Paralogous HOX13 Genes in Human Cancers
Abstract
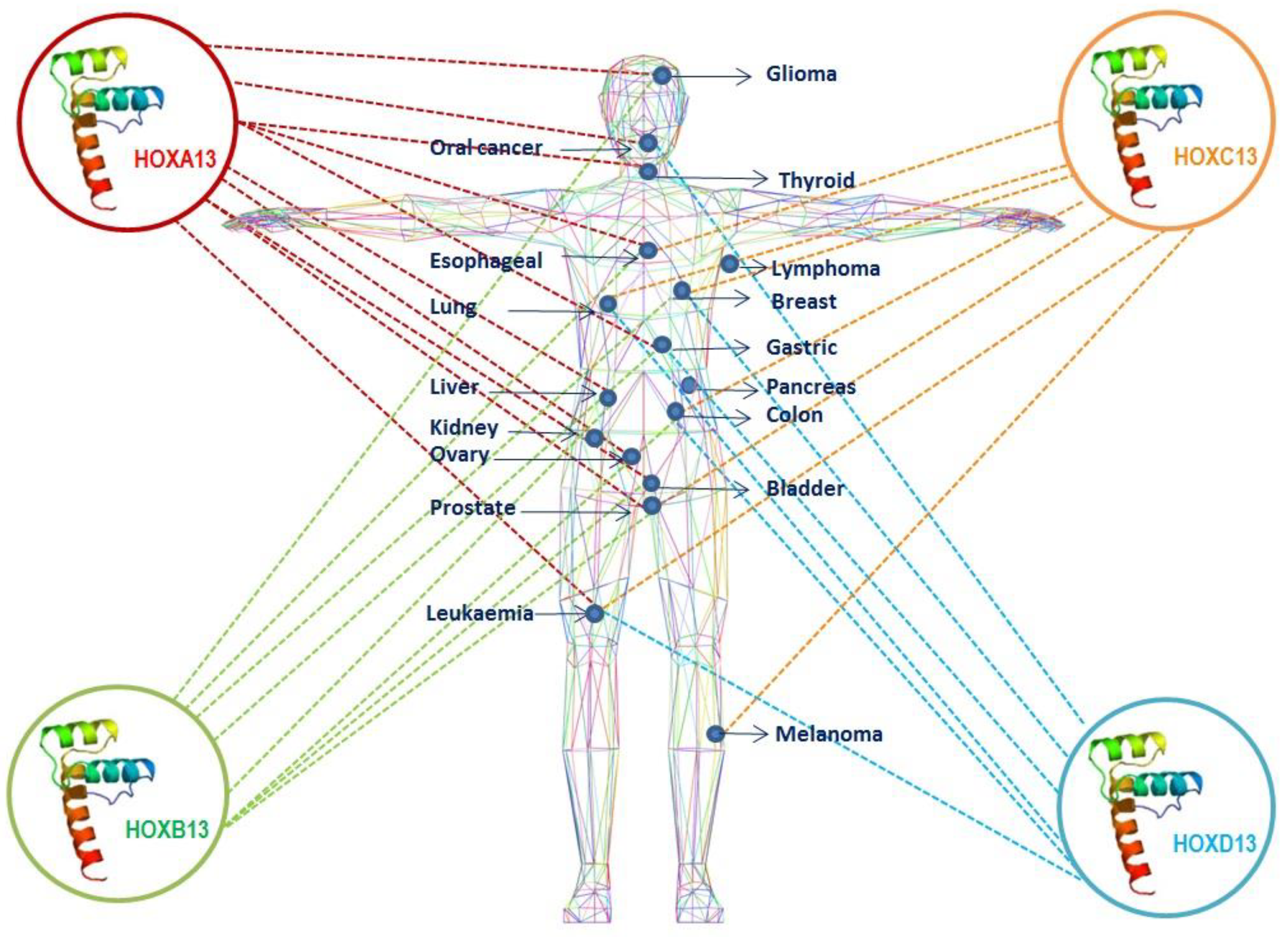
:1. Introduction
2. HOXA13
3. HOXB13
4. HOXC13
5. HOXD13
6. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Gehring, W.; Hiromi, Y. Homeotic genes and the homeobox. Annu. Rev. Genet. 1986, 20, 147–173. [Google Scholar] [CrossRef]
- Krumlauf, R. Hox genes in vertebrate development. Cell 1994, 78, 191–201. [Google Scholar] [CrossRef]
- Apiou, F.; Flagiello, D.; Cillo, C.; Malfoy, B.; Poupon, M.F.; Dutrillaux, B. Fine mapping of human HOX gene clusters. Cytogenet Cell Genet. 1996, 73, 114–115. [Google Scholar] [CrossRef] [PubMed]
- Graham, A.; Papalopulu, N.; Krumlauf, R. The murine and Drosophila homeobox gene complexes have common features of organization and expression. Cell 1989, 57, 367–378. [Google Scholar] [CrossRef]
- Gehring, W.J.; Kloter, U.; Suga, H. Evolution of the Hox gene complex from an evolutionary ground state. Curr. Top. Dev. Biol. 2009, 88, 35–61. [Google Scholar] [PubMed]
- Schuettengruber, B.; Bourbon, H.M.; Di Croce, L.; Cavalli, G. Genome Regulation by Polycomb and Trithorax: 70 Years and Counting. Cell 2017, 171, 34–57. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schuettengruber, B.; Chourrout, D.; Vervoort, M.; Leblanc, B.; Cavalli, G. Genome regulation by polycomb and trithorax proteins. Cell 2007, 128, 735–745. [Google Scholar] [CrossRef]
- Lander, E.S. The new genomics: Global views of biology. Science 1996, 274, 536–539. [Google Scholar] [CrossRef]
- Durston, A.; Wacker, S.; Bardine, N.; Jansen, H. Time space translation: A hox mechanism for vertebrate a-p patterning. Curr. Genom. 2012, 13, 300–307. [Google Scholar] [CrossRef]
- Durston, A.J. Global posterior prevalence is unique to vertebrates: A dance to the music of time? Dev. Dyn. 2012, 241, 1799–1807. [Google Scholar] [CrossRef] [Green Version]
- Luo, Z.; Rhie, S.K.; Farnham, P.J. The Enigmatic HOX Genes: Can We Crack Their Code? Cancers 2019, 11, 323. [Google Scholar] [CrossRef] [PubMed]
- Botti, G.; De Chiara, A.; Di Bonito, M.; Cerrone, M.; Malzone, M.G.; Collina, F.; Cantile, M. Noncoding RNAs within the HOX gene network in tumor pathogenesis and progression. J. Cell. Physiol. 2018, 234, 395–413. [Google Scholar] [CrossRef] [PubMed]
- Burgess, D.J. Non-coding RNA: HOTTIP goes the distance. Nat. Rev. Genet. 2011, 12, 300. [Google Scholar] [CrossRef] [PubMed]
- Quagliata, L.; Matter, M.S.; Piscuoglio, S.; Arabi, L.; Ruiz, C.; Procino, A.; Kovac, M. Long noncoding RNA HOTTIP/HOXA13 expression is associated with disease progression and predicts outcome in hepatocellular carcinoma patients. Hepatology 2014, 59, 911–923. [Google Scholar] [CrossRef]
- Woo, C.J.; Kingston, R.E. HOTAIR lifts noncoding RNAs to new levels. Cell 2007, 129, 1257–1259. [Google Scholar] [CrossRef] [PubMed]
- Cillo, C.; Faiella, A.; Cantile, M.; Boncinelli, E. Homeobox genes and cancer. Exp. Cell Res. 1999, 248, 1–9. [Google Scholar] [CrossRef]
- Cillo, C.; Cantile, M.; Faiella, A.; Boncinelli, E. Homeobox genes in normal and malignant cells. J. Cell. Physiol. 2001, 188, 161–169. [Google Scholar] [CrossRef] [Green Version]
- Lv, X.; Li, L.; Lv, L.; Qu, X.; Li, K.; Deng, X.; Cheng, L.; He, H.; Dong, L. HOXD9 promotes epithelial–mesenchymal transition and cancer metastasis by ZEB1 regulation in hepatocellular carcinoma. J. Exp. Clin. Cancer Res. 2015, 34, 133. [Google Scholar] [CrossRef] [Green Version]
- Cantile, M.; Scognamiglio, G.; Anniciello, A.; Farina, M.; Gentilcore, G.; Santonastaso, C.; Fulciniti, F.; Cillo, C.; Franco, R.; Ascierto, P.A.; et al. Increased HOX C13 expression in metastatic melanoma progression. J. Transl. Med. 2012, 10, 91. [Google Scholar] [CrossRef]
- Haria, D.; Naora, H. Homeobox Gene Deregulation: Impact on the Hallmarks of Cancer. Cancer Hallm. 2013, 1, 67–76. [Google Scholar] [CrossRef] [Green Version]
- Procino, A.; Cillo, C. The HOX genes network in metabolic diseases. Cell Biol. Int. 2013, 37, 1145–1148. [Google Scholar] [CrossRef]
- Sheth, R.; Barozzi, I.; Langlais, D.; Osterwalder, M.; Nemec, S.; Carlson, H.L.; Stadler, H.S.; Visel, A.; Drouin, J.; Kmita, M. Distal Limb Patterning Requires Modulation of cis-Regulatory Activities by HOX13. Cell Rep. 2016, 17, 2913–2926. [Google Scholar] [CrossRef]
- Beck, F. Homeobox genes in gut development. Gut 2002, 51, 450–454. [Google Scholar] [CrossRef] [Green Version]
- de Santa Barbara, P.; Roberts, D.J. Tail gut endoderm and gut/genitourinary/tail development: A new tissue-specific role for Hoxa13. Development 2002, 129, 551–561. [Google Scholar] [PubMed]
- Scott, V.; Morgan, E.A.; Stadler, H.S. Genitourinary functions of Hoxa13 and Hoxd13. J. Biochem. 2005, 137, 671–676. [Google Scholar] [CrossRef]
- Javed, S.; Langley, S.E. Importance of HOX genes in normal prostate gland formation, prostate cancer development and its early detection. BJU Int. 2014, 113, 535–540. [Google Scholar] [CrossRef] [PubMed]
- Aquino, G.; Franco, R.; Sabatino, R.; Mantia, E.L.; Scognamiglio, G.; Collina, F.; Longo, F.; Ionna, F.; Losito, N.S.; Liguori, G.; et al. Deregulation of paralogous 13 HOX genes in oral squamous cell carcinoma. Am. J. Cancer Res. 2015, 5, 3042–3055. [Google Scholar]
- Cantile, M.; Scognamiglio, G.; La Sala, L.; La Mantia, E.; Scaramuzza, V.; Valentino, E.; Tatangelo, F.; Losito, S.; Pezzullo, L.; Chiofalo, M.G.; et al. Aberrant expression of posterior HOX genes in well differentiated histotypes of thyroid cancers. Int. J. Mol. Sci. 2013, 14, 21727–21740. [Google Scholar] [CrossRef]
- Cantile, M.; Franco, R.; Tschan, A.; Baumhoer, D.; Zlobec, I.; Schiavo, G.; Forte, I.; Bihl, M.; Liguori, G.; Botti, G.; et al. HOX D13 expression across 79 tumor tissue types. Int. J. Cancer 2009, 125, 1532–1541. [Google Scholar] [CrossRef] [Green Version]
- Roux, M.; Bouchard, M.; Kmita, M. Multifaceted Hoxa13 function in urogenital development underlies the Hand-Foot-Genital Syndrome. Hum. Mol. Genet. 2019, 28, 1671–1681. [Google Scholar] [CrossRef]
- Mortlock, D.P.; Innis, J.W. Mutation of HOXA13 in hand-foot-genital syndrome. Nat. Genet. 1997, 15, 179–180. [Google Scholar] [CrossRef]
- Wen, Y.; Shu, F.; Chen, Y.; Chen, Y.; Lan, Y.; Duan, X.; Zhao, S.C.; Zeng, G. The prognostic value of HOXA13 in solid tumors: A meta-analysis. Clin. Chim. Acta 2018, 483, 64–68. [Google Scholar] [CrossRef]
- Hu, H.; Chen, Y.; Cheng, S.; Li, G.; Zhang, Z. Dysregulated expression of homeobox gene HOXA13 is correlated with the poor prognosis in bladder cancer. Wien. Klin. Wochenschr. 2017, 129, 391–397. [Google Scholar] [CrossRef]
- Guo, B.; Che, T.; Shi, B.; Guo, L.; Yin, Y.; Li, L.; Wang, J.; Yan, D.; Chen, Y. Screening and identification of specific markers for bladder transitional cell carcinoma from urine urothelial cells with suppressive subtractive hybridization and cDNA microarray. Can. Urol. Assoc. J. 2011, 5, 129–137. [Google Scholar] [CrossRef]
- Guo, B.; Che, T.; Shi, B.; Guo, L.; Zhang, Z.; Li, L.; Cai, C.; Chen, Y. Interaction network analysis of differentially expressed genes and screening of cancer marker in the urine of patients with invasive bladder cancer. Int. J. Clin. Exp. Med. 2015, 8, 3619–3628. [Google Scholar]
- Dong, Y.; Cai, Y.; Liu, B.; Jiao, X.; Li, Z.T.; Guo, D.Y.; Li, X.W.; Wang, Y.J.; Yang, D.K. HOXA13 is associated with unfavorable survival and acts as a novel oncogene in prostate carcinoma. Future Oncol. 2017, 13, 1505–1516. [Google Scholar] [CrossRef]
- Taketani, T.; Taki, T.; Ono, R.; Kobayashi, Y.; Ida, K.; Hayashi, Y. The chromosome translocation t(7;11)(p15;p15) in acute myeloid leukemia results in fusion of the NUP98 gene with a HOXA cluster gene, HOXA13, but not HOXA9. Genes Chromosomes Cancer 2002, 34, 437–443. [Google Scholar] [CrossRef]
- Fujino, T.; Suzuki, A.; Ito, Y.; Ohyashiki, K.; Hatano, Y.; Miura, I.; Nakamura, T. Single-translocation and double-chimeric transcripts: Detection of NUP98-HOXA9 in myeloid leukemias with HOXA11 or HOXA13 breaks of the chromosomal translocation t(7;11)(p15;p15). Blood 2002, 99, 1428–1433. [Google Scholar] [CrossRef]
- Su, X.; Drabkin, H.; Clappier, E.; Morgado, E.; Busson, M.; Romana, S.; Soulier, J.; Berger, R.; Bernard, O.A.; Lavau, C. Transforming potential of the T-cell acute lymphoblastic leukemia-associated homeobox genes HOXA13, TLX1, and TLX3. Genes Chromosomes Cancer 2006, 45, 846–855. [Google Scholar] [CrossRef]
- Han, Y.; Tu, W.W.; Wen, Y.G.; Li, D.P.; Qiu, G.Q.; Tang, H.M.; Peng, Z.H.; Zhou, C.Z. Identification and validation that up-expression of HOXA13 is a novel independent prognostic marker of a worse outcome in gastric cancer based on immunohistochemistry. Med. Oncol. 2013, 30, 564. [Google Scholar] [CrossRef]
- Yang, Y.C.; Wang, S.W.; Wu, I.C.; Chang, C.C.; Huang, Y.L.; Lee, O.K.; Chang, J.G.; Chen, A.; Kuo, F.C.; Wang, W.M.; et al. A tumorigenic homeobox (HOX) gene expressing human gastric cell line derived from putative gastric stem cell. Eur. J. Gastroenterol. Hepatol. 2009, 21, 1016–1023. [Google Scholar] [CrossRef]
- He, Y.X.; Song, X.H.; Zhao, Z.Y.; Zhao, H. HOXA13 upregulation in gastric cancer is associated with enhanced cancer cell invasion and epithelial-to-mesenchymal transition. Eur. Rev. Med. Pharmacol. Sci. 2017, 21, 258–265. [Google Scholar]
- Qu, L.P.; Zhong, Y.M.; Zheng, Z.; Zhao, R.X. CDH17 is a downstream effector of HOXA13 in modulating the Wnt/β-catenin signaling pathway in gastric cancer. Eur. Rev. Med. Pharmacol. Sci. 2017, 21, 1234–1241. [Google Scholar]
- Han, Y.; Song, C.; Wang, J.; Tang, H.; Peng, Z.; Lu, S. HOXA13 contributes to gastric carcinogenesis through DHRS2 interacting with MDM2 and confers 5-FU resistance by a p53-dependent pathway. Mol. Carcinog. 2018, 57, 722–734. [Google Scholar] [CrossRef]
- Yan, W.P.; Shen, L.Y.; Gu, Z.D.; Chen, K.N. Up-regulation of HOXA13 in esophageal squamous cell carcinoma of stage IIa and its effect on the prognosis. Chin. J. Gastrointest. Surg. 2009, 12, 20–23. [Google Scholar]
- Gu, Z.D.; Shen, L.Y.; Wang, H.; Chen, X.M.; Li, Y.; Ning, T.; Chen, K.N. HOXA13 promotes cancer cell growth and predicts poor survival of patients with esophageal squamous cell carcinoma. Cancer Res. 2009, 69, 4969–4973. [Google Scholar] [CrossRef]
- Ma, R.L.; Shen, L.Y.; Chen, K.N. Coexpression of ANXA2, SOD2 and HOXA13 predicts poor prognosis of esophageal squamous cell carcinoma. Oncol. Rep. 2014, 31, 2157–2164. [Google Scholar] [CrossRef]
- Shi, Q.; Shen, L.; Dong, B.; Fu, H.; Kang, X.; Dai, L.; Yang, Y.; Yan, W.; Chen, K.N. Downregulation of HOXA13 sensitizes human esophageal squamous cell carcinoma to chemotherapy. Thorac. Cancer 2018, 9, 836–846. [Google Scholar] [CrossRef] [Green Version]
- Huang, T.; Chesnokov, V.; Yokoyama, K.K.; Carr, B.I.; Itakura, K. Expression of the Hoxa-13 Gene Correlates to Hepatitis B and C Virus Associated HCC. Biochem Biophys Res. Commun. 2001, 281, 1041–1044. [Google Scholar] [CrossRef]
- Cillo, C.; Schiavo, G.; Cantile, M.; Bihl, M.P.; Sorrentino, P.; Carafa, V.; D’Armiento, M.; Roncalli, M.; Sansano, S.; Vecchione, R.; et al. The HOX gene network in hepatocellular carcinoma. Int. J. Cancer 2011, 129, 2577–2587. [Google Scholar] [CrossRef] [Green Version]
- Pan, T.T.; Jia, W.D.; Yao, Q.Y.; Sun, Q.K.; Ren, W.H.; Huang, M.; Ma, J.; Li, J.S.; Ma, J.L.; Yu, J.H.; et al. Overexpression of HOXA13 as a potential marker for diagnosis and poor prognosis of hepatocellular carcinoma. Tohoku J. Exp. Med. 2014, 234, 209–219. [Google Scholar] [CrossRef] [PubMed]
- Quagliata, L.; Quintavalle, C.; Lanzafame, M.; Matter, M.S.; Novello, C.; di Tommaso, L.; Pressiani, T.; Rimassa, L.; Tornillo, L.; Roncalli, M.; et al. High expression of HOXA13 correlates with poorly differentiated hepatocellular carcinomas and modulates sorafenib response in in vitro models. Lab. Investig. 2018, 98, 95–105. [Google Scholar] [CrossRef] [PubMed]
- Deng, Y.; He, R.; Zhang, R.; Gan, B.; Zhang, Y.; Chen, G.; Hu, X. The expression of HOXA13 in lung adenocarcinoma and its clinical significance: A study based on The Cancer Genome Atlas, Oncomine and reverse transcription-quantitative polymerase chain reaction. Oncol. Lett. 2018, 15, 8556–8572. [Google Scholar] [CrossRef] [Green Version]
- Kang, J.U. Characterization of amplification patterns and target genes on the short arm of chromosome 7 in early-stage lung adenocarcinoma. Mol. Med. Rep. 2013, 8, 1373–1378. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kelly, Z.; Moller-Levet, C.; McGrath, S.; Butler-Manuel, S.; Kavitha Madhuri, T.; Kierzek, A.M.; Pandha, H.; Morgan, R.; Michael, A. The prognostic significance of specific HOX gene expression patterns in ovarian cancer. Int. J. Cancer 2016, 139, 1608–1617. [Google Scholar] [CrossRef] [PubMed]
- Duan, R.; Han, L.; Wang, Q.; Wei, J.; Chen, L.; Zhang, J.; Kang, C.; Wang, L. HOXA13 is a potential GBM diagnostic marker and promotes glioma invasion by activating the Wnt and TGF-β pathways. Oncotarget 2015, 6, 27778–27793. [Google Scholar] [CrossRef] [Green Version]
- Wang, K.C.; Yang, Y.W.; Liu, B.; Sanyal, A.; Corces-Zimmerman, R.; Chen, Y.; Lajoie, B.R.; Protacio, A.; Flynn, R.A.; Gupta, R.A.; et al. A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature 2011, 472, 120–124. [Google Scholar] [CrossRef]
- Fu, Z.; Chen, C.; Zhou, Q.; Wang, Y.; Zhao, Y.; Zhao, X.; Li, W.; Zheng, S.; Ye, H.; Wang, L.; et al. LncRNA HOTTIP modulates cancer stem cell properties in human pancreatic cancer by regulating HOXA9. Cancer Lett. 2017, 410, 68–81. [Google Scholar] [CrossRef]
- Chang, S.; Liu, J.; Guo, S.; He, S.; Qiu, G.; Lu, J.; Wang, J.; Fan, L.; Zhao, W.; Che, X. HOTTIP and HOXA13 are oncogenes associated with gastric cancer progression. Oncol. Rep. 2016, 35, 3577–3585. [Google Scholar] [CrossRef]
- Wang, S.S.; Wuputra, K.; Liu, C.J.; Lin, Y.C.; Chen, Y.T.; Chai, C.Y.; Lin, C.S.; Kuo, K.K.; Tsai, M.H.; Wang, S.W.; et al. Oncogenic function of the homeobox A13-long noncoding RNA HOTTIP-insulin growth factor-binding protein 3 axis in human gastric cancer. Oncotarget 2016, 7, 36049–36064. [Google Scholar] [CrossRef] [Green Version]
- Luo, Z.; Rhie, S.K.; Lay, F.D.; Farnham, P.J. A Prostate Cancer Risk Element Functions as a Repressive Loop that Regulates HOXA13. Cell Rep. 2017, 21, 1411–1417. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.R.; Yang, J.K.; Xie, J.K.; Zhao, L.C. Long noncoding RNA HOTTIP contributes to the progression of prostate cancer by regulating HOXA13. Cell Mol. Biol. 2016, 62, 84–88. [Google Scholar]
- Lin, C.; Wang, Y.; Wang, Y.; Zhang, S.; Yu, L.; Guo, C.; Xu, H. Transcriptional and posttranscriptional regulation of HOXA13 by lncRNA HOTTIP facilitates tumorigenesis and metastasis in esophageal squamous carcinoma cells. Oncogene 2017, 36, 5392–5406. [Google Scholar] [CrossRef]
- Sang, Y.; Zhou, F.; Wang, D.; Bi, X.; Liu, X.; Hao, Z.; Li, Q.; Zhang, W. Up-regulation of long non-coding HOTTIP functions as an oncogene by regulating HOXA13 in non-small cell lung cancer. Am. J. Transl. Res. 2016, 8, 2022–2032. [Google Scholar]
- Stelnicki, E.J.; Arbeit, J.; Cass, D.L.; Saner, C.; Harrison, M.; Largman, C. Modulation of the human homeobox genes PRX-2 and HOXB13 in scarless fetal wounds. J. Investig. Dermatol. 1998, 111, 57–63. [Google Scholar] [CrossRef]
- Kömüves, L.G.; Ma, X.K.; Stelnicki, E.; Rozenfeld, S.; Oda, Y.; Largman, C. HOXB13 homeodomain protein is cytoplasmic throughout fetal skin development. Dev. Dyn. 2003, 227, 192–202. [Google Scholar] [CrossRef] [Green Version]
- Economides, K.D.; Capecchi, M.R. Hoxb13 is required for normal differentiation and secretory function of the ventral prostate. Development 2003, 130, 2061–2069. [Google Scholar] [CrossRef] [Green Version]
- Norris, J.D.; Chang, C.Y.; Wittmann, B.M.; Kunder, R.S.; Cui, H.; Fan, D.; Joseph, J.D.; McDonnell, D.P. The homeodomain protein HOXB13 regulates the cellular response to androgens. Mol. Cell 2009, 36, 405–416. [Google Scholar] [CrossRef] [PubMed]
- Cantile, M.; Pettinato, G.; Procino, A.; Feliciello, I.; Cindolo, L.; Cillo, C. In vivo expression of the whole HOX gene network in human breast cancer. Eur. J. Cancer 2003, 39, 257–264. [Google Scholar] [CrossRef]
- Ma, X.J.; Wang, Z.; Ryan, P.D.; Isakoff, S.J.; Barmettler, A.; Fuller, A.; Muir, B.; Mohapatra, G.; Salunga, R.; Tuggle, J.T.; et al. A two-gene expression ratio predicts clinical outcome in breast cancer patients treated with tamoxifen. Cancer Cell 2004, 5, 607–616. [Google Scholar] [CrossRef] [Green Version]
- Goetz, M.P.; Suman, V.J.; Ingle, J.N.; Nibbe, A.M.; Visscher, D.W.; Reynolds, C.A.; Lingle, W.L.; Erlander, M.; Ma, X.J.; Sgroi, D.C.; et al. A two-gene expression ratio of homeobox 13 and interleukin-17B receptor for prediction of recurrence and survival in women receiving adjuvant tamoxifen. Clin. Cancer Res. 2006, 12, 2080–2087. [Google Scholar] [CrossRef]
- Wang, Z.; Dahiya, S.; Provencher, H.; Muir, B.; Carney, E.; Coser, K.; Shioda, T.; Ma, X.J.; Sgroi, D.C. The prognostic biomarkers HOXB13, IL17BR, and CHDH are regulated by estrogen in breast cancer. Clin. Cancer Res. 2007, 13, 6327–6334. [Google Scholar] [CrossRef]
- Jansen, M.P.; Sieuwerts, A.M.; Look, M.P.; Ritstier, K.; Meijer-van Gelder, M.E.; van Staveren, I.L.; Klijn, J.G.; Foekens, J.A.; Berns, E.M. HOXB13-to-IL17BR expression ratio is related with tumor aggressiveness and response to tamoxifen of recurrent breast cancer: A retrospective study. J. Clin. Oncol. 2007, 25, 662–668. [Google Scholar] [CrossRef]
- Jerevall, P.L.; Jansson, A.; Fornander, T.; Skoog, L.; Nordenskjöld, B.; Stål, O. Predictive relevance of HOXB13 protein expression for tamoxifen benefit in breast cancer. Breast Cancer Res. 2010, 12, R53. [Google Scholar] [CrossRef]
- Ma, X.J.; Hilsenbeck, S.G.; Wang, W.; Ding, L.; Sgroi, D.C.; Bender, R.A.; Osborne, C.K.; Allred, D.C.; Erlander, M.G. The HOXB13:IL17BR expression index is a prognostic factor in early-stage breast cancer. J. Clin. Oncol. 2006, 24, 4611–4619. [Google Scholar] [CrossRef]
- Jerevall, P.L.; Brommesson, S.; Strand, C.; Gruvberger-Saal, S.; Malmström, P.; Nordenskjöld, B.; Wingren, S.; Söderkvist, P.; Fernö, M.; Stål, O. Exploring the two-gene ratio in breast cancer--independent roles for HOXB13 and IL17BR in prediction of clinical. Breast Cancer Res. Treat. 2008, 107, 225–234. [Google Scholar] [CrossRef]
- Zhao, L.; Zhu, S.; Gao, Y.; Wang, Y. Two-gene expression ratio as predictor for breast cancer treated with tamoxifen: Evidence from meta-analysis. Tumour Biol. 2014, 35, 3113–3117. [Google Scholar] [CrossRef]
- Ma, X.J.; Salunga, R.; Dahiya, S.; Wang, W.; Carney, E.; Durbecq, V.; Harris, A.; Goss, P.; Sotiriou, C.; Erlander, M.; et al. A five-gene molecular grade index and HOXB13:IL17BR are complementary prognostic factors in early stage breast cancer. Clin. Cancer Res. 2008, 14, 2601–2608. [Google Scholar] [CrossRef]
- Habel, L.A.; Sakoda, L.C.; Achacoso, N.; Ma, X.J.; Erlander, M.G.; Sgroi, D.C.; Fehrenbacher, L.; Greenberg, D.; Quesenberry, C.P., Jr. HOXB13:IL17BR and molecular grade index and risk of breast cancer death among patients with lymph node-negative invasive disease. Breast Cancer Res. 2013, 15, R24. [Google Scholar] [CrossRef]
- Sgroi, D.C.; Chapman, J.A.; Badovinac-Crnjevic, T.; Zarella, E.; Binns, S.; Zhang, Y.; Schnabel, C.A.; Erlander, M.G.; Pritchard, K.I.; Han, L.; et al. Assessment of the prognostic and predictive utility of the Breast Cancer Index (BCI): An NCIC CTG MA.14 study. Breast Cancer Res. 2016, 18, 1. [Google Scholar] [CrossRef]
- Rodriguez, B.A.; Cheng, A.S.; Yan, P.S.; Potter, D.; Agosto-Perez, F.J.; Shapiro, C.L.; Huang, T.H. Epigenetic repression of the estrogen-regulated Homeobox B13 gene in breast cancer. Carcinogenesis 2008, 29, 1459–1465. [Google Scholar] [CrossRef] [Green Version]
- Shah, N.; Jin, K.; Cruz, L.A.; Park, S.; Sadik, H.; Cho, S.; Goswami, C.P.; Nakshatri, H.; Gupta, R.; Chang, H.Y.; et al. HOXB13 mediates tamoxifen resistance and invasiveness in human breast cancer by suppressing ERα and inducing IL-6 expression. Cancer Res. 2013, 73, 5449–5458. [Google Scholar] [CrossRef]
- Liu, B.; Wang, T.; Wang, H.; Zhang, L.; Xu, F.; Fang, R.; Li, L.; Cai, X.; Wu, Y.; Zhang, W.; et al. Oncoprotein HBXIP enhances HOXB13 acetylation and co-activates HOXB13 to confer tamoxifen resistance in breast cancer. J. Hematol. Oncol. 2018, 11, 26. [Google Scholar] [CrossRef] [Green Version]
- Sreenath, T.; Orosz, A.; Fujita, K.; Bieberich, C.J. Androgen-independent expression of hoxb-13 in the mouse prostate. Prostate 1999, 41, 203–207. [Google Scholar] [CrossRef]
- Jung, C.; Kim, R.S.; Lee, S.J.; Wang, C.; Jeng, M.H. HOXB13 homeodomain protein suppresses the growth of prostate cancer cells by the negative regulation of T-cell factor 4. Cancer Res. 2004, 64, 3046–3051. [Google Scholar] [CrossRef]
- Jung, C.; Kim, R.S.; Zhang, H.J.; Lee, S.J.; Jeng, M.H. HOXB13 induces growth suppression of prostate cancer cells as a repressor of hormone-activated androgen receptor signaling. Cancer Res. 2004, 64, 9185–9192. [Google Scholar] [CrossRef]
- Kim, Y.R.; Oh, K.J.; Park, R.Y.; Xuan, N.T.; Kang, T.W.; Kwon, D.D.; Choi, C.; Kim, M.S.; Nam, K.I.; Ahn, K.Y.; et al. HOXB13 promotes androgen independent growth of LNCaP prostate cancer cells by the activation of E2F signaling. Mol. Cancer 2010, 9, 124. [Google Scholar] [CrossRef]
- Jeong, T.O.; Oh, K.J.; Nguyen, X.; Thi, N.; Kim, Y.R.; Kim, M.S.; Lee, S.D.; Ryu, S.B.; Jung, C. Evaluation of HOXB13 as a molecular marker of recurrent prostate cancer. Mol. Med. Rep. 2012, 5, 901–904. [Google Scholar] [CrossRef] [Green Version]
- Varinot, J.; Cussenot, O.; Roupret, M.; Conort, P.; Bitker, M.O.; Chartier-Kastler, E.; Cheng, L.; Compérat, E. HOXB13 is a sensitive and specific marker of prostate cells, useful in distinguishing between carcinomas of prostatic and urothelial origin. Virchows Arch. 2013, 463, 803–809. [Google Scholar] [CrossRef]
- Barresi, V.; Ieni, A.; Cardia, R.; Licata, L.; Vitarelli, E.; Reggiani Bonetti, L.; Tuccari, G. HOXB13 as an immunohistochemical marker of prostatic origin in metastatic tumors. APMIS 2016, 124, 188–193. [Google Scholar] [CrossRef]
- Larnaudie, L.; Compérat, E.; Conort, P.; Varinot, J. HOXB13 a useful marker in pleomorphic giant cell adenocarcinoma of the prostate: A case report and review of the literature. Virchows Arch. 2017, 471, 133–136. [Google Scholar] [CrossRef]
- Zabalza, C.V.; Adam, M.; Burdelski, C.; Wilczak, W.; Wittmer, C.; Kraft, S.; Krech, T.; Steurer, S.; Koop, C.; Hube-Magg, C.; et al. HOXB13 overexpression is an independent predictor of early PSA recurrence in prostate cancer treated by radical prostatectomy. Oncotarget 2015, 6, 12822–12834. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, I.J.; Kang, T.W.; Jeong, T.; Kim, Y.R.; Jung, C. HOXB13 regulates the prostate-derived Ets factor: Implications for prostate cancer cell invasion. Int. J. Oncol. 2014, 45, 869–876. [Google Scholar] [CrossRef]
- Kim, Y.R.; Kang, T.W.; To, P.K.; Xuan Nguyen, N.T.; Cho, Y.S.; Jung, C.; Kim, M.S. HOXB13-mediated suppression of p21WAF1/CIP1 regulates JNK/c-Jun signaling in prostate cancer cells. Oncol. Rep. 2016, 35, 2011–2016. [Google Scholar] [CrossRef]
- Kim, Y.R.; Kim, I.J.; Kang, T.W.; Choi, C.; Kim, K.K.; Kim, M.S.; Nam, K.I.; Jung, C. HOXB13 downregulates intracellular zinc and increases NF-κB signaling to promote prostate cancer metastasis. Oncogene 2014, 33, 4558–4567. [Google Scholar] [CrossRef]
- Johng, D.; Torga, G.; Ewing, C.M.; Jin, K.; Norris, J.D.; McDonnell, D.P.; Isaacs, W.B. HOXB13 interaction with MEIS1 modifies proliferation and gene expression in prostate cancer. Prostate 2019, 79, 414–424. [Google Scholar] [CrossRef] [PubMed]
- Ewing, C.M.; Ray, A.M.; Lange, E.M.; Zuhlke, K.A.; Robbins, C.M.; Tembe, W.D.; Wiley, K.E.; Isaacs, S.D.; Johng, D.; Wang, Y.; et al. Germline mutations in HOXB13 and prostate-cancer risk. N. Engl. J. Med. 2012, 366, 141–149. [Google Scholar] [CrossRef] [PubMed]
- Breyer, J.P.; Avritt, T.G.; McReynolds, K.M.; Dupont, W.D.; Smith, J.R. Confirmation of the HOXB13 G84E germline mutation in familial prostate cancer. Cancer Epidemiol. Biomark. Prev. 2012, 21, 1348–1353. [Google Scholar] [CrossRef]
- Karlsson, R.; Aly, M.; Clements, M.; Zheng, L.; Adolfsson, J.; Xu, J.; Grönberg, H.; Wiklund, F. A population-based assessment of germline HOXB13 G84E mutation and prostate cancer risk. Eur. Urol. 2014, 65, 169–176. [Google Scholar] [CrossRef]
- Lin, X.; Qu, L.; Chen, Z.; Xu, C.; Ye, D.; Shao, Q.; Wang, X.; Qi, J.; Chen, Z.; Zhou, F.; et al. A novel germline mutation in HOXB13 is associated with prostate cancer risk in Chinese men. Prostate 2013, 73, 169–175. [Google Scholar] [CrossRef]
- Akbari, M.R.; Trachtenberg, J.; Lee, J.; Tam, S.; Bristow, R.; Loblaw, A.; Narod, S.A.; Nam, R.K. Association between germline HOXB13 G84E mutation and risk of prostate cancer. J. Natl. Cancer Inst. 2012, 104, 1260–1262. [Google Scholar] [CrossRef]
- Xu, J.; Lange, E.M.; Lu, L.; Zheng, S.L.; Wang, Z.; Thibodeau, S.N.; Cannon-Albright, L.A.; Teerlink, C.C.; Camp, N.J.; Johnson, A.M.; et al. International Consortium for Prostate Cancer Genetics.HOXB13 is a susceptibility gene for prostate cancer: Results from the International Consortium for Prostate Cancer Genetics (ICPCG). Hum. Genet. 2013, 132, 5–14. [Google Scholar] [CrossRef]
- Stott-Miller, M.; Karyadi, D.M.; Smith, T.; Kwon, E.M.; Kolb, S.; Stanford, J.L.; Ostrander, E.A. HOXB13 mutations in a population-based, case-control study of prostate cancer. Prostate 2013, 73, 634–641. [Google Scholar] [CrossRef]
- Smith, S.C.; Palanisamy, N.; Zuhlke, K.A.; Johnson, A.M.; Siddiqui, J.; Chinnaiyan, A.M.; Kunju, L.P.; Cooney, K.A.; Tomlins, S.A. HOXB13 G84E-related familial prostate cancers: A clinical, histologic, and molecular survey. Am. J. Surg. Pathol. 2014, 38, 615–626. [Google Scholar] [CrossRef]
- Maia, S.; Cardoso, M.; Pinto, P.; Pinheiro, M.; Santos, C.; Peixoto, A.; Bento, M.J.; Oliveira, J.; Henrique, R.; Jerónimo, C.; et al. Identification of Two Novel HOXB13 Germline Mutations in Portuguese Prostate Cancer Patients. PLoS ONE 2015, 10, e0132728. [Google Scholar] [CrossRef] [PubMed]
- Jung, C.; Kim, R.S.; Zhang, H.; Lee, S.J.; Sheng, H.; Loehrer, P.J.; Gardner, T.A.; Jeng, M.H.; Kao, C. HOXB13 is downregulated in colorectal cancer to confer TCF4-mediated transactivation. Br. J. Cancer 2005, 92, 2233–2239. [Google Scholar] [CrossRef] [Green Version]
- Ghoshal, K.; Motiwala, T.; Claus, R.; Yan, P.; Kutay, H.; Datta, J.; Majumder, S.; Bai, S.; Majumder, A.; Huang, T.; et al. HOXB13, a target of DNMT3B, is methylated at an upstream CpG island, and functions as a tumor suppressor in primary colorectal tumors. PLoS ONE 2010, 5, e10338. [Google Scholar] [CrossRef] [PubMed]
- Tatangelo, F.; Di Mauro, A.; Scognamiglio, G.; Aquino, G.; Lettiero, A.; Delrio, P.; Avallone, A.; Cantile, M.; Botti, G. Posterior HOX genes and HOTAIR expression in the proximal and distal colon cancer pathogenesis. J. Transl. Med. 2018, 16, 350. [Google Scholar] [CrossRef]
- Akbari, M.R.; Anderson, L.N.; Buchanan, D.D.; Clendenning, M.; Jenkins, M.A.; Win, A.K.; Hopper, J.L.; Giles, G.G.; Nam, R.; Narod, S.; et al. Germline HOXB13 p. Gly84Glu mutation and risk of colorectal cancer. Cancer Epidemiol. 2013, 37, 424–427. [Google Scholar] [CrossRef]
- Yuan, H.; Kajiyama, H.; Ito, S.; Chen, D.; Shibata, K.; Hamaguchi, M.; Kikkawa, F.; Senga, T. HOXB13 promotes ovarian cancer progression, HOXB13 and ALX4 induce SLUG expression for the promotion of EMT and cell invasion in ovarian cancer cells. Oncotarget 2015, 6, 13359–13370. [Google Scholar] [CrossRef] [PubMed]
- Saha, S.S.; Chowdhury, R.R.; Mondal, N.R.; Roy, S.; Sengupta, S. Expression signatures of HOX cluster genes in cervical cancer pathogenesis: Impact of human papillomavirus type 16 oncoprotein E7. Oncotarget 2017, 8, 36591–36602. [Google Scholar] [CrossRef] [Green Version]
- Zhao, Y.; Yamashita, T.; Ishikawa, M. Regulation of tumor invasion by HOXB13 gene overexpressed in human endometrial cancer. Oncol. Rep. 2005, 13, 721–726. [Google Scholar] [CrossRef]
- Marra, L.; Cantile, M.; Scognamiglio, G.; Perdonà, S.; La Mantia, E.; Cerrone, M.; Gigantino, V.; Cillo, C.; Caraglia, M.; Pignata, S.; et al. Deregulation of HOX B13 expression in urinary bladder cancer progression. Curr. Med. Chem. 2013, 20, 833–839. [Google Scholar]
- Okuda, H.; Toyota, M.; Ishida, W.; Furihata, M.; Tsuchiya, M.; Kamada, M.; Tokino, T.; Shuin, T. Epigenetic inactivation of the candidate tumor suppressor gene HOXB13 in human renal cell carcinoma. Oncogene 2006, 25, 1733–1742. [Google Scholar] [CrossRef]
- Sui, B.Q.; Zhang, C.D.; Liu, J.C.; Wang, L.; Dai, D.Q. HOXB13 expression and promoter methylation as a candidate biomarker in gastric cancer. Oncol. Lett. 2018, 15, 8833–8840. [Google Scholar] [CrossRef] [Green Version]
- Zhu, J.Y.; Sun, Q.K.; Wang, W.; Jia, W.D. High-level expression of HOXB13 is closely associated with tumor angiogenesis and poor prognosis of hepatocellular carcinoma. Int. J. Clin. Exp. Pathol. 2014, 7, 2925–2933. [Google Scholar]
- Zhang, E.; Han, L.; Yin, D.; He, X.; Hong, L.; Si, X.; Qiu, M.; Xu, T.; De, W.; Xu, L.; et al. H3K27 acetylation activated-long non-coding RNA CCAT1 affects cell proliferation and migration by regulating SPRY4 and HOXB13 expression in esophageal squamous cell carcinoma. Nucleic Acids Res. 2017, 45, 3086–3101. [Google Scholar] [CrossRef]
- Cazal, C.; Sobral, A.P.; de Almeida, F.C.; das Graças Silva-Valenzuela, M.; Durazzo, M.D.; Nunes, F.D. The homeobox HOXB13 is expressed in human minor salivary gland. Oral Dis. 2006, 12, 424–427. [Google Scholar] [CrossRef]
- Xiong, Y.; Kuang, W.; Lu, S.; Guo, H.; Wu, M.; Ye, M.; Wu, L. Long noncoding RNA HOXB13-AS1 regulates HOXB13 gene methylation by interacting with EZH2 in glioma. Cancer Med. 2018, 7, 4718–4728. [Google Scholar] [CrossRef]
- Liu, X.F.; Olsson, P.; Wolfgang, C.D.; Bera, T.K.; Duray, P.; Lee, B.; Pastan, I. PRAC: A novel small nuclear protein that is specifically expressed in human prostate and colon. Prostate 2001, 47, 125–131. [Google Scholar] [CrossRef]
- Olsson, P.; Motegi, A.; Bera, T.K.; Lee, B.; Pastan, I. PRAC2: A new gene expressed in human prostate and prostate cancer. Prostate 2003, 56, 123–130. [Google Scholar] [CrossRef]
- Jiang, R.; Zhao, C.; Gao, B.; Shao, N.; Wang, S.; Song, W. IL-22 promotes the progression of breast cancer through regulating HOXB-AS5. Oncotarget 2017, 8, 103601–103612. [Google Scholar] [Green Version]
- Jonker, L.; Kist, R.; Aw, A.; Wappler, I.; Peters, H. Pax9 is required for filiform papilla development and suppresses skin-specific differentiation of the mammalian tongue epithelium. Mech. Dev. 2004, 121, 1313–1322. [Google Scholar] [CrossRef]
- Godwin, A.R.; Capecchi, M.R. Hoxc13 mutant mice lack external hair. Genes Dev. 1998, 12, 11–20. [Google Scholar] [CrossRef] [Green Version]
- Magli, M.C.; Barba, P.; Celetti, A.; De Vita, G.; Cillo, C.; Boncinelli, E. Coordinate regulation of HOX genes in human hematopoietic cells. Proc. Natl. Acad. Sci. USA 1991, 88, 6348–6352. [Google Scholar] [CrossRef]
- Jave-Suarez, L.F.; Winter, H.; Langbein, L.; Rogers, M.A.; Schweizer, J. HOXC13 is involved in the regulation of human hair keratin gene expression. J. Biol. Chem. 2002, 277, 3718–3726. [Google Scholar] [CrossRef]
- Jave-Suárez, L.F.; Schweizer, J. The HOXC13-controlled expression of early hair keratin genes in the human hair follicle does not involve TALE proteins MEIS and PREP as cofactors. Arch. Dermatol. Res. 2006, 297, 372–376. [Google Scholar] [CrossRef]
- Luan, L.; Shi, J.; Yu, Z.; Andl, T. The major miR-31 target genes STK40 and LATS2 and their implications in the regulation of keratinocyte growth and hair differentiation. Exp. Dermatol. 2017, 26, 497–504. [Google Scholar] [CrossRef] [PubMed]
- Cribier, B.; Peltre, B.; Grosshans, E.; Langbein, L.; Schweizer, J. On the regulation of hair keratin expression: Lessons from studies in pilomatricomas. J. Investig. Dermatol. 2004, 122, 1078–1083. [Google Scholar] [CrossRef] [PubMed]
- Cribier, B.; Worret, W.I.; Braun-Falco, M.; Peltre, B.; Langbein, L.; Schweizer, J. Expression patterns of hair and epithelial keratins and transcription factors HOXC13, LEF1, and beta-catenin in a malignant pilomatricoma: A histological and immunohistochemical study. J. Cutan Pathol. 2006, 33, 1–9. [Google Scholar] [CrossRef]
- Yoon, S.J.; LeBlanc-Straceski, J.; Ward, D.; Krauter, K.; Kucherlapati, R. Organization of the human keratin type II gene cluster at 12q13. Genomics 1994, 24, 502–508. [Google Scholar] [CrossRef]
- Godwin, A.R.; Capecchi, M.R. Hair defects in Hoxc13 mutant mice. J. Investig. Dermatol. Symp. Proc. 1999, 4, 244–247. [Google Scholar] [CrossRef]
- Kasiri, S.; Ansari, K.I.; Hussain, I.; Bhan, A.; Mandal, S.S. Antisense oligonucleotide mediated knockdown of HOXC13 affects cell growth and induces apoptosis in tumor cells and over expression of HOXC13 induces 3D-colony formation. RSC Adv. 2013, 3, 3260–3269. [Google Scholar] [CrossRef]
- Cillo, C.; Cantile, M.; Mortarini, R.; Barba, P.; Parmiani, G.; Anichini, A. Differential patterns of HOX gene expression are associated with specific integrin and ICAM profiles in clonal populations isolated from a single human melanoma metastasis. Int. J. Cancer 1996, 66, 692–697. [Google Scholar] [CrossRef]
- Maeda, K.; Hamada, J.; Takahashi, Y.; Tada, M.; Yamamoto, Y.; Sugihara, T.; Moriuchi, T. Altered expressions of HOX genes in human cutaneous malignant melanoma. Int. J. Cancer 2005, 114, 436–441. [Google Scholar] [CrossRef] [PubMed]
- Panagopoulos, I.; Isaksson, M.; Billström, R.; Strömbeck, B.; Mitelman, F.; Johansson, B. Fusion of the NUP98 gene and the homeobox gene HOXC13 in acute myeloid leukemia with t(11;12)(p15;q13). Genes Chromosomes Cancer 2003, 36, 107–112. [Google Scholar] [CrossRef] [PubMed]
- La Starza, R.; Trubia, M.; Crescenzi, B.; Matteucci, C.; Negrini, M.; Martelli, M.F.; Pelicci, P.G.; Mecucci, C. Human homeobox gene HOXC13 is the partner of NUP98 in adult acute myeloid leukemia with t(11;12)(p15;q13). Genes Chromosomes Cancer 2003, 36, 420–423. [Google Scholar] [CrossRef]
- Kobzev, Y.N.; Martinez-Climent, J.; Lee, S.; Chen, J.; Rowley, J.D. Analysis of translocations that involve the NUP98 gene in patients with 11p15 chromosomal rearrangements. Genes Chromosomes Cancer 2004, 41, 339–352. [Google Scholar] [CrossRef] [PubMed]
- Tosić, N.; Stojiljković, M.; Colović, N.; Colović, M.; Pavlović, S. Acute myeloid leukemia with NUP98-HOXC13 fusion and FLT3 internal tandem duplication mutation: Case report and literature review. Cancer Genet. Cytogenet. 2009, 193, 98–103. [Google Scholar] [CrossRef] [PubMed]
- La Starza, R.; Brandimarte, L.; Pierini, V.; Nofrini, V.; Gorello, P.; Crescenzi, B.; Berchicci, L.; Matteucci, C.; Romoli, S.; Beacci, D.; et al. A NUP98-positive acute myeloid leukemia with a t(11;12)(p15;q13) without HOXC cluster gene involvement. Cancer Genet. Cytogenet. 2009, 193, 109–111. [Google Scholar] [CrossRef]
- Yamada, T.; Shimizu, T.; Suzuki, M.; Kihara-Negishi, F.; Nanashima, N.; Sakurai, T.; Fan, Y.; Akita, M.; Oikawa, T.; Tsuchida, S. Interaction between the homeodomain protein HOXC13 and ETS family transcription factor PU.1 and its implication in the differentiation of murine erythroleukemia cells. Exp. Cell Res. 2008, 314, 847–858. [Google Scholar] [CrossRef]
- Zhong, M.; Wang, J.; Gong, Y.B.; Li, J.C.; Zhang, B.; Hou, L. Expression of HOXC13 in ameloblastoma. Chin. J. Stomatol. 2007, 42, 43–46. [Google Scholar]
- Schiavo, G.; D’Antò, V.; Cantile, M.; Procino, A.; Di Giovanni, S.; Valletta, R.; Terracciano, L.; Baumhoer, D.; Jundt, G.; Cillo, C. Deregulated HOX genes in ameloblastomas are located in physical contiguity to keratin genes. J. Cell Biochem. 2011, 112, 3206–3215. [Google Scholar] [CrossRef] [PubMed]
- Hong, Y.S.; Wang, J.; Liu, J.; Zhang, B.; Hou, L.; Zhong, M. Expression of HOX C13 in odontogenic tumors. Shanghai J. Stomatol. 2007, 16, 587–591. [Google Scholar]
- Marcinkiewicz, K.M.; Gudas, L.J. Altered epigenetic regulation of homeobox genes in human oral squamous cell carcinoma cells. Exp. Cell Res. 2014, 320, 128–143. [Google Scholar] [CrossRef]
- Marcinkiewicz, K.M.; Gudas, L.J. Altered histone mark deposition and DNA methylation at homeobox genes in human oral squamous cell carcinoma. J. Cell. Physiol. 2014, 229, 1405–1416. [Google Scholar] [CrossRef]
- Luo, J.; Wang, Z.; Huang, J.; Yao, Y.; Sun, Q.; Wang, J.; Shen, Y.; Xu, L.; Ren, B. HOXC13 promotes proliferation of esophageal squamous cell carcinoma via repressing transcription of CASP3. Cancer Sci. 2018, 109, 317–329. [Google Scholar] [CrossRef]
- Chen, F.; Li, Y.; Wang, L.; Hu, L. Knockdown of BMI-1 causes cell-cycle arrest and derepresses p16INK4a, HOXA9 and HOXC13 mRNA expression in HeLa cells. Med. Oncol. 2011, 28, 1201–1209. [Google Scholar] [CrossRef]
- Cantile, M.; Galletta, F.; Franco, R.; Aquino, G.; Scognamiglio, G.; Marra, L.; Cerrone, M.; Malzone, G.; Manna, A.; Apice, G.; et al. Hyperexpression of HOXC13, located in the 12q13 chromosomal region, in well-differentiated and dedifferentiated human liposarcomas. Oncol. Rep. 2013, 30, 2579–2586. [Google Scholar] [CrossRef] [Green Version]
- Komisarof, J.; McCall, M.; Newman, L.; Bshara, W.; Mohler, J.L.; Morrison, C.; Land, H. A four gene signature predictive of recurrent prostate cancer. Oncotarget 2017, 8, 3430–3440. [Google Scholar] [CrossRef]
- Yao, Y.; Luo, J.; Sun, Q.; Xu, T.; Sun, S.; Chen, M.; Lin, X.; Qian, Q.; Zhang, Y.; Cao, L.; et al. HOXC13 promotes proliferation of lung adenocarcinoma via modulation of CCND1 and CCNE1. Am. J. Cancer Res. 2017, 7, 1820–1834. [Google Scholar]
- Alvarado, D.M.; McCall, K.; Hecht, J.T.; Dobbs, M.B.; Gurnett, C.A. Deletions of 5′ HOXC genes are associated with lower extremity malformations including clubfoot and vertical talus. J. Med. Genet. 2016, 53, 250–255. [Google Scholar] [CrossRef]
- Gao, C.; Lu, W.; Lou, W.; Wang, L.; Xu, Q. Long noncoding RNA HOXC13-AS positively affects cell proliferation and invasion in nasopharyngeal carcinoma via modulating miR-383-3p/HMGA2 axis. J. Cell. Physiol. 2018, 234, 12809–12820. [Google Scholar] [CrossRef]
- Rinn, J.L.; Kertesz, M.; Wang, J.K.; Squazzo, S.L.; Xu, X.; Brugmann, S.A.; Goodnough, L.H.; Helms, J.A.; Farnham, P.J.; Segal, E.; et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell 2007, 129, 1311–1323. [Google Scholar] [CrossRef]
- Li, L.; Liu, B.; Wapinski, O.L.; Tsai, M.C.; Qu, K.; Zhang, J.; Carlson, J.C.; Lin, M.; Fang, F.; Gupta, R.A.; et al. Targeted disruption of Hotair leads to homeotic transformation and gene derepression. Cell Rep. 2013, 5, 3–12. [Google Scholar] [CrossRef]
- Tang, Q.; Hann, S.S. HOTAIR: An Oncogenic Long Non-Coding RNA in Human Cancer. Cell Physiol. Biochem. 2018, 47, 893–913. [Google Scholar] [CrossRef]
- Botti, G.; Marra, L.; Malzone, M.G.; Anniciello, A.; Botti, C.; Franco, R.; Cantile, M. LncRNA HOTAIR as Prognostic Circulating Marker and Potential Therapeutic Target in Patients with Tumor Diseases. Curr. Drug Targets 2017, 18, 27–34. [Google Scholar] [CrossRef]
- Davis, A.P.; Capecchi, M.R. A mutational analysis of the 5′ HoxD genes: Dissection of genetic interactions during limb development in the mouse. Development 1996, 122, 1175–1185. [Google Scholar]
- Fromental-Ramain, C.; Warot, X.; Messadecq, N.; LeMeur, M.; Dollé, P.; Chambon, P. Hoxa-13 and Hoxd-13 play a crucial role in the patterning of the limb autopod. Development 1996, 122, 2997–3011. [Google Scholar]
- Zákány, J.; Duboule, D. Hox genes in digit development and evolution. Cell Tissue Res. 1999, 296, 19–25. [Google Scholar] [CrossRef]
- Muragaki, Y.; Mundlos, S.; Upton, J.; Olsen, B.R. Altered growth and branching patterns in synpolydactyly caused by mutations in HOXD13. Science 1996, 272, 548–551. [Google Scholar] [CrossRef]
- Raza-Egilmez, S.Z.; Jani-Sait, S.N.; Grossi, M.; Higgins, M.J.; Shows, T.B.; Aplan, P.D. NUP98-HOXD13 gene fusion in therapy-related acute myelogenous leukemia. Cancer Res. 1998, 58, 4269–4273. [Google Scholar]
- Pineault, N.; Buske, C.; Feuring-Buske, M.; Abramovich, C.; Rosten, P.; Hogge, D.E.; Aplan, P.D.; Humphries, R.K. Induction of acute myeloid leukemia in mice by the human leukemia-specific fusion gene NUP98-HOXD13 in concert with Meis1. Blood 2003, 101, 4529–4538. [Google Scholar] [CrossRef] [Green Version]
- Slape, C.; Hartung, H.; Lin, Y.W.; Bies, J.; Wolff, L.; Aplan, P.D. Retroviral insertional mutagenesis identifies genes that collaborate with NUP98-HOXD13 during leukemic transformation. Cancer Res. 2007, 67, 5148–5155. [Google Scholar] [CrossRef]
- Imren, S.; Heuser, M.; Gasparetto, M.; Beer, P.A.; Norddahl, G.L.; Xiang, P.; Chen, L.; Berg, T.; Rhyasen, G.W.; Rosten, P.; et al. Modeling de novo leukemogenesis from human cord blood with MN1 and NUP98HOXD13. Blood 2014, 124, 3608–3612. [Google Scholar] [CrossRef] [Green Version]
- Monlish, D.A.; Bhatt, S.T.; Duncavage, E.J.; Greenberg, Z.J.; Keller, J.L.; Romine, M.P.; Yang, W.; Aplan, P.D.; Walter, M.J.; Schuettpelz, L.G. Loss of Toll-like receptor 2 results in accelerated leukemogenesis in the NUP98-HOXD13 mouse model of MDS. Blood 2018, 131, 1032–1035. [Google Scholar] [CrossRef]
- Slape, C.; Lin, Y.W.; Hartung, H.; Zhang, Z.; Wolff, L.; Aplan, P.D. NUP98-HOX translocations lead to myelodysplastic syndrome in mice and men. J. Natl. Cancer Inst. Monogr. 2008, 39, 64–68. [Google Scholar] [CrossRef]
- Lin, Y.W.; Slape, C.; Zhang, Z.; Aplan, P.D. NUP98-HOXD13 transgenic mice develop a highly penetrant, severe myelodysplastic syndrome that progresses to acute leukemia. Blood 2005, 106, 287–295. [Google Scholar] [CrossRef] [Green Version]
- Greenblatt, S.; Li, L.; Slape, C.; Nguyen, B.; Novak, R.; Duffield, A.; Huso, D.; Desiderio, S.; Borowitz, M.J.; Aplan, P.; et al. Knock-in of a FLT3/ITD mutation cooperates with a NUP98-HOXD13 fusion to generate acute myeloid leukemia in a mouse model. Blood 2012, 119, 2883–2894. [Google Scholar] [CrossRef] [Green Version]
- Humeniuk, R.; Koller, R.; Bies, J.; Aplan, P.; Wolff, L. Brief report: Loss of p15Ink4b accelerates development of myeloid neoplasms in Nup98-HoxD13 transgenic mice. Stem Cells 2014, 32, 1361–1366. [Google Scholar] [CrossRef]
- Slape, C.; Liu, L.Y.; Beachy, S.; Aplan, P.D. Leukemic transformation in mice expressing a NUP98-HOXD13 transgene is accompanied by spontaneous mutations in Nras, Kras, and Cbl. Blood 2008, 112, 2017–2019. [Google Scholar] [CrossRef] [Green Version]
- Xu, H.; Menendez, S.; Schlegelberger, B.; Bae, N.; Aplan, P.D.; Göhring, G.; Deblasio, T.R.; Nimer, S.D. Loss of p53 accelerates the complications of myelodysplastic syndrome in a NUP98-HOXD13-driven mouse model. Blood 2012, 120, 3089–3097. [Google Scholar] [CrossRef]
- Zhong, Z.B.; Shan, M.; Qian, C.; Liu, T.; Shi, Q.Y.; Wang, J.; Liu, Y.; Liu, Y.; Huang, Y.X.; Pang, D. Prognostic significance of HOXD13 expression in human breast cancer. Int. J. Clin. Exp. Pathol. 2015, 8, 11407–11413. [Google Scholar]
- Ke, D.; Yang, R.; Jing, L. Combined diagnosis of breast cancer in the early stage by MRI and detection of gene expression. Exp. Ther. Med. 2018, 16, 467–472. [Google Scholar] [CrossRef]
- Zhong, Z.; Shan, M.; Wang, J.; Liu, T.; Xia, B.; Niu, M.; Ren, Y.; Pang, D. HOXD13 methylation status is a prognostic indicator in breast cancer. Int. J. Clin. Exp. Pathol. 2015, 8, 10716–10724. [Google Scholar]
- Shan, M.; Yin, H.; Li, J.; Li, X.; Wang, D.; Su, Y.; Niu, M.; Zhong, Z.; Wang, J.; Zhang, X.; et al. Detection of aberrant methylation of a six-gene panel in serum DNA for diagnosis of breast cancer. Oncotarget 2016, 7, 18485–18494. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Han, L.; Xu, G.; Xu, C.; Liu, B.; Liu, D. Potential prognostic biomarkers identified by DNA methylation profiling analysis for patients with lung adenocarcinoma. Oncol. Lett. 2018, 15, 3552–3557. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- von Heyking, K.; Roth, L.; Ertl, M.; Schmidt, O.; Calzada-Wack, J.; Neff, F.; Lawlor, E.R.; Burdach, S.; Richter, G.H. The posterior HOXD locus: Its contribution to phenotype and malignancy of Ewing sarcoma. Oncotarget 2016, 7, 41767–41780. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumon, K.; Kobayashi, H.; Namiki, T.; Tsunematsu, Y.; Miyauchi, J.; Kikuta, A.; Horikoshi, Y.; Komada, Y.; Hatae, Y.; Eguchi, H.; et al. Frequent increase of DNA copy number in the 2q24 chromosomal region and its association with a poor clinical outcome in hepatoblastoma: Cytogenetic and comparative genomic hybridization analysis. Jpn. J. Cancer Res. 2001, 92, 854–862. [Google Scholar] [CrossRef]
- Delpretti, S.; Montavon, T.; Leleu, M.; Joye, E.; Tzika, A.; Milinkovitch, M.; Duboule, D. Multiple enhancers regulate Hoxd genes and the Hotdog LncRNA during cecum budding. Cell Rep. 2013, 5, 137–150. [Google Scholar] [CrossRef]
- Shah, N.; Sukumar, S. The Hox genes and their roles in oncogenesis. Nat. Rev. Cancer 2010, 10, 361–371. [Google Scholar] [CrossRef]
- Alharbi, R.A.; Pettengell, R.; Pandha, H.S.; Morgan, R. The role of HOX genes in normal hematopoiesis and acute leukemia. Leukemia 2013, 27, 1000–1008. [Google Scholar] [CrossRef]
- Cantile, M.; Franco, R.; Schiavo, G.; Procino, A.; Cindolo, L.; Botti, G.; Cillo, C. The HOX Genes Network in Uro-Genital Cancers: Mechanisms and Potential Therapeutic Implications. Curr. Med. Chem. 2011, 18, 4872–4884. [Google Scholar] [CrossRef]
- Plowright, L.; Harrington, K.J.; Pandha, H.S.; Morgan, R. HOX transcription factors are potential therapeutic targets in non-small-cell lung cancer. Br. J. Cancer 2009, 100, 470–475. [Google Scholar] [CrossRef]
- Morgan, R.; El-Tanani, M.; Hunter, K.D.; Harrington, K.J.; Pandha, H.S. Targeting HOX/PBX dimers in cancer. Oncotarget 2017, 8, 32322–32331. [Google Scholar] [CrossRef] [Green Version]
- Li, Z.; Zhao, X.; Zhou, Y.; Liu, Y.; Zhou, Q.; Ye, H.; Wang, Y.; Zeng, J.; Song, Y.; Gao, W.; et al. The long non-coding RNA HOTTIP promotes progression and gemcitabine resistance by regulating HOXA13 in pancreatic cancer. J. Transl. Med. 2015, 13, 84. [Google Scholar] [CrossRef]
- Morgan, R.; El-Tanani, M. HOX Genes as Potential Markers of Circulating Tumour Cells. Curr. Mol. Med. 2016, 16, 322–327. [Google Scholar] [CrossRef]
- Botti, G.; Cantile, M. Circulating long non-coding RNAs: Could they be a useful tool for cancer therapy monitoring? Expert Rev. Anticancer Ther. 2018, 18, 1167–1168. [Google Scholar] [CrossRef]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Botti, G.; Cillo, C.; De Cecio, R.; Malzone, M.G.; Cantile, M. Paralogous HOX13 Genes in Human Cancers. Cancers 2019, 11, 699. https://doi.org/10.3390/cancers11050699
Botti G, Cillo C, De Cecio R, Malzone MG, Cantile M. Paralogous HOX13 Genes in Human Cancers. Cancers. 2019; 11(5):699. https://doi.org/10.3390/cancers11050699
Chicago/Turabian StyleBotti, Gerardo, Clemente Cillo, Rossella De Cecio, Maria Gabriella Malzone, and Monica Cantile. 2019. "Paralogous HOX13 Genes in Human Cancers" Cancers 11, no. 5: 699. https://doi.org/10.3390/cancers11050699
APA StyleBotti, G., Cillo, C., De Cecio, R., Malzone, M. G., & Cantile, M. (2019). Paralogous HOX13 Genes in Human Cancers. Cancers, 11(5), 699. https://doi.org/10.3390/cancers11050699




