Abstract
Expression of DNA repair genes was studied in uveal melanoma (UM) in order to identify genes that may play a role in metastases formation. We searched for genes that are differentially expressed between tumors with a favorable and unfavorable prognosis. Gene-expression profiling was performed on 64 primary UM from the Leiden University Medical Center (LUMC), Leiden, The Netherlands. The expression of 121 genes encoding proteins involved in DNA repair pathways was analyzed: a total of 44 genes differed between disomy 3 and monosomy 3 tumors. Results were validated in a cohort from Genoa and Paris and the The Cancer Genome Atlas (TCGA) cohort. Expression of the PRKDC, WDR48, XPC, and BAP1 genes was significantly associated with clinical outcome after validation. PRKDC was highly expressed in metastasizing UM (p < 0.001), whereas WDR48, XPC, and BAP1 were lowly expressed (p < 0.001, p = 0.006, p = 0.003, respectively). Low expression of WDR48 and XPC was related to a large tumor diameter (p = 0.01 and p = 0.004, respectively), and a mixed/epithelioid cell type (p = 0.007 and p = 0.03, respectively). We conclude that the expression of WDR48, XPC, and BAP1 is significantly lower in UM with an unfavorable prognosis, while these tumors have a significantly higher expression of PRKDC. Pharmacological inhibition of DNA-PKcs resulted in decreased survival of UM cells. PRKDC may be involved in proliferation, invasion and metastasis of UM cells. Unraveling the role of DNA repair genes may enhance our understanding of UM biology and result in the identification of new therapeutic targets.
1. Introduction
Uveal melanoma (UM) is an ocular malignancy that arises from melanocytes residing in the uveal tract, which consists of the iris, ciliary body and choroid. It is the second most common type of melanoma and the most common primary intraocular malignancy in adults, affecting approximately 5.1 individuals per million per year; it is most frequent in Caucasians [1,2], as a fair skin and light eye color have been identified as host susceptibility factors [3,4].
In general, local tumor control is excellent, with large primary ocular melanoma being treated by enucleation, and small- to medium-sized tumors by application of a radioactive plaque, stereotactic irradiation or proton beam therapy [5,6,7,8,9,10]. Despite excellent regional tumor control, UM is still often lethal: up to 50% of patients will develop metastatic disease, for which no effective treatment exists [11]. The liver is involved in approximately 90% of cases with metastasized disease [12]. Metastatic disease may develop at any time from the initial diagnosis of the primary tumor to several years after diagnosis [13].
Several pathological characteristics of the primary tumor are known to be associated with an infaust prognosis. These include a large size, ciliary body involvement, epithelioid cell type, extrascleral invasion and the presence of extravascular matrix loops [14,15,16,17,18]. Furthermore, specific genetic features, such as monosomy 3, amplification of chromosome 8q, and loss of chromosome 1p, correlate with poor survival [19,20,21,22,23]. On the contrary, an additional copy of chromosome 6p is associated with a favorable prognosis [24,25]. Microarray gene expression analyses have resulted in the identification of two classes of UMs: class 1 tumors have low metastatic risk, while class 2 tumors are associated with a high rate of metastatic death [26,27,28].
Recently, mutations in specific genes such as BAP1 (BRCA1-associated protein-1), SF3B1 (splicing factor 3b subunit 1), and EIF1AX (eukaryotic translation initiation factor 1A, X-linked) have been reported to have prognostic value [29,30,31]. Aberrant DNA repair during the evolution of many malignancies and, accordingly, genomic instability is considered a hallmark of cancer cells [32]. Recent research in UM has focused on genetics, with the aim of unraveling UM biology and identifying specific aberrations that underlie the development of UM and may indicate potential targets of therapy [29,30,31,33]. The BAP1 protein, the loss of which correlates to a poor prognosis in UM [29,34,35], has been shown to promote DNA double-strand break repair [36]. Yet, the role of DNA repair in tumor development and progression remains poorly studied. Although counterintuitive, DNA-repair proteins in compensating pathways may be targets for cancer therapeutics [37,38]. Since tumor cells that have lost a repair pathway may (over)rely on them (principle of synthetic lethality), one may try to block DNA-repair proteins to decrease the ability of UM cells to repair DNA damage. This may subsequently help to sensitize tumors to traditional anti-cancer treatment by chemotherapy or radiotherapy [39].
However, it is not yet known whether and how the DNA-repair pathways are involved in the initiation and progression of UM. We, therefore, set out to analyze the expression of genes involved in DNA repair in UM and looked for genes that were associated with prognosis in UM.
To test our hypothesis that genes involved in DNA repair are differentially expressed between tumors with a favorable and unfavorable prognosis, we determined the expression of such genes in 64 UMs and made a comparison between tumors with and without loss of chromosome 3. Additionally, the relation with survival was evaluated for differentially expressed genes. Interesting associations were validated in two other sets of UM and a potential druggable target was explored further.
2. Results
2.1. Population Characteristics
Our cohort included 64 UM patients who had undergone primary enucleation at a median age of 61.6 years and of whom 33 (52%) were males (Table 1). The median largest basal diameter (LBD) was 13.0 mm and the median thickness 8 mm. Most tumors were either classified as American Joint Committee on Cancer (AJCC) tumor size T2 (39%) or T3 (48%). A mixed/epithelioid cell type was recorded in 66% of cases. Monosomy 3 was detected in 63% of the tumors. At last follow-up, 37 (58%) of patients had developed clinical metastases. We validated our data using two other independent cohorts: a set of 110 tumors from Genoa [40] and Paris [41], and the 80 UMs of The Cancer Genome Atlas (TCGA) project [42]. The characteristics of all cohorts are depicted in Table 1.

Table 1.
Baseline characteristics of the test and validation sets. Percentages are rounded and may not equal 100.
2.2. Gene Expression in Relation to Chromosome 3 Status
As loss of one copy of chromosome 3 is a very important prognostic marker in UM, we searched for DNA-repair-related genes that showed differential expression between tumors with and without loss of one chromosome 3.
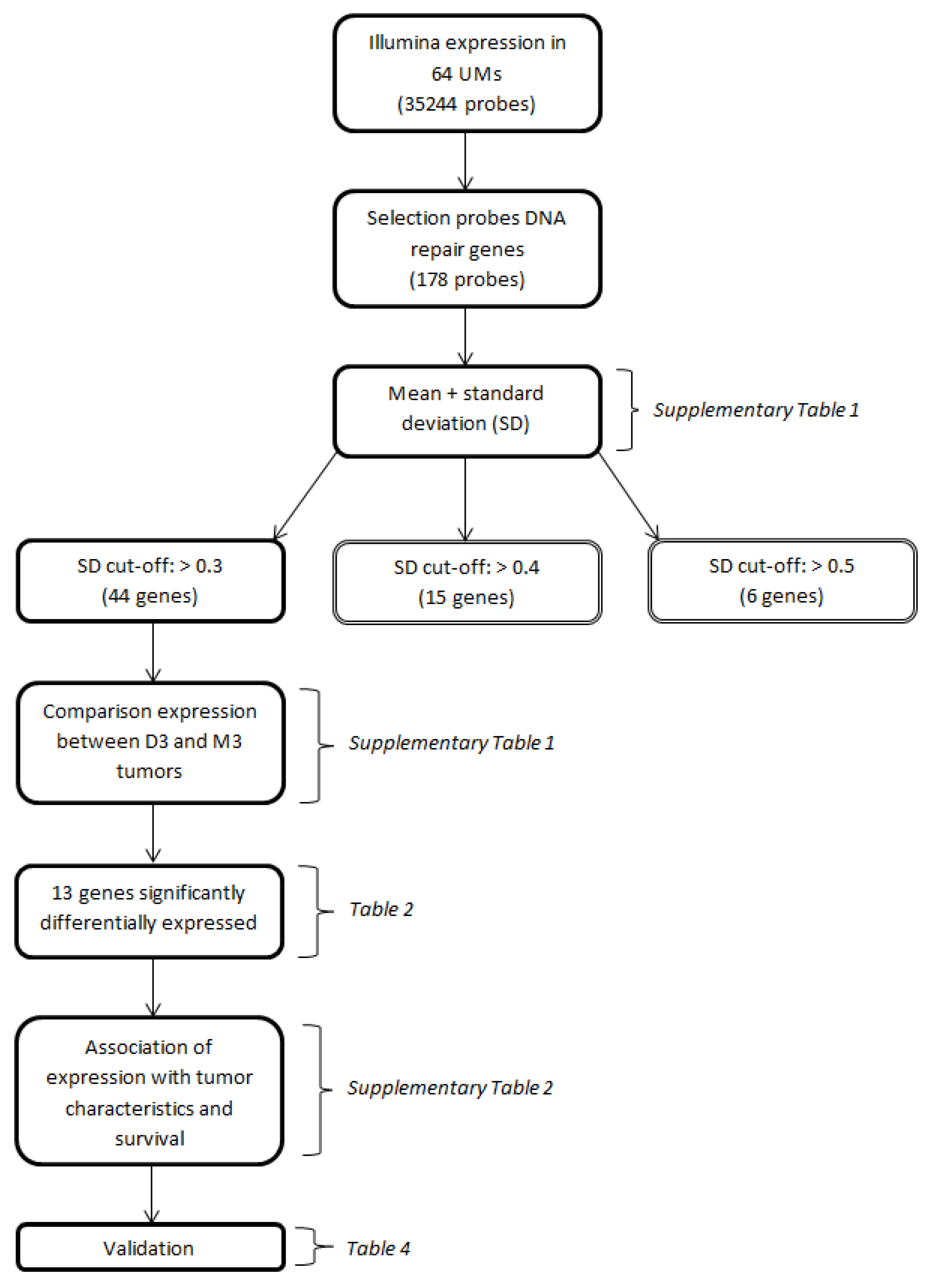
We identified 121 genes encoding proteins involved in DNA repair mechanisms, based on a literature review on DNA repair, using the platforms Gene, OMIM, KEGG and PubMed. As our goal was to identify genes with a variable expression level, we determined the standard deviations of the expression levels of the DNA-repair-gene probes on the Illumina chip (n = 178) (Appendix Table A1). A selection of genes was made based on a cut-off value of the standard deviation of the expression (Figure 1).
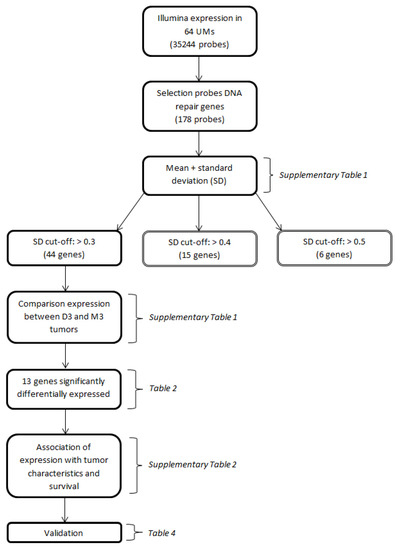
Figure 1.
Flow-chart depicting the major conducted analyses. Parentheses indicate the tables in which the results of the respective analyses are presented.
The median expression of the 44 genes of interest was calculated and compared between disomy 3 and monosomy 3 tumors. Thirteen genes were significantly differentially expressed: three genes (CENPX, DDB1, PRKDC) were significantly higher in monosomy 3 tumors (Table 2A), while ten genes (APEX1, BAP1, CETN2, GTF2H4, MLH1, RMI2, RPA1, SEM1, WDR48, XPC) showed a significant down-modulation in tumors with monosomy 3 (Table 2B).

Table 2.
Differentially-expressed genes in relation to chromosome 3 status. Only significantly differentially expressed genes between monosomic and disomic chromosome 3 UMs are shown. Table 2A shows the genes that had a higher expression in tumors with monosomy 3, and Table 2B shows the tumors with a lower expression in tumors with monosomy 3. The Mann–Whitney U test and Bonferroni correction were applied. BER: base excision repair; DSBR: double-strand break repair; FA: Fanconi Anemia; MMR: mismatch repair; NER: nucleotide excision repair.
2.3. Gene Expression in Relation to Histological Data and Survival
The expression of the 13 genes that were differentially expressed between disomy 3 and monosomy 3 tumors was compared to histopathological data and survival (Appendix Table A2).
With regard to associations between gene expression and tumor diameter, we noticed an association between low expression of WDR48 and XPC and a large LBD (p = 0.01 and p = 0.004, respectively), while a high expression of CENPX correlated with a large LBD (p = 0.02). Although the difference in expression was small, CENPX showed a significantly higher expression in tumors with a mixed/epithelioid cell type (p = 0.04). On the contrary, the expression of the genes WDR48 (p = 0.007) and XPC (p = 0.03) was significantly lower in cases with a mixed/epithelioid cell type. Regarding AJCC size categories, the expression of CENPX (p = 0.01) was significantly higher in tumors with higher AJCC categories, while the expression of the RPA1 gene (p = 0.03) was significantly lower in cases with a higher AJCC category. The genes CENPX and PRKDC were highly expressed in tumors that gave rise to metastases (both p < 0.001). The genes BAP1, CETN2, GTF2H4, MLH1, RMI2, SEM1, WDR48, and XPC, instead, showed a lower expression in the metastasis group (Mann–Whitney U test). Considering survival, CENPX and PRKDC genes were associated with poor survival when highly expressed. A low expression of the genes BAP1, GTF2H4, RMI2, SEM1, WDR48, and XPC was instead associated with an unfavorable prognosis (log-rank test).
2.4. Chromosome Dose Effect and Expression Levels
As previously noticed for other genes, the loss or gain of chromosomal material might influence gene expression levels [43,44]. Therefore, we combined gene expression levels of all the 44 genes of interest with the SNP copy number value of the chromosome region harboring the gene (Table 3). We divided tumors into three groups: no aberration in the specified chromosome area, duplication in the specified region or deletion of the region of interest. The analysis was only reliable for genes located on chromosomes 3, 6 or 8, since SNP analyses of other chromosomes showed no aberrant copy number in most tumors.

Table 3.
Relationship between chromosome dose and gene expression for all 44 genes of interest. The analysis was reliable only for genes located on chromosomes 3, 6 or 8, since SNP analyses of other chromosomes showed no aberrant copy number in most tumors.
As we expected, the four genes located on chromosome 3p (BAP1, MLH1, WDR48, XPC) showed an association between a decreased expression and presence of monosomy 3, while a trend towards decreased expression was noted for MBD4 (chromosome 3q) (Table 3). The genes FANCE and GTF2H4 (chromosome 6p) showed a significantly increased expression in tumors with a gain of 6p, while for GTF2H5 (chromosome 6q), a significantly lower expression was found in tumors with loss of 6q. The expression of POLB (chromosome 8p) was significantly decreased in tumors with loss of 8p, while an increased expression of NBN and PRKDC, which are located on the long arm of chromosome 8, was related to a gain of genetic material in that chromosome region.
2.5. Validation
The 13 genes statistically differentially expressed were validated on the datasets of two other UMs: a set of 110 tumors from Genoa and Paris and another set of 80 UMs of The Cancer Genome Atlas (TCGA) project [42]. In each validation set, median expression levels for every gene were calculated to establish two groups of tumors for Kaplan–Meier analyses. The occurrence of metastases was the event of interest in the tumors from Genoa and Paris (taken together), while death due to UM metastases was the endpoint of analysis for the TCGA tumors. In Genoa and Paris sets, more than one p-value is presented for some genes, since several probes were available for these genes.
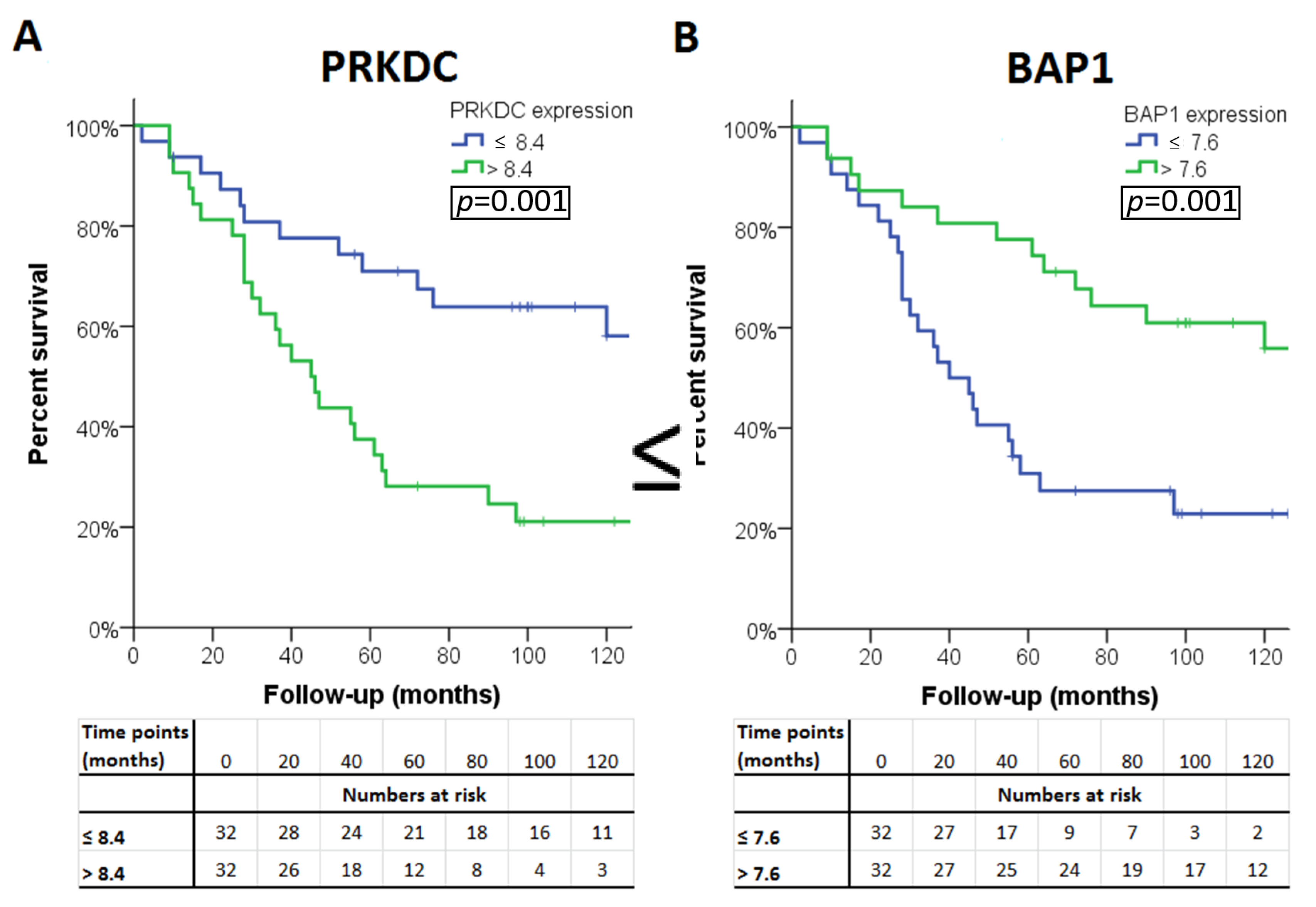
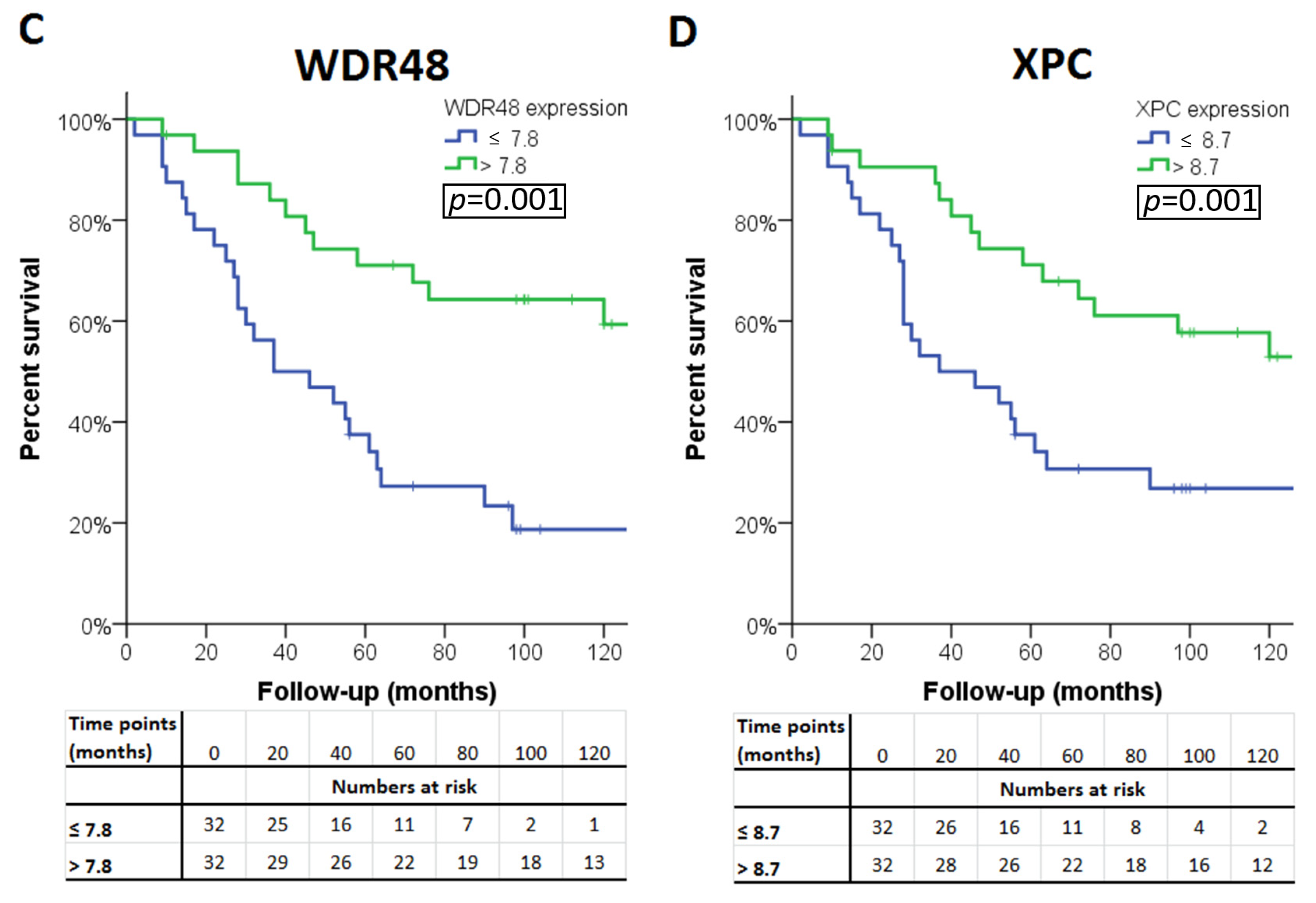
The association of the expression of a gene with survival was considered ‘validated’ if a significant association was observed in all three sets (LUMC set and the two validation sets). Four of the 13 genes were significantly associated with survival in all three cohorts. A high expression of PRKDC was associated with poor survival, as well as a low expression of BAP1, WDR48, and XPC (Table 4). Survival curves for these genes in patients from the LUMC cohort are shown in Figure 2. As cut-off value, we used the median expression of each gene.

Table 4.
Validation of the 13 significantly differentially expressed genes between disomy 3 and monosomy 3 tumors in the LUMC cohort. Validation was performed in an independent cohort of 110 tumors (Genoa + Paris) and in the TCGA cohort of 80 tumors. p-values of the log-rank test are shown. Significant p-values are in bold. Genes that are significantly associated with survival in all cohorts are depicted in the last column. Abbreviations: BER: base excision repair; DSBR: double-strand break repair; FA: Fanconi Anemia; MMR: mismatch repair; NER: nucleotide excision repair.
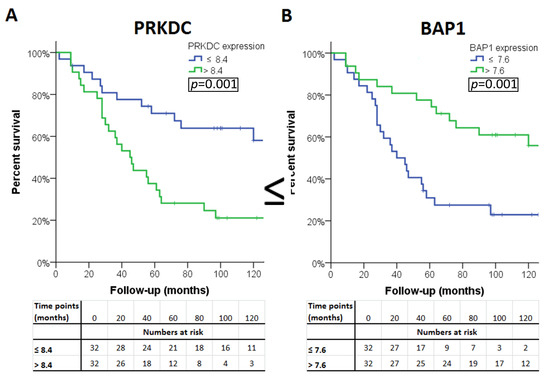
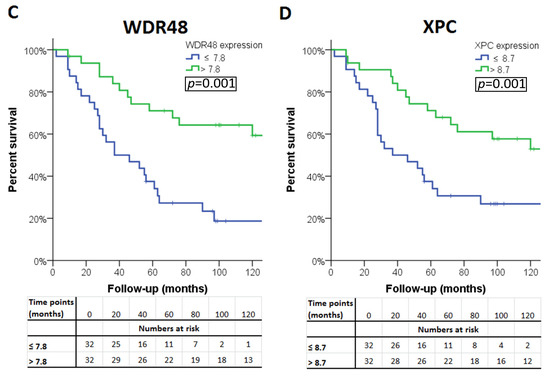
Figure 2.
Kaplan–Meier survival curves of the four genes: (A) PAKDC, (B) BAP1 (C) WDR48 and (D) XPC significantly associated with clinical outcome in all three cohorts.
2.6. PRKDC
Considering that a high expression of the PRKDC gene located on chromosome 8q is related to an unfavorable prognosis and the fact that gain of material of chromosome 8q predicts an adverse clinical outcome, we decided to perform further (experimental) analyses to study the biological significance of the PRKDC gene in UM. Our decision to focus on PKRDC was furthermore fueled by the finding that the DNA-PKcs protein encoded by PRKDC has been shown to modulate cell survival, proliferation, invasion and migration in other cancers [45,46].
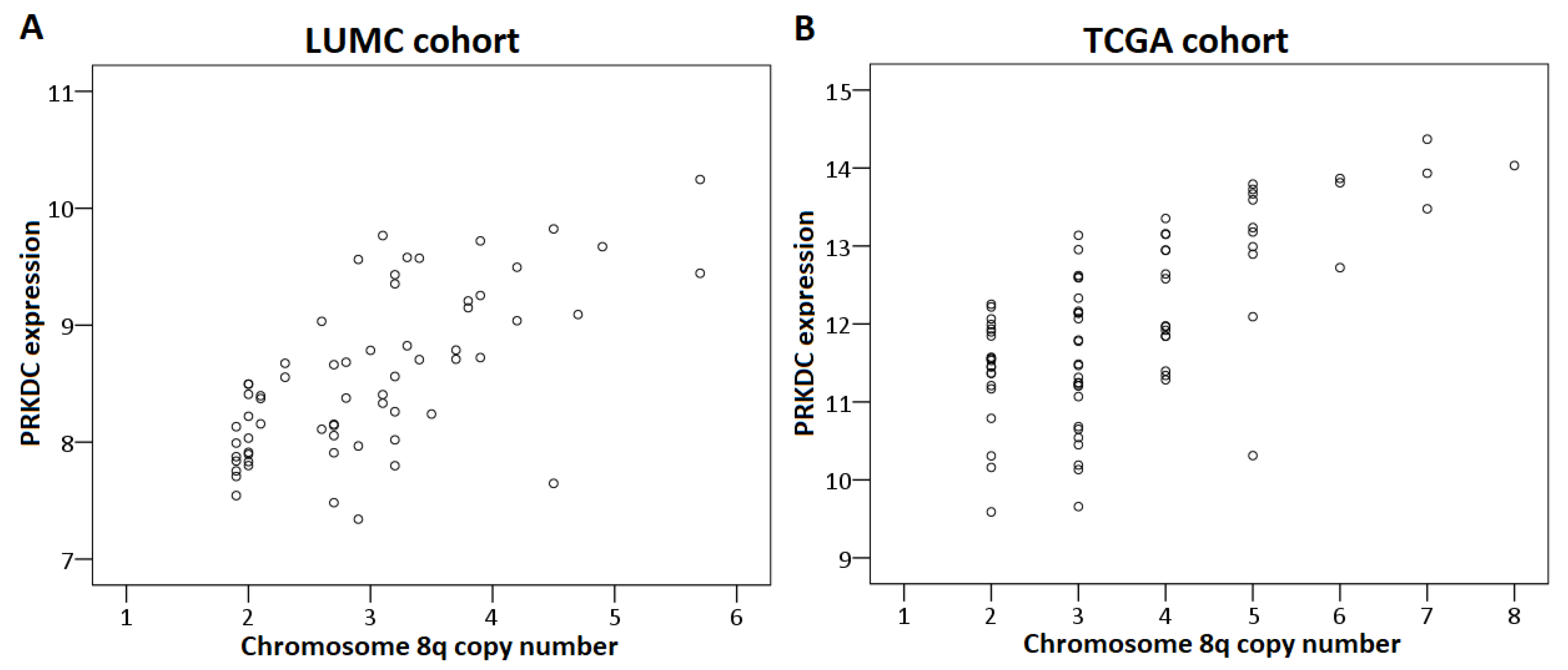
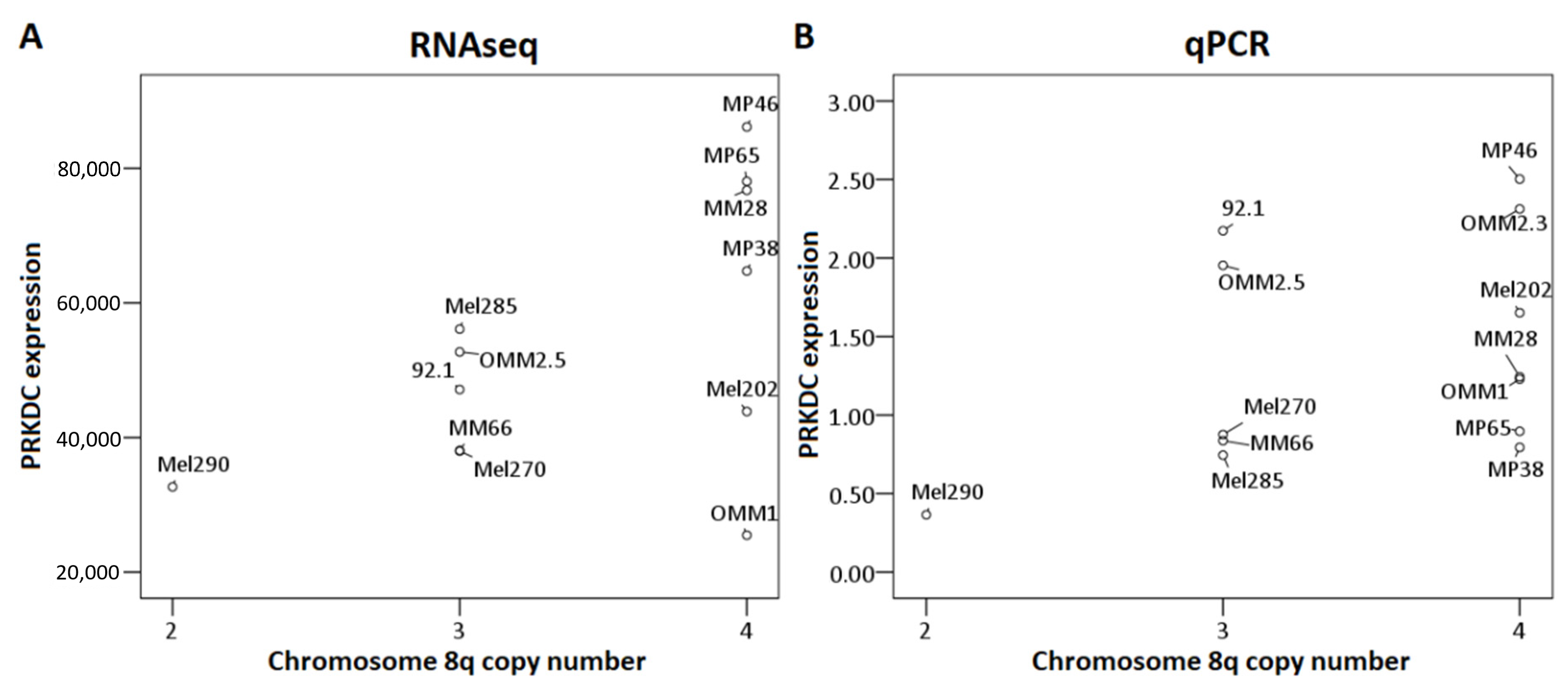
First, we analyzed the relation between chromosome 8q copy number variation and PRKDC expression in the LUMC and the TCGA cohort. This analysis could not be performed for the Genoa and Paris cohort because the chromosome 8q status of these tumors was unknown. A higher chromosome 8q copy number was significantly correlated to a higher expression of PRKDC in the LUMC cohort (correlation coefficient: 0.67, p < 0.001) as well as the TCGA cohort (correlation coefficient: 0.61, p < 0.001) (Figure 3).
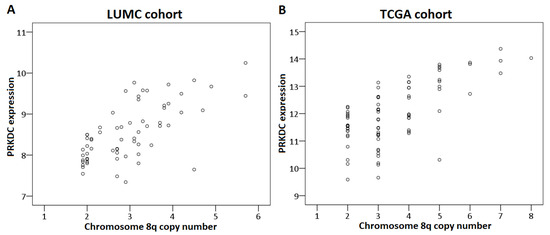
Figure 3.
Correlation between PRKDC expression and chromosome 8q copy number in primary UM. The Spearman’s correlation test was performed. (A) LUMC cohort, (B) TCGA cohort.
We also analyzed the association between 8q copy number and PRKDC expression, determined by RNAseq in 12 UM cell lines, and by qPCR in 13 UM cell lines (Figure 4). Although the association was not significant (RNAseq: p = 0.23, qPCR: p = 0.2 [Kruskal–Wallis test]), we observed a trend towards higher expression of PRKDC in cell lines with more copies of 8q, which was in agreement with our findings in primary tumors (Figure 3). However, this association was less evident than in primary tumors, due to the lower number of cases and the lack of cell lines with two copies of chromosome 8q or more than four copies of 8q. The correlation was most pronounced in the RNAseq analysis (Figure 4A) and less clear in the qPCR analysis (Figure 4B), where the correlation was slightly distorted by cell lines 92.1 and OMM2.5, which have three copies of chromosome 8q but show a PRKDC expression that is comparable to cell lines with four copies. However, there was a subpopulation of cells having four copies of chromosome 8q in cell line 92.1, indicating mosaicism.
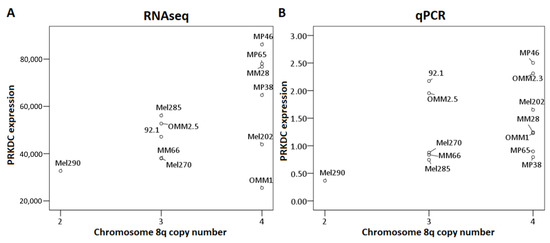
Figure 4.
Association between PRKDC expression and chromosome 8q copy number in UM cell lines. The Kruskal–Wallis test was performed. (A): RNAseq, (B): qPCR.
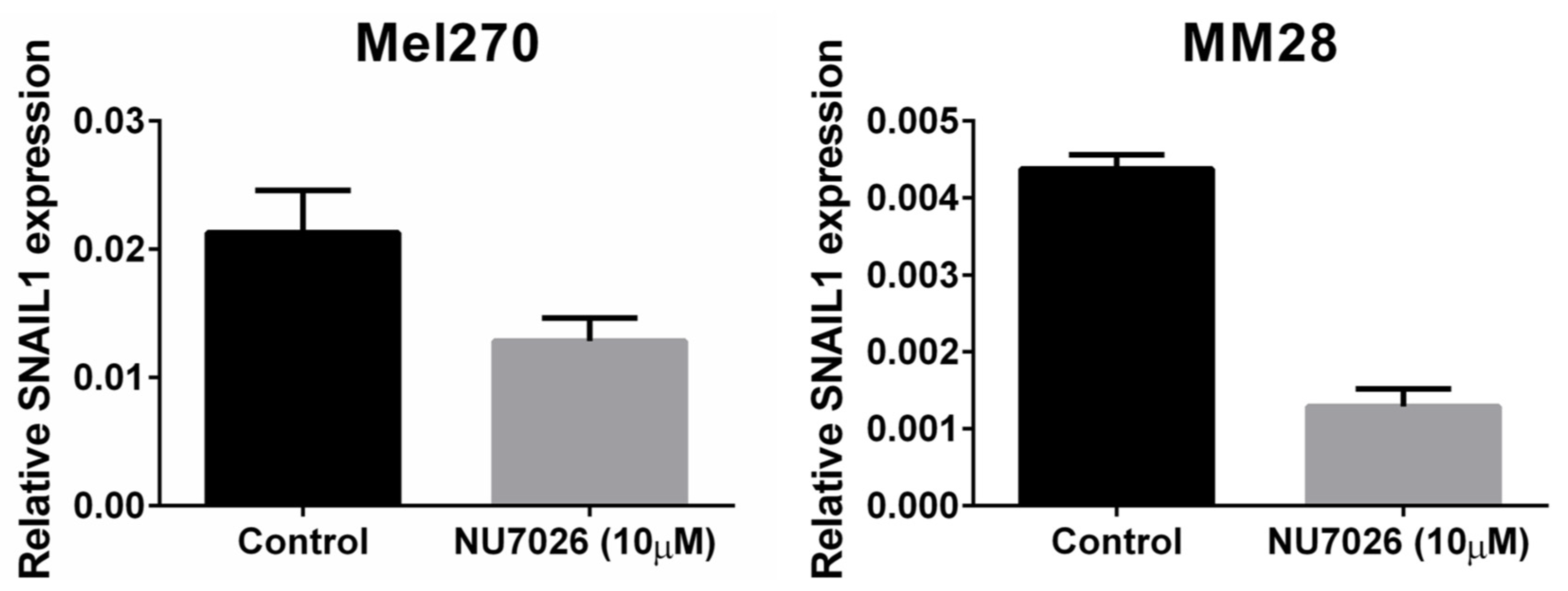
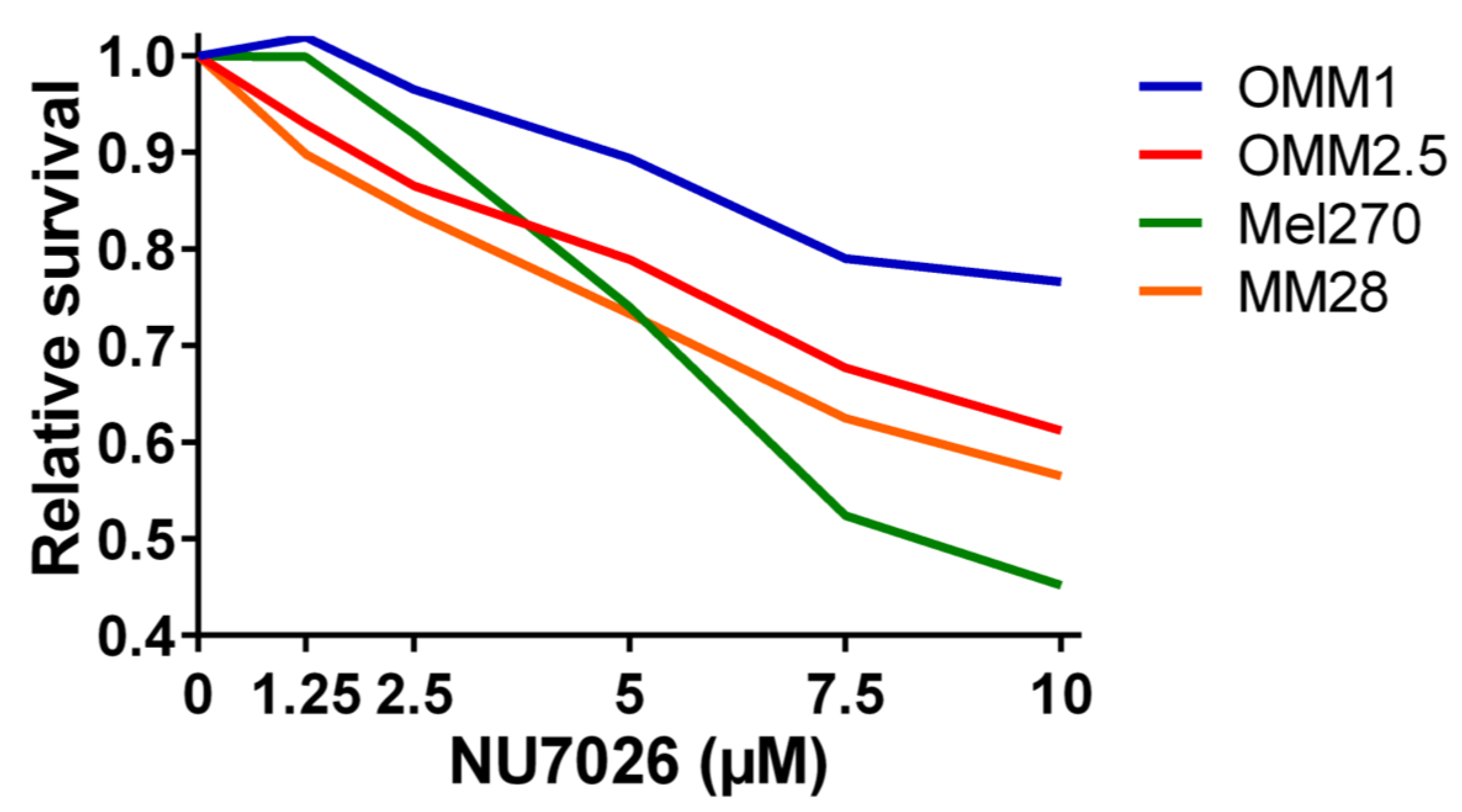
To test our hypothesis that PRKDC is a possible driver of metastasis in UM, we wondered in which ways PRKDC could be involved in invasion and migration of UM cells. A study in prostate cancer showed that transcriptional regulation by the DNA-PKcs protein encoded by the PRKDC gene promotes invasion, migration and metastasis [45]. As the expression of ZEB1, TWIST1 and SNAIL1 have been proposed to play a role in invasion of UM cells, [47] we evaluated whether inhibition of DNA-PKcs with NU7026 would influence the expression of these genes. NU7026 is an inhibitor of DNA-dependent protein kinase (DNA-PK), an enzyme involved in the non-homologous end-joining (NHEJ) DNA-repair pathway [48]. NU7026 sensitizes cells to radiation and has potential for use in anticancer therapies [49,50]. The expression of ZEB1, TWIST1 and SNAIL1 was evaluated in a primary UM cell line (Mel270) and in a metastatic UM cell line (MM28) before and after treating the cells with 10μM NU7026 for 5 days. The basal expression level of these genes was low in both cell lines. Inhibition of DNA-PKcs by NU7026 led to a downregulation of SNAIL1 in Mel270 as well as MM28 cells (Figure 5). ZEB1 and TWIST1 expression were not affected. To analyze the effect of DNA-PKcs inhibition on cell proliferation, we treated four cell lines (OMM1, OMM2.5, Mel270, MM28) with increasing doses of NU7026 up to 10 µM for a period of 5 days (Figure 6). The proliferaton of all cell lines was affected by the DNA-PKcs inhibitor. The strongest growth inhibitory effect was noted in cell lines Mel270 and MM28, showing a 55% and 43% inhibition, respectively.
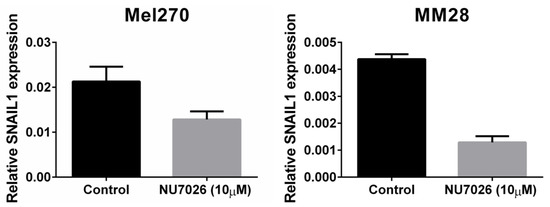
Figure 5.
The effect of DNA-PKcs inhibition on the mRNA expression of SNAIL1 in cell lines Mel270 and MM28. Cells were treated with 10 µM of the DNA-PKcs inhibitor NU7026 for 5 days.
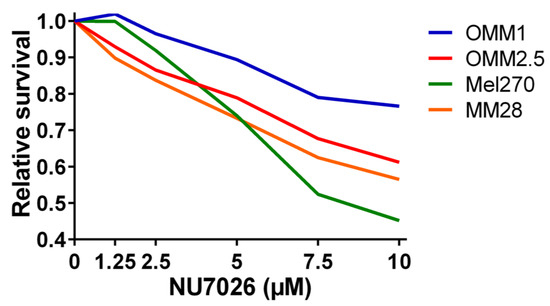
Figure 6.
The relative survival in UM cell lines OMM1, OMM2.5, Mel270, and MM28 upon treatment with increasing doses of the DNA-PKcs inhibitor NU7026 for 5 days.
3. Discussion
Biological cellular responses following DNA damage include DNA damage repair, damage tolerance, cell-cycle checkpoint control and apoptosis. These mechanisms are tightly regulated and which pathway becomes activated depends on the type and severity of the DNA damage. In case of severe damage, the complex signaling pathways may eventually lead to cell cycle arrest (providing the cell more time for repair and tolerance mechanisms) or to apoptosis [51,52]. The recognition of expression patterns of the genes involved in DNA repair in UM is the first step in understanding the way these genes might play a role in UM development and may help in identifying new targets for therapy. We evaluated the expression of DNA-repair-related genes in the Leiden cohort of 64 UMs and aimed to identify genes with a variable expression between prognostically favorable and prognostically unfavorable UM. After validation in two other independent cohorts, we identified four genes which were associated with the degree of malignancy in UM: three genes (BAP1, WDR48, and XPC1) showed an association between a low expression and poor survival, while PRKDC was highly expressed in cases with an unfavorable prognosis. The genes BAP1, WDR48, and XPC1 are all located on chromosome 3p and showed a significantly lower expression in monosomy 3 tumors. A lower expression of the MLH1 gene, which is also located on chromosome 3p, was significantly related to prognosis in one cohort and showed a near-significant effect in the other cohorts. Since these four genes play a role in DNA repair, we can expect that impaired DNA repair is one of the results of the loss of a copy of chromosome 3. Sustained DNA damage as a result of deficient DNA repair mechanisms may lead to the accumulation of chromosomal abnormalities and gene mutations, which may promote cell growth and proliferation. Chromosome 3 loss does not occur in a single step since small tumors with partial monosomy have been observed [53], but apparently, loss of the entire chromosome confers a selective advantage that might be mediated by the DNA-repair genes identified here.
BAP1 (BRCA1-associated protein 1) is a gene located on chromosome 3p21.1. The BAP1 gene encodes a nuclear ubiquitin carboxy-terminal hydrolase, which is a deubiquitinating enzyme [54]. It has been described to be a tumor suppressor gene in the BRCA-1 control pathway. The BAP1 protein contains binding domains for BRCA1 and BARD1, enzymes that form a heterodimeric complex that functions as a tumor suppressor [55]. Loss of BAP1 has been shown to be related to a poor clinical outcome in UM [29]. Similarly, a lower gene expression of BAP1 in our study corresponded to a poor survival.
Ubiquination and deubiquination regulate essential biological processes such as DNA replication and DNA repair [42,55]. In accordance, BAP1 has been shown to play a role in the repair of DNA double-strand breaks by homologous recombination [36,56]. It has been suggested that the DNA-repair function of BAP1 may be the molecular basis for its tumor suppressor role in UM [36].
Another DNA-repair-related gene involved in deubiquitination, which in our study showed a low expression in metastasizing uveal melanoma, is WDR48. It is also known as UAF1 and is located in close proximity (on 3p22.2) to BAP1. UAF1 and USP1, a deubiquitinating enzyme, form the UAF1/USP1 complex, which regulates the Fanconi Anemia DNA-repair pathway [57]. UAF1 activates USP1, and USP1 regulates the Fanconi Anemia repair pathway by deubiquitinating FANCD2, one of the most important players in this pathway. Fanconi Anemia is an inherited genomic instability disorder that led to the discovery of a novel DNA-repair pathway. The Fanconi Anemia repair pathway plays a role in the repair of DNA cross-links and can be activated after various types of DNA damage, such as ionizing radiation and ultraviolet light [58,59]. Accurate deubiquitination of the FANCD2 protein by the USP1/UAF1 complex is essential for an intact Fanconi Anemia pathway and proper DNA damage repair [60,61]. Because of this crucial role of the WDR48 gene, and the association that we found of a low expression of WDR48 with poor prognosis, a defective Fanconi Anemia repair pathway may play a role in the malignant transformation of UM. Murine fibroblasts deficient in UAF1 have been shown to exhibit profound chromosomal instability [62].
XPC (Xeroderma Pigmentosum, complementation group C) is the third gene located on chromosome 3p. Its low expression was associated with poor survival in our study. The XPC gene, located in the region 3p25.1, encodes a protein that helps to form the XPC repair complex and is involved in the early steps of the DNA Nucleotide Excision Repair (NER) pathway. Mutations in XPC that impair the production of the XPC protein are related to Xeroderma Pigmentosum (XP), a rare recessive disorder, which makes patients extremely sensitive to ultraviolet light. This results in the frequent development of skin tumors, mainly in areas of the body exposed to the sun. The XPC protein acts a sensor detecting DNA damage [63,64,65,66]. The association of the low expression of XPC with poor survival in UM is interesting, since evidence for the association of ultraviolet light exposure and UM development is inconclusive. However, XPC may play a role that is independent of its direct function related to UV-damage, as evidenced by the association of epigenetic silencing of XPC with shorter survival in bladder cancer [67]. The XPC repair complex contains the CETN2 protein, which shows a significantly lower expression in metastasizing UMs in the two validation cohorts of our study (Table 4) [68]. Xeroderma Pigmentosum is associated with a higher risk for ocular malignancies [69].
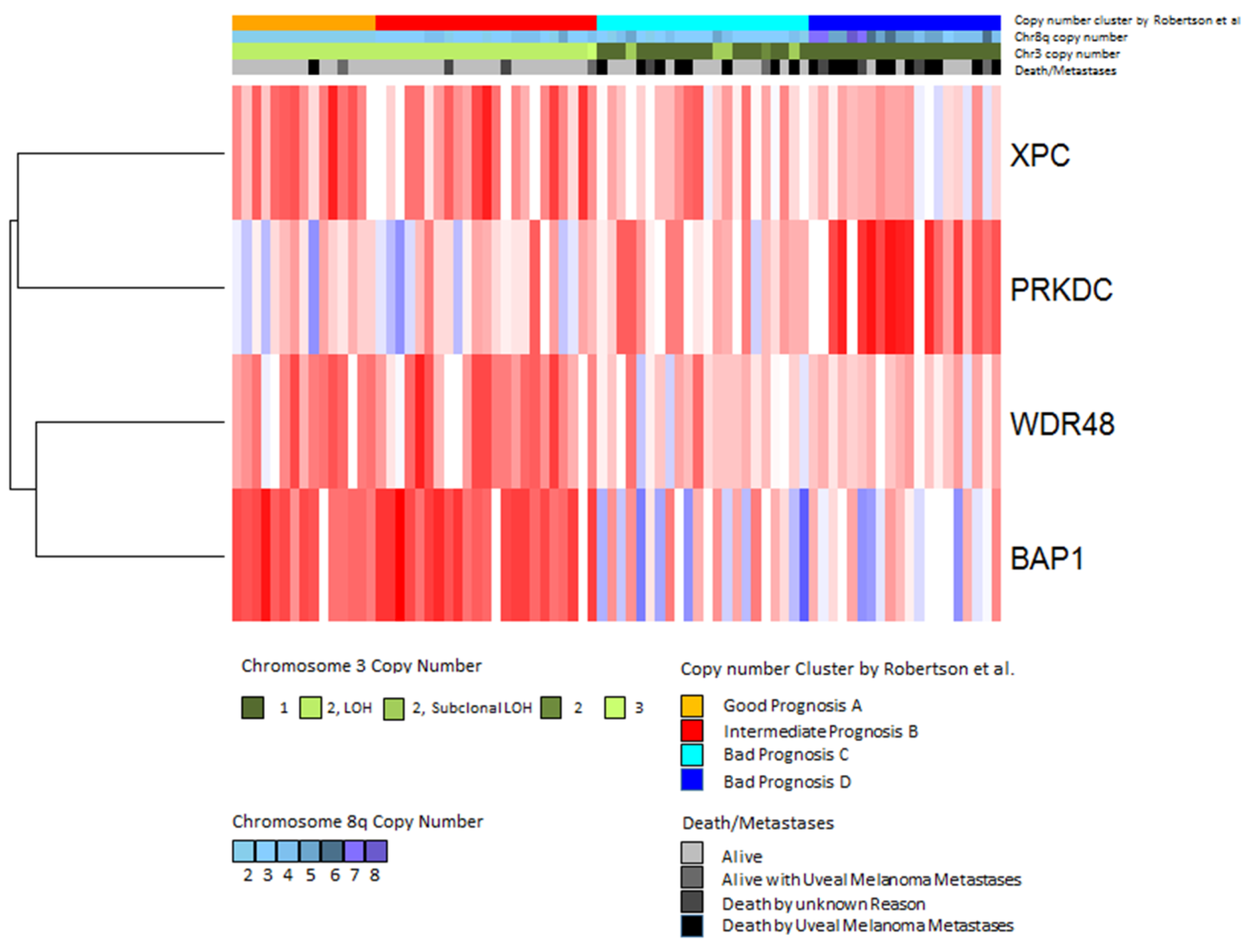
In contrast to the genes discussed above, the PRKDC gene that is located on chromosome 8q11.21 was found to be associated with worse survival when highly expressed [70]. A heatmap showing the patients that developed metastases makes it clear that a low BAP1 expression (blue) is associated with a high PRKDC expression (Figure 7). PRKDC encodes the catalytic subunit of DNA-dependent serine/threonine protein kinase (DNA-PKcs). DNA-PK is involved in the repair of double-strand breaks (DSBs) by non-homologous end-joining (NHEJ) [71,72,73]. DSBs can develop due to the effects of reactive oxygen intermediates or by exogenous agents such as ionizing radiation and anticancer chemotherapeutic drugs [74].
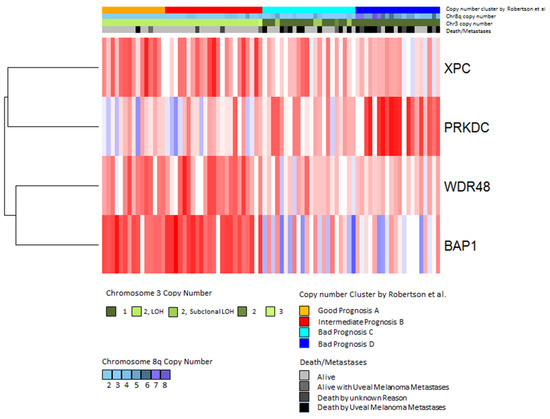
Figure 7.
Expression of DNA-repair genes in the TCGA dataset. Gene expression values of the four significantly differentially expressed genes were analyzed by hierarchical clustering using Euclidean distance and average linkage. Each column shows one tumor sample and each row shows one gene. Expression values are shown according to the mean value for each gene (blue = expression below the mean, red = expression above the mean, white = expression at the mean). Subtypes defined by Robertson et al. [42] are indicated in the upper bar above the heatmap as follows: good prognosis (orange), intermediate prognosis (red), bad prognosis group C (cyan) and bad prognosis D (blue) (Jager et al. [75]).
High expression of DNA-repair proteins such as DNA-PKcs may increase the ability of tumor cells to withstand damage caused by chemotherapy or irradiation. Accordingly, increased DNA-PKcs activity was related to glioma resistance to cisplatin chemotherapy [76]. Moreover, upregulation of DNA-PKcs was detected after irradiation of oral squamous cell carcinoma (OSCC) cells that were resistant to radiotherapy. Targeting DNA-PKcs has been suggested as a novel sensitization therapy of OSCC, and it has been shown to increase anticancer drug sensitivity in osteosarcoma cell lines [77,78]. Since the majority of primary UMs is treated by radiotherapy and certain chemotherapeutic targets are being tested for their effectiveness in killing UM metastases, elucidating the role of DNA-PKcs in UM may pave the way for sensitization therapy in UM by inhibiting DNA-PKcs. While some preliminary results indicate that inhibition of DNA-PKcs by NU7026 sensitizes UM cell lines for the topoisomerase I inhibitor, Topotecan, studies on cervical and breast cancer cells, as well as on lung cancer cells, have shown that this treatment sensitizes tumor cells to radiation treatment [79,80]. As far as we know, this combination has not been tried on UM cells. Van Oorschot et al. showed that the combination of hyperthermia and treatment with NU7441 led to an even better sensitization [79].
We demonstrate that gain of chromosome 8q is related to a higher expression of PRKDC in our cases, as well as in the TCGA cohort and UM cell lines. It is known that amplification of chromosome 8q is associated with an adverse clinical outcome in UM [25,81]. Although the exact mechanisms by which gain of chromosome 8q confers its malignant effect has not yet been elucidated, overexpression of DDEF1 has been suggested as one potential mechanism [82]. A recent study in prostate cancer has shown that the DNA-PKcs protein encoded by PRKDC modulates cell invasion and migration and acts as a strong driver of tumor progression and metastasis [45]. In addition, activated DNA-PKcs has been correlated with increased proliferation, decreased apoptosis and poor survival in hepatocellular carcinoma [46]. In accordance, DNA-PKcs has been shown to be involved in normal cell cycle progression by controlling proper chromosome segregation and cytokinesis [83].
In this study, we show that inhibition of DNA-PKcs results in decreased proliferation of UM cells. A recent study by Kotula et al. in the cutaneous melanoma cell line SK28 demonstrated that DNA-PKcs has pro-metastatic activity by modulating the tumor microenvironment through controlling the secretion of, e.g., matrix metalloproteinases (MMPs) and tissue inhibitors of matrix metalloproteinases (TIMPs) [84]. We found a low and variable expression of MMPs and TIMPs in the majority of UM cell lines we analyzed and we did not observe an evident regulatory effect following DNA-PKcs inhibition. Since DNA-PKcs is postulated to be a driver of invasion and metastasis, we analyzed the effect of DNA-PKcs inhibition on the expression of an epithelial-to-mesenchymal transformation (EMT)—associated factors that have been shown to play a role in the invasiveness of UM cells (ZEB1, TWIST1, SNAIL1) [47]. Although the basal expression of these factors was low in the UM cell lines, we observed a decrease in the expression of the pro-metastatic SNAIL1 upon DNA-PKcs inhibition. The inhibition of the protein interaction between DNA-PKCs and Snail1 has been suggested to be an effective strategy for inhibiting tumor migration [85].
Considering the suggested pro-metastatic functions of DNA-PKcs, it is conceivable that an increased expression of PRKDC, as a result of amplification of 8q, may contribute to the malignant progression in UM. This would imply that DNA-PKcs could be a potential target for therapy in UM. Furthermore, the use of inhibitors of DNA-repair proteins is a promising option for treating metastases, since cancer cells only retain some DNA-repair modules and are dependent on these for survival [86].
4. Materials and Methods
4.1. Study Population
Our ‘training set’ contained 64 UMs obtained by primary enucleation at the Leiden University Medical Center (LUMC), Leiden, The Netherlands, between 1999 and 2008. Patient and tumor characteristics are shown in Table 1. Sufficient frozen material of these tumors was available and DNA of adequate quality could be retrieved. Survival data was retrieved from the patients’ charts and from the Netherlands Comprehensive Cancer Organisation (https://iknl.nl/over-iknl/about-iknl), and updated in March 2017. In The Netherlands, general physicians report every cancer patient to the Netherlands Comprehensive Cancer Organisation, which collects and registers information on the survival status by contacting the general physicians yearly. The follow-up in The Netherlands is not intensive because of a lack of effective treatments for UM metastases and patients are often referred back to their general physician after treatment of the primary tumor. The median follow-up time was 62 months and no patient was lost to follow-up.
Validation of the data was performed using two independent cohorts of post enucleation surgery patients: microarray datasets from Genoa and Paris, and RNAseq data of The Cancer Genome Atlas (TCGA) project [42,75]. Sixty-three untreated uveal melanoma provided by the Biological Resource Centre of Institut Curie (GSE2213840) [86] and 48 UM samples from the Genoa cohort (GSE2783141 and GSE5188042) [41,87] were obtained from the Gene Expression Omnibus (www.ncbi.nlm.nih.gov/geo/). The datasets were combined and normalized as described.
The study followed the tenets of the Declaration of Helsinki (World Medical Association of Declaration 1964; ethical principles for medical research involving human subjects) and the Medical Ethics Committee of the LUMC, Leiden, The Netherlands, had no objection regarding this research (G16.076/NV/gk).
4.2. Histologic Examination
After opening the enucleated bulbus, a part of the tumor was retrieved and snap frozen at −80 °C. The remaining tumor tissue was formalin fixed (4% neutral-buffered) and embedded in paraffin. A conventional histologic evaluation by an ophthalmic pathologist for confirmation of diagnosis and determination of characteristics was done. Parameters such as largest basal diameter (LBD, in millimeters), thickness (in millimeters), mitotic count (per 2mm2 at 40× magnification, 8 high-power fields), tumor location, cell type (assessed according to the Armed Forces Institute of Pathology atlas) [88] were evaluated on 4 μm-thick hematoxylin and eosin-stained sections. The 8th edition of the AJCC Cancer Staging Manual [89] was used to stage tumors according to the TNM classification system.
4.3. Genetic Analyses
DNA and RNA were isolated from fresh-frozen tissue. DNA for single nucleotide polymorphism (SNP) analysis was extracted with the QIAmp DNA Mini kit and RNA for gene-expression profiling with the RNeasy Mini Kit (both from Qiagen, Venlo, The Netherlands). SNP array analysis to determine the chromosome copy number was performed with the Affymetrix 250K_NSP microarray chip (Affymetrix, Santa Clara, CA, USA) on all 64 UMs and with the Affymetrix Cytoscan HD chip (Affymetrix) on the cell lines. The Chromosome Analysis Suite (ChAS, version 2.0225) from Affymetrix was used to determine chromosome copy numbers. Gene-expression profiling at the transcriptional level was carried out on RNA of 64 UMs using 35,244 probes from the Illumina HT-12v4 chip (Illumina, San Diego, CA, USA).
RNA for real-time PCR analysis in cell lines was isolated using the SV total RNA isolation kit (Promega, Madison, WI, USA), then cDNA was synthesized using the reverse transcriptase reaction mixture, as indicated by Promega. qPCR was performed using SYBR green mix (Roche Diagnostics, IN, USA) in a C1000 touch Thermal Cycler (Bio-Rad laboratories, Hercules, CA, USA). Relative expression of PRKDC and SNAIL1 was determined compared to housekeeping genes CAPNS1 and SRPR. The untreated samples average was set at 1.
RNAseq analysis in the cell lines was conducted at Institut Curie (Paris, France) after isolation of total RNA using a NucleoSpin Kit (Macherey-Nagel, Düren, Germany). cDNA synthesis was conducted with MuLV Reverse Transcriptase in accordance with the manufacturers’ instructions (Invitrogen, Carlsbad, CA, USA), with quality assessments conducted on an Agilent (Santa Clara, CA, USA) 2100 Bioanalyzer. Libraries were constructed using the TruSeq Stranded mRNA Sample Preparation Kit (Illumina) and sequenced on an Illumina HiSeq 2500 platform using a 100 bp paired-end sequencing strategy. TopHat (v2.0.6) was used to align the reads against the human reference genome Hg19 RefSeq (RNA sequences, GRCh37) downloaded from the UCSC Genome Browser (http://genome.ucsc.edu). Gene expression was determined by featureCounts and normalized using DESeq2.
Heatmaps and hierarchical clustering were performed in R (heatmap.plus package) using Euclidean distance and average linkage. Gene expression data were also correlated with Uveal Melanoma subtypes according to Robertson et al. [42].
4.4. Gene Selection Procedure
We identified 121 genes encoding proteins involved in DNA repair mechanisms, based on a literature review on DNA repair, using the platforms Gene, Online Inheritance in Man (OMIM), Kyoto Encyclopedia of Genes and Genomes (KEGG) and PubMed. As our goal was to identify genes with a variable expression level, we determined the standard deviations of the expression levels of the DNA repair gene probes on the Illumina chip (n = 178) (Appendix Table A1). Certain genes were analyzed multiple times because they are encoded by different Illumina probes (in that case, the distinction between probes is made by placing letters in alphabetic order at the end of the gene name), while 18 genes were not analyzed since they were not on the Illumina chip. A selection of genes was made based on a cut-off value of the standard deviation of the expression (Figure 1). A cut-off value of >0.5 would result in 6 genes, of >0.4 in 15 genes, and a cut-off value of >0.3 would lead to a total of 44 genes (encoded by 49 probes). A cut-off value of >0.3 was chosen to have a reasonably-sized group of genes with an acceptable level of variation in expression. The median expression of the probes of these 44 genes was compared between disomy 3 (D3) and monosomy 3 (M3) tumors and corrected for multiple testing using the Bonferroni method. A total of 13 Genes which were significantly differentially expressed after Bonferroni correction were selected for further analysis.
4.5. Cell Lines, DNA-PKcs Inhibition, and Proliferation Assay
Cell lines OMM2.5 (originally called OMM1.5 derived from a liver metastasis) and Mel270, which are derived from the same patient, were obtained from Dr. Bruce Ksander [90] and maintained in RPMI supplemented with 10% FBS (fetal bovine serum) and antibiotics. MM28 was obtained from Dr. Sergio Roman-Roman [91] and grown in IMDM supplemented with 20% FBS and antibiotics. The OMM1 cell line, maintained in RPMI supplemented with 10% FBS and antibiotics was established by Dr. Gré Luyten [92]. Cell line 92.1 was developed in Leiden by Dr. Martine Jager [93]. MM28 cells lack BAP1 expression, whereas Mel270, OMM2.5, and OMM1 cells are BAP1-positive.
To evaluate the effect of DNA-PKcs inhibition on the expression of pro-metastatic factors, the expression of these factors was evaluated in a primary UM cell line (Mel270) and in a metastatic UM cell line (MM28) before and after treating the cells with 10 µM NU7026 (#13308, Cayman Chemical, Ann Arbor, MI, USA, stock concentration 20 mM in DMSO) for 5 days. In order to analyze the effect of the DNA-PKcs inhibitor on the growth of these UM cell lines, the cells were seeded in triplicate in 96-well plates. Treatment with NU7026 was started the next day. Cells were replenished with fresh medium with or without drugs after three days. Relative survival was determined after five days with the use of the CellTitre-Blue cell viability assay (Promega) according to the manufacturer’s protocol.
4.6. Statistical Analysis
For data analysis, we used the statistical programming language R version 3.0.1 (R: A Language and Environment for Statistical Computing, R Core Team, R foundation for Statistical Computing, Vienna, Austria, 2014, http://www.R-project.org) supplemented with specialized packages for SNP and RNA analysis. The main package used for SNP analysis was aroma.affymetrix, supported by ‘DNAcopy’ (Venkatraman E. Seshan and Adam Olshen, DNAcopy: DNA copy number data analysis. R package version 1.34.0), ‘sfit’ (Henrik Bengtsson and Pratyaksha Wirapati (2013), sfit: Multidimensional simplex fitting. R package version 0.3.0/r185, http://R-Forge.R-project.org/projects/matrixstats/), and ‘R.utils’ (Henrik Bengtsson (2014), R.utils: Various programming utilities, R package version 1.29.8, http://CRAN.R-project.org/package=R.utils). The ‘Aroma.Affymetrix’ package made it possible to use the information from the SNP microarrays to determine copy number values [94,95].
The packages used for RNA microarray analysis were ‘limma’ version 3.16.8, and the specific packages for Illumina microarrays: ‘lumi’ version 2.12.0, ‘annotate’ (R. Gentleman, annotate: Annotation for microarrays, R package version 1.38.0), and the database package ‘IlluminaHumanv4.db’ (Mark Dunning, Andy Lynch and Matthew Eldridge, IlluminaHumanv4.db: Illumina HumanHT12v4 annotation data (chip IlluminaHumanv4), R package version 1.18.0).
The statistical software package SPSS v.20.0.0 (IBM SPSS Statistics for Windows, IBM Corp., Armonk, NY, USA) was used for data analysis. Population characteristics were described using medians and percentages. The Mann–Whitney U test was performed to analyze numerical variables between two groups, and the Kruskal–Wallis test in case more than two groups were compared. Kaplan–Meier survival curves were made and the log rank test was used to analyze significance. Differences were considered to be significant if p < 0.05 after correction for multiple testing.
5. Conclusions
We show that several important DNA-repair molecules are differentially expressed between tumors with good and adverse prognosis. Furthermore, we report on the effects of DNA-PKcs inhibition on cell survival and expression of pro-metastatic genes in UM cell lines. We suggest that DNA-PKcs, encoded by the PRKDC gene on chromosome 8q, may be involved in proliferation, invasion, and metastasis of UM cells and should be investigated further. An increased insight of factors involved in DNA repair mechanisms in uveal melanoma will hopefully enhance our understanding of the pathogenesis of this disease and may eventually result in the identification of new targets of therapy.
Author Contributions
Conceptualization, M.D., A.R.T., U.P., A.G.J., M.J.J.; methodology, M.D., A.R.T., J.C., S.I.v.P., P.A.v.d.V., U.P., A.G.J., M.J.J.; software, M.D., S.I.v.P.; validation, S.A., A.A., U.P.; formal analysis, M.D., S.I.v.P., A.A.; investigation, M.D., A.R.T., J.C., S.C., S.G.v.D., P.A.v.d.V., A.A., U.P., A.G.J., M.J.J.; resources, S.C., W.G.M.K., A.F.A.S.T., S.A., S.G.v.D., G.P.M.L., P.A.v.d.V., A.A., U.P., and A.G.J.; data curation, X.X.; writing—original draft preparation, X.X.; writing—review and editing, M.D., A.R.T., J.C., S.C., W.G.M.K., A.F.A.S.T., S.A., S.G.v.D., G.P.M.L., P.A.v.d.V., A.A., U.P., A.G.J., and M.J.J.; visualization, M.D., A.A..; supervision, U.P., A.G.J., M.J.J.; project administration, U.P., A.G.J., M.J.J.; funding acquisition, A.R.T., U.P., A.G.J., and M.J.J.
Funding
The research was funded by ‘Funding Program Science Without Borders, CNPq/CAPES, Brazilian Government’, The Eye Cancer Foundation (New York, USA), ‘Horizon2020 UM CURE grant #667787’, and by a grant from the Associazione Italiana per la Ricerca sul Cancro, IG 17103, and a grant from the Compagnia di San Paolo (#20067) to UP.
Conflicts of Interest
The authors declare no conflict of interest.
Appendix A

Table A1.
Alphabetic list of all DNA repair genes (n = 121, encoded by 178 probes) evaluated in our cohort. The expression of genes with a standard deviation >0.3 (n = 44, encoded by 49 probes) was compared between disomy 3 (n = 24) and monosomy (n = 40) tumors. Bonferroni correction was applied to the unrounded p-values. Significant corrected p-values and corresponding probes are in bold.
Table A1.
Alphabetic list of all DNA repair genes (n = 121, encoded by 178 probes) evaluated in our cohort. The expression of genes with a standard deviation >0.3 (n = 44, encoded by 49 probes) was compared between disomy 3 (n = 24) and monosomy (n = 40) tumors. Bonferroni correction was applied to the unrounded p-values. Significant corrected p-values and corresponding probes are in bold.
| GENE | MEAN EXPRESSION | STANDARD DEVIATION | DISOMY 3 (n = 24) Median (Range) | MONOSOMY 3 (n = 40) Median (Range) | p-VALUE | CORRECTED p-VALUE |
|---|---|---|---|---|---|---|
| APEX1 | 9.76 | 0.58 | 10.2 (8.3–10.8) | 9.6 (8.5–10.7) | 0.001 | 0.07 |
| APEX1a | 10.69 | 0.42 | 11.0 (9.9–11.4) | 10.5 (9.6–11.4) | <0.001 | 0.004 |
| APEX2 | 7.87 | 0.21 | ||||
| APITD1 | Not in Illumina | Not in Illumina | ||||
| ATR | Not in Illumina | Not in Illumina | ||||
| ATRIP | 7.33 | 0.22 | ||||
| ATRIPa | 7.12 | 0.21 | ||||
| BAP1 | 7.52 | 0.53 | 8.0 (6.6–8.5) | 7.4 (6.4–8.1) | <0.001 | <0.001 |
| BIVM-ERCC5 | 7.46 | 0.28 | ||||
| BIVM-ERCC5a | 7.82 | 0.31 | 7.9 (7.4–8.3) | 7.8 (7.1–8.5) | 0.05 | 1 |
| BLM | 6.46 | 0.11 | ||||
| BRCA1 | 6.49 | 0.15 | ||||
| BRCA1a | 6.73 | 0.16 | ||||
| BRCA2 | Not in Illumina | Not in Illumina | ||||
| BRIP1 | 6.47 | 0.1 | ||||
| C17orf70 | 9.1 | 0.39 | 9.0 (8.3–9.8) | 9.2 (8.3–10.4) | 0.02 | 0.84 |
| C19orf40 | 7.06 | 0.19 | ||||
| CCNH | 7.54 | 0.34 | 7.4 (6.8–8.2) | 7.6 (7.0–8.4) | 0.008 | 0.38 |
| CCNHa | 7.66 | 0.22 | ||||
| CDK7 | 8.89 | 0.39 | 8.8 (7.7–9.7) | 8.9 (8.4–10.3) | 0.21 | 1 |
| CENPX | 9.61 | 0.38 | 9.3 (8.9–10.3) | 9.7 (9.0–10.6) | <0.001 | <0.001 |
| CETN2 | 10.02 | 0.38 | 10.2 (9.7–11.2) | 9.9 (9.3–10.7) | <0.001 | 0.002 |
| CUL4B | 7.71 | 0.27 | ||||
| CUL4Ba | 6.53 | 0.17 | ||||
| DCLRE1C | 6.7 | 0.14 | ||||
| DCLRE1Ca | 7.28 | 0.32 | 7.2 (6.7–8.4) | 7.3 (6.9–8.3) | 0.31 | 1 |
| DDB1 | 12.33 | 0.37 | 12.1 (11.3–13.0) | 12.4 (11.7–13.0) | 0.001 | 0.04 |
| DDB1a | 6.63 | 0.18 | ||||
| DDB1b | 6.9 | 0.18 | ||||
| DDB1c | 8.27 | 0.28 | ||||
| DDB2 | 6.95 | 0.23 | ||||
| DNTT | Not in Illumina | Not in Illumina | ||||
| EME1 | 6.54 | 0.1 | ||||
| EME2 | Not in Illumina | Not in Illumina | ||||
| ERCC1 | 9.52 | 0.27 | ||||
| ERCC1a | 7.38 | 0.3 | ||||
| ERCC1b | 9.74 | 0.29 | ||||
| ERCC1c | 6.53 | 0.11 | ||||
| ERCC2 | 7.79 | 0.28 | ||||
| ERCC3 | 7.98 | 0.17 | ||||
| ERCC3a | 6.6 | 0.11 | ||||
| ERCC4 | 6.47 | 0.09 | ||||
| ERCC6 | 6.48 | 0.11 | ||||
| ERCC8 | 6.62 | 0.13 | ||||
| ERCC8a | 6.52 | 0.12 | ||||
| EXO1 | 6.62 | 0.25 | ||||
| EXO1a | 6.39 | 0.13 | ||||
| FAN1 | 8.71 | 0.28 | ||||
| FAN1a | 7.1 | 0.16 | ||||
| FANCA | 6.5 | 0.12 | ||||
| FANCAa | 6.52 | 0.11 | ||||
| FANCAb | 6.36 | 0.13 | ||||
| FANCB | 6.4 | 0.12 | ||||
| FANCC | Not in Illumina | Not in Illumina | ||||
| FANCD2 | 6.74 | 0.2 | ||||
| FANCE | 7.59 | 0.36 | 7.7 (6.8–8.6) | 7.4 (7.1–8.3) | 0.003 | 0.13 |
| FANCF | Not in Illumina | Not in Illumina | ||||
| FANCG | 7.48 | 0.34 | 7.4 (7.0–8.5) | 7.4 (6.9–8.4) | 0.69 | 1 |
| FANCI | 6.59 | 0.18 | ||||
| FANCL | 6.42 | 0.14 | ||||
| FANCLa | 7.78 | 0.4 | 7.9 (7.1–8.6) | 7.6 (6.9–8.9) | 0.06 | 1 |
| FANCM | Not in Illumina | Not in Illumina | ||||
| FEN1 | 7.09 | 0.19 | ||||
| FEN1a | 8.97 | 0.32 | 9.0 (8.4–9.7) | 9.0 (8.1–9.6) | 0.98 | 1 |
| GTF2H1 | 6.91 | 0.19 | ||||
| GTF2H1a | 7.42 | 0.25 | ||||
| GTF2H2 | 6.27 | 0.13 | ||||
| GTF2H2B | 7.13 | 0.32 | 7.0 (6.6–7.9) | 7.2 (6.5–8.1) | 0.37 | 1 |
| GTF2H3 | 7.12 | 0.19 | ||||
| GTF2H4 | 8.09 | 0.47 | 8.5 (6.9–9.4) | 7.9 (7.2–9.3) | <0.001 | <0.001 |
| GTF2H5 | 10.19 | 0.43 | 10.1 (9.3–11.0) | 10.3 (9.3–11.2) | 0.01 | 0.62 |
| LIG1 | 8.27 | 0.31 | 8.2 (7.6–8.7) | 8.3 (7.8–9.0) | 0.09 | 1 |
| LIG3 | 6.85 | 0.12 | ||||
| LIG3a | 7.18 | 0.25 | ||||
| LIG4 | 6.63 | 0.08 | ||||
| LIG4a | 6.47 | 0.11 | ||||
| MBD4 | 8.26 | 0.46 | 8.4 (7.5–9.9) | 8.1 (7.5–9.3) | 0.33 | 1 |
| MGMT | 9.34 | 0.43 | 9.6 (8.7–10.3) | 9.2 (8.3–10.1) | 1 | |
| MLH1 | 7.94 | 0.33 | 8.2 (7.5–8.8) | 7.8 (7.1–8.3) | <0.001 | <0.001 |
| MLH3 | 6.44 | 0.11 | ||||
| MLH3a | 6.63 | 0.15 | ||||
| MNAT1 | 6.8 | 0.2 | ||||
| MPG | 6.61 | 0.09 | ||||
| MPGa | 6.31 | 0.12 | ||||
| MRE11A | 6.59 | 0.08 | ||||
| MRE11Aa | 6.74 | 0.17 | ||||
| MSH2 | 6.46 | 0.12 | ||||
| MSH2a | 6.68 | 0.12 | ||||
| MSH3 | 7.72 | 0.26 | ||||
| MSH3a | 13.39 | 0.65 | 13.3 (12.1–14.7) | 13.5 (12.3–15.0) | 0.13 | 1 |
| MSH6 | 8.8 | 0.3 | ||||
| MUS81 | 7.76 | 0.22 | ||||
| MUTYH | 6.61 | 0.12 | ||||
| MUTYHa | 7.85 | 0.38 | 7.9 (7.3–8.4) | 7.9 (6.9–8.5) | 0.39 | 1 |
| MUTYHb | 6.78 | 0.19 | ||||
| NBN | 8.1 | 0.42 | 7.9 (7.4–8.5) | 8.2 (7.3–9.2) | 0.005 | 0.24 |
| NBNa | 7.01 | 0.25 | ||||
| NEIL1 | 6.58 | 0.11 | ||||
| NEIL2 | 8.06 | 0.45 | 8.3 (7.4–8.8) | 7.9 (6.9–9.1) | 0.002 | 0.1 |
| NHEJ1 | Not in Illumina | Not in Illumina | ||||
| NEIL3 | Not in Illumina | Not in Illumina | ||||
| NTHL1 | 7.76 | 0.31 | 7.8 (7.1–8.5) | 7.7 (7.2–8.3) | 0.01 | 0.64 |
| OGG1 | 6.48 | 0.14 | ||||
| OGG1a | 6.94 | 0.2 | ||||
| PALB2 | 7.21 | 0.19 | ||||
| PARP2 | 7.43 | 0.22 | ||||
| PARP2a | 6.89 | 0.16 | ||||
| PCNA | 6.58 | 0.16 | ||||
| PCNAa | 8.53 | 0.51 | 8.3 (7.7–9.7) | 8.6 (7.5–9.9) | 0.002 | 0.12 |
| PMS2 | 6.97 | 0.21 | ||||
| PMS2a | 6.44 | 0.15 | ||||
| PMS2CL | 6.79 | 0.14 | ||||
| PMS2CLa | 6.48 | 0.11 | ||||
| POLB | 9.72 | 0.63 | 9.9 (9.1–10.7) | 9.7 (8.1–10.9) | 0.06 | 1 |
| POLD3 | 6.77 | 0.13 | ||||
| POLE3 | 9.62 | 0.31 | 9.7 (8.8–10.3) | 9.6 (9.1–10.3) | 0.24 | 1 |
| POLH | 6.52 | 0.13 | ||||
| POLHa | 6.55 | 0.09 | ||||
| POLI | Not in Illumina | Not in Illumina | ||||
| POLK | Not in Illumina | Not in Illumina | ||||
| POLL | 6.89 | 0.2 | ||||
| POLM | 7.14 | 0.22 | ||||
| POLN | 6.53 | 0.22 | ||||
| PRKDC | 6.64 | 0.21 | ||||
| PRKDCa | 8.55 | 0.68 | 8.0 (7.3–8.6) | 8.8 (7.8–10.2) | <0.001 | <0.001 |
| PRKDCb | 6.64 | 0.15 | ||||
| PRKDCc | 6.54 | 0.12 | ||||
| PRKDCd | 6.56 | 0.1 | ||||
| RAD23A | 9.77 | 0.24 | ||||
| RAD50 | 7.59 | 0.19 | ||||
| RAD51 | 6.97 | 0.26 | ||||
| RAD51a | 6.81 | 0.13 | ||||
| RAD51C | Not in Illumina | Not in Illumina | ||||
| RAD52 | Not in Illumina | Not in Illumina | ||||
| RAD54B | 6.52 | 0.1 | ||||
| RAD54Ba | 6.81 | 0.16 | ||||
| RBX1 | 10.17 | 0.39 | 10.0 (9.4–10.8) | 10.2 (9.6–11.1) | 0.01 | 0.55 |
| REV1 | 7.78 | 0.17 | ||||
| REV1a | 7.75 | 0.19 | ||||
| REV3L | 6.66 | 0.2 | ||||
| RFC1 | 7.37 | 0.19 | ||||
| RFC1a | 8.66 | 0.28 | ||||
| RMI1 | 6.84 | 0.14 | ||||
| RMI1a | 7.27 | 0.2 | ||||
| RMI2 | 7.07 | 0.32 | 7.2 (6.7–7.7) | 6.9 (6.5–7.7) | <0.001 | 0.02 |
| RPA1 | 8.2 | 0.34 | 8.5 (7.7–8.9) | 8.1 (7.4–8.9) | 0.006 | 0.28 |
| RPA1a | 9.74 | 0.36 | 9.9 (8.7–10.4) | 9.7 (8.9–10.5) | 0.02 | 1 |
| RPA1b | 8.39 | 0.36 | 8.7 (7.7–9.2) | 8.3 (7.4–8.9) | 0.001 | 0.04 |
| RPA2 | 9.95 | 0.38 | 10.1 (9.3–10.9) | 10.0 (9.2–10.7) | 0.22 | 1 |
| RPA3 | 9 | 0.41 | 9.1 (8.5–10.1) | 8.9 (8.0–9.9) | 0.09 | 1 |
| RPA4 | 6.38 | 0.13 | ||||
| SEM1 | 11.32 | 0.24 | ||||
| SEM1a | 7.53 | 0.32 | 7.7 (7.3–8.4) | 7.4 (6.8–8.0) | <0.001 | 0.01 |
| SLX1A | Not in Illumina | Not in Illumina | ||||
| SLX1B | Not in Illumina | Not in Illumina | ||||
| SLX4 | 6.96 | 0.19 | ||||
| SMUG1 | 9.04 | 0.23 | ||||
| TELO2 | 6.96 | 0.29 | ||||
| TDG | Not in Illumina | Not in Illumina | ||||
| TOP3A | 7.39 | 0.36 | 7.3 (6.7–8.6) | 7.4 (6.9–8.3) | 0.22 | 1 |
| TOP3Aa | 6.78 | 0.15 | ||||
| TOP3B | 7.89 | 0.26 | ||||
| UBE2T | 7.32 | 0.37 | 7.3 (6.8–8.2) | 7.3 (6.7–8.5) | 0.49 | 1 |
| UNG | 6.31 | 0.12 | ||||
| UNGa | 9.6 | 0.41 | 9.6 (8.8–10.5) | 9.7 (8.7–10.3) | 0.53 | 1 |
| UNGb | 6.54 | 0.11 | ||||
| USP1 | 7.57 | 0.29 | ||||
| USP1a | 6.48 | 0.12 | ||||
| WDR48 | 7.82 | 0.36 | 8.2 (7.4–8.6) | 7.6 (7.2–8.2) | <0.001 | <0.001 |
| XPA | 6.99 | 0.14 | ||||
| XPC | 8.76 | 0.42 | 9.2 (8.3–9.7) | 8.6 (8.0–9.3) | <0.001 | <0.001 |
| XRCC1 | 7.88 | 0.3 | 7.9 (7.3–8.7) | 7.9 (7.3–8.5) | 0.57 | 1 |
| XRCC4 | Not in Illumina | Not in Illumina | ||||
| XRCC5 | 8.94 | 0.32 | 9.0 (8.2–9.6) | 9.0 (8.1–9.6) | 0.95 | 1 |
| XRCC6 | 8.5 | 0.31 | 8.3 (7.8–8.8) | 8.6 (7.7–9.3) | 0.005 | 0.26 |
| XRCC6a | 6.73 | 0.13 | ||||
| XRCC6b | 8.75 | 0.38 | 8.7 (7.9–9.2) | 8.8 (7.9–10.0) | 0.65 | 1 |
| XRCC6c | 10.61 | 0.37 | 10.5 (9.5–11.2) | 10.7 (9.8–11.2) | 0.02 | 0.87 |
Appendix B

Table A2.
Association of the 13 significantly differentially expressed genes between disomy 3 and monosomy 3 tumors, with clinicopathologic parameters and survival. Significant p-values are indicated in bold.
Table A2.
Association of the 13 significantly differentially expressed genes between disomy 3 and monosomy 3 tumors, with clinicopathologic parameters and survival. Significant p-values are indicated in bold.
| A: Highly expressed genes in monosomy 3 tumors. | ||||||||||
| CHARACTERISTIC | GENE | |||||||||
| CENPX (17q25.3) | DDB1 (11q12) | PRKDC (8q11.21) | ||||||||
| PATHWAY | ||||||||||
| FA | NER | DSBR | ||||||||
| Largest Basal Diameter | ||||||||||
| ≤13 mm (n = 34) median(range) | 9.4 (9.0–10.6) | 12.3 (11.3–13.0) | 8.4 (7.5–9.8) | |||||||
| >13 mm (n = 30) median(range) | 9.7 (8.9–10.5) | 12.3 (11.7–12.8) | 8.7 (7.3–10.2) | |||||||
| p-value * | 0.02 | 0.93 | 0.11 | |||||||
| Cell type | ||||||||||
| Spindle (n = 22) median (range) | 9.5 (8.9–10.4) | 12.3 (11.7–13.0) | 8.2 (7.3–9.7) | |||||||
| Mixed/epithelioid (n = 42) median(range) | 9.6 (9.0–10.6) | 12.3 (11.3–13.0) | 8.6 (7.5–10.2) | |||||||
| p-value * | 0.04 | 0.59 | 0.12 | |||||||
| AJCC Stage | ||||||||||
| Stage I (n = 5) median (range) | 9.3 (9.0–9.9) | 12.6 (12.2–13.0) | 8.2 (7.7–9.6) | |||||||
| Stage II (n = 36) median (range) | 9.5 (8.9–10.6) | 12.3 (11.3–13.0) | 8.4 (7.5–9.8) | |||||||
| Stage III (n = 23) median (range) | 9.7 (9.3–10.5) | 12.3 (11.8–12.9) | 8.7 (7.3–10.2) | |||||||
| p-value † | 0.01 | 0.1 | 0.19 | |||||||
| Presence of metastases | ||||||||||
| No (n = 27) median (range) | 9.3 (9.0–10.4) | 12.3 (11.2–13.0) | 8.1 (7.5–9.4) | |||||||
| Yes (n = 37) median (range) | 9.8 (8.9–10.6) | 12.4 (11.7–13.0) | 8.8 (7.3–10.2) | |||||||
| p-value * | <0.001 | 0.3 | <0.001 | |||||||
| Survival analysis | ||||||||||
| Expression lower than or equal to median | ≤9.6 | ≤12.3 | ≤8.4 | |||||||
| Expression higher than median | >9.6 § | >12.3 | >8.4 § | |||||||
| p-value ‡ | <0.001 | 0.48 | 0.001 | |||||||
| B: Lowly expressed genes in monosomy 3 tumors. | ||||||||||
| CHARACTERISTIC | GENE | |||||||||
| APEX1 (14q11.2) | BAP1 (3p21.1) | CETN2 (Xq28) | GTF2H4 (6p21.33) | MLH1 (3p22.2) | RMI2 (16p13.13) | RPA1 (17p13.3) | SEM1 (7q21.3) | WDR48 (3p22.2) | XPC (3p25.1) | |
| PATHWAY | ||||||||||
| BER | DSBR | NER | NER | MMR/FA | DSBR | DSBR/MMR/NER | DSBR | FA | NER | |
| LBD | ||||||||||
| ≤13 mm (n = 34) median(range) | 10.7 (9.9–11.4) | 7.7 (6.6–8.5) | 10.0 (9.3–11.2) | 8.0(6.9–9.1) | 8.0(7.5–8.8) | 7.09 (6.6–7.7) | 8.4 (7.7–9.2) | 7.5(7.0–8.4) | 8.0(7.2–8.5) | 8.9 (8.0–9.6) |
| >13 mm (n = 30) median(range) | 10.7 (9.6–11.4) | 7.4 (6.4–8.5) | 9.9 (9.3–10.9) | 8.0(7.2–9.4) | 7.9 (7.1–8.6) | 7.0 (6.5–7.7) | 8.4 (7.4–8.8) | 7.4 (6.8–8.0) | 7.7 (7.2–8.6) | 8.6 (8.0–9.7) |
| p-value * | 0.75 | 0.17 | 0.24 | 0.86 | 0.16 | 0.11 | 0.2 | 0.11 | 0.01 | 0.004 |
| Cell type | ||||||||||
| Spindle (n = 22) median (range) | 10.8 (9.6–11.4) | 7.7 (6.6–8.5) | 10.1 (9.6–10.9) | 8.2 (7.2–9.4) | 7.9 (7.1–8.8) | 7.1 (6.5–7.7) | 8.6 (7.4–9.2) | 7.5 (6.8–8.0) | 8.0(7.3–8.6) | 8.9 (8.0–9.7) |
| Mixed/epithelioid (n = 42) median(range) | 10.6 (9.6–11.4) | 7.6 (6.4–8.5) | 10.0 (9.3–11.2) | 8.0 (6.9–9.1) | 7.9 (7.2–8.8) | 6.9 (6.6–7.7) | 8.4(7.5–9.1) | 7.5 (6.9–8.4) | 7.7 (7.2–8.5) | 8.7 (8.0–9.3) |
| p-value * | 0.13 | 0.15 | 0.1 | 0.12 | 0.97 | 0.15 | 0.06 | 0.09 | 0.007 | 0.03 |
| AJCC Stage | ||||||||||
| Stage I (n = 5) median (range) | 10.7 (10.4–11.0) | 7.7 (7.5–8.1) | 10.1 (9.3–11.2) | 8.1 (7.9–8.5) | 7.8 (7.6–8.4) | 7.1 (6.9–7.6) | 8.7 (7.9–9.1) | 7.6 (7.1–8.2) | 7.9 (7.5–8.5) | 8.8 (8.6–9.2) |
| Stage II (n = 36) median (range) | 10.8 (9.7–11.4) | 7.7 (6.4–8.5) | 10.0(9.6–10.7) | 8.0 (6.9–9.4) | 8.0 (7.5–8.8) | 7.0 (6.6–7.7) | 8.5 (7.7–9.2) | 7.5 (7.0–8.4) | 8.1 (7.3–8.6) | 8.8 (8.0–9.7) |
| Stage III (n = 23)median (range) | 10.5 (9.6–11.3) | 7.4 (6.6–8.5) | 9.9 (9.3–10.9) | 8.0 (7.5–9.1) | 7.8 (7.1–8.6) | 7.1 (6.5–7.7) | 8.3 (7.4–8.8) | 7.4 (6.8–8.0) | 7.7 (7.2–8.3) | 8.6 (8.0–9.3) |
| p-value † | 0.1 | 0.35 | 0.16 | 0.76 | 0.05 | 0.44 | 0.03 | 0.27 | 0.08 | 0.14 |
| Presence of metastases | ||||||||||
| No (n = 27) median (range) | 10.8 (9.9–11.4) | 7.8 (6.6–8.5) | 10.1 (9.7–11.2) | 8.3 (6.9–9.3) | 8.1 (7.5–8.8) | 7.1(6.7–7.7) | 8.4(7.7–9.2) | 7.6 (7.3–8.4) | 8.1(7.2–8.6) | 9.0 (8.0–9.6) |
| Yes (n = 37) median (range) | 10.6 (9.6–11.4) | 7.4(6.4–8.3) | 9.9 (9.3–10.9) | 7.9 (7.2–9.4) | 7.9 (7.1–8.6) | 6.9 (6.5–7.7) | 8.4 (7.4–8.9) | 7.4 (6.8–8.01) | 7.7 (7.2–8.3) | 8.6 (8.0–9.7) |
| p-value * | 0.11 | 0.003 | 0.03 | 0.005 | 0.02 | 0.003 | 0.09 | 0.006 | <0.001 | 0.006 |
| Survival analysis | ||||||||||
| Expression ≤ median | ≤10.7 | ≤7.6 § | ≤10 | ≤8 § | ≤7.9 | ≤7.1 § | ≤8.4 | ≤7.5 § | ≤7.8 § | ≤8.7 § |
| Expression higher than median | >10.7 | >7.6 | >10 | >8 | >7.9 | >7.1 | >8.4 | >7.5 | >7.8 | >8.7 |
| p-value ‡ | 0.05 | 0.001 | 0.18 | 0.001 | 0.07 | 0.02 | 0.41 | 0.006 | <0.001 | 0.005 |
Abbreviations: BER: base excision repair; DSBR: double-strand break repair; FA: fanconi anemia; MMR: mismatch repair; NER: nucleotide excision repair. Symbols: *: Mann–Whitney U test, †: Kruskal–Wallis test, ‡: Log-rank test, §: worse survival.
References
- Chang, A.E.; Karnell, L.H.; Menck, H.R. The National Cancer Data Base report on cutaneous and noncutaneous melanoma: A summary of 84,836 cases from the past decade. The American College of Surgeons Commission on Cancer and the American Cancer Society. Cancer 1998, 83, 1664–1678. [Google Scholar] [CrossRef]
- Singh, A.D.; Turell, M.E.; Topham, A.K. Uveal melanoma: Trends in incidence, treatment, and survival. Ophthalmology 2011, 118, 1881–1885. [Google Scholar] [CrossRef] [PubMed]
- Holly, E.A.; Aston, D.A.; Char, D.H.; Kristiansen, J.J.; Ahn, D.K. Uveal melanoma in relation to ultraviolet light exposure and host factors. Cancer Res. 1990, 50, 5773–5777. [Google Scholar] [PubMed]
- Weis, E.; Shah, C.P.; Lajous, M.; Shields, J.A.; Shields, C.L. The association between host susceptibility factors and uveal melanoma: A meta-analysis. Arch. Ophthalmol. 2006, 124, 54–60. [Google Scholar] [CrossRef] [PubMed]
- Diener-West, M.; Hawkins, B.S.; Markowitz, J.A.; Schachat, A.P. A review of mortality from choroidal melanoma. II. A meta-analysis of 5-year mortality rates following enucleation, 1966 through 1988. Arch. Ophthalmol. 1992, 110, 245–250. [Google Scholar] [CrossRef] [PubMed]
- Jampol, L.M.; Moy, C.S.; Murray, T.G.; Reynolds, S.M.; Albert, D.M.; Schachat, A.P.; Diddie, K.R.; Engstrom, R.E., Jr.; Finger, P.T.; Hovland, K.R.; et al. The COMS randomized trial of iodine 125 brachytherapy for choroidal melanoma: IV. Local treatment failure and enucleation in the first 5 years after brachytherapy. COMS report no. 19. Ophthalmology 2002, 109, 2197–2206. [Google Scholar] [CrossRef]
- Krohn, J.; Monge, O.R.; Skorpen, T.N.; Mork, S.J.; Dahl, O. Posterior uveal melanoma treated with I-125 brachytherapy or primary enucleation. Eye 2008, 22, 1398–1403. [Google Scholar] [CrossRef]
- Tarmann, L.; Wackernagel, W.; Avian, A.; Mayer, C.; Schneider, M.; Winkler, P.; Langmann, G. Ruthenium-106 plaque brachytherapy for uveal melanoma. Br. J. Ophthalmol. 2015, 99, 1644–1649. [Google Scholar] [CrossRef]
- Riechardt, A.I.; Cordini, D.; Willerding, G.D.; Georgieva, I.; Weber, A.; Seibel, I.; Lakotka, N.; Bechrakis, N.E.; Foerster, M.H.; Moser, L.; et al. Proton beam therapy of parapapillary choroidal melanoma. Am. J. Ophthalmol. 2014, 157, 1258–1265. [Google Scholar] [CrossRef]
- Seddon, J.M.; Gragoudas, E.S.; Egan, K.M.; Glynn, R.J.; Howard, S.; Fante, R.G.; Albert, D.M. Relative survival rates after alternative therapies for uveal melanoma. Ophthalmology 1990, 97, 769–777. [Google Scholar] [CrossRef]
- Augsburger, J.J.; Correa, Z.M.; Shaikh, A.H. Effectiveness of treatments for metastatic uveal melanoma. Am. J. Ophthalmol. 2009, 148, 119–127. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.D.; Bergman, L.; Seregard, S. Uveal melanoma: Epidemiologic aspects. Ophthalmol. Clin. N. Am. 2005, 18, 75–84. [Google Scholar] [CrossRef] [PubMed]
- Kujala, E.; Makitie, T.; Kivela, T. Very long-term prognosis of patients with malignant uveal melanoma. Investig. Ophthalmol. Vis. Sci. 2003, 44, 4651–4659. [Google Scholar] [CrossRef] [PubMed]
- Kujala, E.; Damato, B.; Coupland, S.E.; Desjardins, L.; Bechrakis, N.E.; Grange, J.D.; Kivela, T. Staging of ciliary body and choroidal melanomas based on anatomic extent. J. Clin. Oncol. 2013, 31, 2825–2831. [Google Scholar] [CrossRef] [PubMed]
- Seddon, J.M.; Albert, D.M.; Lavin, P.T.; Robinson, N. A prognostic factor study of disease-free interval and survival following enucleation for uveal melanoma. Arch. Ophthalmol. 1983, 101, 1894–1899. [Google Scholar] [CrossRef] [PubMed]
- Affeldt, J.C.; Minckler, D.S.; Azen, S.P.; Yeh, L. Prognosis in uveal melanoma with extrascleral extension. Arch. Ophthalmol. 1980, 98, 1975–1979. [Google Scholar] [CrossRef] [PubMed]
- Makitie, T.; Summanen, P.; Tarkkanen, A.; Kivela, T. Microvascular loops and networks as prognostic indicators in choroidal and ciliary body melanomas. J. Nat. Cancer Inst. 1999, 91, 359–367. [Google Scholar] [CrossRef] [PubMed]
- Shields, C.L.; Furuta, M.; Thangappan, A.; Nagori, S.; Mashayekhi, A.; Lally, D.R.; Kelly, C.C.; Rudich, D.S.; Nagori, A.V.; Wakade, O.A.; et al. Metastasis of uveal melanoma millimeter-by-millimeter in 8033 consecutive eyes. Arch. Ophthalmol. 2009, 127, 989–998. [Google Scholar] [CrossRef] [PubMed]
- Horsman, D.E.; Sroka, H.; Rootman, J.; White, V.A. Monosomy 3 and isochromosome 8q in a uveal melanoma. Cancer Genet. Cytogenet. 1990, 45, 249–253. [Google Scholar] [CrossRef]
- Prescher, G.; Bornfeld, N.; Hirche, H.; Horsthemke, B.; Jockel, K.H.; Becher, R. Prognostic implications of monosomy 3 in uveal melanoma. Lancet 1996, 347, 1222–1225. [Google Scholar]
- Sisley, K.; Rennie, I.G.; Parsons, M.A.; Jacques, R.; Hammond, D.W.; Bell, S.M.; Potter, A.M.; Rees, R.C. Abnormalities of chromosomes 3 and 8 in posterior uveal melanoma correlate with prognosis. Genes Chromosomes Cancer 1997, 19, 22–28. [Google Scholar] [CrossRef]
- Kilic, E.; Naus, N.C.; van Gils, W.; Klaver, C.C.; van Til, M.E.; Verbiest, M.M.; Stijnen, T.; Mooy, C.M.; Paridaens, D.; Beverloo, H.B.; et al. Concurrent loss of chromosome arm 1p and chromosome 3 predicts a decreased disease-free survival in uveal melanoma patients. Investig. Ophthalmol. Vis. Sci. 2005, 46, 2253–2257. [Google Scholar] [CrossRef]
- Dogrusöz, M.; Jager, M.J. Genetic prognostication in uveal melanoma. Acta Ophthalmol. 2018, 96, 331–347. [Google Scholar] [CrossRef]
- White, V.A.; Chambers, J.D.; Courtright, P.D.; Chang, W.Y.; Horsman, D.E. Correlation of cytogenetic abnormalities with the outcome of patients with uveal melanoma. Cancer 1998, 83, 354–359. [Google Scholar] [CrossRef]
- Damato, B.; Dopierala, J.; Klaasen, A.; van Dijk, M.; Sibbring, J.; Coupland, S.E. Multiplex ligation-dependent probe amplification of uveal melanoma: Correlation with metastatic death. Investig. Ophthalmol. Vis. Sci. 2009, 50, 3048–3055. [Google Scholar] [CrossRef]
- Onken, M.D.; Worley, L.A.; Ehlers, J.P.; Harbour, J.W. Gene expression profiling in uveal melanoma reveals two molecular classes and predicts metastatic death. Cancer Res. 2004, 64, 7205–7209. [Google Scholar] [CrossRef]
- Onken, M.D.; Worley, L.A.; Tuscan, M.D.; Harbour, J.W. An accurate, clinically feasible multi-gene expression assay for predicting metastasis in uveal melanoma. J. Mol. Diagn. 2010, 12, 461–468. [Google Scholar] [CrossRef]
- Onken, M.D.; Worley, L.A.; Char, D.H.; Augsburger, J.J.; Correa, Z.M.; Nudleman, E.; Aaberg, T.M., Jr.; Altaweel, M.M.; Bardenstein, D.S.; Finger, P.T.; et al. Collaborative Ocular Oncology Group report number 1: Prospective validation of a multi-gene prognostic assay in uveal melanoma. Ophthalmology 2012, 119, 1596–1603. [Google Scholar] [CrossRef]
- Harbour, J.W.; Onken, M.D.; Roberson, E.D.; Duan, S.; Cao, L.; Worley, L.A.; Council, M.L.; Matatall, K.A.; Helms, C.; Bowcock, A.M. Frequent mutation of BAP1 in metastasizing uveal melanomas. Science 2010, 330, 1410–1413. [Google Scholar] [CrossRef]
- Yavuzyigitoglu, S.; Koopmans, A.E.; Verdijk, R.M.; Vaarwater, J.; Eussen, B.; van Bodegom, A.; Paridaens, D.; Kilic, E.; de Klein, A. Rotterdam Ocular Melanoma Study Group. Uveal Melanomas with SF3B1 Mutations: A Distinct Subclass Associated with Late-Onset Metastases. Ophthalmology 2016, 123, 1118–1128. [Google Scholar] [CrossRef]
- Ewens, K.G.; Kanetsky, P.A.; Richards-Yutz, J.; Purrazzella, J.; Shields, C.L.; Ganguly, T.; Ganguly, A. Chromosome 3 Status Combined with BAP1 and EIF1AX Mutation Profiles Are Associated with Metastasis in Uveal Melanoma. Investig. Ophthalmol. Vis. Sci. 2014, 55, 5160–5167. [Google Scholar] [CrossRef]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef]
- Amaro, A.; Gangemi, R.; Piaggio, F.; Angelini, G.; Barisione, G.; Ferrini, S.; Pfeffer, U. The biology of uveal melanoma. Cancer Metastasis Rev. 2017, 36, 109–140. [Google Scholar] [CrossRef]
- van Essen, T.H.; van Pelt, S.I.; Versluis, M.; Bronkhorst, I.H.; van Duinen, S.G.; Marinkovic, M.; Kroes, W.G.; Ruivenkamp, C.A.; Shukla, S.; de Klein, A.; et al. Prognostic parameters in uveal melanoma and their association with BAP1 expression. Br. J. Ophthalmol. 2014, 98, 1738–1743. [Google Scholar] [CrossRef]
- Shah, A.A.; Bourne, T.D.; Murali, R. BAP1 protein loss by immunohistochemistry: A potentially useful tool for prognostic prediction in patients with uveal melanoma. Pathology 2013, 45, 651–656. [Google Scholar] [CrossRef]
- Yu, H.; Pak, H.; Hammond-Martel, I.; Ghram, M.; Rodrigue, A.; Daou, S.; Barbour, H.; Corbeil, L.; Hebert, J.; Drobetsky, E.; et al. Tumor suppressor and deubiquitinase BAP1 promotes DNA double-strand break repair. Proc. Nat. Acad. Sci. USA 2014, 111, 285–290. [Google Scholar] [CrossRef]
- Helleday, T.; Petermann, E.; Lundin, C.; Hodgson, B.; Sharma, R.A. DNA repair pathways as targets for cancer therapy. Nat. Rev. Cancer 2008, 8, 193–204. [Google Scholar] [CrossRef]
- Lieberman, H.B. DNA damage repair and response proteins as targets for cancer therapy. Curr. Med. Chem. 2008, 15, 360–367. [Google Scholar] [CrossRef]
- Kelley, M.R.; Fishel, M.L. DNA repair proteins as molecular targets for cancer therapeutics. Anti Cancer Agents Med. Chem. 2008, 8, 417–425. [Google Scholar] [CrossRef]
- Gangemi, R.; Mirisola, V.; Barisione, G.; Fabbi, M.; Brizzolara, A.; Lanza, F.; Mosci, C.; Salvi, S.; Gualco, M.; Truini, M.; et al. Mda-9/syntenin is expressed in uveal melanoma and correlates with metastatic progression. PLoS ONE 2012, 7, e29989. [Google Scholar] [CrossRef]
- Laurent, C.; Valet, F.; Planque, N.; Silveri, L.; Maacha, S.; Anezo, O.; Hupe, P.; Plancher, C.; Reyes, C.; Albaud, B.; et al. High PTP4A3 phosphatase expression correlates with metastatic risk in uveal melanoma patients. Cancer Res. 2011, 71, 666–674. [Google Scholar] [CrossRef]
- Robertson, A.G.; Shih, J.; Yau, C.; Gibb, E.A.; Oba, J.; Mungall, K.L.; Hess, J.M.; Uzunangelov, V.; Walter, V.; Danilova, L.; et al. Integrative Analysis Identifies Four Molecular and Clinical Subsets in Uveal Melanoma. Cancer Cell 2018, 33, 151. [Google Scholar] [CrossRef]
- Tschentscher, F.; Husing, J.; Holter, T.; Kruse, E.; Dresen, I.G.; Jockel, K.H.; Anastassiou, G.; Schilling, H.; Bornfeld, N.; Horsthemke, B.; et al. Tumor classification based on gene expression profiling shows that uveal melanomas with and without monosomy 3 represent two distinct entities. Cancer Res. 2003, 63, 2578–2584. [Google Scholar]
- Van Gils, W.; Lodder, E.M.; Mensink, H.W.; Kilic, E.; Naus, N.C.; Bruggenwirth, H.T.; van Ijcken, W.; Paridaens, D.; Luyten, G.P.; de Klein, A. Gene expression profiling in uveal melanoma: Two regions on 3p related to prognosis. Investig. Ophthalmol. Vis. Sci. 2008, 49, 4254–4262. [Google Scholar] [CrossRef]
- Goodwin, J.F.; Kothari, V.; Drake, J.M.; Zhao, S.; Dylgjeri, E.; Dean, J.L.; Schiewer, M.J.; McNair, C.; Jones, J.K.; Aytes, A.; et al. DNA-PKcs-Mediated Transcriptional Regulation Drives Prostate Cancer Progression and Metastasis. Cancer Cell 2015, 28, 97–113. [Google Scholar] [CrossRef]
- Evert, M.; Frau, M.; Tomasi, M.L.; Latte, G.; Simile, M.M.; Seddaiu, M.A.; Zimmermann, A.; Ladu, S.; Staniscia, T.; Brozzetti, S.; et al. Deregulation of DNA-dependent protein kinase catalytic subunit contributes to human hepatocarcinogenesis development and has a putative prognostic value. Br. J. Cancer 2013, 109, 2654–2664. [Google Scholar] [CrossRef]
- Asnaghi, L.; Gezgin, G.; Tripathy, A.; Handa, J.T.; Merbs, S.L.; van der Velden, P.A.; Jager, M.J.; Harbour, J.W.; Eberhart, C.G. EMT-associated factors promote invasive properties of uveal melanoma cells. Mol. Vis. 2015, 21, 919–929. [Google Scholar]
- Ma, H.; Takahashi, A.; Yoshida, Y.; Adachi, A.; Kanai, T.; Ohno, T.; Nakano, T. Combining carbon ion irradiation and non-homologous end-joining repair inhibitor NU7026 efficiently kills cancer cells. Radiat. Oncol. 2015, 10, 255. [Google Scholar] [CrossRef]
- Fontana, A.O.; Augsburger, M.A.; Grosse, N.; Guckenberger, M.; Lomax, A.J.; Sartori, A.A.; Pruschy, M.N. Differential DNA repair pathway choice in cancer cells after proton- and photon-irradiation. Radiother. Oncol. 2015, 116, 374–380. [Google Scholar] [CrossRef]
- Dolman, M.E.; van der Ploeg, I.; Koster, J.; Bate-Eya, L.T.; Versteeg, R.; Caron, H.N.; Molenaar, J.J. DNA-Dependent Protein Kinase as Molecular Target for Radiosensitization of Neuroblastoma Cells. PLoS One 2015, 10, e0145744. [Google Scholar] [CrossRef]
- Friedberg, E.C. DNA damage and repair. Nature 2003, 421, 436–440. [Google Scholar] [CrossRef]
- Zhou, B.B.; Elledge, S.J. The DNA damage response: Putting checkpoints in perspective. Nature 2000, 408, 433–439. [Google Scholar] [CrossRef]
- Shields, C.L.; Ganguly, A.; Bianciotto, C.G.; Turaka, K.; Tavallali, A.; Shields, J.A. Prognosis of Uveal Melanoma in 500 Cases Using Genetic Testing of Fine-Needle Aspiration Biopsy Specimens. Ophthalmology 2011, 118, 396–401. [Google Scholar] [CrossRef]
- Jensen, D.E.; Proctor, M.; Marquis, S.T.; Gardner, H.P.; Ha, S.I.; Chodosh, L.A.; Ishov, A.M.; Tommerup, N.; Vissing, H.; Sekido, Y.; et al. BAP1: A novel ubiquitin hydrolase which binds to the BRCA1 RING finger and enhances BRCA1-mediated cell growth suppression. Oncogene 1998, 16, 1097–1112. [Google Scholar] [CrossRef]
- Nishikawa, H.; Wu, W.; Koike, A.; Kojima, R.; Gomi, H.; Fukuda, M.; Ohta, T. BRCA1-associated protein 1 interferes with BRCA1/BARD1 RING heterodimer activity. Cancer Res. 2009, 69, 111–119. [Google Scholar] [CrossRef]
- Ismail, I.H.; Davidson, R.; Gagne, J.P.; Xu, Z.Z.; Poirier, G.G.; Hendzel, M.J. Germline mutations in BAP1 impair its function in DNA double-strand break repair. Cancer Res. 2014, 74, 4282–4294. [Google Scholar] [CrossRef]
- Cohn, M.A.; Kee, Y.; Haas, W.; Gygi, S.P.; D’Andrea, A.D. UAF1 is a subunit of multiple deubiquitinating enzyme complexes. J. Biol. Chem. 2009, 284, 5343–5351. [Google Scholar] [CrossRef]
- Kennedy, R.D.; D’Andrea, A.D. The Fanconi Anemia/BRCA pathway: New faces in the crowd. Genes Dev. 2005, 19, 2925–2940. [Google Scholar] [CrossRef]
- Dunn, J.; Potter, M.; Rees, A.; Runger, T.M. Activation of the Fanconi anemia/BRCA pathway and recombination repair in the cellular response to solar ultraviolet light. Cancer Res. 2006, 66, 11140–11147. [Google Scholar] [CrossRef]
- Nijman, S.M.; Huang, T.T.; Dirac, A.M.; Brummelkamp, T.R.; Kerkhoven, R.M.; D’Andrea, A.D.; Bernards, R. The deubiquitinating enzyme USP1 regulates the Fanconi anemia pathway. Mol. Cell 2005, 17, 331–339. [Google Scholar] [CrossRef]
- Cohn, M.A.; Kowal, P.; Yang, K.; Haas, W.; Huang, T.T.; Gygi, S.P.; D’Andrea, A.D. A UAF1-containing multisubunit protein complex regulates the Fanconi anemia pathway. Mol. Cell 2007, 28, 786–797. [Google Scholar] [CrossRef]
- Park, E.; Kim, J.M.; Primack, B.; Weinstock, D.M.; Moreau, L.A.; Parmar, K.; D’Andrea, A.D. Inactivation of Uaf1 causes defective homologous recombination and early embryonic lethality in mice. Mol. Cell. Biol. 2013, 33, 4360–4370. [Google Scholar] [CrossRef]
- Sands, A.T.; Abuin, A.; Sanchez, A.; Conti, C.J.; Bradley, A. High susceptibility to ultraviolet-induced carcinogenesis in mice lacking XPC. Nature 1995, 377, 162–165. [Google Scholar] [CrossRef]
- De Laat, W.L.; Jaspers, N.G.; Hoeijmakers, J.H. Molecular mechanism of nucleotide excision repair. Genes Dev. 1999, 13, 768–785. [Google Scholar] [CrossRef]
- Sugasawa, K.; Ng, J.M.; Masutani, C.; Iwai, S.; van der Spek, P.J.; Eker, A.P.; Hanaoka, F.; Bootsma, D.; Hoeijmakers, J.H. Xeroderma pigmentosum group C protein complex is the initiator of global genome nucleotide excision repair. Mol. Cell 1998, 2, 223–232. [Google Scholar] [CrossRef]
- Volker, M.; Mone, M.J.; Karmakar, P.; van Hoffen, A.; Schul, W.; Vermeulen, W.; Hoeijmakers, J.H.; van Driel, R.; van Zeeland, A.A.; Mullenders, L.H. Sequential assembly of the nucleotide excision repair factors in vivo. Mol. Cell 2001, 8, 213–224. [Google Scholar] [CrossRef]
- Yang, J.; Xu, Z.; Li, J.; Zhang, R.; Zhang, G.; Ji, H.; Song, B.; Chen, Z. XPC epigenetic silence coupled with p53 alteration has a significant impact on bladder cancer outcome. J. Urol. 2010, 184, 336–343. [Google Scholar] [CrossRef]
- Araki, M.; Masutani, C.; Takemura, M.; Uchida, A.; Sugasawa, K.; Kondoh, J.; Ohkuma, Y.; Hanaoka, F. Centrosome protein centrin 2/caltractin 1 is part of the xeroderma pigmentosum group C complex that initiates global genome nucleotide excision repair. J. Biol. Chem. 2001, 276, 18665–18672. [Google Scholar] [CrossRef]
- Ramkumar, H.L.; Brooks, B.P.; Cao, X.; Tamura, D.; Digiovanna, J.J.; Kraemer, K.H.; Chan, C.C. Ophthalmic manifestations and histopathology of xeroderma pigmentosum: Two clinicopathological cases and a review of the literature. Surv. Ophthalmol. 2011, 56, 348–361. [Google Scholar] [CrossRef]
- Sipley, J.D.; Menninger, J.C.; Hartley, K.O.; Ward, D.C.; Jackson, S.P.; Anderson, C.W. Gene for the catalytic subunit of the human DNA-activated protein kinase maps to the site of the XRCC7 gene on chromosome 8. Proc. Nat. Acad. Sci. USA 1995, 92, 7515–7519. [Google Scholar] [CrossRef]
- Jackson, S.P.; Jeggo, P.A. DNA double-strand break repair and V(D)J recombination: Involvement of DNA-PK. Trends Biochem. Sci. 1995, 20, 412–415. [Google Scholar] [CrossRef]
- Yoo, S.; Dynan, W.S. Geometry of a complex formed by double strand break repair proteins at a single DNA end: Recruitment of DNA-PKcs induces inward translocation of Ku protein. Nucleic Acids Res. 1999, 27, 4679–4686. [Google Scholar] [CrossRef]
- An, J.; Huang, Y.C.; Xu, Q.Z.; Zhou, L.J.; Shang, Z.F.; Huang, B.; Wang, Y.; Liu, X.D.; Wu, D.C.; Zhou, P.K. DNA-PKcs plays a dominant role in the regulation of H2AX phosphorylation in response to DNA damage and cell cycle progression. BMC Mol. Biol. 2010, 11, 18. [Google Scholar] [CrossRef]
- Mehta, A.; Haber, J.E. Sources of DNA Double-Strand Breaks and Models of Recombinational DNA Repair. Cold Spring Harb. Perspect. Biol. 2014, 6, a016428. [Google Scholar] [CrossRef]
- Jager, M.J.; Brouwer, N.J.; Esmaeli, B. The Cancer Genome Atlas Project: An integrated molecular view of uveal melanoma. Ophthalmology 2018, 125, 1139–1142. [Google Scholar] [CrossRef]
- Mukherjee, B.; McEllin, B.; Camacho, C.V.; Tomimatsu, N.; Sirasanagandala, S.; Nannepaga, S.; Hatanpaa, K.J.; Mickey, B.; Madden, C.; Maher, E.; et al. EGFRvIII and DNA double-strand break repair: A molecular mechanism for radioresistance in glioblastoma. Cancer Res. 2009, 69, 4252–4259. [Google Scholar] [CrossRef]
- Shintani, S.; Mihara, M.; Li, C.; Nakahara, Y.; Hino, S.; Nakashiro, K.; Hamakawa, H. Up-regulation of DNA-dependent protein kinase correlates with radiation resistance in oral squamous cell carcinoma. Cancer Sci. 2003, 94, 894–900. [Google Scholar] [CrossRef]
- Li, X.; Tian, J.; Bo, Q.; Li, K.; Wang, H.; Liu, T.; Li, J. Targeting DNA-PKcs increased anticancer drug sensitivity by suppressing DNA damage repair in osteosarcoma cell line MG63. Tumour Biol. 2015, 36, 9365–9372. [Google Scholar] [CrossRef]
- Van Oorschot, B.; Granata, G.; Di Franco, S.; ten Cate, R.; Rodermond, H.M.; Todaro, M.; Medema, J.P.; Franken, N.A.P. Targeting DNA double strand break repair with hyperthermia and DNA-PKcs inhibition to enhance the effect of radiation treatment. Oncotarget 2016, 7, 65504–65513. [Google Scholar] [CrossRef]
- Sunada, S.; Kanai, H.; Lee, Y.; Yasuda, T.; Hirakawa, H.; Liu, C.; Fujimori, A.; Uesaka, M.; Okayasu, R. Nontoxic concentration of DNA-PK inhibitor NU7441 radio-sensitizes lung tumor cells with little effect on double strand break repair. Cancer Sci. 2016, 107, 1250–1255. [Google Scholar] [CrossRef]
- Cassoux, N.; Rodrigues, M.J.; Plancher, C.; Asselain, B.; Levy-Gabriel, C.; Lumbroso-Le Rouic, L.; Piperno-Neumann, S.; Dendale, R.; Sastre, X.; Desjardins, L.; et al. Genome-wide profiling is a clinically relevant and affordable prognostic test in posterior uveal melanoma. Br. J. Ophthalmol. 2014, 98, 769–774. [Google Scholar] [CrossRef]
- Ehlers, J.P.; Worley, L.; Onken, M.D.; Harbour, J.W. DDEF1 is located in an amplified region of chromosome 8q and is overexpressed in uveal melanoma. Clin. Cancer Res. 2005, 11, 3609–3613. [Google Scholar] [CrossRef]
- Huang, B.; Shang, Z.F.; Li, B.; Wang, Y.; Liu, X.D.; Zhang, S.M.; Guan, H.; Rang, W.Q.; Hu, J.A.; Zhou, P.K. DNA-PKcs associates with PLK1 and is involved in proper chromosome segregation and cytokinesis. J. Cell. Biochem. 2014, 115, 1077–1088. [Google Scholar] [CrossRef]
- Kotula, E.; Berthault, N.; Agrario, C.; Lienafa, M.C.; Simon, A.; Dingli, F.; Loew, D.; Sibut, V.; Saule, S.; Dutreix, M. DNA-PKcs plays role in cancer metastasis through regulation of secreted proteins involved in migration and invasion. Cell Cycle 2015, 14, 1961–1972. [Google Scholar] [CrossRef]
- Kang, G.Y.; Pyun, B.J.; Seo, H.R.; Jin, Y.B.; Lee, H.J.; Lee, Y.J.; Lee, Y.S. Inhibition of Snail1-DNA-PKcs protein-protein interface sensitizes cancer cells and inhibits tumor metastasis. J. Biol. Chem. 2013, 288, 32506–32516. [Google Scholar] [CrossRef]
- Velic, D.; Couturier, A.M.; Ferreira, M.T.; Rodrigue, A.; Poirier, G.G.; Fleury, F.; Masson, J.Y. DNA Damage Signalling and Repair Inhibitors: The Long-Sought-After Achilles’ Heel of Cancer. Biomolecules 2015, 5, 3204–3259. [Google Scholar] [CrossRef]
- Amaro, A.; Mirisola, V.; Angelini, G.; Musso, A.; Tosetti, F.; Esposito, A.I.; Perri, P.; Lanza, F.; Nasciuti, F.; Mosci, C.; et al. Evidence of epidermal growth factor receptor expression in uveal melanoma: Inhibition of epidermal growth factor-mediated signalling by Gefitinib and Cetuximab triggered antibody-dependent cellular cytotoxicity. Eur. J. Cancer 2013, 49, 3353–3365. [Google Scholar] [CrossRef]
- McLean, I.W.; Foster, W.D.; Zimmerman, L.E.; Gamel, J.W. Modifications of Callender’s Classification of Uveal Melanoma at the Armed Forces Institute of Pathology. Am. J. Ophthalmol. 2018, 195, lvi–lx. [Google Scholar] [CrossRef]
- Amin, M.B. The American Joint Committee on Cancer: AJCC Cancer Staging Manual, 8th ed.; Springer International Publishing: New York, NY, USA, 2017; pp. 805–818. [Google Scholar] [CrossRef]
- Chen, P.W.; Murray, T.G.; Uno, T.; Salgaller, M.L.; Reddy, R.; Ksander, B.R. Expression of MAGE genes in ocular melanoma during progression from primary to metastatic disease. Clin. Exp. Metastasis 1997, 15, 509–1890. [Google Scholar] [CrossRef]
- Amirouchene-Angelozzi, N.; Nemati, F.; Gentien, D.; Nicolas, A.; Dumont, A.; Carita, G.; Camonis, J.; Desjardins, L.; Cassoux, N.; Piperno-Neumann, S.; et al. Establishment of novel cell lines recapitulating the genetic landscape of uveal melanoma and preclinical validation of mTOR as a therapeutic target. Mol. Oncol. 2014, 8, 1508–1520. [Google Scholar] [CrossRef]
- Luyten, G.P.; Naus, N.C.; Mooy, C.M.; Hagemeijer, A.; Kan-Mitchell, J.; Van Drunen, E.; Vuzevski, V.; De Jong, P.T.; Luider, T.M. Establishment and characterization of primary and metastatic uveal melanoma cell lines. Int. J. Cancer 1996, 66, 380–387. [Google Scholar] [CrossRef]
- De Waard-Siebinga, I.; Blom, D.J.; Griffioen, M.; Schrier, P.I.; Hoogendoorn, E.; Beverstock, G.; Danen, E.H.; Jager, M.J. Establishment and characterization of an uveal melanoma cell line. Int. J. Cancer 1995, 62, 155–161. [Google Scholar] [CrossRef]
- Bengtsson, H.; Irizarry, R.; Carvalho, B.; Speed, T.P. Estimation and assessment of raw copy numbers at the single locus level. Bioinformatics 2008, 24, 759–767. [Google Scholar] [CrossRef]
- Bengtsson, H.; Wirapati, P.; Speed, T.P. A single-array preprocessing method for estimating full-resolution raw copy numbers from all Affymetrix genotyping arrays including GenomeWideSNP 5 & 6. Bioinformatics 2009, 25, 2149–2156. [Google Scholar]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).