid tumors including TNBC | No standard therapy available | NCT03538028 | Not yet recruiting | |
| Sym022 | No | I | Advanced solid tumors | No standard therapy available | NCT03489369 | Recruiting | |
| + Sym021 (anti-PD1) or Sym023 (anti-TIM3) | I | Advanced solid tumors | No standard therapy available | NCT03311412 | Recruiting | ||
| MGD013 (Anti- LAG3 + Anti-PD1) | No | I | Advanced solid tumors | No standard therapy available | NCT03219268 | Recruiting | |
| FS118 (Anti-LAG3 + Anti-PDL1) | No | I | Advanced solid tumors that progressed on anti-PD1/PDL-1 therapy | ≥ 1 line | NCT03440437 | Recruiting | |
| XmAb®22841 (Anti- LAG3 + Anti-CTLA4) | No | I | Advanced solid tumors including TNBC | No standard therapy available | NCT03849469 | Not yet recruiting | |
| TIM-3 | MBG453 | +/− PDR001 (anti-PD1) | I-Ib/II | Advanced solid tumors (phase I) | No standard therapy available | NCT02608268 | Recruiting |
| TSR-022 | No | I | Advanced solid tumors | No standard therapy available | NCT02817633 | Recruiting | |
| + Carboplatin + Nab-paclitaxel + TSR-042 (anti-PD1) | I | Advanced solid tumors | ≤ 1 line (part B) ≤ 4 lines (part A) | NCT03307785 | Recruiting | ||
| LY3321367 | +/− LY3300054 (anti-PDL1) | Ia/Ib | Advanced solid tumors | No standard therapy available | NCT03099109 | Recruiting | |
| INCAGN02390 | No | I | Advanced solid tumors including TNBC | No standard therapy availaible | NCT03652077 | Recruiting | |
| Sym023 | No | I | Advanced solid tumors | No standard therapy availaible | NCT03489343 | Recruiting | |
| + Sym021 (anti-PD1) or Sym022 (anti-LAG3) | I | Advanced solid tumors | No standard therapy available | NCT03311412 | Recruiting | ||
| LY3321367 | +/− LY3300054 (anti-PDL1) | I | Advanced solid tumors | Any line | NCT03099109 | Recruiting | |
| BGB-A425 | +/− Tislelizumab (anti-PD1) for phase II | I/II | Advanced solid tumors | No standard therapy available | NCT03744468 | Recruiting | |
| LY3415244 (Anti-TIM3 + Anti-PDL1) | No | Ia/Ib | Advanced solid tumors | Any line (phase Ia) ≥ 1 line with anti-PD1 or anti-PDL1 therapy (phase Ib) | NCT03752177 | Recruiting | |
| MBG453 | + PDR001 (anti-PD1) | I/II | Advanced solid tumors | No standard therapy available and no prior anti-PD1/PDL1 therapy | NCT02608268 | Recruiting | |
| VISTA | CA-170 | No | I | Advanced solid tumors including TNBC | No standard therapy availaible | NCT02812875 | Recruiting |
| TIGIT | AB154 | +/− AB122 (anti-PD1) | I | Advanced solid tumors | No standard therapy availaible | NCT03628677 | Recruiting |
| OMP-313M32 (Etigilimab) | +/− Nivolumab (anti-PD1) | Ia/Ib | Advanced solid tumors | No standard therapy availaible | NCT03119428 | Active, not recruiting | |
| BMS-986207 | +/− Nivolumab (anti-PD1) | I/II | Advanced solid tumors | No standard therapy availaible | NCT02913313 | Recruiting | |
| GITR | MK-4166 | +/− Pembrolizumab (anti-PD1) | I | Advanced solid tumors | No standard therapy availaible | NCT02132754 | Active, not recruiting |
| INCAGN01876 | +/− Epacadostat (IDO1 inhibitor) +/− Pembrolizumab (anti-PD1) | I/II | Advanced solid tumors (phase I) | No standard therapy availaible | NCT03277352 | Active, not recruiting | |
| +/− Nivolumab (anti-PD1) +/− Ipilimumab (anti-CTLA4) | I/II | Advanced solid tumors (phase I) | No standard therapy availaible | NCT03126110 | Recruiting | ||
| No | I/II | Advanced solid tumors (phase I) | No standard therapy availaible | NCT02697591 | Recuiting | ||
| TRX518 | +/− Gemcitabine +/− Pembrolizumab (anti-PD1) +/− Nivolumab (anti-PD1) | I | Advanced solid tumors (monotherapy and association with Gemcitabine) | No standard therapy availaible or indication for Gemcitabine | NCT02628574 | Recruiting | |
| No | I | Advanced solid tumors | No standard therapy availaible | NCT01239134 | Recruiting, safety results published [81] | ||
| + Cyclophosphamide and/or Avelumab (anti-PDL1) | I/II | Advanced solid tumors including TNBC and hormone receptor positive refractory BC | TNBC: 2nd or 3rd line Hormone receptor positive BC: ≥ 1 line with aromatase inhibitor | NCT03861403 | Not yet recruiting | ||
| BMS-986156 | +/− Nivolumab (anti-PD1) | I/Iia | Advanced solid tumors | No standard therapy availaible | NCT02598960 | Active, not recruiting preliminary results [82] | |
| +/− Nivolumab (anti-PD1) | I | Advanced solid tumors | ≥ 2 lines | NCT03335540 | Recruiting | ||
| GWN323 | +/− PDR001 (anti-PD1) | I/Ib | Advanced solid tumors | Not specified | NCT02740270 | Active, not recruiting | |
| MEDI1873 | No | I | Advanced solid tumors | Not specified | NCT02583165 | Completed, no published results | |
| OMP-336B11 | No | Ia | Advanced solid tumors | No standard therapy availaible | NCT03295942 | Active, not recruiting | |
| B7-H3 | MGA271 (Enoblituzumab) | +/− Pembrolizumab (anti-PD1) | I | Advanced solid tumors including TNBC | No standard therapy available | NCT02475213 | Active, not recruiting |
| + Ipilimumab (anti-CTLA4) | I | Advanced solid tumors including TNBC | No standard therapy available | NCT02381314 | Active, not recruiting | ||
| MGD009 (Orlotamab) | No | I | Advanced solid tumors including TNBC | ≥ 1 prior line | NCT02628535 | Recruiting | |
| MGA012 (anti-PD1) | I | Advanced solid tumors expressing B7-H3 | No standard therapy available | NCT03406949 | Recruiting | ||
| MGC018 | +/− MGA012 (anti-PD1) | I/II | Advanced solid tumors | No standard therapy available | NCT03729596 | Recruiting | |
| ICOS | JTX-2011 | +/− Nivolumab (anti-PD1) +/− Ipilimumab (anti-CTLA4) +/− Pembrolizumab (anti-PD1) | I/II | Advanced solid tumors | No standard therapy availaible | NCT02904226 | Recruiting, safety results published [83] |
| BMS-986226 | +/− Nivolumab (anti-PD1) or Ipilimumab (anti-CTLA4) | I/II | Advanced solid tumors | ≥ 1 prior line | NCT03251924 | Recruiting | |
| 4-1BB | PF-05082566 (Utolimumab) | + Trastuzumab – Vinorelbine – Avelumab (anti-PDL1) + Trastuzumab – Avelumab (anti-PDL1) | II | Advanced Her2+ BC | ≥ 1 prior line with progression under Trastuzumab - Pertuzumab | NCT03414658 | Recruiting |
| Cohort 1: + Trastuzumab – Emtansine Cohort 2: + Trastuzumab | IB | Advanced Her2+ BC | Cohort 1: ≥ 1 prior line with taxane and trastuzumab Cohort 2: ≥ 2 prior lines | NCT03364348 | Recruiting | ||
| + Avelumab (anti-PDL1) | IB/II | Advanced solid tumors including TNBC | Any line | NCT02554812 | Recruiting | ||
| Arm A: + Avelumab (Anti-PD-L1) Arm C: + Avemulmab (anti-PD-L1) and PF-04518600 (anti-OX40) | I/II | Advanced solid tumors | No strandard therapy available | NCT03217747 | Recruiting | ||
| BMS-663513 (Urelumab) | +/− Nivolumab (anti-PD1) | I/II | Advanced solid tumors | Any line | NCT02253992 | Active, not recruiting | |
| + SBRT – Nivolumab (anti-PD1) | I | Advanced solid tumors | Any line | NCT03431948 | Recruiting | ||
| No | I | Advanced solid tumors | No strandard therapy available | NCT01471210 | Completed, preliminary safety results published [84] | ||
| + Nivolumab (anti-PD1) | I | Advanced solid tumors | No strandard therapy available | NCT02534506 | Active, not recruiting | ||
| + Nivolumab (anti-PD1) | I/II | Advanced solid tumors | No strandard therapy available | NCT03792724 | Not yet recruiting | ||
| PRS-343 | + Atezolizumab (anti-PDL1) | IB | Advanced solid tumors including Her2+ BC | ≥ 2nd line | NCT03650348 | Recruiting | |
| No | I | Advanced solid tumors including Her2+ BC | No strandard therapy available | NCT03330561 | Recruiting | ||
| ADG106 | No | I | Advanced solid tumors | No strandard therapy available | NCT03802955 | Recruiting | |
| No | I | Advanced solid tumors | No strandard therapy available | NCT03707093 | Recruiting | ||
| CD27/CD70 | Anti-hCD70 CAR PBL | + Aldeskeukin (IL-2) | I/II | Advanced solid tumors expressing CD70 | ≥ 2nd line | NCT02830724 | Recruiting |
| ARGX-110 (Cusatuzumab) | No | I/II | Advanced solid tumors expressing CD70 | No standard therapy available | NCT01813539 | Active, not recruiting Safety results published [85] | |
| CDX-1127 (Varlilumab) | + ONT-10 (Immunovaccine) | IB | Advanced BC | ≥ 2nd line | NCT02270372 | Completed, no published results | |
| OX40/OX40L | MOXR0916 (Vonlerolizumab) | No | I | Advanced solid tumors | No standard therapy available | NCT02219724 | Active, not recruiting |
| + Atezolizumab (anti-PDL1) | IB | Advanced solid tumors | No standard therapy available | NCT02410512 | Active, not recruiting Preliminary safety results published [86] | ||
| PF-04518600 | + Avelumab (anti-PDL1) Or + Utolilumab (Anti-4-1BB) and Avelumab (anti-PDL1) +/− Radiation | I/II | Advanced solid tumors | No standard therapy available | NCT03217747 | Recruiting | |
| MEDI6383 | +/− MEDI4736 (anti-PDL1) | I | Advanced solid tumors | No standard therapy available ≤ 5 prior lines | NCT02221960 | Completed, no published results | |
| MEDI0562 | +/− MEDI4736 (anti-PDL1) Or +/− Tremelilumab (anti-CTLA4) | I | Advanced solid tumors | No standard therapy available ≤ 3 prior lines | NCT02705482 | Active, not recruiting | |
| INCAGN01949 | No | I/II | Advanced solid tumors | No standard therapy available | NCT02923349 | Active, not recruiting | |
| +/− Nivolumab (anti-PD1) +/− Ipilimumab (anti-CTLA4) | I/II | Advanced solid tumors (phase I) | No standard therapy available | NCT03241173 | Active, not recruiting | ||
| GSK3174998 | +/− Pembrolizumab (anti-PD1) | I | Advanced solid tumors | No standard therapy available ≤ 5 prior lines | NCT02528357 | Recruiting | |
| + GSK1795091 (TLR4 agonist) | I | Advanced solid tumors including BC but not TNBC | No standard therapy available | NCT03447314 | Recruiting | ||
| MEDI6469 | + SBRT to liver or lung metastases | I/II | Advanced BC | ≥ 1 prior line | NCT01862900 | Completed, no published results | |
| mRNA 2416 | No | I | Advanced solid tumors | No standard therapy available | NCT03323398 | Recruiting | |
| BMS-986178 | + intra-tumoral SD-101 (TLR9 agonist) | I | Advanced solid tumors | ≥ 1 prior line | NCT03831295 | Recruiting | |
| +/− Nivolumab (anti-PD1) and/or Ipilimumab (anti-CTLA4) | I/IIa | Advanced solid tumors | ≥ 1 prior line | NCT02737475 | Recruiting | ||
| BTLA | NA | NA | NA | NA | NA | NA | NA |
| TLR9 | IMO-2125 (Tilsotolomid) Intra-tumoral | No | Ib | Advanced solid tumors | Any line (previously treated with anti-PDL1 therapy if indicated) | NCT03052205 | Active, not recruiting Preliminary safety results published [87] |
| Agatolimod (CPG 7909; PF-3512676) | + Trastuzumab | I/II | Advanced Her2+ BC | ≤ 3 lines | NCT00043394 | Completed, no published results | |
| + Trastuzumab | I/II | Advanced Her2+ BC | Not specified | NCT00031278 | Completed, no published results | ||
| + Montanide® ISA-51 (immune modulator) + NY-ESO-l protein (therapeutic vaccine) | I | Localised solid tumors | Neo-adjuvant or adjuvant chemotherapy authorised | NCT00299728 | Completed, no published results | ||
| + Montanide ISA 720 (immune modulator) + Cyclophosphamide + NY-ESO-1-derived Peptides or Protein (therapeutic vaccine) | I | Advanced solid tumors expressing NY-ESO-1 | ≥ 2nd line | NCT00819806 | Completed, no results published | ||
| MGN1703 | + Ipilimumab (anti-CTLA4) | I | Advanced solid tumors | No standard therapy available | NCT02668770 | Recruiting | |
| SD-101 | + BMS 986178 (anti-OX40) | I | Advanced solid tumors | ≥ 1 prior line | NCT03831295 | Recruiting | |
| + Pembrolizumab (anti-PD1) | II | Stage II or III BC | No prior treatment | NCT01042379 | Recruiting | ||
| Adenosine pathway | |||||||
| A2aR | NIR178 | +/− NZV930 (anti-CD73) +/− PDR001 (anti-PD1) | I/IB | Advanced solid tumors including TNBC | No standard therapy available | NCT03549000 | Recruiting |
| + PDR001 (anti-PD1) and LAG525 (anti-LAG3) | I | Advanced TNBC | ≤ 2 prior lines | NCT03742349 | Recruiting | ||
| AZD4635 | +/− Durvalumab (Anti-PDL1) | I | Advanced solid tumors | No standard therapy available | NCT02740985 | Recruiting | |
| AB928 | + AB122 (anti-PD1) | I | Advanced solid tumors | No standard therapy available | NCT03629756 | Recruiting | |
| +/− Pegylated liposomal doxorubicin | I/Ib | Advanced TNBC | No standard therapy available | NCT03719326 | Recruiting | ||
| CPI-444 | +/− Atezolizumab (anti-PDL1) | I | Advanced solid tumors including TNBC | ≥ 1 and ≤ 5 prior lines | NCT02655822 | Recruiting | |
| +/− CPI-006 (anti-CD73) | I/IB | Advanced solid tumors including TNBC | ≥ 1 and ≤ 5 prior lines | NCT03454451 | Recruiting | ||
| CD73 | SRF373 (NZV930) | +/− PDR001 (anti-PD1) +/− NIR178 (A2aR antagonist) | I/IB | Advanced solid tumors including TNBC | No standard therapy available | NCT03549000 | Recruiting |
| CPI-006 | +/− CPI-444 (A2aR antagonist) +/− Pembrolizumab (anti-PD1) | I/IB | Advanced solid tumors including TNBC | ≥ 1 and ≤ 5 prior lines | NCT03454451 | Recruiting | |
| BMS-986179 | +/− Nivolumab (anti-PD1) +/− rHuPH20 (Recombinant human hyaluronidase) | I/IIA | Advanced solid tumors | Any line | NCT02754141 | Recruiting, preliminary results published [88] | |
| MEDI9447 (Oleclumab) | +/− MEDI4736 (anti-PDL1) | I | Advanced solid tumors | Any line | NCT02503774 | Recruiting | |
| + Paclitaxel – Carboplatin – Durvalumab (anti-PDL1) | I/II | Advanced TNBC | 1st line | NCT03616886 | Recruiting | ||
| No | I | Advanced solid tumors (Japanese population) | No standard therapy available | NCT03736473 | Active, not recruiting | ||
| + NACT + pre-operative surgery + Durvalumab (anti-PDL1) | II | Luminal B BC (neo-adjuvant setting) | Neo-adjuvant setting | NCT03875573 | Not yet recruiting | ||
| + Paclitaxel + Durvalumab (anti-PDL1) | I/II | Advanced TNBC | 1st line | NCT03742102 | Recruiting | ||
| CD39 | NA | NA | NA | NA | NA | NA | NA |
| Marker | BC Subtype | Number of Patients | Method | Positive/Overexpressing Cases | Prognostic/Predictive value | Comments | Reference |
|---|---|---|---|---|---|---|---|
| Macrophage-related | |||||||
| CSF-1/CSF-1R | All | 581 (301 node-negative, 280 node-positive) | IHC | Positive cases: node-negative 114/301 (38.9%) node-positive 189/280 (67.5%) | Positivity in node negative: worse OS (not in node positive patients) | [264] | |
| All | 196 | IHC in situ RNA detection | 74% CSF-1+ and 58% CSF-1R+ tumors | CSF-1+ tumor cells: poor survival | CSF-1+ tumor cells: more frequent metastases | [263] | |
| All | 572 | ELISA (circulating CSF1 levels) | NA | logCSF1: worse BCCS high CSF-1: worse outcome in post-menopausal patients | Cut-off: median serum CSF-1 expression | [262] | |
| All | 68 | IHC | NA | High CSF-1: worse DSS | High CSF-1R: marginally correlated to worse DSS | [261] | |
| CCL2/CCR2 | All | 137 | IHC | CCL2+ tumor cells: 30.7% in PTs vs 39.4% in paired recurrences CCL2+ stromal cells: 18.2% in PTs vs 22.6% in paired recurrences | No correlation | Significantly higher CCL2 expression in tumor cells of recurrences (especially the early ones) compared to PTs | [267] |
| All | 427 | IHC | NA | Stromal but not epithelial CCL2 expression: worse RFS in basal-like subtype | Stromal CCL2 remained an independent factor of worse prognosis in basal-like subtype | [268] | |
| All | 63 | IHC | NA | CCR2 expression in tumor cells: worse DFS, MFS and OS | CCR2 expression in tumor cells and CCL2 expression in stromal cells associated with higher risk of metastasis. CCR2 expression in tumor cells remained an independent factor of worse MFS | [269] | |
| All | 151 (135 evaluable) | IHC | CCL2 high: 65/135 (48.1%) CCL2 low: 70/135 (51.9%) | CCL2 high: worse RFS | High combined CCL2/VEGF expression was independently associated with worse RFS | [270] | |
| All | 3554 (TCGA and kmplot.com) | RNA-seq | NA | High mRNA CCL2 expression: better RFS in basal-like, HER2-enriched and luminal-B subtypes (median cutoff of mRNA expression) | No significant association between RFS and expression of CCL2 mRNA in the whole cohort and in luminal-A subtype | [271] | |
| CD40 | All | 181 | IHC | Cytoplasmic tumor cell expression: 53% Membrane tumor cell expression: 7.7% Nuclear tumor cell expression: 81% | CD40 cytoplasmic positivity: better OS | Positive association of CD40 cytoplasmic expression in HR+ breast tumors | [272] |
| NK cell-related | |||||||
| CD94/NKG2A | All | 28 (TDLN) | Flow cytometry | NA | NA | High expression of NKG2A in NK cells of tumor-draining lymph nodes described NKG2A+ NK cells correlated to locally advanced disease | [273] |
| NKG2D ligands (MICBAB, ULBP1-5) | All | 677 | IHC | Tumor cell expression: MIC-AB: 50% ULBP-1: 90% ULBP-2: 99% ULBP-3: 100% ULBP-4: 26% ULBP-5: 90% | High MIC-AB and ULBP-2 expression better RFS | Combined low expression of MIC-AB and ULBP-2 correlated to worse RFS | [274] |
| Dendritic cell-related | |||||||
| IDO | All (Pakistani population) | 100 | IHC | 100% positive 24/100 low IDO (24%) 27/100 medium IDO (27%) 49/100 high IDO (49%) | Medium and high IDO: worse OS | IDO expression correlated to TNBC | [275] |
| All | 203 | IHC | 100% positive 108/203 low IDO (53.2%) 95/203 intermediate and high IDO (46.8%) | General population: no difference in OS ER+ IDO intermediate/high: better OS Node-positive IDO intermediate/high: better DSS | IDO expression correlated to ER+ | [276] | |
| All | 26 primary tumor + TDLN 10 benign lesions | IHC | IDO positivity: PT: 12/26 (46.15%) TDLN: 19/26 (73.08%) Benign lesions: 1/10 (10%) | IDO expression: statistically not significant worse OS and TTP | IDO expression correlated to advanced stages, lymph-node metastasis and Treg infiltration No expression in healthy adjacent tissue | [277] | |
| All | 155 | IHC | Stromal positivity (>5%): 49/155 (31%) Epithelial positivity (>10%) 24/155 (15%) | IDO positivity: better OS | IDO positivity correlated to absence of lymph-node metastasis, ER- and TNBC | [278] | |
| All | 242 primary tumor 20 TDLN 19 metastasis | IHC | IDO positivity: PT: 34/242 (14%) TDLN: 1/20 (5%) Metastasis: 0/19 (0%) | NA | IDO positivity correlated to high grade and TNBC Co-expression of IDO in 70% of PDL-1+ cases | [279] | |
| All | 65 | IHC | IDO positivity: 42/65 (64.6%) | IDO expression: worse OS and PFS in UA but not MA | IDO expression correlated to high grade, lymph-node metastastasis | [280] | |
| All | 54 | IHC | IDO positivity: 27/54 (68.5%) | IDO expression: worse response to NACT and statistically not significant worse PFS and OS | IDO expression correlated to advanced stages, lymph-node metastasis | [281] | |
| All | 129 PT 10 normal LN 17 metastatic LN | IHC | IDO expression: PT: NA Normal lymph-nodes 80% Metastatic lymph nodes 88.2% | NA | IDO expression correlated to lymph-node metastasis, ER-, TNBC and PD-1 expression | [282] | |
| All | 54 PT 11 healthy controls | qRT-PCR | NA | NA | IDO expression reduced in tumor compared to healthy tissue IDO expression in tumor correlated to advanced stage | [283] | |
| All | 46 | IHC | IDO high: 26/46 (56.5%) | IDO high: worse response to NACT and worse PFS and OS | IDO high correlated to advanced stage and lymph-node metastasis | [284] | |
| HR+ | 362 | IHC | IDO expression 276/362 (76.2%) | IDO expression: worse OS | IDO expression not correlated to clinico-pathological characteristics IDO expression negatively correlated to B-cell infiltration | [285] | |
| All | 202 | IHC | NA | IDO high (expression by CAFs): worse DSS and MFS | [286] | ||
| All | 91 PT 21 benign lesions 10 healthy controls | IHC | IDO expression: PT: 55/91 (60%) Benign lesions 9/21 (43%) Healthy controls 2/10 (20%) | NA | IDO expression correlated to advanced stage | [287] | |
| All | 85 | IHC | NA | NA | IDO expression correlated to Treg infiltration and lymph-node metastasis | [288] | |
| All | 5 | IHC | IDO expression 5/5 (100%) | NA | [289] | ||
| Target | Drug | Other Agent(s) | Phase | Disease | Line | NCT Identifier | Trial Status |
|---|---|---|---|---|---|---|---|
| TAM-stimulatory markers | |||||||
| CSF-1/CSF-1R | |||||||
| CSF-1R/CSF-1 inhibitors | PLX 3397 (Pexidartinib) | + Eribulin | Ib/II | Metastatic breast cancer | ≥ 1 prior line | NCT01596751 | Active, not recruiting |
| No | I | Advanced solid tumors | No standard therapy available | NCT01004861 | Active, not recruiting | ||
| +/− Paclitaxel | Ib | Advanced solid tumors | Not specified | NCT01525602 | Completed, no published results | ||
| ARRY-382 | +/− Pembrolizumab (anti-PD1) | Ib/II | Advanced solid tumors including TNBC (phase Ib) | No standard therapy available | NCT02880371 | Recruiting | |
| No | I | Advanced or metastatic solid tumors | No standard therapy available | NCT01316822 | Completed, no published results | ||
| BLZ945 | +/− PDR001 (anti-PD1) | I | Advanced solid tumors including TNBC | Not specified | NCT02829723 | Recruiting | |
| Anti CSF-1R antibodies | LY3022855 (IMC-CS4) | No | I | Advanced BC | ≥ 1 prior line | NCT02265536 | Completed, no published results |
| + Durvalumab (anti-PDL1) or Tremelimumab (anti-CTLA4) | I | Advanced solid tumors | Not specified | NCT02718911 | Completed, no published results | ||
| No | I | Advanced solid tumors | No standard therapy available | NCT01346358 | Completed, safety results published [290] | ||
| RO5509554 (Emactuzumab) | + Atezolizumab (anti-PDL1) | I | Advanced solid tumors including TNBC | Not specified | NCT02323191 | Recruiting | |
| +/− Paclitaxel | I | Advanced solid tumors | No standard therapy available | NCT01494688 | Completed, preliminary safety and activity results published [291] | ||
| + RO7009789 (CD40 agonist) | Ib | Advanced solid tumors including TNBC | No standard therapy available | NCT02760797 | Completed, no published results | ||
| AMG820 | No | I | Advanced solid tumors | Not specified | NCT01444404 | Completed, no published results | |
| SNDX-6352 | Phase Ia: SNDX-6352 monotherapy Phase Ib: + Durvalumab (anti-PDL1) | I | Advanced solid tumors | ≥ 1 prior line and no standard therapy available | NCT03238027 | Recruiting | |
| Cabiralizumab (BMS-986227, FPA008) | +/− Nivolumab (anti-PD1) | I | Advanced malignancies | No standard therapy available | NCT03158272 | Recruiting | |
| + Nivolumab (anti-PD1) and SBRT | I | Advanced malignancies | No standard therapy available | NCT03431948 | Recruiting | ||
| PD 0360324 (M-CSF mAb) | + Avelumab (anti-PDL1) | Ib/II | Advanced solid tumors including TNBC | No standard therapy available | NCT02554812 | Recruiting | |
| CCL2/CCR2 | |||||||
| CCR2 antagonist | PF-04136309 | + Avelumab (anti-PDL1) +Utomilumab (anti-4-1BB) | Ib/II | Advanced solid tumors including TNBC | No standard therapy available | NCT02554812 | Recruiting |
| CD47 – SIRPα | |||||||
| Anti-CD47 antibodies | Hu5F9-G4 | + Cetuximab (anti-EGFR) | Ib/II | Advanced solid tumors including BC (phase Ib) | ≥ 1 prior line | NCT02953782 | Recruiting |
| +Avelumab (anti-PDL1) | Ib | Advanced solid tumors | Not specified | NCT03558139 | Recruiting | ||
| CC-90002 | No | I | Advanced solid tumors | No standard therapy available | NCT02367196 | Recruiting | |
| IBI188 | No | Ia | Advanced solid tumors | No standard therapy available | NCT03763149 | Recruiting | |
| No | I | Advanced solid tumors | No standard therapy available | NCT03717103 | Recruiting | ||
| AO-176 | No | I | Advanced solid tumors | No standard therapy available | NCT03834948 | Recruiting | |
| SRF231 | No | I/Ib | Advanced solid tumors | No standard therapy available | NCT03512340 | Recruiting | |
| SIRPα-IgG1-Fc | TTI-621 (intra-tumoral injection) | +/− PD1/PDL1 Inhibitor | I | Advanced solid tumors with percutaneously accessible lesions | No standard therapy available | NCT02890368 | Recruiting |
| ALX148 | +/− Trastuzumab or Pembrolizumab (anti-PD1) or Rituximab (anti-CD20) | I | Advanced solid tumors | No standard therapy available | NCT03013218 | Recruiting, preliminary safety results published [292] | |
| TAM-inhibitory markers | |||||||
| CD40 (agonists) | CP-870,893 | No | I | Advanced solid tumors | No standard therapy available | NCT02225002 | Completed, no published results |
| No | I | Advanced solid tumors | Patients who had clinical benefit following a single infusion of CP-870, 893 | NCT02157831 | Completed | ||
| RO7009789 Selicrelumab | + Atezolizumab (anti PDL1) | Ib | Advanced solid tumors | No standard therapy available | NCT02304393 | Recruiting | |
| + Emactuzumab (anti-CSF-1R) | I | Advanced solid tumors including TNBC | No standard therapy available | NCT02760797 | Completed, no published results | ||
| + Vanucizumab (anti-VEGF-A and angiopoietin-2) | I | Metastatic solid tumors | Not specified | NCT02665416 | Recruiting | ||
| ADC-1013 (intra-tumoral or intra-venous injection) | No | I | Advanced solid tumors | Not specified | NCT02379741 | Completed, no published results | |
| JNJ-64457107 | No | I | Advanced solid tumors | Not specified | NCT0282909 | Recruiting | |
| TLR7 (agonists) | Imiquimod | + Cyclophosphamide and Radiotherapy | I/II | Advanced BC with skin metastases | Any line | NCT01421017 | Completed, no published results |
| NK cell-inhibitory markers | |||||||
| CD94/ NKG2A | IPH2201 | + Durvalumab (anti-PDL1) | I/II | Advanced solid tumors | Any line | NCT02671435 | Recruiting |
| KIR family | Lirilumab (anti-KIR2DL1,2,3 antibody) | +Nivolumab (anti-PD1) Or + Nivolumab (anti-PD1) and Ipilimumab (anti-CTLA4) | I | Advanced and/or metastatic solid tumors | Not specified | NCT03203876 | Active, not recruiting |
| +Nivolumab (anti-PD1) | I/II | Advanced solid tumors | ≥ 1 and ≤ 5 prior lines | NCT01714739 | Active, not recruiting | ||
| IDO | |||||||
| Small-molecule inhibitor of IDO-1 | Epacadostat (INCB024360) | + INCB01158 (arginase inhibitor) +/− Pembrolizumab (anti-PD1) | I/II | Advanced solid tumors | No standard therapy available | NCT03361228 | Active, not recruiting |
| + Pembrolizumab (anti-PD1) | I/II | Advanced or metastatic solid tumors including TNBC (phase I) | ≥ 1 prior line | NCT02178722 | Active, not recruiting Preliminary safety and efficacy results published [293] | ||
| + Sirolimus (mTOR inhinitor) | I | Advanced solid tumors | ≥ 1 prior line and no standard therapy available | NCT03217669 | Recruiting | ||
| +Nivolumab (anti-PD1) and Ipilimumab (anti-CTLA4) (group A) + Nivolumab (anti-PD1) + lirilumab (anti-KIR) (group B) | I/II | Advanced solid tumors | No standard therapy available (phase I) ≥ 1 prior line (phase II) | NCT03347123 | Active, not recruiting | ||
| + Durvalumab (anti-PDL1) | I/II | Advanced solid tumors | ≥ 1 prior line | NCT02318277 | Active, not recruiting | ||
| + Pembrolizumab (anti-PD1) And mFOLFOX6 Or (anti-PD1) Gemcitabine and Nab-Paclitaxel Or Carboplatin and Paclitaxel Or Pemetrexed, and Platinium agent Or Cyclophosphamide Or Gemcitabine and Platinium agent Or Platinium agent and 5-Fu | I/II | Advanced solid tumors | Not specified | NCT03085914 | Active, not recruiting | ||
| +/− Pembrolizumab (anti-PD1) Or +/−Pembrolizumab (anti-PD1) and Carboplatin or Cisplatin and Paclitaxel Or +/− Pembrolizumab (anti-PD1) and Carboplatin and Paclitaxel | I | Advanced solid tumors (Japanese population) | No standard therapy available | NCT02862457 | Active, not recruiting Preliminary safety and efficacy results published [294] | ||
| + Pembrolizumab (anti-PD1) and Azacitidine (DNA methyl transferase inhibitor) Or + INCB057643 (BET inhibitor) + Pembrolizumab (anti-PD1) Or + INCB059872 (LSD1 inhibitor) and Pembrolizumab (anti-PD1) | I/II | Advanced solid tumors | No standard therapy available | NCT02959437 | Active, not recruiting | ||
| No | I | Advanced solid tumors | No standard therapy available | NCT01195311 | Completed, safety results published [295] | ||
| + Pembrolizumab (anti-PD1) and INCAGN01876 (anti-GITR) | I/II | Advanced solid tumors | No standard therapy avaialble | NCT03277352 | Active, not recruiting | ||
| + Itacitinib (JAK inhibitor) | I | Advanced solid tumors including TNBC | No standard therapy available | NCT02559492 | Active, not recruiting | ||
| No | Ib | Resectable metastatic solid tumors | Eligible for surgical resection and no standard therapy available | NCT03471286 | Recruiting | ||
| GDC-0919 (navoximod) | + Atezolizumab (anti-PD-1) | Ib | Advanced solid tumors | ≥ 1 prior line | NCT02471846 | Active, not recruiting | |
| NLG802 | No | I | Advanced solid tumors | Not specified | NCT03164603 | Recruiting | |
| Marker | Types of Cells Expressed | Function on Anti-tumor Immunity |
|---|---|---|
| LAG-3 | Effector T-cells, Tregs, NK-cells, B-cells, dendritic cells (DC) | Co-inhibitory |
| TIM-3 | CD8+, CD4+ T helper 1 cells (Th1 cells), Tregs, NK cells, DC, monocytes, macrophages | Co-inhibitory |
| VISTA | CD8+, CD4+ T-cells, Tregs, NK cells, DC, monocytes, macrophages, granulocytes | Co-inhibitory |
| TIGIT | Effector, memory, follicular helper (Tfh) T-cells, Tregs, NK-cells | Co-inhibitory |
| GITR | T-cells | Co-stimulatory |
| B7-H3 | T-cells, antigen-presenting cells (APC), NK-cells | Co-stimulatory Co-inhibitory |
| ICOS | T-cells | Co-stimulatory Co-inhibitory |
| 4-1BB | Effector, helper T-cells, Tregs, B-cells, NK-cells, DC, neutrophils, eosinophils, mast cells, monocytes, macrophages | Co-stimulatory |
| CD27 | T-cells, B-cells, NK-cells | Co-stimulatory |
| OX40 | Tregs, neutrophils, NK-cells and NKT-cells, CD4+ and CD8+ T-cells (upon TCR stimulation) | Co-stimulatory |
| BTLA | T-cells, B-cells, Tfh cells, macrophages, DC, NKT-cells, NK-cells | Co-inhibitory |
| A2aR | T-cells, NKT-cells, B-cells, monocytes, macrophages, DC, NK-cells, mast cells, eosinophils, platelets | Co-inhibitory |
| CD73 | B-cells, CD8+, CD4+ T-cells, Tregs, neutrophils, MDSC, monocytes, macrophages, DC, NK-cells, endothelial cells, cancer cells | Co-inhibitory |
| CD39 | Platelets, endothelial cells, cancer cells | Co-inhibitory |
| CCR2 | Monocytes, macrophages | Co-inhibitory |
| CD47 | Cancer cells | Co-inhibitory |
| CD40 | APC, macrophages | Co-stimulatory |
| CD94/NKG2A | NK-cells, CD8+ T-cells | Co-inhibitory |
| NKG2D | NK-cells | Co-stimulatory |
| IDO | Cancer cells, stromal dendritic-like cells, myoepithelial cells | Co-inhibitory |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chrétien, S.; Zerdes, I.; Bergh, J.; Matikas, A.; Foukakis, T. Beyond PD-1/PD-L1 Inhibition: What the Future Holds for Breast Cancer Immunotherapy. Cancers 2019, 11, 628. https://doi.org/10.3390/cancers11050628
Chrétien S, Zerdes I, Bergh J, Matikas A, Foukakis T. Beyond PD-1/PD-L1 Inhibition: What the Future Holds for Breast Cancer Immunotherapy. Cancers. 2019; 11(5):628. https://doi.org/10.3390/cancers11050628
Chicago/Turabian StyleChrétien, Sebastian, Ioannis Zerdes, Jonas Bergh, Alexios Matikas, and Theodoros Foukakis. 2019. "Beyond PD-1/PD-L1 Inhibition: What the Future Holds for Breast Cancer Immunotherapy" Cancers 11, no. 5: 628. https://doi.org/10.3390/cancers11050628
APA StyleChrétien, S., Zerdes, I., Bergh, J., Matikas, A., & Foukakis, T. (2019). Beyond PD-1/PD-L1 Inhibition: What the Future Holds for Breast Cancer Immunotherapy. Cancers, 11(5), 628. https://doi.org/10.3390/cancers11050628





